Abstract
Listeria spp. isolated from different food products and collected from 12 Brazilian states were sent to the Laboratory of Bacterial Zoonoses (Oswaldo Cruz Institute, Brazil) for identification. The aims of this study were to characterize these isolates, from 1990 to 2012, by using biochemical, morphological, and serotyping tests, and to analyze the distribution of L. monocytogenes serotypes on different food products and geographical locations. Serotyping was performed using polyclonal somatic and flagellar antisera. Of 5953 isolates, 5770 were identified as Listeria spp., from which 3429 (59.4%) were L. innocua, 2248 (38.9%) were L. monocytogenes, and 93 (1.6%) were other Listeria spp. L. innocua was predominantly isolated from 1990 to 2000, while L. monocytogenes was from 2001 to 2012. Regarding the serotype distribution in the foods, serotypes 1/2a and 4b were most common in processed meat and ready-to-eat products, respectively; serotypes 1/2a, 1/2b, and 4b were the most common in nonprocessed meat. The results above confirm the presence of the main serotypes of L. monocytogenes in different parts of the food chain from three regions of the country and emphasize the importance of improving the control measures, as tolerance zero policy and microbiological surveillance in Brazil.
1. Introduction
The genus Listeria includes pathogenic species as Listeria monocytogenes and Listeria ivanovii, the latter is common in warm-blooded animals, and it is widespread in nature. Its ubiquity is evident by the isolation of this microorganism in fecal specimens from healthy hosts, soil, water, waste, vegetables, and processed silage or food (as well as their factories) [1].
Listeria monocytogenes differs from most bacterial food pathogens due its ability to survive harsh environment conditions, grow over a wide temperature range (1–45°C), and survive under wide pH range (4.5–9.6), high salt concentration (10 to 15% NaCl), and very low water activity (aw = 0.94). Therefore it can grow in different types of food products [2].
From a public health perspective, L. monocytogenes (mainly the serotypes 1/2a, 1/2b, and 4b) is responsible for severe syndromes in humans, such as meningitis, septicemia, abortion, and febrile enteritis. Its main impact is in immunocompromised individuals, with a high fatality rate of 20% to 30% [3]. As it is a foodborne pathogen, individuals are infected predominantly through contaminated food consumption [4].
Although sporadic cases of listeriosis have been reported in Brazil [5–8], there is no information on foodborne outbreaks involving L. monocytogenes [9, 10]. On the other hand, different foods have been recognized as potential sources of this pathogen [6–8, 11–18], but there is no systematic evaluation of these products over time.
In Brazil the Normative Ruling Number 09, promulgated on 8 April 2009 by the Ministry of Agriculture, Cattle Raising and Supply, established criteria and procedures for the implementation of L. monocytogenes Control Procedures in ready-to-eat products of animal origin in order to monitor and ensure the safety of these products. These procedures are supervised by a national program called Federal Inspection Service (SIF). The National Agency of Sanitary Surveillance also has the RDC (Board Resolution) Number 12 on 2 January 2001, which establishes the absence of this pathogen in 25 g sample of cheese [19, 20].
Since there is no compulsory notification for cases of Listeriosis in Brazil, Sanitary Surveillance and Public Health has trouble to identify the occurrence of outbreaks, and only isolated cases are reported, which may explain the few listeriosis cases in the literature and the absence of outbreaks reports in our country.
The aims of this study were to (i) identify the species and serotypes of Listeria isolated from different food products and regions of Brazil and (ii) analyze the presence of main serotypes of L. monocytogenes in different foods and geographical locations of Brazil in an attempt to present a more detailed view of the distribution of Listeria spp. in the country.
These isolates were received by the Laboratory of Bacterial Zoonoses (LABZOO) at Oswaldo Cruz Institute (IOC), FIOCRUZ, Rio de Janeiro, Brazil, for identification and serotyping.
2. Materials and Methods
2.1. Bacterial Cultures
From 1990 to 2012 the laboratory received, from different sources (industries, public universities, research institutes, and other public institutions), 5953 isolates suspected of being Listeria spp. for identification, isolated from several food products of 12 different Brazilian states. All these isolates were received in nutrient agar.
2.2. Identification of Listeria spp
All isolates were screened through hemolysis and colony morphology on Columbia agar containing 5% defibrinated sheep blood, evaluation of the mobility to 25–28°C by stab-inoculation in semisolid agar (tryptose broth containing 0,4% agar), and Gram staining. In this initial screening, 183 (3.07%) isolates were discarded which includes 156 nonmotile Gram positive cocci and 27 motile Gram negative rods. Consequently, 5770 (96.9%) isolates went to further identification and serotyping. The identification was performed using previously published morphological and biochemical tests characterized by catalase, motility, and biochemical tests including acid production from D-xylose, D-mannitol, L-rhamnose, and α-methyl-D-mannoside. When necessary, L. monocytogenes was differentiated from other species of Listeria by using API Listeria (BioMeriéux) kit. The final confirmation was provided by Christie-Atkins-Munch-Peterson (CAMP) Test [21].
