Abstract
This study compared microRNA (miRNA) expression profiles between rheumatic heart disease (RHD) patients and healthy controls to investigate their differential expression and help elucidate their mechanisms of action. Microarray analysis was used to measure miRNA expression, and a total of 133 miRNAs were shown to be significantly upregulated in RHD patients compared with controls, including miR-1183 and miR-1299. A total of 137 miRNAs, including miR-4423-3p and miR-218-1-3p, were significantly downregulated in RHD patients. Quantitative real-time-PCR confirmed microarray findings for miR-1183 and miR-1299 in both tissue and plasma. Bioinformatic predictions were also made of differentially expressed miRNAs as biomarkers in RHD by databases and GO/pathway analysis. Furthermore, we investigated miR-1183 and miR-1299 expression in RHD patients with secondary pulmonary hypertension (PAH). Our findings identified an important role for miR-1299 as a direct regulator of RHD, while the observed difference in expression of miR-1183 between RHD-PAH patients with high or low pulmonary artery pressure suggests that miR-1183 overexpression may reflect pulmonary artery remodeling. miR-1183 and miR-1299 appear to play distinct roles in RHD pathogenesis accompanied by secondary PAH and could be used as potential biological markers for disease development.
1. Introduction
MicroRNAs (miRNAs) are a class of single-stranded endogenous noncoding RNA molecules [1, 2], approximately 22 nucleotides (nt) in length, that negatively regulate gene expression by targeting the 3′-untranslated region of specific mRNAs for degradation or translational repression [3, 4]. The rapid growth in miRNA studies has demonstrated that they play an important role in a range of biological processes and are viewed as critical regulators in immune cell lineage commitment, differentiation, maturation, and immune signaling pathways [5, 6]. Additionally, deregulated miRNA expression patterns have been documented in many human diseases including inflammatory and autoimmune diseases [7–9].
Early miRNA studies focused on their role in cancer [10–13]; however, more recently, there has been a shift of attention to their possible impact on cardiovascular development and diseases [14]. Indeed, they are important in tissue development and influence the pathological processes of many cardiovascular diseases, including acute myocardial infarction, heart failure, coronary artery disease, stroke, and hypertension [1, 2]. To the best of our knowledge, the effects of miRNAs on the development of rheumatic heart disease (RHD) in either healthy tissues or circulating plasma have not yet been investigated.
RHD is primarily an autoimmune sequela of an acute rheumatic fever [15, 16], which occurs as a result of beta-hemolytic streptococcal infection [17, 18]. RHD can cause chronic inflammation of the endocardium and myocardium, leading to valvular dysfunction and hemodynamic changes [19] and, commonly, heart failure, stroke, or other serious related complications. Unfortunately, because of the lack of a specific method of detecting RHD, many patients have been diagnosed with irreversible valvular dysfunction, for which valvular surgery is one of the main treatments. RHD continues to be a burden in several developing countries such as India and China, though it is reasonably rare in western countries—probably because of the widespread use of antibiotics [20–22]. Therefore, the identification of a biomarker of characteristic RHD pathophysiology will be valuable to aid early detection and enable patients to avoid surgery by starting effective treatment at an early stage.
Pulmonary arterial hypertension (PAH) is a common complication of many RHD patients. Because PAH is characterized by the enhanced proliferation and reduced apoptosis of pulmonary artery smooth muscle cells [23], and as some miRNAs are also associated with the regulation of cell proliferation and apoptosis, it is hypothesized that they might be implicated in the etiology of PAH [22–26].
Therefore, in this study, we analyzed the miRNA expression profiles of RHD patients using microarray and confirmed our findings using quantitative real-time- (qRT-) PCR. We screened the roles of differentially expressed miRNAs in RHD with secondary PAH and used bioinformatics to predict and analyze their target genes as potential biomarkers of RHD. We also propose new directions for their potential therapeutic use in RHD.
2. Materials and Methods
2.1. Patients
A total of 100 subjects were selected for the study from the Inpatient Clinic of Ningbo Medical Center, Lihuili Hospital (Ningbo, China), between March 2012 and October 2013. Of these subjects, 50 were RHD patients (case group), and the remaining 50 were normal healthy adults (control group) with no medical history of congenital heart disease, cardiomyopathy, or liver or renal diseases. The inclusion criteria of the RHD group are as follows: (i) every patient diagnosed with mitral valve prolapse because of mitral chordae tendineae fracture and mitral insufficiency and scheduled for mitral valve replacement; (ii) left ventricular ejection fraction (EF) > 50%; (iii) left ventricular end-diastolic diameter (LVEDD) < 55 mm. RHD cases and their controls were well matched based on the following details: (iv) same gender; (v) difference of age < 5 years; (vi) other physiological indexes from physical check in close. All human materials were obtained in accordance with the hospital's regulations and hence were approved by the Ethics Committee of Lihuili Hospital. Written informed consent was also obtained from all subjects in advance.
2.2. Sample Collection
Blood samples were collected in EDTA tubes for plasma collection from the 50 RHD cases and stored at −80°C. All blood samples of cases and controls were collected by the same investigators. Left ventricular papillary muscles were obtained from 12 cases of resected mitral valves from RHD cases. They were later transferred into a physiological saline solution and then into liquid nitrogen and stored at −80°C. Six normal tissues for comparison were obtained from donors who had died from trauma.
2.3. RNA Isolation and Characterization
Peripheral blood was coagulated at room temperature for 30 min then centrifuged at 3000 rpm for 15 min to completely remove cell debris. It was stored at −80°C until required for miRNA detection. Total RNA was extracted from 625 μL of plasma using the mirVana PARIS kit (Ambion, USA) according to the manufacturer's instructions. Total RNA was also extracted from 10–50 mg left ventricular papillary muscles using the mirVana isolation kit according to the manufacturer's protocol (Ambion) [27–29]. The final elution volume of all RNA samples was 100 μL, and concentrations were determined by the ultramicro nucleic acid ultraviolet tester (NANODROP 1000, Wilmington, USA). RNA was reverse-transcribed into cDNA using the TaqMan microRNA reverse transcription kit (Applied Biosystems, Foster City, CA) using miRNA-specific primers provided by the manufacturer in Applied BioSystems 9700 Thermocycler. All cDNAs were stored at −20°C.
