Abstract
Insights into inflammatory bowel disease (IBD) are advancing rapidly owing to immunologic investigations of a plethora of animal models of intestinal inflammation, ground-breaking advances in the interrogation of diseases that are inherited as complex genetic traits, and the development of culture-independent methods to define the composition of the intestinal microbiota. These advances are bringing a deeper understanding to the genetically determined interplay between the commensal microbiota, intestinal epithelial cells, and the immune system and the manner in which this interplay might be modified by relevant environmental factors in the pathogenesis of IBD. This review examines these interactions and, where possible, potential lessons from IBD-directed, biologic therapies that may allow for elucidation of pathways that are central to disease pathogenesis in humans.
Keywords: Crohn’s disease, ulcerative colitis, intestinal inflammation, genetics, microbiota
INTRODUCTION
The two major clinically defined forms of inflammatory bowel disease (IBD), Crohn’s disease (CD) and ulcerative colitis (UC), are chronic remittent or progressive inflammatory conditions that may affect the entire gastrointestinal tract and the colonic mucosa, respectively, and are associated with an increased risk for colon cancer. IBD has long been appreciated to have a genetic basis and likely involves a response of the immune system to some environmental agent(s). The discordance of IBD among monozygotic twins (1) and the development of IBD in immigrants to high-prevalence countries (2) and in countries undergoing rapid Westernization also highlights the importance of environmental factors in disease pathogenesis (3).
The discovery that interleukin-2 (IL-2), IL-10, or T cell receptor (TCR) (4–6) mutant mice develop IBD-like enterocolitis, and the success of tumor necrosis factor (TNF)-α blockade in treating patients with CD, stimulated a new era of investigation in the early 1990s. Mechanisms deduced from numerous animal models (7) could be tested for relevance in human IBD by target-specific biologics (8). Since the recent dramatic expansion of studies into the genetic basis of complex diseases such as IBD (9), and the possibility to study the intestinal microbiome by sequencing (10), the pace of pathophysiologic discovery has further quickened. Genetically based interactions between the human intestinal microbiome and mucosal immune system and the manner in which environmental factors modify these relationships appear particularly relevant for the development of IBD. Among the insights that have emerged is the central role played by the innate immune system and its relationship to the commensal microbiota and adaptive immune system in the initiation and perpetuation of IBD. This review aims to integrate recent discoveries in the genetics, microbiology, and immunobiology of IBD together with lessons learned from the application of biologic therapies to emphasize the dynamic relationships of each of these components and the importance of considering them in their totality in order to understand the pathogenesis of these disorders.
GENETIC BASIS OF IBD
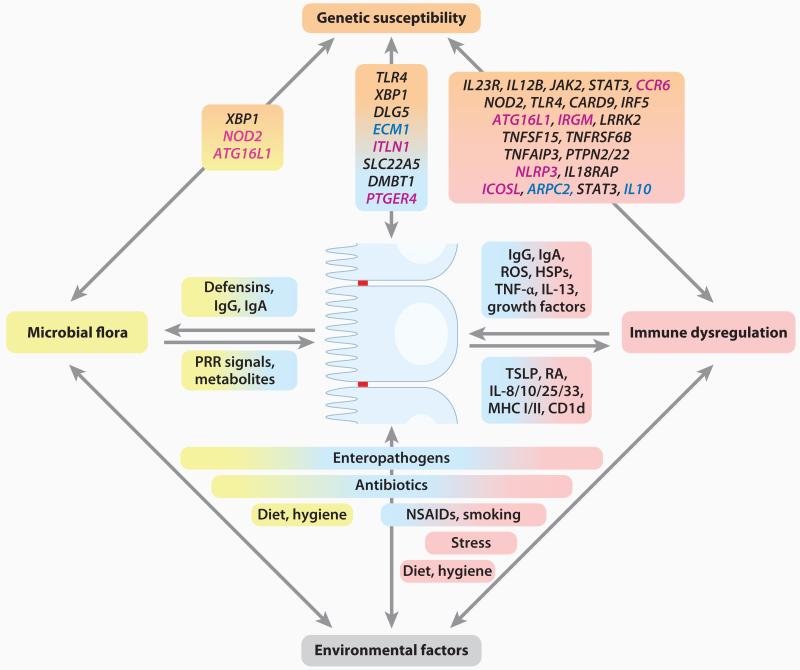
Both types of IBD occur in genetically susceptible individuals through interplay with poorly understood environmental factors. IBD, considered a polygenic disorder, is familial in 5–10% of individuals and sporadic in the remainder (1). Monozygotic twins exhibit phenotypic concordance in 50–75% of CD patients, and the relative risk of developing CD is 800-fold greater compared to the general population (1). In UC, phenotypic concordance in monozygotic twins is less frequent (10–20%), suggesting that heritability is less important in UC, that the relevant environmental exposure(s) is less common, or that copy number variations and/or epigenetic differences between twin pairs are more frequent, thus limiting the possibility for true concordance (1). Genetic studies, including candidate gene approaches, linkage mapping studies, and in particular genome-wide association studies (GWASs), have significantly advanced our understanding on the importance of genetic susceptibility in IBD (11). The GWASs performed to date together with a meta-analysis of several GWASs involving CD have identified more than 30 risk-conferring loci (see References 9, 11, and 12 for recent reviews of IBD genetics). These studies highlight pathways previously identified through immunologic studies [e.g., IL-23 and T helper (Th) 17 cells (13)], but have also discovered previously unappreciated pathways such as autophagy (14), raising novel hypotheses about disease pathogenesis (summarized in Figure 1 and in Supplemental Table 1; follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org).
Figure 1.
Inflammatory bowel disease (IBD) as a multifactorial disorder. The development and course of IBD are affected by several factors, including genetic susceptibility of the host, the intestinal microbiota, other environmental factors, and the host immune system. In addition, these factors cross-regulate each other in multiple ways, as shown. IBD-associated genes are summarized by molecular pathways with genes belonging to the same pathway arranged next to each other in one line. Polymorphisms in genes specific for Crohn’s disease (CD) are shown in magenta text, whereas those specific for ulcerative colitis (UC) are shown in dark blue text. Genetic associations shared between both diseases are shown in black text. Abbreviations: HSPs, heat shock proteins; MHC, major histocompatibility complex; NSAIDs, nonsteroidal anti-inflammatory drugs; PRR, pattern-recognition receptor; RA, retinoic acid; ROS, reactive oxygen species; TSLP, thymic stromal lymphopoietin.
Interestingly, GWASs have also revealed a substantial overlap in genetic risk factors between CD and UC (15, 16). However, it is possible that these similarities are not shared at the level of structurally or functionally relevant polymorphisms because causal variants are mostly unknown. The wide phenotypic diversity of cystic fibrosis associated with diverse CFTR variants might serve as precedent (17). However, some loci are quite unique for CD or UC. For example, autophagy genes (e.g., ATG16L1, IRGM), NOD-like receptors (e.g., NOD2), and intelectins (ITLN1) are highly specific for CD, whereas loci related to regulatory pathways (IL10 and ARPC2), intestinal epithelial cell (IEC) function (e.g., ECM1), and an E3 ubiquitin ligase (e.g., HERC2) appear to be specific for UC (Supplemental Table 1). Moreover, associations within the HLA/MHC region are stronger with UC compared to CD, a genetic trait of IBD shared with a number of other autoimmune diseases (9, 15). Although IBD is classified as an immune-mediated disease, there is no evidence to date that autoimmunity plays a direct pathogenic role in either UC or CD despite the existence of detectable autoantibodies that are cross-reactive with bacterial antigens (18). These genetic associations within the HLA/MHC region raise the possibility, however, that autoimmunity may ultimately be defined as another pathogenetic mechanism.
In addition to the HLA/MHC region, a number of other IBD risk loci are also associated with a diverse set of immune-related diseases. These include type 1 diabetes mellitus (e.g., PTPN2 and PTPN22), type 2 diabetes mellitus (e.g., CDKAL1), asthma (e.g., ORMDL3), psoriasis (e.g., CDKAL1 and GCKR), systemic lupus erythematosus (e.g., PTPN22), Graves’ disease (e.g., PTPN22), and rheumatoid arthritis (e.g., PTPN22) (9, 15, 19). The sharing of associations among different diseases implies that a general inherited propensity to develop immune-related diseases may exist and that environmental (or epigenetic) factors may determine not only disease phenotype but also the specific immune-mediated disease that develops. Moreover, IBD risk loci vary remarkably between different populations. For example, NOD2 and autophagy genes, the major risk loci in the Caucasian population, are not susceptibility factors in the Asian population (3). Hence, despite commonalities in the genetic basis of CD and UC (15), substantial genetic heterogeneity exists within and between populations. However, despite genotypic differences among various populations, the clinicopathologic phenotype is largely similar, as is the overall response to various therapies (3). This might predict that a large variety of genotypes converge on a limited set of phenotypic pathways that are responsible for initiating disease and are amenable to therapeutic manipulation.