All isolates identified as Listeria were stored in Brain Heart Infusion (Difco) with 20% (v/v) glycerol at −80°C. All the Listeria isolates were deposited in the Culture Collection of Listeria (CLIST/LABZOO).
2.3. Listeria spp. Serotyping
The isolates were serotyped using polyclonal antisera produced against Listeria somatic and flagellar antigens previously manufactured in the LABZOO according to the method described by Seeliger and Höhne [22].
2.4. Statistical Analysis
All the isolates were tabulated using Excel 2007 software. Subsequently, the distribution of all continuous variables, means, medians, and interquartile values were studied. The frequency of all the categorical variables was described. Bivariate analysis of continuous variables was performed using Student's t-test or Mann-Whitney test if the variable did not follow normal distribution. Bivariate analysis of categorical variables was performed using Fisher exact test. Statistical analysis was performed using the statistical package STATA version 13.0, Texas, USA.
3. Results
The sources of Listeria isolates sent to the lab are geographically depicted in Table 1. Of those, 4714 (81.7%) were meat products (processed or/and not processed). Most of the strains (1474, 31.27%) were from poultry products and originated from Goiás, a state in the Middle-West of Brazil.
Table 1.
Geographical distribution of Listeria spp. strains by food type in Brazil. 1990–2012.
| State | Region∗ | Milk | Cheese | Meat | Ready-to-eat | Vegetables | Unknow source∗∗ | Total | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Nonprocessed | Processed | N | % | |||||||
| Alagoas | NE | 34 | 34 | 0.58 | ||||||
| Bahia | NE | 4 | 1 | 39 | 19 | 2 | 65 | 1.12 | ||
| Paraíba | NE | 25 | 8 | 33 | 0.57 | |||||
| Pernambuco | NE | 28 | 2 | 2 | 32 | 0.55 | ||||
| Goiás | MW | 1474 | 60 | 1534 | 26.58 | |||||
| Mato Grosso | MW | 17 | 43 | 223 | 283 | 4.9 | ||||
| Minas Gerais | SE | 87 | 346 | 472 | 17 | 922 | 15.97 | |||
| Rio de Janeiro | SE | 2 | 36 | 474 | 57 | 8 | 26 | 603 | 10.45 | |
| São Paulo | SE | 92 | 95 | 352 | 428 | 114 | 56 | 49 | 1186 | 20.55 |
| Paraná | S | 12 | 29 | 4 | 45 | 0.77 | ||||
| Santa Catarina | S | 42 | 380 | 4 | 79 | 505 | 8.75 | |||
| Rio Grande do Sul | S | 22 | 200 | 171 | 135 | 528 | 9.15 | |||
|
| ||||||||||
| Total | 145 | 560 | 3310 | 1404 | 203 | 99 | 49 | 5770 | ||
∗NE, Northeast; MW, Middle-West; SE, Southeast; S, South.
∗∗Food source not informed by the senders.
A total of 1404 (24.3%) strains were isolated from processed meat products (cooked, cured, or smoked products), with the most frequent (900 strains) being mixed sausages (chicken and pork) from two Southeastern Brazilian states. As for dairy products, the home-made cheese from the Southern Brazilian state of Rio Grande do Sul was the main origin.
As can be seen in Table 1, there was a predominance of the southeastern region (states of Minas Gerais, Rio de Janeiro, and São Paulo), with 2711 strains (46.9%), followed by the Midwestern region (states of Mato Grosso and Goiás) with 1817 strains (31.4%), mainly from meat products. These two regions contributed a total of 4528 strains of Listeria (78.3%). On a smaller scale, the South and Northeast regions totalized 1078 (18.6%) and 164 strains (2.82%), respectively, mainly from meat and dairy products. Among the states with isolated Listeria spp. in dairy products, São Paulo and Rio Grande do Sul contributed with more strains. While São Paulo had a similar number of Listeria strains from milk and cheese, in Rio Grande do Sul cheese was the major source (OR = 8.8, 95% CI = 5.2–14.9, p < 0.01).