2.4. Quantitative Real-Time-PCR
Quantitative real-time-PCR (qRT-PCR) was performed as previously described [30]. Each reaction was performed in a final volume of 10 μL containing 4.5 μL cDNA, 5 μL TaqMan Universal PCR Master Mix (No AmpErase), and 0.5 μL TaqMan miRNA Assay (Applied Biosystems). The thermal cycle was set as start with 10 min template denaturation at 95°C, 40 cycles of denaturation at 95°C for 15 s, and combined primer annealing/elongation at 60°C for 1 min. Each sample was run in triplicate, and noncoding small RNA RNU6B was used as the internal control gene, according to the Applied Biosystems Application Note. RNU6B has previously been demonstrated to have both abundant and stable expression across 38 different human tissues and organs. It is regarded as one of the control genes with the least variability for miRNA assays and has been widely used in different fields, including cardiovascular research. We used Taqman microRNA assay for the qRT-PCR. The primer sequences were searched on the ABI official website, shown as follows.
The assay ID of miR-1183 assay is 002841; hsa-miR-1183 mature miRNA sequence is CACUGUAGGUGAUGGUGAGAGUGGGCA. The assay ID of miR-1299 assay is 241065_mat; hsa-miR-1299 mature miRNA sequence is UUCUGGAAUUCUGUGUGAGGGA. The assay ID of RNU6B assay is 001093; control RNU6b sequence is CGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTTT.
2.5. miRNA Microarray Analysis
To study the differential expression of miRNAs in RHD patients, we performed miRNA expression profiling on plasma samples using the miRCURY LNA Array system because of its comprehensive profiling and high sensitivity and specificity (see S2_Fig in Supplementary Material available online at http://dx.doi.org/10.1155/2015/524519). Here we selected 3 patients named numbers 1, 2, and N1-3 for microarray assay from the above 50 RHD patients. Meanwhile, we also selected 3 healthy controls named numbers 3, 4, and 5 from 50 control groups, which are also evaluated for microarray assay. Microarray analysis was performed by Kangcheng Bio-tech Inc. (Shanghai, China). miRNAs selected for investigation in our study were further filtered on the basis of expression levels and previously published data [31]. The RNA of each individual was analyzed on a separate chip. Data were analyzed in Genepix Pro 6.0 and saved as EXCEL files. GeneSpring 7.2 was used for further data analysis. Fold changes in miRNAs between groups, either twofold greater or less, were considered to represent differential expression.
2.6. Statistical Analysis
Experimental data were analyzed using SPSS19.0 statistical software. RNA concentration and Ct value levels were presented as means ± SD.
3. Results
3.1. RNA Concentration of RHD Patients in Plasma and Tissue
First, we validated the feasibility of miRNA detection from plasma in all subjects. No differences in RNA concentrations were observed among the different groups (plasma RHD group: 17.74 ± 5.59 ng/μL; plasma control group: 16.43 ± 4.32 ng/μL; tissue RHD group: 40.95 ± 27.90 ng/μL; tissue control group: 52.28 ± 28.21 ng/μL) (S1_Fig). However, the lower RNA yield from plasma hampered the actual RNA quantification compared with that from tissue (S1_Fig).
3.2. Genome-Wide Expression Profiling of Plasma miRNAs by Microarray Analysis
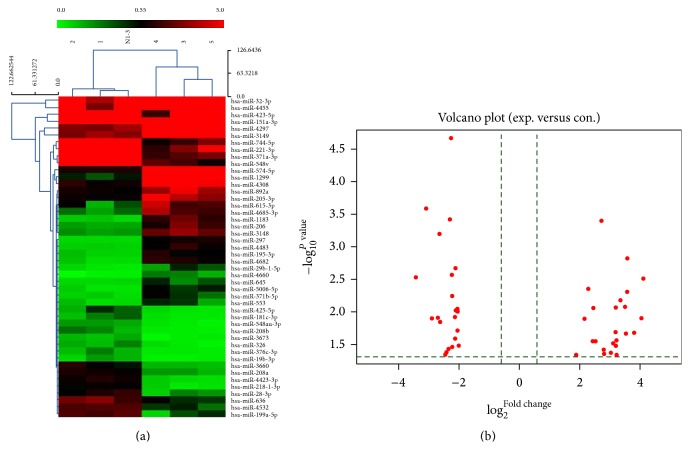
Hundreds of miRNAs showed differential expression between RHD cases and controls (S2_Fig). A total of 133 miRNAs, including miR-1299 and miR-1183, were significantly upregulated (>2-fold), while 137 were significantly downregulated (>2-fold), including miR-4423-3p and miR-218-1-3p (S3_Fig). We selected several significantly differentially expressed miRNAs for further study. Gene chips represents different fold changes from many upregulated miRNAs and downregulated miRNAs, either by heat map of microRNA microarray expression data (Figure 1(a)) or volcano plot (Figure 1(b)). Chip results suggested that miR-1299 and miR-1183 expressed in RHD were significantly upregulated (>10-fold), whereas miR-4423-3p and miR-218-1-3p expressed in RHD were significantly downregulated (>5-fold) in RHD cases compared with controls (Table 1).
Figure 1.
MicroRNA microarray expression data from plasma samples of rheumatic heart disease (n = 3) and healthy control subjects (n = 3). (a) Heat map of microRNA microarray expression data from plasma samples of rheumatic heart disease (n = 3) and healthy control subjects (n = 3). The expression of miRNA is hierarchically clustered on the y-axis, and RHD plasma samples or healthy control plasma samples or healthy control plasma samples are hierarchically clustered on the x-axis. The legend on the right indicates the miRNA represented in the corresponding row. The relative miRNA expression is depicted according to the color scale shown on the right. Red indicated upregulation; blue indicated downregulation; numbers 1, 2, and N1-3 indicate healthy control samples; numbers 3, 4, and 5 indicate RHD plasma samples. (b) Volcano plot from gene chips represents different fold changes from many upregulated miRNAs and downregulated miRNAs.
Table 1.