A striking but potentially instructive outcome of GWASs is that the vast majority of identified loci individually confer extremely modest risk [odds-ratios (ORs), mostly between 1.11 and 1.29]. Collectively, the loci identified to date represent ≈10–20% of the overall variance of potential disease risk; a dominant contribution is provided by the three common NOD2 variants (20). Moreover, for most of the confirmed loci the causal gene(s) or variant(s) (ranging from rare to common) is not yet known (9, 11, 12). This “missing heritability” has led to at least two potential interpretations. The genetic basis for common phenotypic traits such as sporadic IBD may be due to the cumulative effect of interactions between an unknown quantity of potentially hundreds or thousands of common single nucleotide polymorphisms (SNPs) of minor individual biologic impact (21) and/or that IBD, especially the familial form, may be due to the effects of rare variants with profound impact that may be modified by more common variants (21, 22). In this latter model, at least a subset of IBD, such as those with a familial pattern of inheritance, may potentially be due to a “more” Mendelian form of heredity, which is supported by several lines of evidence. First, multiple rare primary genetic syndromes with Mendelian inheritance may develop IBD as a part of the syndrome (e.g., Wiskott-Aldrich Syndrome, Hermansky-Pudlak Syndrome, glycogen storage disease type 1b, and immunodeficiency polyendocrinopathy with eczema and X-linked, or IPEX). Second, a familial form of early-onset CD has been recently identified as a monogenic disorder due to homozygous mutations in either IL10RA or IL10RB, which encode subunits of the IL-10 receptor (23). Moreover, these IL-10R variants appear to functionally map to hematopoietic cells, as cure was observed after allogeneic hematopoietic stem cell transplantation, a modality recognized as of potential utility in a select subset of IBD subjects (24). It is therefore interesting that Il10−/− (5) and Il10rb−/− (25) mice as well as mice with Stat3 deletion in macrophages (26) all develop spontaneous intestinal inflammation. Although IL-10R1 (encoded by IL10RA) is unique to IL-10R, IL-10R2 (encoded by IL10RB) is shared with other receptors such as IL-22, which may protect against colitis via goblet cells (27). Thus, rare variants with strong biologic effects and common variants may reside within a functional pathway, as may be the case for IL-10R, IL-10, and STAT3. As spontaneous IBD rarely develops in animal models targeted at IBD susceptibility loci, it may be speculated that those cases that do develop disease in rodents might be monogenically inherited in humans (e.g., IL-10).
ROLE OF THE MICROBIOTA AS A MAJOR ENVIRONMENTAL DRIVER OF IBD
Insight into the genetic basis of IBD has focused attention on the relationships between the immune system and the intestinal microbiota. The intestinal microbiota profoundly affects host immune composition under physiologic conditions and is likely the most important environmental factor in IBD as the target of the inflammatory response (28). This is supported by a wide variety of observations in humans and mouse models, as recently reviewed elsewhere (7, 29). Perhaps most important among these are the observations that numerous genetic mouse models of intestinal inflammation do not develop disease after germ-free rederivation (30), and T cell lines specific for bacterial antigens, but not when nonspecifically activated, can induce intestinal inflammation (31, 32). IBD may represent an inappropriate immune response to the commensal microbiota in a genetically predisposed host. This finding has led to an intense interest in the composition of the intestinal microbiota, its regulation by the host and environmental factors, and the interactions between the microbiota and host.
Regulation of Mucosal Immune Functions by the Commensal Microbiota
Humans (and experimental animals) associate with numerous microorganisms at environmentally exposed surfaces of the body (10). The gastrointestinal tract harbors more than 1014 microorganisms of more than 1000 species (33, 34), mostly contained within the colon and not accessible to conventional culture techniques (10). Most (>90%) belong to two different phyla that account for the majority of gram-negative bacteria (Bacteroidetes) and gram-positive bacteria (Firmicutes); the remainder belong to rarer phyla such as Proteobacteria (containing genuses such as Escherichia and Helicobacter) and Actinobacteria as well as viruses, protists, and fungi (10). Collectively, the microbiota carries out many physiological functions important in mammalian biology (35). In fact, the microbiota is required for the development and differentiation of local and systemic immune and nonimmune components (28). As an example, Bacteroides thetaiotaomicron affects innate immune capabilities by regulating antimicrobial peptide (e.g., angiogenin) expression within the intestinal epithelium through direct activation of Toll-like receptors (TLR) on Paneth cells (36). Similarly, adaptive immune functions within the intestines related to TCRαβ intraepithelial lymphocytes (37), T regulatory cells (Tregs) (38), and Th17 cells (39–41) are determined by specific bacteria, although the mechanisms behind these effects cannot yet be explained by simple rules. Systemic immune responses are also impaired in germ-free mice, including the development of adequate Treg responses leading to increased systemic autoimmunity (38, 42), which may have implications for the development of extraintestinal manifestations in IBD. An example of a microbial mechanism that affects host inflammatory responses and that is also affected by dietary intake, i.e., environmental factors, is that associated with short-chain fatty acids (SCFA). SCFA derive from microbial fermentation of dietary fiber, bind to G protein–coupled receptor 43 (GPR43), and play a profound role in various inflammatory conditions, such as colitis, arthritis, and asthma (43). Consequently, Gpr43−/− mice, similar to germ-free mice that lack SCFA, exhibit a profound impairment in the resolution of inflammation (43). Overall, the commensal microbiota has major effects on the composition and function of innate and adaptive immune pathways as they may relate to IBD.
Determinants of Commensal Microbiota Composition: Nature and/or Nurture?
Large throughput and next generation sequencing of variable regions of microbial 16S rRNA have allowed for detailed insights into the composition of the intestinal microbiota and their functional genes (i.e., microbiome) in animal models and humans (10). These studies reveal an astounding degree of complexity as defined by phylotypes, with remarkable interindividual differences observed even in healthy subjects (34) and an important role for environmental as well as genetic factors in determining the microbial niche. Specifically, twin studies revealed only a slightly decreased similarity of community structure in dizygotic compared to monozygotic twins, while exhibiting substantial similarity with their mothers (34). This finding suggests that microbial commensalism is largely “inherited” from the mothers and modified by genetic and other environmental (e.g., dietary) factors. Despite the significant interindividual differences at the phylotype level, there is an apparent effort on the part of the host and microbial communities to achieve the existence of a defined “core microbiome” of predicted metabolic functionality that is shared under disparate genotypes (34). This is perhaps the most important virtue of the microbiota in exerting its influences on homeostasis and/or disease. How this attribute relates to IBD has not yet been studied (10).
Animal studies do support the notion that host factors affect microbial commensalism, however. For example, Cd1d−/− mice exhibit an overgrowth of commensal bacteria upon monocolonization from the germ-free state and an alteration of the overall architecture of their microbial communities (44). This effect might be mediated by Paneth cells given their altered morphology and function in Cd1d−/− mice. Thus, host innate immune factors that act through Paneth cells (44–47) or other (potentially genetically defined) factors affecting the composition of mucins (48) or microbial adherence to IECs (49) may also be determinants of the microbial niche. One example of altered colonization is the upregulation of carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) on the cell surface of IECs in IBD, which serves as ligand for certain Proteobacteria such as enteroadherent Escherichia coli that may bloom in a subset of individuals with intestinal inflammation (50). CEACAM6 on IECs likely accounts for the localization of enteroadherent E. coli on inflamed epithelium adjacent to ulcerated areas, implying an important secondary factor in further promoting intestinal inflammation (50). Similarly, adaptive immune factors such as secretory IgA may also affect commensalism, and the commensals in turn drive the generation of secretory IgA (51).
Composition of Commensal Microbiota in Intestinal Inflammation and IBD
Recent studies have sought to determine whether specific alterations can be identified in the intestinal microbiota in IBD. 16S rRNA sequencing revealed a detectable difference between the intestinal microbiota in CD and UC compared to healthy controls (52). This difference in microbial phylotypes largely arises from a distinct subset of CD and UC patients (so-called IBD subset) with the remaining IBD patients being similar to healthy controls, although this awaits further more extensive metagenomic analyses. This IBD subset is characterized by depletion of commensal bacteria with a tenfold lower bacterial load and affects both major classes of commensal phyla, Firmicutes and Bacteroidetes (52). Whether this “dysbiosis” in the IBD subset is associated with particular genotypes (and hence a primary effect antecedent to inflammation) or is a consequence of inflammation per se is unknown (see Table 1). Several genetic loci associated with IBD (e.g., NOD2, ATG16L1, XBP1) affect or have been predicted to affect Paneth cells (e.g., ITLN1 or intelectin 1) (20, 45–47, 53), which secrete abundant quantities of antimicrobial factors. Hence, the function of a number of genes could affect microbial community structure and predispose to inflammation. For example, NOD2 expression is regulated by the microbiota and, in turn, regulates the quantity of bacteria within the intestines, perhaps via Paneth cells (54). However, dysbiosis in the absence of NOD2 appears neither inflammatory in its own right nor secondary to inflammation given the absence of spontaneous inflammation in Nod2−/− mice (47, 55). It is interesting to note that NOD2 polymorphisms were recently identified as risk factors for Mycobacterium paratuberculosis (MAP) infection in cattle (56), which phenocopies IBD of the small intestine (29). However, the major NOD2 polymorphisms linked to CD are not associated with the presence of MAP in the peripheral blood as detected by PCR (29). Nonetheless, these insights suggest that, in the context of inflammation, an overgrowth of certain organisms (e.g., enteroadherent E. coli or MAP) together with altered interactions between these organisms with the host (e.g., increased CEACAM6) may be a relevant secondary factor in driving inflammation in IBD.