The characterization of the species in the period of 1990–2012 (Figure 1) revealed the prevalence of L. innocua (3429, 59.4%) and L. monocytogenes (2248, 38.9%) and discrete occurrences of L. welshimeri, L. seeligeri, and L. grayi subsp. murrayi (93, 1.6%). However, L. ivanovii was not isolated from any of the food materials.
Figure 1.
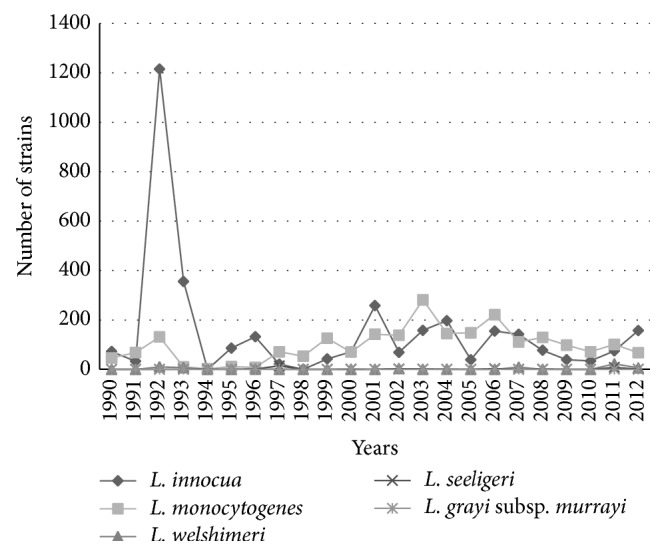
Temporal distribution of Listeria spp. isolated from food in Brazil from 1990 to 2012.
The analysis of the trend isolation of L. monocytogenes and L. innocua during two periods, 1990–2000 and 2001–2012, (number of strains in the period/total strains for the species) showed that L. innocua in the first period amounted to 58.9% (2029/3429) and to 40.8% (1400/3429) in the second period. As for L. monocytogenes, it amounted to 26.5% (596/2248) in the first period and reached 73.4% (1652/2248) in the second (OR = 3.92, 95% CI = 3.50–4.39, p < 0.01).
In species distribution by geographical origin (Table 2), four states, Paraíba, Mato Grosso, São Paulo, and Rio Grande do Sul, had higher levels of L. monocytogenes strains (1456 of 2030 strains, 71.7%), surpassing the other Listeria spp. isolated, while this was not observed in the other states (792 of 3740 strains, 21.2%) (OR = 9.4, 95% CI = 8.3–10.7, p < 0.01). In contrast, in the other states, the prevalence of L. innocua (2876 of 3740 strains, 76.9%) was higher (OR = 8.9, 95% CI = 8.9–10.1, p < 0.01).
Table 2.
Geographical distribution of Listeria spp. isolates, in Brazil. 1990–2012.
| States | Species | Total | |||||
|---|---|---|---|---|---|---|---|
| L. innocua | L. monocytogenes | L. welshimeri | L. seeligeri | L. grayi subsp. murrayi | N | % | |
| Alagoas | 22 | 12 | 34 | 0.58 | |||
| Bahia | 57 | 6 | 2 | 65 | 1.12 | ||
| Paraíba | 6 | 27 | 33 | 0.57 | |||
| Pernambuco | 28 | 4 | 32 | 0.55 | |||
| Goiás | 1350 | 169 | 15 | 1534 | 26.58 | ||
| Mato Grosso | 64 | 216 | 3 | 283 | 4.9 | ||
| Minas Gerais | 763 | 154 | 3 | 2 | 922 | 15.97 | |
| Rio de Janeiro | 330 | 259 | 14 | 603 | 10.45 | ||
| São Paulo | 334 | 837 | 13 | 2 | 1186 | 20.55 | |
| Paraná | 28 | 14 | 3 | 45 | 0.77 | ||
| Santa Catarina | 298 | 174 | 25 | 8 | 505 | 8.75 | |
| Rio Grande do Sul | 149 | 376 | 3 | 528 | 9.15 | ||
|
| |||||||
| Total | |||||||
| N | 3429 | 2248 | 59 | 32 | 2 | 5770 | 99.8 |
| % | 59.42 | 38.96 | 1.02 | 0.55 | 0.03 | ||
Based on the regional distribution of the species, of the 4692 strains from Northeast, Midwest, and Southeast regions, 2954 (62.9%) were identified as L. innocua, while 1684 (35.8%) were identified as L. monocytogenes. In the South region, with 1078 strains, L. monocytogenes was more frequent (564-52.3%) than L. innocua (475-44%) (OR = 2.1, 95% CI = 1.8–2.4, p < 0.01), a result biased by the sampling from Rio Grande do Sul.