List of the hsa-miRNAs with at least 2-fold changes.
| Upregulated miRNA (>10 folds) | Downregulated miRNAs (>5 folds) | ||||
|---|---|---|---|---|---|
| miRNA | Folds | P value | miRNA | Folds | P value |
| miR-1299 | 17.13272 | 0.003132 | miR-4423-3p | 0.092672 | 0.002951 |
| miR-1183 | 16.5075 | 0.012476 | miR-218-1-3p | 0.117514 | 0.000261 |
| miR-4455 | 13.98099 | 0.020973 | miR-744-5p | 0.13412 | 0.01252 |
| miR-3148 | 11.95398 | 0.001515 | miR-4666a-5p | 0.139837 | 0.018463 |
| miR-4660 | 11.9266 | 0.004957 | miR-208b | 0.154499 | 0.012267 |
| miR-3149 | 11.56086 | 0.021548 | miR-199a-5p | 0.159151 | 0.000643 |
| miR-4682 | 11.37554 | 0.008373 | miR-548v | 0.162247 | 0.014474 |
| miR-297 | 10.12601 | 0.006702 | miR-3660 | 0.184096 | 0.045429 |
| miR-206 | 9.313578 | 0.045881 | miR-28-3p | 0.188257 | 0.041912 |
| miR-32-3p | 9.312469 | 0.027754 | miR-425-5p | 0.196498 | 0.037533 |
| miR-4308 | 9.191765 | 0.020532 | miR-548-3p | 0.203344 | 0.000383 |
| miR-4483 | 9.155266 | 0.033305 | miR-4532 | 0.209924 | 0.000217 |
| miR-574-5p | 9.132102 | 0.008565 | miR-3673 | 0.211119 | 0.0027 |
| miR-4685-3p | 8.583969 | 0.03023 | miR-221-3p | 0.215457 | 0.005767 |
| miR-4297 | 8.120556 | 0.042287 | miR-181c-3p | 0.216058 | 0.034691 |
| miR-4722-5p | 7.657944 | 0.002658 | miR-636 | 0.226805 | 0.011922 |
| miR-3591-5p | 7.405006 | 0.020074 | miR-376c-3p | 0.227146 | 0.025902 |
| miR-615-3p | 7.082924 | 0.044276 | miR-4798-3p | 0.228792 | 0.038073 |
| miR-195-3p | 6.976838 | 0.039299 | miR-208a | 0.231132 | 0.002143 |
| miR-4657 | 6.682702 | 0.00013 | miR-371a-3p | 0.233856 | 0.009664 |
| miR-3657 | 6.671498 | 0.024803 | miR-4708-5p | 0.23388 | 0.010049 |
| miR-5006-5p | 6.639262 | 0.000398 | miR-151a-3p | 0.239967 | 0.00974 |
| miR-4458 | 6.188047 | 4.05E − 06 | miR-4678 | 0.240886 | 0.004481 |
| miR-29b-1-5p | 5.735279 | 0.028064 | miR-423-5p | 0.240937 | 0.009271 |
| miR-205-3p | 5.521096 | 0.008749 | miR-326 | 0.243204 | 0.019519 |
| miR-498 | 5.497149 | 0.017617 | miR-4703-3p | 0.244857 | 0.021779 |
| miR-4450 | 5.491613 | 0.026159 | miR-19b-3p | 0.24777 | 0.033327 |
| miR-371b-5p | 5.378559 | 0.028034 | miR-3612 | 0.250328 | 0.017766 |
| miR-2113 | 5.371673 | 0.025333 | miR-4748 | 0.252973 | 0.026891 |
| miR-3689a-5p | 5.342335 | 0.015381 | miR-186-5p | 0.253236 | 0.045289 |
| miR-3120-5p | 5.221742 | 0.00234 | miR-320b | 0.253705 | 0.029743 |
| miR-4681 | 5.200976 | 0.01057 | miR-550b-2-5p | 0.253829 | 0.045176 |
| miR-3690 | 5.145123 | 0.015582 | miR-346 | 0.254841 | 0.021562 |
| miR-4804-3p | 5.03806 | 0.013518 | miR-130a-3p | 0.256733 | 0.04919 |
| miR-4447 | 4.964015 | 0.001608 | miR-4650-5p | 0.256936 | 0.02673 |
| miR-892a | 4.895473 | 0.004484 | miR-2682-5p | 0.257933 | 0.008486 |
| miR-4795-5p | 4.698387 | 0.000309 | miR-219-5p | 0.263898 | 0.011573 |
| miR-3158-3p | 4.618894 | 0.003654 | miR-337-3p | 0.265439 | 0.016578 |
3.3. Plasma and Tissue miR-1183 and miR-1299 Expressions Are Potential Biochemical Marker for RHD by Quantitative Real-Time-PCR
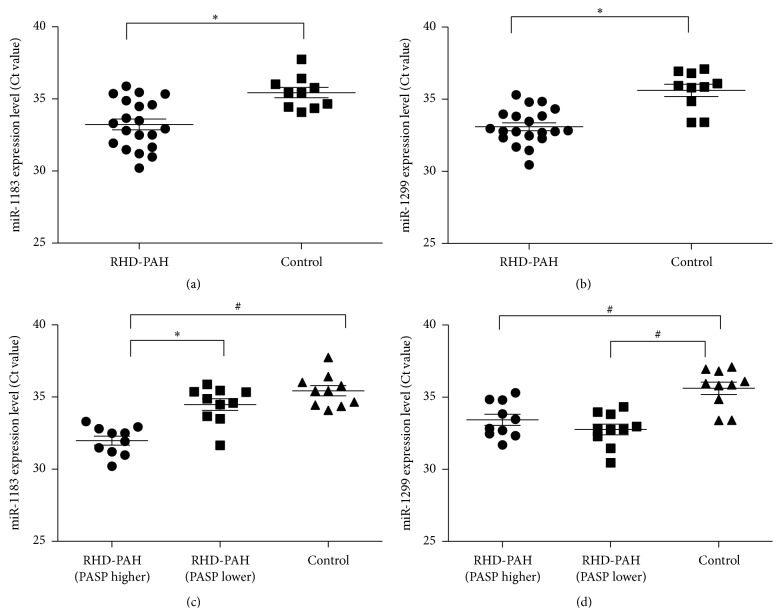
To verify the accuracy of the microarray-based miRNA measurements, expression levels of hsa-miR-1183, hsa-miR-1299, and RNU6B were assessed using qRT-PCR [32]. All miRNAs including RNU6B showed reliable Ct values in most samples (Figure 2), and fluorescent signals failed to reach the set threshold after 40 cycles in very few assays. Results are shown in Figures 2 and 3; qRT-PCR revealed significant differences in the expression of miRNA-1183, miRNA-1299, and RNU6B in both tissue and plasma samples of RHD patients compared with healthy controls, which was consistent with microarray data. Ct values in RHD tissue for miR-1183 were 34.62 ± 2.23, 31.81 ± 2.46 for miR-1299, and 28.33 ± 3.48 for RNU6B.