Table 1. Changes in the microbial flora in selected human diseases and mouse models of diseasea.
| Decreased abundance | Increased abundance | Total bacterial amount | Sample origin | ||
|---|---|---|---|---|---|
| Human | IBD | Bacteroidetes including Bacteroides thetaiotaomicron (52) Clostridia class of Firmicutes including Faecalibacterium prausnitzii (52, 71, 352-354) and butyrate-producing spp. (52) Reduced diversity (353, 355) |
Proteobacteria including Enterobacteriaceae (relative, not absolute increase) (52, 352, 354, 356) Bacilli class of Firmicutes (52) Increase in mucosal adherent bacteria (particularly in adjacent uninflamed mucosa) (104) |
Decreased in recent 16S rRNA studies (52, 352) Increased in DGGE and FISH studies (104, 357) |
Intestinal tissue |
| Indeterminate colitis | Bacteroidetes (358) | Increase in mucosal adherent bacteria (104, 358) |
Intestinal tissue | ||
| Obesity (359) | Bacteroidetes (increase upon calorie restriction) |
Firmicutes (decrease upon calorie restriction) |
Luminal content | ||
| Mouse | DSS colitis (58) | Bacteroidetes (twofold) | Firmicutes (twofold; includes Lachnospiraceae, Lactobacillaceae families) |
Reduced (0.7-fold) | Colon tissue and luminal content |
| Il10−/− (58, 360) | Bacteroidetes (0.7-fold) | Firmicutes (twofold; includes Lachnospiraceae family) Enterobacteriaceae (Proteobacteria) |
Unchanged | Colon tissue and luminal content |
|
|
Citrobacter rodentium
infection (58) |
Bacteroidetes (threefold) | Enterobacteriaceae (Proteobacteria) | Reduced (fourfold) | Colon tissue and luminal content |
|
| ob/ob mice (361, 362) | Bacteroidetes (0.5-fold, division-wide) |
Firmicutes (division-wide) | Cecal content | ||
|
High fat diet (C57BL/6
versus Relmb−/−) (363) |
Bacteroidetes (diet-induced, independent of obesity) |
Firmicutes (Clostridia class) Proteobacteria (Deltaproteobacteria) (both diet-induced, independent of obesity) |
Fecal pellets | ||
|
Myd88−/− on NOD
background (42) |
Reduced Firmicutes/ Bacteroidetes ratio |
Bacteroidetes (Rikenellaceae and Porphyromonadaceae families) Firmicutes (Lactobacillaceae) |
Cecal content | ||
Abbreviations: DGGE, denaturing gradient gel electrophoresis; DSS, dextran sodium sulfate; FISH, fluorescence in situ hybridization.
A recent study on the transcription factor T-bet (encoded by Tbx21) has provided strong support for the possibility that dysbiosis could contribute to intestinal inflammation. Specifically, T-bet deficiency in the innate immune system together with an absence of Tregs led to spontaneous colitis, which was abrogated by antibiotics, supporting a role for the commensal flora (57). Although colonic dendritic cell (DC)-derived TNF was a critical mediator of the induction of colitis and induced IEC apoptosis via TNFR1, colitis was strikingly vertically (mother-pup) and horizontally (adult-adult) transmitted via the intestinal microbiota from Rag2−/−Tbx21−/− mice to T-bet intact immunodeficient and immunocompetent mice (57). Hence, the colonic environment in Rag2−/−Tbx21−/− mice indeed created a milieu that supported the development of a colitogenic microbial community (57). It is therefore intriguing to speculate that TNF could be a decisive factor in regulating microbial community structure including its colitogenic nature, which has important ramifications for both forms of IBD given the responsiveness of both CD and UC to anti-TNF therapies (see below and in Supplemental Table 2). Although there is little evidence that human IBD is a transmissible disease, these observations suggest that the host can primarily (perhaps through TNF) or secondarily through inflammatory mediators and their consequences (e.g., altered mucin content or antimicrobial peptides) affect the intestinal microbiota and its relationship with the host in a manner that induces or perpetuates inflammation. Regardless of the mechanism that creates this microbial niche, these studies show that the commensal microbiota can assume an overall structure that is inflammatory.
Host inflammation per se, induced by a pathogen (e.g., the model pathogen Citrobacter rodentium), chemically (dextran sodium sulfate, DSS), or genetically through Il10 deletion, leads to profound alterations of colonic microbial community structure (58) (see Table 1). In each of these models, inflammation was associated with a decrease in Bacteroidetes and with maintenance, and thus relative increase, of Proteobacteria, in particular aerobes within Enterobacteriaceae (58). Upon pathogenic invasion, C. rodentium filled the void in the microbial niche normally occupied by commensals (58) and appeared to co-opt the inflammatory response to gain a foothold in the intestinal microenvironment. Such a pathogen-induced dysbiosis could be inflammatory in its own right because it would enable the pathogen to maintain its control of the local environment. If dysbiosis includes the overgrowth of other proinflammatory species such as pathogenic E. coli, it is possible that inflammation is perpetuated through these other inflammatory allies of the invading pathogen until homeostasis is restored upon removal of the inciting agent (50). Because chemically or genetically induced inflammation in animal models and human IBD (52) seemingly phenocopies these pathogen-induced changes in the microbiota, inflammation per se may cause dysbiosis with the aforementioned consequences. These studies also support the idea that the genetically susceptible host with IBD responds to the commensal microbiota as if it were a pathogen.
These observations predict that more severe inflammation might be associated with more profound changes in the microbiota, which in turn would increase the quantity of pathogenic bacteria (i.e., commensal microbiota with pathogenic tendencies), thus perpetuating inflammation. It is interesting to speculate whether the IBD subset described by Frank et al. (52) is indeed associated with more severe inflammation together with a more robust host immune response to the commensal microbiota (e.g., robust IgG response to specific microbial antigens including flagellin or outer membrane protein C) (59). Altogether, these observations raise questions concerning the mechanisms whereby inflammation affects microbial composition.
Mechanisms of Host-Commensal Interactions and Their Relationship to Inflammation
The host mechanisms that provide the niche for the gut microbiota and how these change with inflammation are largely unknown. In Drosophila, five commensal species dominate the gut microbiota, making Drosophila more amenable for study than mice or humans (60). In Drosophila, inhibition of the intestinal homeobox gene Caudal increases NF-κB-dependent antimicrobial peptide expression, which in turn alters the commensal populations in the intestine (60). The consequential dominance of one particular gut microbe results in gut cell apoptosis and host mortality, whereas reintroduction of the Caudal gene restores a healthy microbiota and normal host survival (60). Thus, NF-κB-regulated antimicrobial peptides secreted by IECs including Paneth cells could represent a mechanism whereby specific microorganisms may bloom (34) during inflammation.
The ability of microorganisms to control NF-κB may therefore be critical. Probiotic bacteria, for example, may stabilize IκB or promote peroxisome proliferator-activated receptor γ (PPARγ), which diminishes NF-κB (RelA) retention in the nucleus (61, 62). Another example is NADPH oxidase (or dual oxidase) of Drosophila [functionally homologous to NCF4, a genetic risk factor for CD (63) that encodes the human neutrophil NADPH oxidase factor 4], which regulates the quantity of bacteria in the gut (64, 65).
Thus, a proper balance of commensal community architecture and antimicrobial activities of the epithelium (including goblet cells, absorptive epithelial cells, and Paneth cells), innate immune cells (e.g., neutrophils, macrophages), and the adaptive immune system (e.g., IgA) are critical in maintaining the proper composition of the “metagenome,” the expressed genetic composition of the commensal bacteria and host (33, 34).
The Role of Environmental Factors in Regulating Commensalism and Intestinal Inflammation
These observations on the alterations of microbial composition that are observed in humans with IBD or in experimental model systems may also provide a window into an understanding of the role of certain modifying environmental factors in the pathogenesis of IBD such as diet (66), antibiotics, and most importantly pathogenic infections.