Regarding the distribution of L. monocytogenes serotypes in food products, Table 3 shows that those referred to as the most potentially pathogenic isolates to human consumers, 1/2a, 1/2b, and 4b, were more prominent at 30.3% (1751/5770), rising to 36.8% when serotype 1/2c samples (2129/5770) were included in the calculation. Moreover, the predominance of serotype 4b on most products is evident, except in processed meat, where serotype 1/2a was predominant. When we considered all products, the serotypes 1/2a and 4b presented a similar contribution (10,97% and 11,29%, resp.).
Table 3.
Species and serovar frequency of Listeria spp. isolates from food, in Brazil. Period 1990–2012.
| Food type | L. innocua | L. monocytogenes | L. welshimeri | L. seeligeri | L. grayi | Total | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6a | 6b | NT∗ | 1/2a | 1/2b | 1/2c | 3a | 3b | 3c | 4b | 4c | 4e | 7 | NT∗ | R∗∗ | N | % | ||||
| Meat: | ||||||||||||||||||||
| Nonprocessed | 1939 | 90 | 233 | 244 | 250 | 140 | 6 | 20 | 2 | 275 | 28 | 1 | 2 | 4 | 51 | 25 | 3310 | 57.37 | ||
| Processed | 615 | 53 | 27 | 239 | 113 | 144 | 5 | 8 | 163 | 2 | 4 | 18 | 8 | 5 | 1404 | 24.33 | ||||
| Ready-to-eat | 387 | 38 | 45 | 139 | 90 | 80 | 1 | 195 | 13 | 1 | 4 | 2 | 2 | 997 | 17.28 | |||||
| Unknown | 2 | 11 | 13 | 14 | 19 | 59 | 1.02 | |||||||||||||
|
| ||||||||||||||||||||
| Total | ||||||||||||||||||||
| N | 2943 | 181 | 305 | 633 | 466 | 378 | 11 | 29 | 2 | 652 | 2 | 45 | 2 | 24 | 4 | 59 | 32 | 2 | 5770 | 100 |
| % | 51.0 | 3.14 | 5.29 | 10.97 | 8.08 | 6.55 | 0.19 | 0.51 | 0.04 | 11.29 | 0.04 | 0.78 | 0.04 | 0.41 | 0.06 | 1.02 | 0.55 | 0.04 | ||
∗NT = nontypable (smooth form).
∗∗R = rough form.
The serotypes 3a, 3b, 3c, 4c, 4e, and 7 and nontypable samples, with rough R-forms, reached a minimum level (119–2.06%), and the isolation of the serotype 4e in milk samples from São Paulo and unprocessed meats (beef and chicken) from Rio Grande do Sul is also noteworthy.
L. innocua constituted the largest number of strains (3429 of 5770 strains, 59.4%), of which 2943 were identified as serotype 6a, isolated mainly in unprocessed (poultry) and processed meat (sausages), totaling 2554 of 2943 strains (86,8%).
4. Discussion
In the period of 1990–2000, several human listeriosis outbreaks were described in the world with food products as their source; consequently, an intense policy with the implementation of control measures in food industries by Hazard Analysis and Critical Control Points (HACCP) and risk analysis were adopted mainly in Europe and North America. In addition, there are specific recommendations for persons at higher risk for listeriosis. There was also an effort to strengthen quantitative microbiological criteria, with a zero tolerance policy for foods that support and favor the multiplication of bacteria [3, 23]. It should be noted that the policy of zero tolerance has been criticized on the basis that the level of contamination by L. monocytogenes in marketed products is often very low and in some developed countries this measure revealed no significant change on listeriosis incidents [24].
In the present study, the laboratory received suspected isolates of Listeria spp. However, the sender did not inform if they had established a standard microbiological acceptability of the product or product batches based on it or the presence or number of bacterial masses per unit area or batch. There was also no information about where the products came from, if they were from other geographical areas of Brazil, or if they were imported or would be exported.
The data in Table 1 shows how common members of the genus Listeria isolated from foods are in the four regions of Brazil. The lack of samples from the North of Brazil does not indicate that this microorganism is not present in this area. In fact, [25] reported the isolation of Listeria spp. in beef in the city of Belém, state of Pará (one of the main cities from Northern Brazil).
Quantitative variations occurred mainly due to the higher concentration of industries that processed food products of animal origin, in particular meat products, mainly from Middle-West, Southeast, and South regions and these are sold to domestic and international markets. On a smaller scale, but present in all regions examined, are dairy products; their marketing is more restricted to the area surrounding the manufacturing site and many products are identified as artisanal.