Figure 2.
Plasma level of representative differentially expressed miR-1183 and miR-1299 in 20 RHD patients and 10 normal controls (∗ P < 0.05, # P < 0.01).
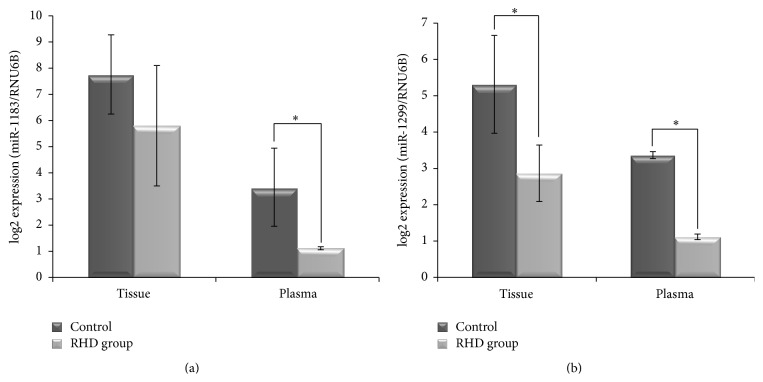
Figure 3.
Upregulation of microRNA-1183 and microRNA-1299 expression in rheumatic heart disease tissues and plasma by real-time RT-PCR. (a) The differential expression of miRNA-1183 in the tissue and plasma of RHD patients and normal controls. (b) The differential expression of miRNA-1299 in the tissue and plasma of RHD patients and normal controls (∗ P < 0.05).
Pulmonary artery systolic pressure (PASP) is a key factor to evaluate the severity of PAH, so we next divided RHD patients with secondary PAH into two groups: those with high PASP (RHD-PAH [PASP higher >40 mmHg]) and those with low PASP (RHD-PAH [PASP lower <40 mmHg]) (Table 2). The expression of miR-1183 was significantly upregulated in both RHD plasma samples (Figure 2(a), P = 0.012) and the subset of RHD cases with high PASP (Figure 2(b), P = 0.021). By contrast, although the expression of miR-1299 was significantly upregulated in RHD plasma samples (Figure 2(c), P = 0.011), no significant difference was observed between the subsets with high and low PASP (Figure 2(d), P = 0.566). Moreover, the overexpression of miR-1183 in high versus low PASP cases could reflect the pulmonary artery remodeling of PAH secondary to RHD, meaning that it plays a more important role in secondary PAH complications than primary disease. Although the same trend was also observed when comparing miRNA-1183 in tissue samples between the two groups by qRT-PCR (Figure 3(a)), the difference was not significant. Interestingly, the expression of miRNA-1299 is significantly higher in the RHD group compared with the non-RHD group for both tissue and plasma (Figure 3(b), P < 0.05).
Table 2.
Baseline characteristics of different severe pulmonary arterial hypertension complication on RHD patients and healthy control subjects.
| Groups | Group 1: high pulmonary artery pressure group in RHD patients (n = 10) |
Group 2: low pulmonary artery pressure group in RHD patients (n = 10) |
Group 3: control (n = 10) |
|---|---|---|---|
| Age, years | 54.2 ± 5.73 | 59.2 ± 6.27 | 49.6 ± 7.0 |
| Sex, males/females | 3/7 | 4/6 | 5/5 |
| NYHA grades | II~III | IV | / |
| PASP (mmHg) | 88.3 ± 5.376 | 33.1 ± 4.55 | / |
3.4. Bioinformatic Analysis and Predicted miRNA Molecular Targets Identified in RHD Patients
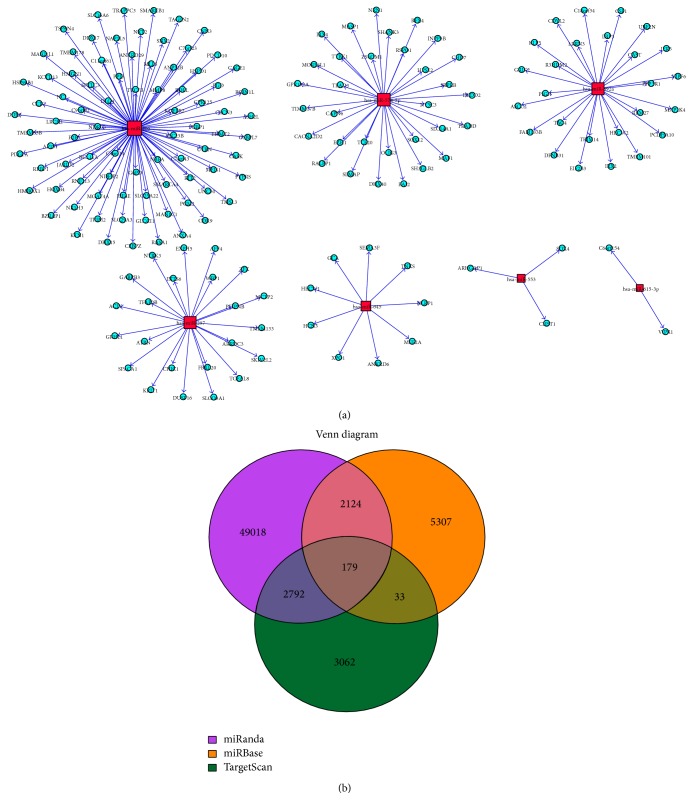
Those miRNAs showing significantly differential expression by microarray were analyzed by bioinformatics, including target gene prediction, gene ontology (GO) analysis, and pathway analysis, with the aim of investigating target genes and regulatory mechanisms. Target genes of differential miRNA expression were predicted using three algorithms: miRBase (http://www.mirbase.org/), miRanda (http://www.microrna.org/), and TargetScan (http://www.targetscan.org/) (Figure 4). Only genes identified by all three algorithms were considered to be the predicted targets for each miRNA.