Support for this concept comes from studies with Helicobacter hepaticus, a commensal bacterium with pathogenic potential (38, 67). Although colonization of wild-type mice with H. hepaticus does not result in inflammation, H. hepaticus induces colitis in Il10−/− (68) or scid/Rag2−/− hosts that received naive CD4+CD45RBhigh T cells (67). This colitis is driven by T cells, including those specific for the flagellar hook protein (FlgE) of H. hepaticus (69). In both of these cases, H. hepaticus–induced colitis requires aggressive T cells in the absence of Tregs, similar to the original observations of Powrie and colleagues (70). Notably, this colitis is prevented by cocolonization with the symbiont Bacteroides fragilis, suggesting that, although not specifically shown, this organism may have been depleted by H. hepaticus infection (38). Notably, protection by B. fragilis is dependent on a single microbial molecule (polysaccharide A, PSA) and involves decreased colonic TNF and increased IL-10 from CD4+ T cells (38). Hence, a symbiotic bacterial molecule might network with the immune system to coordinate anti-inflammatory responses required for homeostasis. Such a symbiont and its protective factor may be lost during infection or in a host genetically susceptible for IBD (38). Along similar lines, decreased Faecalibacterium prausnitzii, a major commensal Firmicute, is associated with post-operative recurrence of CD. F. prausnitzii exerts anti-inflammatory properties and induces IL-10 in hematopoietic cells (71). In summary, these observations suggest that initiation of chronic intestinal inflammation requires perturbations of both the commensal microbiota and host immune system; in other words, a two-hit hypothesis for the initiation of IBD (72).
Commensal Microbiota, Innate Immunity, and Adaptive Immunity: A Continuum
Although an adaptive immune system is not necessary for development of colitis in Rag2−/−Tbx21−/− mice (57), it is no doubt a critical factor with the involvement of both bacterial antigen–specific T and B cells. Transfer of flagellin-specific CD4+ T cells into immunodeficient scid mice can induce colitis (73). Similarly, in human IBD there is a notable serologic switch from a homeostatic IgA-dominant to an IgG-dominant response within the intestines that is largely directed at bacterial antigens (74). This excessive production of IgG in IBD is likely to be inflammatory, as the neonatal Fc receptor (FcRn) within hematopoietic cells promotes intestinal inflammation in response to bacterial flagellins in the presence of anti-flagellin IgG (75). However, mice deficient in innate immune responses (i.e., Myd88−/−Trif−/−) do not develop intestinal inflammation despite increased bacterial translocation into the spleen and a dramatic increase in antibacterial IgG responses (76). Hence, though primary alterations in innate immunity may be antecedent to abnormal adaptive immune responses to the commensal microbiota in IBD, they do not necessarily transform into intestinal inflammation.
In this context, one of the most dominant bacterial antigens inducing IgG responses in human CD and mouse models of colitis are flagellins. Although flagellins are expressed by many intestinal commensals, this IgG response is mainly directed at Clostridia-related species expressing specific flagellins (CBir1–15) (73). The flagellar antigen recognized by anti-CBir1 is notably present in colitic and noncolitic mouse strains (73), indicating that the presence of the antigen (and the IgG response) itself does not correlate with colitis. Consistent with this, neither Myd88−/−Trif−/− mice (with elevated antibacterial IgG) (76) nor mice transgenic for a TCR specific for a CBir1 flagellin develop intestinal inflammation (77). To the contrary, the flagellin-specific TCR transgene results in enhanced Treg responses that appear to promote IgA production with all of its beneficial consequences (77). Consistent with this, Tlr5−/− mice develop spontaneous colitis (78), which suggests that an innate inability to respond to flagellin induces a loss of Treg responses, increased bacterial specific IgG, and bacterial dysbiosis due to a loss of commensal specific IgA; altogether these effects culminate in a loss of tolerance to the microbiota and intestinal inflammation.
Taken together, this suggests that the presence of inflammatory bacterial antigen(s) (e.g., flagellin) within the microbiota is inadequate to induce intestinal inflammation even in the presence of a broad loss of innate immune function, despite an adaptive immune system geared toward inflammation as revealed by the presence of an IgG-dominated B cell response or even an absent adaptive immune response as observed in Rag2−/− mice. Hence, a critical accumulation of irregularities in the intestinal microenvironment is necessary for inflammation to occur; albeit these irregularities may evolve from single critical perturbations, as discussed throughout this review.
THE CENTRAL ROLE OF INNATE IMMUNITY IN IBD AND ITS RELATIONSHIP TO THE COMMENSAL MICROBIOTA
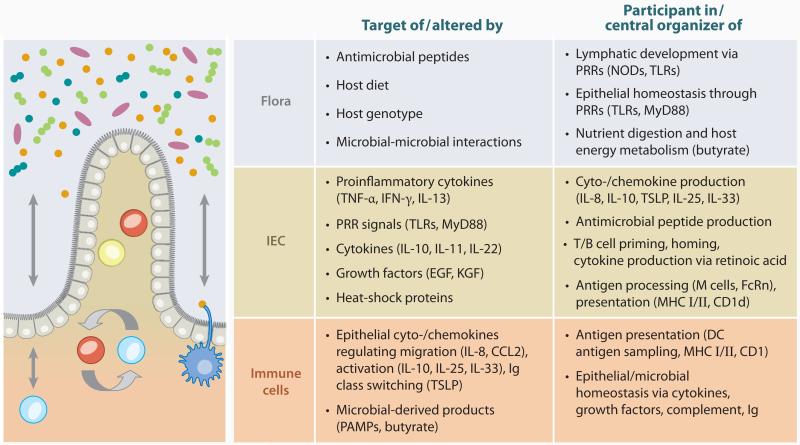
Genetic studies, animal models, and the apparent superior efficacy of biologic agents directed at innate, as opposed to adaptive, immune factors in the treatment of IBD (see Supplemental Table 2) suggest abnormal innate immune responses toward the microbiota as a central underlying theme of IBD (Figure 2).
Figure 2.
The microbial flora, intestinal epithelial cells, and lamina propria immune cells as targets, participants, and central organizers in intestinal immune responses. Abbreviations: EGF, epithelial growth factor; KGF, keratinocyte growth factor; PAMPs, pathogen-associated molecular patterns; PRR, pattern-recognition receptor.
NOD2 and Pattern Recognition of the Microbiota
NOD2 polymorphisms were the first firm genetic association between an individual gene and a polygenic disease (79–81). Between 30% and 40% of patients with CD in the Western hemisphere (compared to ≈10% in healthy controls) carry NOD2 polymorphisms on at least one allele. NOD2 is not a genetic risk factor for UC, but other NOD2 polymorphisms have been linked to Blau syndrome (82). NOD2 is structured into two N-terminal caspase-activation and recruitment domains, a central nucleotide-binding and oligomerization domain, and a C-terminal leucine-rich repeat ligand-binding domain (83). Three individually uncommon (minor allele frequency <5%) polymorphisms that affect protein structure, R702W (NOD2C2104T, “SNP8”), G908R (NOD2G2722C, “SNP12”), and 1007fs (NOD23020insC, “SNP13”), account for 32%, 18%, and 31% of CD-associated variants, respectively, whereas additional rare variants cumulatively account for 19% of the risk associated with NOD2 (84). Altogether, 93% of the variants are located in the leucine-rich repeat region (84), which is involved in binding of N-acetyl muramyl dipeptide (MDP) derived from bacterial peptidoglycan (83) and N-glycolyl MDP from mycobacteria (85), as well as viral ssRNA (86). The OR for CD for simple NOD2 heterozygotes is 2.4, and for homozygotes or compound heterozygotes 17.1 in Caucasians (79, 84), rendering NOD2 as the locus with the strongest effect size among all currently known IBD-associated loci (20).
NOD2 is expressed intracellularly, including in myeloid cells, IECs, Paneth cells (87, 88), and, as recently reported, T cells (89). NOD2 activation by MDP results in binding to receptor-interacting serine-threonine kinase 2 (RIP2; also known as RICK, CARDIAK), which in turn results in NF-κB essential modulator (NEMO; also known as IκB kinase γ) ubiquitination via the E3 ubiquitin ligase TRAF6 (90–92). Ubiquitinated NEMO recruits the TGF-β-activated kinase (TAK1) complex to phosphorylate the IκB kinases (IKK), which promote IκB degradation and consequently release NF-κB into the nucleus to transduce expression of chemokines (e.g., CXCL3/CXCL14 and CCL2) and cytokines (e.g., TNF and IL-6) (92). TLRs signal via a similar pathway with which NOD2 may synergize (92). ssRNA binding to NOD2 results in interaction with mitochondrial antiviral signaling protein, consequent IRF3 activation, and IFN-β production (86). Neither Nod2−/− nor Nod22939insC mice (knock-in of human NOD23020insC) develop enteritis or colitis (47, 55), and a comprehensive pathway to disease has not yet been elucidated. In the search for NOD2 functions in CD, several lines of evidence suggest hypomorphic function of the CD-associated variants, which are related to bacterial innate immune recognition, as detailed below.