From an epidemiological point of view, the movement of food carriers is probably an important route for L. monocytogenes spread to domestic consumers and those from other regions and/or countries [26].
As for the temporal distribution of isolates characterized into species in the two periods (1990–2000 and 2001–2012) (Figure 1), L. innocua had marked predominance in the first period when compared to L. monocytogenes. In the second period, the opposite was seen, and we hypothesized that a possible cause for the difference was the improvement of isolation technique by the senders of the samples, as a result of implemented methods for analysis of foods such as the introduction of new selective culture media [27, 28]. Another important detail was that the technical guidance stating that a number of 5 or more suspected isolated colonies in a selective media should be screened for Listeria spp. decreased the number of false-positives. In the confirmatory phenotypic analysis, the hemolysis test would frequently yield false-negative results, and this is a critical test in the differentiation between L. monocytogenes and L. innocua. To overcome this problem, the CAMP test, the alanyl peptidase detection, or molecular typing [28] is recommended.
As for the other species (Figure 1 and Table 2), there was a low incidence of L. welshimeri, L. seeligeri, and L. grayi subsp. murrayi (93 strains, 1.6%) and absence of L. ivanovii isolates, according to the findings of Gianfranceschi et al., 2003 [29]. In Table 2 it was observed that four states located in each region revealed the prevalence of L. monocytogenes, in opposition to the other areas where L. innocua had a higher incidence. Analyzing the results, it can be assumed that there exists a correlation with the type of product, especially meat products (processed and unprocessed) of avian origin, particularly composed of poultry meat sausages. This situation is depicted in isolates from Bahia, Goiás, Minas Gerais, Rio de Janeiro, and Santa Catarina. It was observed that in isolates from unprocessed meat products (cattle, sheep, and pigs) the occurrence of L. innocua was not as relevant and in some situations L. monocytogenes had a higher incidence in meat products from avian sources in states of Mato Grosso, São Paulo, and Rio Grande do Sul. A hypothesis for the variations in prevalence of the species, whatever the food analyzed is, is that L. innocua, when present in equal or greater numbers, tends to overgrow L. monocytogenes during the stages of selective enrichment, resulting in a smaller number of L. monocytogenes colonies and greater difficulty in viewing them in isolation medium [28].
The presence of L. monocytogenes serotypes isolated from various types of food (Table 3) reproduces previous observations from both national [6–8, 11, 16, 17] and international investigations [3, 21, 30] demonstrating the predominant and cosmopolitan nature of the 1/2a, 1/2b, and 4b serotypes, but with increased detection of serotype 1/2c. Overall, these serotypes were found in all products analyzed, with serotype 4b isolates prevailing in ready-to-eat products, serotype 1/2a prevailing in processed meat, and a similar distribution of the serotypes 1/2a, 1/2b, and 4b in nonprocessed meat.
From an epidemiological standpoint, serotyping has a very limited discriminatory level, and only three serotypes (1/2a, 1/2b, and 4b) have a predominant role in human disease processes [31], although presumably serotyping could allow investigators to understand changes in the temporal occurrence of serotypes by geographical region and a particular food type. Nevertheless, serotyping information is necessary to establish the association of serotypes and outbreaks [32].
The observations of Parihar et al. [32] are supported by numerous references about the predominance of serotype 1/2a in clinical cases in Sweden, perhaps as a result of its predominance in many foods, including processed foods. This situation was also reported in other European countries and in the U.S., with serotype 1/2a isolates from food and clinical cases reported more frequently than 4b [23], in gastrointestinal listeriosis [3], or non-outbreak-associated cases of L. monocytogenes infection [28]. In our research, the difference between the two serotypes (4b with 652 isolates and 1/2a with 633 isolates) is not significant, even when the isolates related to food without identification are not considered. The frequency of serotypes 1/2b and 1/2c was consistent with previous findings with meat products as the main source of isolation [6, 8, 29]. The antigenic characterization of the predominant species in isolates, L. innocua, demonstrated the clear predominance of serotype 6a, mainly in isolates from unprocessed meat products (poultry) and processed meat (sausages containing beef and poultry) as well as in cheeses. The prevalence of other species, identified as L. welshimeri, L. seeligeri, and L. grayi subsp. murrayi, was very low; they were mainly found in nonprocessed meat products.
The association of the resistance of Listeria spp. to the conditions imposed by the environment and its wide dissemination in animal sources is a serious problem for the food industry. This situation is a result of the high level of contamination of raw materials originating from animals and vegetables, as these microorganisms may persist for variable periods in the industrial environment. Of particular importance is the ability of L. monocytogenes to adhere to surfaces and form biofilm [33]. In the food markets and home environments, a flaw in the cold chain constitutes a risk factor for the growth and dissemination of Listeria [3, 34].