Figure 4.
Target genes of differential miRNA expression predicted. (a) MicroRNA–mRNA-Gene-Network of several representative miRNAs with their predicted target genes. (b) Overlapping data of three databases from the target summary by miRBase (http://www.mirbase.org/), miRanda (http://www.microrna.org/), and TargetScan (http://www.targetscan.org/).
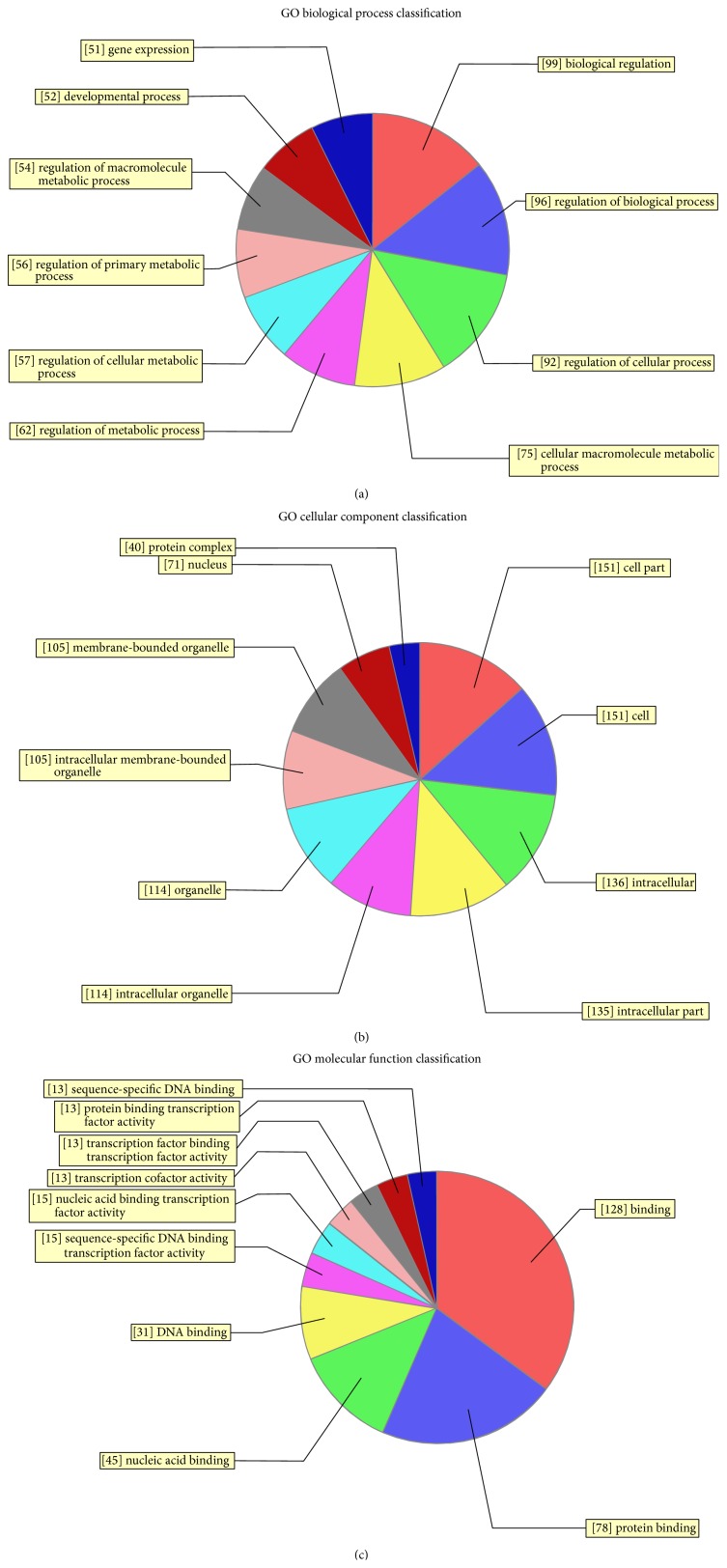
Targets were found to be predominantly involved in the regulation of cellular and biological processes, including SOX family members and MEF2A. Hundreds of target genes were predicted and were mainly involved in transcription coactivator activity, RNA polymerase II transcription coactivator activity, histone-lysine N-methyl transferase activity, protein-lysine N-methyl transferase activity (GO molecular function), the positive regulation of biological processes, histone-lysine methylation, protein alkylation (GO biology process), and intracellular roles (GO cellular component) (Figure 5, S4_Fig). To assess the possible biological impact of the differentially expressed miRNAs, we undertook pathway analyses of the predicted target genes, revealing that the gene set was mostly involved in biological pathways including the pentose phosphate pathway, glutamatergic synapse, and the MAPK signaling pathway (Figure 6).
Figure 5.
GO analysis classifications of predicted target genes regulated by differentially expressed microRNAs (miRNAs) in rheumatic heart disease. GO analysis was performed on genes predicted to be targets of differentially expressed miRNAs.
Figure 6.
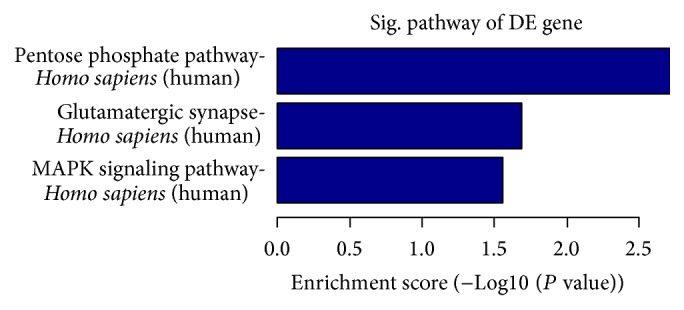
Pathway classifications of predicted target genes regulated by differentially expressed microRNAs (miRNAs) in rheumatic heart disease. Pathway analysis was performed on genes predicted to be targets of differentially expressed miRNAs. The negative log of the P value (log10 P) is plotted on the x-axis.