The three common CD-associated NOD2 polymorphisms abrogate RIP2 binding and NEMO ubiquitination (90) and result in decreased NF-κB transactivation (93), implying hypomorphic NOD2 function. Consequently, PBMCs from patients homozygous for these variants exhibit decreased inflammatory cytokine secretion upon MDP stimulation or MDP-stimulated TLR ligation (94). Accordingly, macrophages from Nod2−/− mice exhibit diminished IκBα phosphorylation upon MDP stimulation and decreased IL-6 and IL-12p40 secretion upon costimulation with TLR ligands (47). However, chronic stimulation of NOD2 via MDP, as predicted to occur in the context of the intestinal microbiota, “tolerizes” against subsequent stimulation through TLRs, which is lost in CD patients homozygous for the NOD23020insC allele (95). Consistent with this, NOD2 signaling may even be inhibitory to TLR2-mediated activation of NF-κB in antigen-presenting cells (APC), which is lost with the murine homolog of the NOD23020insC variant (96). Increased NF-κB activation upon MDP stimulation has also been observed in mice engineered to express the mouse homolog of NOD23020insC (Nod22939insC). These mice were more susceptible to DSS colitis, which could be prevented by recombinant IL-1Ra (55). Along the same lines, Nod2−/− APCs induced height-ened IFN-γ responses in antigen-specific CD4+ T cells, and transfer of OVA-TCR transgenic CD4+ T cells into recipient mice and subsequent exposure to OVA expressed by orally administered E. coli resulted in more severe colitis compared to Nod2+/+ recipient mice (97). However, Nod2−/− mice also exhibit diminished humoral adaptive immune responses to a model antigen in vivo (47), highlighting the complexity of NOD2 functions.
CD-associated NOD2 variants also exhibit a gain of function through active inhibition of (anti-inflammatory and regulatory) IL10 transcription via blockade of p38 interactions with nuclear ribonucleoprotein hnRNP-A1 (98), consistent with decreased TLR ligand–induced IL-10 production of monocytes from patients homozygous for NOD23020insC (98, 99). Altogether, these aspects of NOD2 function on NF-κB, TLRs, and IL-10 predict impairment of the normal innate response toward commensal flora required for the maintenance of tolerance.
Nod2−/− mice also exhibit decreased α-defensin expression in Paneth cells and increased systemic translocation of the orally infected model pathogen Listeria monocytogenes (47), as well as increased overall bacterial load in the intestinal lumen (54). Similarly, Paneth cells from NOD23020insC homozygous patients exhibit decreased α-defensin HD4 and HD5 expression (53), though inflammation could also contribute to downregulation (100). Furthermore, IEC-expressed NOD2 may provide protection against intracellular bacteria like Salmonella typhimurium, a function lost with the NOD23020insC variant (88, 101). NOD2 is also involved in the autophagic response to invasive bacteria (see Autophagy and IBD section, below) as it induces the recruitment of the autophagy protein ATG16L1 to the entry site of bacteria at the plasma membrane (102, 103). Notably, the major CD-associated NOD2 variants fail to induce autophagy via ATG16L1, and consequently autophagic wrapping of invading bacteria is impaired (102, 103). These altered NOD2 functions could contribute to the increased association of intestinal bacteria with the epithelium, which has been observed in IBD (104), as well as lead to an inability to manage pathogens and the commensal flora, setting the stage for an inflammatory environment, as described above (47).
NF-κB and Its Regulation in IBD
As detailed above, there is an intricate relationship between NOD2 and NF-κB, with underlying hypomorphic induction of NF-κB by CD-associated NOD2 variants and its complex outcomes. This relationship draws specific attention to the complex role of the NF-κB pathway (105) in the mucosal immune system with vastly divergent effects in different cellular compartments.
Increased NF-κB activation, associated with increased IL-1β, TNF-α, and IL-6 expression (106), in macrophages and IECs has been reported in CD, UC, nonspecific colitis, and diverticulitis, but not in uninflamed mucosa, and correlates with inflammatory activity (107). NF-κB p65 (RelA) antisense oligonucleotides administered intravenously or rectally ameliorate trinitrobenzene sulfonic acid (TNBS)-induced colitis and colitis in Il10−/− mice (106). Similarly, administration of BMS-345541, a pharmacological inhibitor of IκB destruction, ameliorates DSS colitis (108). Also similarly, deletion of IKKβ (Ikbkb) in macrophages and neutrophils improves colitis in Il10−/− mice (109). In contrast, genetic deletion of IKKβ specifically in IECs results in increased severity of DSS colitis (109, 110), which appears secondary to decreased recruitment of inflammatory cells that contribute to production of barrier protective mediators such as IL-11, IL-22, and heat shock protein 70 (109–111). Consistent with this, IEC-specific Ikbkb deletion did not affect chronic colitis in Il10−/− mice, in contrast to IKKβ deletion in myeloid cells (109), supporting a protective role for IKKβ in the epithelium in contrast to an inflammatory role in myeloid cells (109). Overall, these divergent outcomes resemble TLR4 signaling in the mucosa (promotion of mucosal healing versus promotion of inflammation) (112, 113).
In addition, IKKβ within IECs can direct adaptive immune functions in the lamina propria via distal effects on DCs (114). Specifically, the intestinal microbiota drives expression of thymic stromal lymphopoietin (TSLP) in IECs via an IKKβ-dependent pathway (115, 116), which renders mucosal DCs noninflammatory, characterized by IL-10 and IL-6, but not IL-12, secretion (117). IEC-specific IKKβ (IkbkbVillin-Cre) deletion results in decreased TSLP expression in IECs and a consequent inability to eradicate Trichuris muris infestation secondary to a failure to develop a protective Th2 response (116, 118). Instead, mucosal DCs in IkbkbVillin-Cre or TSLP-receptor (Crlf2)-deficient mice exhibit increased IL-12/23p40 and TNF expression and CD4+ T cells deviated to IFN-γ and IL-17 secretion. Consequently, these mice develop severe intestinal inflammation (116). Indeed, neutralization of IL-12/23p40 and IFN-γ in Trichuris-infested IkbkbVillin-Cre or Crlf2−/− mice results in decreased IFN-γ and IL-17 expression and a concomitant increase in IL-13 expression, restored goblet cell function via increased RELMβ expression, and worm expulsion (116, 118).
Another important aspect of IECs lies in their roles as physical barrier and source of antimicrobial peptides, both in defense against pathogenic invasions and in the maintenance of bacterial commensalism (36, 44, 76). Deletion of NEMO (Ikbkg), or both IKKα (Ikbka) and IKKβ, in IECs results in severe spontaneous colitis secondary to apoptosis of colonic IECs (119). This is associated with decreased production of antimicrobial peptides and translocation of bacteria into the mucosa, which triggers a spontaneous MyD88- and TNFR1-dependent chronic inflammatory response in the colon (119).
A20 (Tnfaip3) is a potent inhibitor of NF-κB signaling by restricting TNF and TLR signals via ubiquitin editing of RIP (120) and TNF receptor-associated factor 6 (TRAF6) (121), respectively. A20 also restricts MyD88-independent TLR signals by inhibiting Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF)-dependent NF-κB signals (122) and MDP-induced NOD2 signaling (123). In a direct feed-back loop, A20 is phosphorylated by IKKβ, which increases its ability to inhibit NF-κB activation. Altogether, these properties render A20 a critical negative regulator of microbially derived signals, and Tnfaip3−/− mice succumb to severe inflammation in various organs, including the intestine (124). Notably, a polymorphism at rs7753394, with the closest gene being TNFAIP3, has been associated with CD (and other immune-related diseases) (125), implying a more general role of A20 in disease pathogenesis in addition to its role in restricting NOD2 signals.
METABOLIC ABNORMALITIES AND IMMUNE FUNCTION OF THE INTESTINAL EPITHELIUM IN IBD
The studies on NOD2 and NF-κB summarized above have furthered the interest in the intestinal epithelium in IBD as an immunophysiologic barrier rather than simply as a structural barrier whose “leakiness” might represent the sole factor antecedent to the development of IBD (Figure 2).