The situation depicted by this study demonstrates how easy the transmission of L. monocytogenes to the consumer is and the underlying reasons for sporadic cases and outbreaks of listeriosis [3, 35]. Indeed, individuals affected by Listeria monocytogenes must encounter the conditions recognized as risk factors [36]. Based on this, attention should be focused on the analysis of risk assessments for ready-to-eat foods, including the establishment of quantitative microbiological criteria as adopted in the U.S. and Europe [3, 35, 37, 38].
Given the results related to ready-to-eat products (Table 3), there is no doubt that consumers are exposed to potential risks of infection, especially the most susceptible population: immune suppressed patients, individuals on extreme age groups, and pregnant women. Furthermore, it is important to take into account the ingested dose of viable bacteria, in addition to the characteristics of the host. This is influenced by the period that the products are kept under refrigeration and the ability of the bacteria to grow in these products [37]. The adoption of a program to reduce the salt concentration in food products (to reduce hypertension) in France raised the hypothesis that this public health measure may have increased the chance of L. monocytogenes contamination in foods (mostly meat and fish) and increased the disease incidence [38, 39].
Although the results presented here point to the presence of the main serovars of L. monocytogenes in different foods, there are no reports of outbreaks of listeriosis in the country, only a few sporadic cases. This fact can be explained by the absence of mandatory reporting in cases of listeriosis, since there is no compulsory notification for cases of listeriosis in Brazil, and also brings up the possibility that those responsible for the diagnosis do not have proper training to identify this pathogen.
In summary, our work depicts a current overview of the genus Listeria distribution in primary foods used as raw material in large national food industry that remain in the final product even after processing.
The eradication of L. monocytogenes in foods and processing environments is a difficult task due to the ability of this pathogen to adapt to different harsh conditions. Therefore, it is fundamental the application of HACCP and GMP (Good Manufacturing Practices) programs in industries, a stronger microbiological surveillance and greater dissemination of information about listeriosis (especially for risk groups). Furthermore a network bringing together the public health and food surveillance is necessary in order to gather more efforts to monitor and reduce the risks of food contamination by L. monocytogenes in Brazil.
Acknowledgments
The authors thank all the microbiology laboratories in Brazil for sending the Listeria isolates and Evaldo Soares da Silva and Sérgio Alves de Azevedo from the Bacterial Zoonosis Laboratory for their technical collaboration. Partial financial support was obtained from the Brazilian National Scientific and Technological Research Council (CNPq), process 301545/2006-5.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Velge P., Roche S. M. Variability of Listeria monocytogenes virulence: a result of the evolution between saprophytism and virulence? Future Microbiology. 2010;5(12):1799–1821. doi: 10.2217/fmb.10.134. [DOI] [PubMed] [Google Scholar]
- 2.Farber J. M., Peterkin P. I. Listeria monocytogenes, a food-borne pathogen. Microbiological Reviews. 1991;55(3):476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swaminathan B., Gerner-Smidt P. The epidemiology of human listeriosis. Microbes and Infection. 2007;9(10):1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Mead P. S., Slutsker L., Dietz V., et al. Food-related illness and death in the United States. Emerging Infectious Diseases. 1999;5(5):607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esper M. R. N. R., Pess G. V. A., Hofer E., et al. Meningite por Listeria monocytogenes em São Paulo, Brasil. Revista do Instuto Adolfo Lutz. 1978;38:37–41. [Google Scholar]