Because of the differences in miR-1183 and miR-1299 expression, we separately predicted the target genes for these two miRNAs. Bcl-2 was predicted to be an important target gene of miR-1299, which may influence cardiomyocyte apoptosis. Considering RHD as an autoimmune sequela of an acute rheumatic fever, it can cause chronic inflammation. PBMC and THP-1 cells as classical immune cells are studied to dig deep into the mechanism study including both miR-1183 and miR-1299 mimic and inhibitor study. Series of studies are in progress including identifying the changes of target genes Bcl-2 and EGFR mRNA expression and related cytokines (data not shown). Up to now, we have already further studied the relationship of miR-1299 mimic, inhibitor, and Bcl-2 expression levels. It seems that miR-1299 mimics can upregulate miRNA level and induce cardiomyocyte apoptosis (data not shown). Meanwhile, CXCR4, EGF, and EGFR were also predicted to be important target genes of miR-1183. However, the following mechanism study including miR mimic was still in progress.
4. Discussion
Several studies have shown that a variety of miRNAs are implicated in cardiovascular diseases, so exploratory research has been conducted into the possibility of using them as biological markers based on their expression in plasma or tissue [33–35]. However, this has not been tested in RHD. The present miRNA array results suggested that miR-1299, miR-1183, and so forth are significantly upregulated, while miR-4423-3p, miR-218-1-3p, and so forth are significantly downregulated in RHD. qRT-PCR confirmed that the expression of miR-1299 and miR-1183 in RHD tissue and plasma was significantly higher in RHD cases than healthy controls. Moreover, the enhanced levels of miR-1183 and miR-1299 expression in plasma are consistent with those in tissues, suggesting that they could be used as potential biological markers in RHD. Besides, some other miRNAs determined to be expressed differently in RHD compared to healthy controls can be dug deeper, like miR206, miR208a, miR208b, and miR574-5p which have already been studied in the regions of other cardiovascular diseases [36–38]. To explore the possible functions on RHD of these miRNAs may provide us ideas whether they are potent therapeutic targets for cardiac hypertrophy, fibrosis, dysfunction, and so forth.
Pulmonary arterial hypertension is a debilitating condition with progressive remodeling of the pulmonary resistance vessels [22]. It is characterized by excessive vascular resistance and smooth muscle cell proliferation in small pulmonary arteries and finally causes elevation of pulmonary vascular resistance, right ventricular failure, and death [39, 40]. Variations in PASP in RHD patients are likely to influence the expression of different miRNAs in RHD-PAH.
We identified an important role for miR-1299 as a direct regulator of RHD. Meanwhile, the difference in expression of miR-1183 between RHD cases with high and low PASP suggests that its overexpression is caused by the pulmonary artery remodeling of PAH secondary to RHD, meaning that it plays a more important role in secondary complications than primary disease. miR-1183 and miR-1299 may therefore have an independent effect on the disease processes of RHD and PAH. We searched the literature for mechanistic articles on miR-1183 or miR-1299 and found them to be extremely limited. miR-1183 was previously reported to be downregulated in tick salivary glands [41], while another study reported that it has a relationship with a functional polymorphism in the EpCAM gene [42]. miR-1183 is also known to be upregulated in locally advanced rectal cancer patients [43]. Information about miR-1299 is even more limited, so further detailed studies are required for a greater understanding of the molecular mechanisms of miR-1183 and miR-1299.
Based on the work which we have done for screening differentially expressed miRNAs (e.g., miR-1183 and miR-1299) of RHD, most importantly, evaluation of the stability and effect of miRNA-based therapeutics is of great importance for the comprehensive understanding of the miR-1183 and miR-1299 functions in rheumatic heart disease. The bioinformatics analysis is often used to further study the mechanism of the differential expression of miRNAs [44–46] and to predict and analyze their target genes with the aim of understanding regulatory mechanisms in the future [47, 48]. Our analysis predicted hundreds of target genes, and pathway and GO analyses demonstrated that the gene set was mostly involved in biological pathways and cellular processes such as the pentose phosphate pathway, glutamatergic synapse, and MAPK signaling pathway. Our findings also suggest that the regulation of cellular and metabolic processes may influence the release of miR-1299 and miR-1183 into the peripheral blood following their overexpression in the tissue. The Bcl-2 gene and its influence on cardiomyocyte apoptosis may be an important factor in association with miR-1299 expression. However, more detailed studies are needed for a greater understanding of this process.
5. Conclusion
Taken together, our data revealed the differential expression of specific miRNAs in RHD accompanied by secondary PAH. miR-1183 and miR-1299 appear to play a distinct role in disease pathology and so could be potential biological markers for both RHD and PAH. Future functional and mechanistic studies on the dynamic changes of miRNA expression in RHD may improve our understanding of the regulatory role of miRNAs in RHD.
Supplementary Material
Genome-wide miRNA profiling using miChip.
Acknowledgments
The research was supported by the grants from Advanced key Scientific and Technological Programs of Ningbo (2012C5017), Zhejiang Provincial Natural Science Foundation (LY12H16002), Science and Technology Innovation Team of Ningbo (2011B82015), Advanced Key Scientific and Technological Programs of Ningbo (2011C51001), Natural Science Foundation of Ningbo (2014A610272), and Clinical medical Research Fund of Zhejiang Province (2013ZYC-A67).
Abbreviation
- RHD:
Rheumatic heart disease
- miR:
MicroRNAs
- PAH:
Pulmonary hypertension
- QRT-PCR:
Quantitative real-time-PCR
- PASP:
Pulmonary artery systolic pressure
- nt:
Nucleotides.