Endoplasmic Reticulum Stress of the Intestinal Epithelium
The unfolded protein response (UPR) is activated upon accumulation of misfolded proteins, which cause endoplasmic reticulum (ER) stress (126). Among three proximal effector pathways, inositol-requiring enzyme 1 (IRE1)/X-box binding protein 1 (XBP1) is the evolutionarily most conserved (126). UPR molecules are ubiquitously expressed, and the relative contribution of individual pathways in various cell types varies profoundly (126, 127). The intestinal epithelium is unique in that it selectively expresses an additional isoform of IRE1α, IRE1β, predicting critical dependency on an efficient UPR (128). Indeed, IRE1β−/− (Ern2) mice are more susceptible to DSS colitis in association with increased ER stress (128). IRE1 activates XBP1 via an unconventional splicing mechanism by excising a 26-nucleotide sequence from the unspliced XBP1 mRNA, resulting in a frameshift and consequent production of an active transcription factor (XBP1s) that contains a DNA transactivating domain at the C terminus (126). Xbp1 deletion in the intestinal epithelium results in unabated ER stress in the epithelium, spontaneous enteritis in the small intestine, and increased susceptibility to DSS colitis (46). Deletion of only one allele is sufficient to induce profound overactivation of IRE1 and enteritis in approximately one-third of mice (46). XBP1 regulates Paneth cell function, with a consequent defect in handling oral Listeria monocytogenes infection (46), similar to Nod2−/− mice (47). Deletion of Xbp1 results in apoptotic depletion of Paneth cells and reduction in goblet cells, whereas IECs exhibit increased inflammatory responsiveness to TLR and cytokine signals (46). A candidate gene study revealed significant associations of the complex XBP1 locus with both CD and UC (46). Three-fold more rare SNPs in CD and UC compared to healthy controls were found by deep sequencing, and CD-/UC-associated variants exhibit hypomorphic induction of UPR target genes (46).
A forward-genetic approach recently yielded the Winnie and Eeyore mouse models with spontaneous colitis resembling UC, which mapped to missense mutations in the Muc2 gene (129). These variants led to aberrant MUC2 oligomerization and induction of ER stress in goblet cells, and goblet cells in UC exhibited a similar phenotype (129). The woodrat (wrt) forward-genetic model with a missense mutation in the membrane-bound transcription factor peptidase site 1 (S1P)-encoding gene (Mbtps1) provides another example of a link between the UPR and intestinal inflammation (130). S1P activates ATF6α, another proximal UPR mediator, upon ER stress (126). Mbtps1wrt/wrt mice exhibited increased sensitivity to DSS colitis, with abnormal Mbtps1 function mapping to nonhematopoietic cells (130). Moreover, administration of the ER stress inducer tunicamycin results in severe colitis in Mbtps1wrt/wrt but not in wild-type mice (130).
The HLA-B27 transgenic rat model of spontaneous colitis may serve as a final example of the association between ER stress and intestinal inflammation (30). The human HLA-B27 heavy chain is remarkably unstable, suggesting that its misfolding induces ER stress in certain tissues (e.g., stomach, intestines, joints, liver, skin), which in turn correlates closely with the extent of colitis (131), presumably through mechanisms as described above. This observation could also explain the high prevalence of (asymptomatic) ileitis in patients with HLA-B27-associated spondyloarthropathies.
In summary, a proper ER stress response in the intestinal epithelium appears to be necessary to maintain homeostasis, with the most highly secretory cell types (Paneth and goblet cells) most vulnerable to these effects. XBP1 and ER stress in general may regulate the ability of the intestinal epithelium to both regulate and sense the composition of the luminal microbiota, which sets the inflammatory tone of the IEC. These studies also suggest that alterations in IECs may be a primordial factor in the development of IBD. Environmental and microbial factors can modulate ER stress in beneficial and detrimental ways (72), and increased ER stress may be a common occurrence in human IBD (46, 129, 132). Thus, primary (genetic) or secondary (environmental) pathways (and their interactions) that lead to ER stress within environmentally exposed and highly secretory cells appear to be an important pathway for development of IBD (133).
Autophagy and IBD
Macroautophagy, a fundamental and evolutionary highly conserved response to fasting, is a lysosomal pathway that is involved in the turnover of cellular macromolecules and organelles and plays an important role in a variety of biological processes as diverse as infection, immunity, cancer, and aging (134). Autophagy is activated by a variety of conditions of cellular stress including ER stress (135) and involves formation of double-membraned autophagosomes engulfing cellular contents that later fuse with lysosomes. The connection between ER stress and autophagy involves several levels and likely differs between cell types (135). ER stress may activate autophagy through the ability of IRE1 to associate with TRAF2 and activate JNK or through PERK-mediated inhibition of eIF2α (135).
It is thus interesting that a GWAS discovered ATG16L1 as a genetic risk factor that is specific for CD, but not for UC (14). Virtually all the risk of this locus was exerted by rs2241880, which codes for a T300A substitution (14). rs2241880 was also one of the main associations reported in another GWAS on CD (63). Together with the identification of polymorphisms close to another gene involved in autophagy, IRGM (125), these studies together revealed a previously unanticipated role for autophagy in the pathogenesis of CD.
Insight into the potential mechanism of ATG16L1 in CD stems from studies in Atg16l1 hypomorphic and deficient mice. ATG16L1 deficiency disrupts the recruitment of the ATG12-ATG5 conjugate to the isolation membrane, and a consequence is severe impairment in autophagosome formation and degradation of long-lived proteins (136). Stimulation of Atg16l1−/− macrophages with LPS resulted in high production of IL-1β and IL-18 via TRIF-dependent activation of caspase-1, showing that ATG16L1 regulates LPS-induced inflammasome activation (136). Deficiency of ATG16L1 in bone marrow resulted in increased susceptibility to DSS colitis, which could be alleviated by IL-1β and IL-18 blockade (136). Hypomorphic ATG16L1 variants and IEC-specific Atg5 deletion revealed abnormalities in the granule exocytosis pathway of Paneth cells, including disorganized granules and decreased granule numbers (45). Similar alterations were found in CD patients homozygous for the ATG16L1 risk allele (45). Despite these abnormalities, ATG16L1 hypomorphic mice—in contrast to Nod2−/− (47) and Xbp1Villin-Cre (46) mice, which also exhibit Paneth cell defects—exhibited no impairment upon oral Listeria monocytogenes infection. Paneth cells in ATG16L1-hypomorphic mice also revealed altered expression of genes involved in PPAR signaling and lipid metabolism together with increased production of the adipocytokines, leptin, and adiponectin (45), implicating them in the regulation of intestinal inflammation. Mice deficient in ATG16L1 in the bone marrow—or ATG16L1 hypomorphic mice—do not, however, develop spontaneous enteritis (45, 136). A knock-down/reconstitution strategy in vitro revealed that the ATG16L1T300A variant resembles NOD2 function (88, 101) by exhibiting impaired capture of Salmonella typhimurium within autophagosomes with no effect on basal autophagy (137). Consistent with this, the autophagic response can be triggered by NOD1 or NOD2 upon intracellular infection with invasive bacteria, which function in inducing the recruitment of ATG16L1 to the plasma membrane (102, 103). The ATG16L1T300A is associated with impaired NOD2-dependent induction of autophagy upon stimulation with MDP (102, 103). Thus, these two genetic risk factors (NOD2 and ATG16L1) function in a common pathway that involves bacterially induced autophagy and the consequent induction of antigen-specific T cells, and this pathway is also impaired in the absence of normal NOD2 function (103).
A 20-kb deletion polymorphism immediately upstream of IRGM, resulting in an altered expression pattern (138), was identified as the potential causal variant of the second autophagy gene discovered in association with CD (125, 139). IRGM belongs to the IFN-γ-induced p47 immunity-related GTPase family (140). Its mouse homolog, LRG-47 (encoded by Irgm1), controls intracellular pathogens by autophagy (141), and Irgm1−/− mice exhibit increased susceptibility to Toxoplasma gondii, Listeria monocytogenes, and Mycobacterium tuberculosis infection (142) due to decreased bacterial killing in Irgm1−/− macrophages (142).
These studies reveal a convergence of several genetic risk factors for IBD (NOD2, XBP1, and ATG16L1) on the function of the intestinal epithelium and especially Paneth cells and concurrently on the regulation of inflammatory pathways in both the epithelium and myeloid cells. Hence, these epithelial cell functions, which are likely susceptible to environmental modification, may be important determinants of the propensity to develop IBD. Although Paneth cell dysfunction may lead to dysbiosis or altered adherence of bacteria to the epithelium (36), isolated Paneth cell deletion (143) and inability to activate their antimicrobial function (144) are not associated with spontaneous intestinal inflammation. It might therefore be hypothesized that dysbiosis must be coupled with immune hypersensitivity to the commensal microbiota to develop intestinal inflammation.