- 6.Hofer E., Ribeiro R., Feitosa D. P. Species and serovars of the genus Listeria isolated from different sources in Brazil from 1971 to 1997. Memórias do Instituto Oswaldo Cruz. 2000;95(5):615–620. doi: 10.1590/s0074-02762000000500005. [DOI] [PubMed] [Google Scholar]
- 7.Lemes-Marques E. G., Cruz C. D., Destro M. T. Pheno- and genotypic characterization of Listeria monocytogenes clinical isolates from the Southwestern region of the State of São Paulo, Brazil. Brazilian Journal of Microbiology. 2007;38(2):287–292. doi: 10.1590/s1517-83822007000200019. [DOI] [Google Scholar]
- 8.Bueno V. F., Banerjee P., Banada P. P., José de Mesquita A., Lemes-Marques E. G., Bhunia A. K. Characterization of Listeria monocytogenes isolates of food and human origins from Brazil using molecular typing procedures and in vitro cell culture assays. International Journal of Environmental Health Research. 2010;20(1):43–59. doi: 10.1080/09603120903281283. [DOI] [PubMed] [Google Scholar]
- 9.Landgraf I. M., Kobata A. M. M., Jakabi M., Kirschbaum C. R. A., Marchi C. R. Surto de meningite neonatal por Listeria monocytogenes . Revista do Instituto Adolfo Lutz. 1999;58:63–67. [Google Scholar]
- 10.Martins I. S., Faria F. C. C., Miguel M. A. L., et al. A cluster of Listeria monocytogenes infections in hospitalized adults. The American Journal of Infection Control. 2010;38(9):31–36. doi: 10.1016/j.ajic.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Destro M. T., de Melo Serrano A., Kabuki D. Y. Isolation of Listeria species from some Brazilian meat and dairy products. Food Control. 1991;2(2):110–112. doi: 10.1016/0956-7135(91)90147-o. [DOI] [Google Scholar]
- 12.da Silva M. C. D., Hofer E., Tibana A. Incidence of Listeria monocytogenes in cheese produced in Rio de Janeiro, Brazil. Journal of Food Protection. 1998;61(3):354–356. doi: 10.4315/0362-028x-61.3.354. [DOI] [PubMed] [Google Scholar]
- 13.Catão R. M. R., Ceballos B. S. O. Pesquisa de Listeria spp., coliformes totais e fecais e Escherichia coli no leite cru e pasteurizado de uma indústria de laticínios, no estado da Paraíba (Brasil) Ciência e Tecnologia de Alimentos. 2001;21(3):281–287. [Google Scholar]
- 14.Barbalho T. C. F., Almeida P. F., Almeida R. C. C., Hofer E. Prevalence of Listeria spp. at a poultry processing plant in Brazil and a phage test for rapid confirmation of suspect colonies. Food Control. 2005;16(3):211–216. doi: 10.1016/j.foodcont.2004.01.014. [DOI] [Google Scholar]
- 15.Chiarini E., Tyler K., Farber J. M., Pagotto F., Destro M. T. Listeria monocytogenes in two different poultry facilities: manual and automatic evisceration. Poultry Science. 2009;88(4):791–797. doi: 10.3382/ps.2008-00396. [DOI] [PubMed] [Google Scholar]
- 16.Barancelli G. V., Camargo T. M., Reis C. M. F., Porto E., Hofer E., Oliveira C. A. F. Incidence of Listeria monocytogenes in cheese manufacturing plants from the northeast region of São Paulo, Brazil. Journal of Food Protection. 2011;74(5):816–819. doi: 10.4315/0362-028x.jfp-10-489. [DOI] [PubMed] [Google Scholar]
- 17.Sant'Ana A. S., Igarashi M. C., Landgraf M., Destro M. T., Franco B. D. G. M. Prevalence, populations and pheno- and genotypic characteristics of Listeria monocytogenes isolated from ready-to-eat vegetables marketed in São Paulo, Brazil. International Journal of Food Microbiology. 2012;155(1-2):1–9. doi: 10.1016/j.ijfoodmicro.2011.12.036. [DOI] [PubMed] [Google Scholar]
- 18.Moreno L. Z., Paixão R., Gobbi D. D., et al. Characterization of atypical Listeria innocua isolated from swine slaughterhouses and meat markets. Research in Microbiology. 2012;163(4):268–271. doi: 10.1016/j.resmic.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 19.RDC. Resolution-RDC. 12. Brasília, Brazil: National Agency of Sanitary Surveillance (ANVISA); 2001. Technical regulation on microbiological standards for foods. [Google Scholar]
- 20. Instrução Normativa no. 09, de 8 de abril de 2009. Ministério da Agricultura Pecuária e Abastecimento (MAPA) http://www.agricultura.gov.br/arq_editor/Programa_de_Listeria%281%29.pdf.