Disclosure
Ni Li and Jiangfang Lian are co-first authors of this work.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.van Empel V. P. M., de Windt L. J., da Costa Martins P. A. Circulating miRNAs: reflecting or affecting cardiovascular disease. Current Hypertension Reports. 2012;14(6):498–509. doi: 10.1007/s11906-012-0310-7. [DOI] [PubMed] [Google Scholar]
- 2.Egea V., Schober A., Weber C. Circulating miRNAs: messengers on the move in cardiovascular disease. Thrombosis and Haemostasis. 2012;108(4):590–591. doi: 10.1160/th12-08-0613. [DOI] [PubMed] [Google Scholar]
- 3.Donzelli S., Strano S., Blandino G. microRNAs: short non-coding bullets of gain of function mutant p53 proteins. Oncoscience. 2014;1:427–433. doi: 10.18632/oncoscience.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellinger J., Müller S. C. MicroRNAs: a novel non-invasive biomarker for patients with urological malignancies. Current Pharmaceutical Biotechnology. 2014;15(5):486–491. doi: 10.2174/1389201015666140519124909. [DOI] [PubMed] [Google Scholar]
- 5.Zhu S., Pan W., Qian Y. MicroRNA in immunity and autoimmunity. Journal of Molecular Medicine. 2013;91(9):1039–1050. doi: 10.1007/s00109-013-1043-z. [DOI] [PubMed] [Google Scholar]
- 6.Zibert J. R., Løvendorf M. B., Litman T., Olsen J., Kaczkowski B., Skov L. MicroRNAs and potential target interactions in psoriasis. Journal of Dermatological Science. 2010;58(3):177–185. doi: 10.1016/j.jdermsci.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Li Y., Xu Z., Wang K., Wang N., Zhu M. Network analysis of microRNAs, genes and their regulation in human bladder cancer. Biomedical Reports. 2013;1:918–924. doi: 10.3892/br.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Guire V., Robitaille R., Tétreault N., et al. Circulating miRNAs as sensitive and specific biomarkers for the diagnosis and monitoring of human diseases: promises and challenges. Clinical Biochemistry. 2013;46(10-11):846–860. doi: 10.1016/j.clinbiochem.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 9.Van Roosbroeck K., Pollet J., Calin G. A. miRNAs and long noncoding RNAs as biomarkers in human diseases. Expert Review of Molecular Diagnostics. 2013;13(2):183–204. doi: 10.1586/erm.12.134. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh W. J., Lin F.-M., Huang H.-D., Wang H. Investigating microRNA-target interaction-supported tissues in human cancer tissues based on miRNA and target gene expression profiling. PLoS ONE. 2014;9(4) doi: 10.1371/journal.pone.0095697.e95697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vrba L., Muñoz-Rodríguez J. L., Stampfer M. R., Futscher B. W. miRNA gene promoters are frequent targets of aberrant DNA methylation in human breast cancer. PLoS ONE. 2013;8(1) doi: 10.1371/journal.pone.0054398.e54398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Zhao J., Huang J., Tang H., Yu S., Chen Y. The regulatory roles of miRNA and methylation on oncogene and tumor suppressor gene expression in pancreatic cancer cells. Biochemical and Biophysical Research Communications. 2012;425(1):51–57. doi: 10.1016/j.bbrc.2012.07.047. [DOI] [PubMed] [Google Scholar]
- 13.Tong A. W., Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Therapy. 2008;15(6):341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- 14.Chen J., Wang D.-Z. MicroRNAs in cardiovascular development. Journal of Molecular and Cellular Cardiology. 2012;52(5):949–957. doi: 10.1016/j.yjmcc.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah B., Sharma M., Kumar R., Brahmadathan K. N., Abraham V. J., Tandon R. Rheumatic heart disease: progress and challenges in India. Indian Journal of Pediatrics. 2013;80(supplement 1):S77–S86. doi: 10.1007/s12098-012-0853-2. [DOI] [PubMed] [Google Scholar]
- 16.Toor D., Vohra H. Immune responsiveness during disease progression from acute rheumatic fever to chronic rheumatic heart disease. Microbes and Infection. 2012;14(12):1111–1117. doi: 10.1016/j.micinf.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Zheng D., Xu L., Sun L., et al. Comparison of the ventricle muscle proteome between patients with rheumatic heart disease and controls with mitral valve prolapse: HSP 60 may be a specific protein in RHD. BioMed Research International. 2014;2014:9. doi: 10.1155/2014/151726.151726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guilherme L., Köhler K. F., Kalil J. Rheumatic heart disease: mediation by complex immune events. Advances in Clinical Chemistry. 2011;53:31–50. doi: 10.1016/b978-0-12-385855-9.00002-3. [DOI] [PubMed] [Google Scholar]
- 19.Guilherme L., Kalil J. Rheumatic heart disease: molecules involved in valve tissue inflammation leading to the autoimmune process and anti-S. pyogenes vaccine. Frontiers in Immunology. 2013;4, article 352 doi: 10.3389/fimmu.2013.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhardwaj R., Kandoria A., Marwah R., et al. Prevalence of rheumatic fever and rheumatic heart disease in rural population of Himachal—a population based study. Journal of Association of Physicians of India. 2012;60(5):13–14. [PubMed] [Google Scholar]
- 21.Nordet P., Lopez R., Dueñas A., Sarmiento L. Prevention and control of rheumatic fever and rheumatic heart disease: the Cuban experience (1986-1996-2002) Cardiovascular Journal of Africa. 2008;19(3):135–140. [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Xue X.-Y., Liu Y.-X., et al. Pulmonary arterial hypertension and microRNAs—an ever-growing partnership. Archives of Medical Research. 2013;44(7):483–487. doi: 10.1016/j.arcmed.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Courboulin A., Paulin R., Giguère N. J., et al. Role for miR-204 in human pulmonary arterial hypertension. The Journal of Experimental Medicine. 2011;208(3):535–548. doi: 10.1084/jem.20101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millard-Bullock D. The rheumatic fever and rheumatic heart disease control programme—Jamaica. The West Indian Medical Journal. 2012;61(4):361–364. doi: 10.7727/wimj.2012.134. [DOI] [PubMed] [Google Scholar]