Organic Carnitine and Cation Transporters and β-oxidation
The UPR and autophagy pathways regulate important cellular “metabolic” functions associated with diet (145, 146). Given the complex metabolic environment at the host-microbiota interface, and the potential contribution of environmental/nutritional factors to IBD [e.g., increasing incidence upon “Westernization” (3, 147)], it is notable that another genuine metabolic function, β-oxidation, is implicated in CD pathogenesis. Expression quantitative trait locus (eQTL) analysis suggests that SLC22A5 (20), encoding OCTN2, is the associated gene at the IBD5 locus (148). OCTN2 is a Na+-dependent, high-affinity L-carnitine transporter and a polyspecific Na+-independent cation transporter (149). Carnitine has an obligatory role for transport of long-chain fatty acids into mitochondria for β-oxidation, which is of particular importance to the energy metabolism of IECs and liver (150). Gastrointestinal carnitine content in Slc22a5−/− mice is reduced to 5–10% of normal (151). This is associated with increased IEC apoptosis, abnormal villus structure, and inflammatory infiltration in the mucosa with the spontaneous development of small intestinal perforations and (micro) abscesses (151). Similarly, pharmacological inhibition of gut fatty acid β-oxidation also results in experimental colitis (152). Genetically decreased SLC22A5 expression in CD (20) may become particularly relevant in metabolically “challenged” IECs owing to alterations in the microbiota or, in the context of inflammation, when energy needs are increased, due to increased catabolism.
Role of CD1d-Restricted Natural Killer T Cells in IBD
In concluding a discussion of metabolic factors and immune function, we briefly consider the biology of natural killer T (NKT) cells in relationship to IBD. NKT cells respond to phospholipids or glycolipids that are presented by CD1d on an APC leading to an “innate-like” rapid response through secretion of abundant numbers of Th1, Th2, and Th17 cytokines that subsequently trigger almost all branches of the innate and adaptive immune systems (153). NKT cells can be activated by various mechanisms, including direct activation by presentation of self- or microbial-derived lipids by the nonclassical MHC class I molecule CD1d and indirect cytokine-mediated activation mainly through IL-12 and IL-18 (153). The inflamed lamina propria of UC but not of CD patients contains increased numbers of T cells expressing the NK marker CD161, which respond to CD1d with increased secretion of IL-13 (154). However, in humans these NKT cells do not react with CD1d tetramers loaded with the invariant (i) NKT cell ligand α-galactosylceramide and therefore must be considered as type II or noninvariant NKT cells (154). Consistent with this, mice deficient in CD1d and NKT cells are resistant to oxazolone colitis, a murine model of ulcerative colitis (155). However, in contrast to human UC, invariant NKT cells were observed to be the main effectors in oxazolone colitis (155).
The mechanism(s) by which CD1d and NKT cells may be involved in UC pathogenesis remains to be established, but several possibilities exist. Many different cell types that are present in the intestines express CD1d, including DCs, macrophages, B cells, and IECs (156). This raises the questions of whether a particular cell type is responsible for NKT cell activation in colitis and whether the various CD1d-expressing cells in the intestines play differential, protective, or pathogenic roles in colitis as previously described for NF-κB (109, 157) and TLR4 (112). Interestingly, IEC-specific deletion of the microsomal triglyceride transfer protein that normally lipidates apolipoprotein-B during absorption of dietary lipids, and that also assists in loading nascent lipid antigens onto CD1d within the ER and is necessary for CD1d-restricted antigen presentation (158), leads to increased mortality upon oxazolone challenge, which can be prevented by systemic antibody-mediated blockade of CD1d (T. Olszack, S. Zeissig, A. Kaser, and R.S. Blumberg, unpublished observations). These findings suggest a protective role of CD1d on IECs in murine oxazolone colitis in contrast to a pathogenic role of CD1d on hematopoietic cells. The importance of these findings is high-lighted by the fact that IECs of IBD subjects exhibit decreased CD1d expression (159) while overall CD1d expression in the lamina propria is increased, presumably owing to mononuclear cell infiltration (160). This proinflammatory effect of CD1d on hematopoietic cells may be mediated by IL-23 and the IL-23R, both of which are genetic risk factors for both CD and UC (see below). IL-23R is expressed on NKT cells and regulates IL-17 expression by NKT cells (161). CD1d-restricted NKT cells have also been linked to the pathogenesis of asthma, which, similar to UC, is associated with increased secretion of IL-13 (162).
INNATE IMMUNE CYTOKINE PATHWAYS AND IBD
Abnormalities of innate immune function and their relationship to the commensal microbiota have been identified to be key properties that characterize the immunogenetic profile of human IBD and animal models of intestinal inflammation, as described in detail above. Another line of evidence that supports a central role of innate immune functional abnormalities in IBD pathogenesis is the cytokine environment that is observed, as well as the efficacy of therapies that are directed at the specific cytokines and the cells that are responsible for their production. The experience with biologic therapies in humans with IBD is particularly instructive in furthering our understanding of the immunogenetic pathogenesis of these disorders and in assigning relevance to potential functional pathways (see Supplemental Table 2).
TNF and TNF-Related Cytokines (TL1A)
The currently most efficacious treatment for IBD is anti-TNF antibodies (8). Surprisingly, the mechanistic basis of their effectiveness remains enigmatic, as does the specific relationship of TNF to the genetic underpinning of IBD. It is noteworthy that TNF is located 1 MB apart from the MHC locus, which has been associated with UC more so than with CD (163). The dramatic efficacy of anti-TNF antibodies predicts it is a major factor on which many pathways associated with IBD converge.
TNF acts as a transmembrane or soluble protein by transducing signals ranging from cellular activation and proliferation to cytotoxicity and apoptosis through two distinct TNF receptors, TNFR1 (p55) and TNFR2 (p75) (164). NF-κB and NF-AT control Tnf transcription, and AU-rich elements (ARE) in the 5′ UTR control mRNA destabilization and translational repression (165). TnfΔARE mice overproduce TNF and develop inflammatory polyarthritis and spondyloarthritis and CD-like deep transmural intestinal inflammation with granulomas primarily in the terminal ileum (165, 166). Intestinal pathology is attenuated in Tnfr2−/−TnfΔARE double mutants and is absent in Tnfr1−/−TnfΔARE mice (165). Rag1−/−TnfΔARE mice exhibit only mild inflammation confined to the intestinal mucosa (165). This suggests that TNF can inflict superficial injury in the absence of mature B and T cells, but such are required for transmural inflammation typical of CD and severe disease observed in TnfΔARE mice. The superficial inflammation observed in Rag1−/−TnfΔARE mice is reminiscent of TNFR1-dependent inflammation observed in Rag2−/−Tbx21−/− mice (57). Bone marrow and parenchymal cells are equally responsive to the pathogenic effects of TNF in the development of intestinal inflammation in the TnfΔARE model (166, 167). Interestingly, TNFR1 on mesenchymal cells adjacent to IECs appears to be particularly important for disease in TnfΔARE mice, which may regulate the balance of matrix metalloproteinases (MMP) and inhibitor of MMP (TIMP) expression, leading to leukocyte recruitment and tissue destruction (166, 168). Thus, one mechanism of anti-TNF therapies may be through blockade of a superficial pathway of tissue injury that is common to both UC and CD and involves TNF production by parenchymal cells or innate immune cells that act upon TNFR1 within superficial cellular structures of the gut. Consistent with this, human colon IECs express TNF (along with GM-CSF, CXCL8, CCL2, and IL-6) ex vivo upon exposure to inflammatory stimuli or pathogenic bacteria (169). Moreover, IEC apoptosis is increased in CD and reduced upon anti-TNF antibody treatment (170). In a similar manner, blockade of TNF reduces IEC apoptosis in the Rag2−/−Tbx21−/− (57), IkbkgΔIEC (119), and SAMP1/YitFc (171) models.
Transmural, CD-like inflammation in TnfΔARE mice requires CD8+ T cells, is dependent on IL-12/23p40 and IFN-γ, and is regulated by CD4+ T cells (167). IFN-γ-secreting CD8+ effector T cells are also involved in a hapten-mediated colitis model (172), suggesting that CD8 effectors are more important in IBD than currently appreciated. Myeloid cells or T (Th1) lymphocytes may be sources of TNF (167). However, numerous other cell types, including Paneth cells and adipocytes, exist that may promote inflammation within their specific intestinal microenvironments (173). TNF also plays an important role in other models of intestinal inflammation, like TNBS-induced models (174), in Il10−/− mice (175), and in the spontaneous UC-like disease in the cotton-top tamarin model (176). Notably, nonlymphocyte-derived TNF was sufficient for the development of colitis in the CD45RBhigh transfer model with TNF found in macrophages (localized close to epithelial erosions) and colonic epithelial cells, especially during early phases of disease (177). In the CD45RBhigh transfer model of colitis, continuous anti-TNF antibody treatment was required to decrease disease severity (178). Expression of noncleavable membrane-bound TNF in Rag2−/−Tnf−/− recipients that lack soluble TNF is sufficient to cause colitis upon transfer of TNF-deficient CD4+CD45RBhigh T cells (179). TNF+ cells are increased in the lamina propria of both UC and CD, with localization confined to subepithelial macrophages (UC) or evenly distributed throughout the lamina propria (CD) (180). Overall, these studies suggest that in transmural colitis, TNF is likely derived from many different cell types and that pathogenic T cells may create a permissive environment for the inflammatory effects of TNF to occur. Moreover, they highlight the pathologic potential of TNF in both UC and CD and the importance of the membrane-bound TNF in its own right, which is consistent with the role of anti-TNF-induced apoptosis as a therapeutic pathway in CD (181). Specifically, retrograde signaling via transmembrane TNF has been suggested to distinguish effective (anti-TNF) and ineffective (TNFR2-Fc) TNF-targeted therapies in CD (8, 181). However, it should be noted that in oxazolone-induced colitis (155, 182), a TNFR1-IgG1-fusion protein led to more extensive disease (183), which was associated with decreased TGF-β1. TGF-β1 usually limits the extent of disease to the distal part of the colon (184). Because TNFR1-Fc (and TNFR2-Fc) may bind both soluble TNF and lymphotoxin-α, this may reflect a role for lymphotoxin-α in regulating TGF-β production and intestinal inflammation.