- 21.Hitchins A. D. Bacteriological Analytical Manual. chapter 10. US Food and Drug Administration (FDA); 2013. Detection and enumeration of Listeria monocytogenes in foods. http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm071400.htm. [Google Scholar]
- 22.Seeliger H. P. R., Höhne K. Serotyping of Listeria monocytogenes and related species. Methods in Microbiology. 1979;13:31–49. doi: 10.1016/s0580-9517(08)70372-6. [DOI] [Google Scholar]
- 23.Voetsch A. C., Angulo F. J., Jones T. F., et al. Reduction in the incidence of invasive listeriosis in foodborne diseases active surveillance network sites, 1996–2003. Clinical Infections Diseases. 2007;44:513–520. doi: 10.1086/511006. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y., Ross W. H., Scott V. N., Gombas D. E. Listeria monocytogenes: low levels equal low risk. Journal of Food Protection. 2003;66(4):570–577. doi: 10.4315/0362-028x-66.4.570. [DOI] [PubMed] [Google Scholar]
- 25.Maroja O. M., Abreu A. C. V. V., Mendonça W. B. Encontro de um organismo identificado com Listeria monocytogenes em bovinos do Pará. Revista do Serviço. Especializado em Saúde Pública. 1962;12:179–184. [Google Scholar]
- 26.Varma J. K., Samuel M. C., Marcus R., et al. Listeria monocytogenes infection from foods prepared in a commercial establishment: a case-control study of potential sources of sporadic illness in the United States. Clinical Infectious Diseases. 2007;44(4):521–528. doi: 10.1086/509920. [DOI] [PubMed] [Google Scholar]
- 27.Vlaemynck G., Lafarge V., Scotter S. Improvement of the detection of Listeria monocytogenes by the application of ALOA, a diagnostic, chromogenic isolation medium. Journal of Applied Microbiology. 2000;88(3):430–441. doi: 10.1046/j.1365-2672.2000.00978.x. [DOI] [PubMed] [Google Scholar]
- 28.Scotter S. L., Langton S., Lombard B., et al. Validation of ISO method 11290. Part 2. Enumeration of Listeria monocytogenes in foods. International Journal of Food Microbiology. 2001;70(1-2):121–129. doi: 10.1016/s0168-1605(01)00530-x. [DOI] [PubMed] [Google Scholar]
- 29.Gianfranceschi M., Gattuso A., Tartaro S., Aureli P. Incidence of Listeria monocytogenes in food and environmental samples in Italy between 1990 and 1999: serotype distribution in food, environmental and clinical samples. European Journal of Epidemiology. 2003;18(10):1001–1006. doi: 10.1023/a:1025849532417. [DOI] [PubMed] [Google Scholar]
- 30.Rocourt J. Listeriosis 1985–1995: microbiologic and epidemiologic aspects. Bulletin de l'Académie Nationale de Médecine. 1995;179(8):1613–1624. [PubMed] [Google Scholar]
- 31.Norton D. M., Scarlett J. M., Horton K., et al. Characterization and pathogenic potential of Listeria monocytogenes isolates from the smoked fish industry. Applied and Environmental Microbiology. 2001;67(2):646–653. doi: 10.1128/aem.67.2.646-653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parihar V. S., Lopez-Valladares G., Danielsson-Tham M.-L., et al. Characterization of human invasive isolates of Listeria monocytogenes in Sweden 1986–2007. Foodborne Pathogens and Disease. 2008;5(6):755–761. doi: 10.1089/fpd.2008.0123. [DOI] [PubMed] [Google Scholar]
- 33.Borucki M. K., Peppin J. D., White D., Loge F., Call D. R. Variation in biofilm formation among strains of Listeria monocytogenes . Applied and Environmental Microbiology. 2003;69(12):7336–7342. doi: 10.1128/aem.69.12.7336-7342.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salvat G., Fravalo P. Risk assessment strategies for Europe: integrated safety strategy or final product control: example of Listeria monocytogenes in processed products from pork meat industry. Deutsche Tierarztliche Wochenschrift. 2004;111(8):331–334. [PubMed] [Google Scholar]
- 35.Le Monnier A., Leclercq A. Listeria et listériose: des animaux d'élevage à nos assiettes. Pathologie Biologie. 2009;57(1):17–22. doi: 10.1016/j.patbio.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 36.FAO. Risk characterization of Salmonella spp. in eggs and broiler chickens and Listeria monocytogenes in ready-to-eat foods. Proceedings of the Joint FAO/WHO Expert Consultation on Risk Assessment of Microbiological Hazards in Foods; April-May 2001; Rome, Italy. FAO; FAO Food and Nutrition Paper. [Google Scholar]
- 37.Rocourt J., BenEmbarek P., Toyofuku H., Schlundt J. Quantitative risk assessment of Listeria monocytogenes in ready-to-eat foods: the FAO/WHO approach. FEMS Immunology and Medical Microbiology. 2003;35(3):263–267. doi: 10.1016/s0928-8244(02)00468-6. [DOI] [PubMed] [Google Scholar]
- 38.Goulet V., Hedberg C., Le Monnier A., de Valk H. Increasing incidence of listeriosis in France and other European countries. Emerging Infectious Diseases. 2008;14(5):734–740. doi: 10.3201/eid1405.071395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goulet V. What can we do to prevent listeriosis in 2006? Clinical Infectious Diseases. 2007;44(4):529–530. doi: 10.1086/509932. [DOI] [PubMed] [Google Scholar]