- 25.Mirrakhimov A. E., Ali A. M., Barbaryan A., Prueksaritanond S. Human immunodeficiency virus and pulmonary arterial hypertension. ISRN Cardiology. 2013;2013:11. doi: 10.1155/2013/903454.903454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bockmeyer C. L., Maegel L., Janciauskiene S., et al. Plexiform vasculopathy of severe pulmonary arterial hypertension and microRNA expression. The Journal of Heart and Lung Transplantation. 2012;31(7):764–772. doi: 10.1016/j.healun.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Kim J., Kang Y., Kojima Y., et al. An endothelial apelin-FGF link mediated by miR-424 and miR-503 is disrupted in pulmonary arterial hypertension. Nature Medicine. 2013;19(1):74–82. doi: 10.1038/nm.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Podolska A., Kaczkowski B., Litman T., Fredholm M., Cirera S. How the RNA isolation method can affect microRNA microarray results. Acta Biochimica Polonica. 2011;58(4):535–540. [PubMed] [Google Scholar]
- 29.Kong L., Zhu J., Han W., et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetologica. 2011;48(1):61–69. doi: 10.1007/s00592-010-0226-0. [DOI] [PubMed] [Google Scholar]
- 30.Camarillo C., Swerdel M., Hart R. P. Comparison of microarray and quantitative real-time PCR methods for measuring MicroRNA levels in MSC cultures. Methods in Molecular Biology. 2011;698:419–429. doi: 10.1007/978-1-60761-999-4_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callari M., Tiberio P., De Cecco L., et al. Feasibility of circulating miRNA microarray analysis from archival plasma samples. Analytical Biochemistry. 2013;437(2):123–125. doi: 10.1016/j.ab.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Pan X., Murashov A. K., Stellwag E. J., Zhang B. Monitoring microRNA expression during embryonic stem-cell differentiation using quantitative real-time PCR (qRT-PCR) Methods in Molecular Biology. 2010;650:213–224. doi: 10.1007/978-1-60761-769-3_16. [DOI] [PubMed] [Google Scholar]
- 33.Ferreira L. R. P., Frade A. F., Santos R. H. B., et al. microRNAs miR-1, miR-133a, miR-133b, miR-208a and miR-208b are dysregulated in Chronic Chagas disease Cardiomyopathy. International Journal of Cardiology. 2014;175(3):409–417. doi: 10.1016/j.ijcard.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 34.Elton T. S., Khan M., Terentyev D. MicroRNAs in cardiovascular disease. F1000 Medicine Reports. 2011;3(1, article 10) doi: 10.3410/m3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.da Costa Martins P. A., Leptidis S., Salic K., de Windt L. D. Microrna regulation in cardiovascular disease. Current Drug Targets. 2010;11(8):900–906. doi: 10.2174/138945010791591322. [DOI] [PubMed] [Google Scholar]
- 36.Boštjančič E., Zidar N., Štajer D., Glavač D. MicroRNAs miR-1, miR-133a, miR-133b and miR-208 are dysregulated in human myocardial infarction. Cardiology. 2010;115(3):163–169. doi: 10.1159/000268088. [DOI] [PubMed] [Google Scholar]
- 37.Huang Y., Li J. MicroRNA208 family in cardiovascular diseases: therapeutic implication and potential biomarker. Journal of Physiology and Biochemistry. 2015;71(3):479–486. doi: 10.1007/s13105-015-0409-9. [DOI] [PubMed] [Google Scholar]
- 38.Novák J., Kružliak P., Bienertová-Vaškü J., Slabý O., Novák M. MicroRNA-206: a promising theranostic marker. Theranostics. 2014;4(2):119–133. doi: 10.7150/thno.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou G., Chen T., Raj J. U. MicroRNAs in pulmonary arterial hypertension. American Journal of Respiratory Cell and Molecular Biology. 2015;52(2):139–151. doi: 10.1165/rcmb.2014-0166tr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bienertova-Vasku J., Novak J., Vasku A. MicroRNAs in pulmonary arterial hypertension: pathogenesis, diagnosis and treatment. Journal of the American Society of Hypertension. 2015;9(3):221–234. doi: 10.1016/j.jash.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 41.Zhou J., Zhou Y., Cao J., Zhang H., Yu Y. Distinctive microRNA profiles in the salivary glands of Haemaphysalis longicornis related to tick blood-feeding. Experimental and Applied Acarology. 2013;59(3):339–349. doi: 10.1007/s10493-012-9604-3. [DOI] [PubMed] [Google Scholar]
- 42.Hu M., Jian L., Zhang L., et al. Functional polymorphism in the EpCAM gene is associated with occurrence and advanced disease status of cervical cancer in Chinese population. Molecular Biology Reports. 2012;39(7):7303–7309. doi: 10.1007/s11033-012-1560-9. [DOI] [PubMed] [Google Scholar]
- 43.Della Vittoria Scarpati G., Falcetta F., Carlomagno C., et al. A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. International Journal of Radiation Oncology Biology Physics. 2012;83(4):1113–1119. doi: 10.1016/j.ijrobp.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y., Kan Q., Zhang P., Zhang X.-Q., Zhou X.-G., Zhou X.-Y. Bioinformatic analysis of target gene prediction and related signaling pathways of miRNA-126∗ . Chinese Journal of Contemporary Pediatrics. 2013;15(3):227–232. doi: 10.7499/j.issn.1008-8830.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 45.Zhao C., Tian F., Yu Y., et al. miRNA-dysregulation associated with tenderness variation induced by acute stress in Angus cattle. Journal of Animal Science and Biotechnology. 2012;3(1, article 12) doi: 10.1186/2049-1891-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W., Edwards A., Fan W., Flemington E. K., Zhang K. MiRNA-mRNA correlation-network modules in human prostate cancer and the differences between primary and metastatic tumor subtypes. PLoS ONE. 2012;7(6) doi: 10.1371/journal.pone.0040130.e40130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Juhila J., Sipilä T., Icay K., et al. MicroRNA expression profiling reveals MiRNA families regulating specific biological pathways in mouse frontal cortex and hippocampus. PLoS ONE. 2011;6(6) doi: 10.1371/journal.pone.0021495.e21495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai E. C. miRNAs: Whys and wherefores of miRNA-mediated regulation. Current Biology. 2005;15(12):R458–R460. doi: 10.1016/j.cub.2005.06.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genome-wide miRNA profiling using miChip.