Polymorphisms in TNFSF15 (encoding TL1A, TNF ligand–related molecule 1A), another TNF-related family member, confer substantial risk for CD in Japanese and Korean cohorts with ORs of up to 2.40 and 3.49, respectively (185, 186), as compared to the relatively modest OR of 1.22 exhibited in the Caucasian GWAS meta-analysis (20). TL1A is a TNF-like cytokine that is increased in CD and interacts with the death domain receptor (DR3), which signals through NF-κB (187). TL1A is produced by human DCs and monocytes and enhances IFN-γ production by T and NK cells (187). Both DSS-induced colitis and the spontaneous colitis in SAMP1/Yit mice exhibit increased expression of TL1A, DR3, and Th1 and Th17 cytokines, which are decreased together with mucosal inflammation by neutralization of TL1A (188, 189). This highlights TL1A as another example of an innate immune-derived (TNF-related) molecule that drives adaptive immune-mediated, intestinal inflammation, and it is tantalizing to speculate that this might be a particularly interesting therapeutic target in Asians with IBD.
IL-6, gp130, JAK2, and STAT3
IL-6 signals through the IL-6R expressed on the cell surface and through soluble IL-6R (sIL-6R) via binding of the sIL-6R/IL-6 complex to the transmembrane receptor β subunit gp130 (trans signaling) (190). Redundancy within the IL-6 family, which comprises IL-6, IL-11, leukemia inhibitory factor, oncostatin M, ciliary neurotrophic factor, and cardiotrophin-1, is attributed to the common use of gp130. gp130 signals via two distinct pathways: The first is Janus kinase (JAK) 1, JAK2, and tyrosine kinase 2 (Tyk2) and consequent signal transducer and activator of transcription 3 (STAT3) activation. The second pathway is through STAT1 leading to activation of NF-κB. The second pathway involves engagement of src-homology tyrosine phosphatase (SHP2) and subsequent activation of the Ras-ERK pathway.
IL-6 and sIL-6 secretion is increased in both CD and UC mucosa and likely derives predominantly from non-T cells (191). Phosphorylated STAT3 expression indicates T cells and macrophages as the major targets of IL-6 signaling (191). IL-6 trans signaling is important for the survival of CD4+ T cells, and possibly macrophages, and for their production of inflammatory cytokines such as TNF, IFN-γ, and IL-1β (191, 192). IL-6 plays an important role in immune-deviating T cells from a Treg fate toward an inflammatory (i.e., Th17) phenotype (193–195). Consistent with this, blockade of IL-6 trans signaling via a gp130-Fc decoy receptor or complete IL-6R signaling via an anti-IL-6R antibody ameliorates colitis in the CD45RBhigh transfer model, in the TNBS model, and in Il10−/− mice (191, 192). This is associated with increased apoptosis of lamina propria mononuclear cells implying that excess IL-6 secretion by innate immune cells promotes the survival and activity of proinflammatory T cells (and possibly macrophages), which drive inflammation (191, 192).
Accordingly, a placebo-controlled pilot trial reported benefit of an anti-IL-6R in active CD (Supplemental Table 2). Polymorphisms close to STAT3 and within STAT3 have been associated with CD (20, 125) and UC (196). Moreover, a polymorphism in the JAK2 promoter region also associates with CD (20) and UC (197), highlighting the importance of IL-6-gp130-JAK2-STAT3 related pathways in both forms of IBD. However, the pleiotropic relationships that JAK2 (e.g., gp130 family members, IFN-γ, IL-12) and STAT3 (e.g., gp130 family members; IL-10; leptin; IL-12; and γc family members such as IL-2, IL-7, IL-9, IL-15, and IL-21) have with cytokine signaling pathways are considerable such that it cannot be concluded that the associations identified between JAK2 and STAT3 and IBD are definitively related to the biology of IL-6-mediated signaling.
Additional complexity stems from the fact that gp130 signaling involves two cascades with distinct biologic consequences (STAT1/STAT3-NF-κB mediated and SHP2-Ras-ERK mediated) that have unique effects on the intestinal epithelium. Deletion of the STAT binding domain (gp130ΔSTAT) leads to spontaneous ulcerations of the rectum (198) and augments DSS-induced colitis (199), implying a cytoprotective role for gp130-induced STAT3 (and STAT1) signaling in IECs (200, 201). In line with this, IEC-specific Stat3 deletion results in augmented DSS-induced epithelial erosions and subsequent mucosal inflammation, whereas STAT3 overactivation confers protection (202). Gp130ΔSTAT mice also lack intestinal epithelial trefoil factor (TFF) 3 expression (199), a cytoprotective molecule associated with mucins, and Tff3−/− mice consequently phenocopy the sensitivity to DSS administration (203) observed in gp130ΔSTAT mice. IECs also express the IL-6R on the basal surface, and its ligation activates NF-κB (200, 201), which, as discussed above, provides important protective signals to the epithelium.
Mice with disabled gp130-related SHP2-Ras-ERK activation, however, do not exhibit spontaneous mucosal ulcerations, are protected from DSS colitis, and exhibit increased levels of TFF3 expression and increased STAT1/STAT3 activation (199). If one takes together (a) the genetic studies that find an association of JAK2 and STAT3 with IBD, (b) the amelioration of colitis in mouse models through blockade of IL-6 signaling (191, 192), and (c) the potential benefit of anti-IL-6R therapy in human CD (Supplemental Table 2), then in IBD there may be loss of epithelial cytoprotective function of IL-6. This loss of function is due to disabled STAT1/STAT3-mediated, gp130-associated signaling together with excess promotion of inflammatory pathways by IL-6, derived predominantly from innate immune cells within the lamina propria.
Apart from IECs, STAT3 activation by IL-6, IL-10, and other cytokines (mentioned above) in myeloid cells also has important implications for IBD. Deletion of floxed exon 22 of Stat3 in macrophages and neutrophils via LysM-Cre results in spontaneous transmural enterocolitis with depletion of goblet cells (26), along with augmented LPS-induced expression of inflammatory cytokines (26). A comparable phenotype is observed upon deletion of exons 18–20 of Stat3 in bone marrow and endothelial cells by Tie2-Cre (204), which is associated with the formation of granuloma-like structures and crypt abscesses. Similar to Stat3Lys M-Cre, LPS-stimulation of Stat3Tie2-Cre myeloid cells leads to increased IκBα phosphorylation along with increased NF-κB DNA binding activity (204).
These studies suggest that, in the context of a specific inability of macrophages in the intestines to respond to STAT3-mediated signals as may be delivered by IL-10, augmented responses to microbial signals and consequently intestinal inflammation are observed. Accordingly, enterocolitis is improved in Tlr4−/−Stat3Lys M-Cre mice. Moreover, this macrophage-induced inflammation in the absence of STAT3 requires IL-12p40 and lymphocytes because inflammation is ameliorated in Il12b [p40]−/−Stat3Lys M-Cre and Rag2−/−Stat3Lys M-Cre mice, respectively (205). Because inflammation is not ameliorated by loss of STAT1 expression, which is essential for IFN-γ signaling, it might be surmised that the critical cytokine derived from aggressive STAT3-deficient macrophages leading to inflammation is IL-23, although this needs to be directly tested.
NLRP3 and the IL-1 Cytokine Family
Although the prototypical innate proinflammatory mediator IL-1β has been known for decades as an important contributor to mucosal inflammation (206, 207), recent evidence has reinvigorated interest in the relationship of IL-1β to IBD. Specifically, polymorphisms in NLRP3, encoding NALP3/cryopyrin, have been associated with CD in a candidate-gene study (208). NALP3 within the inflammasome directs the conversion of procaspase-1 to caspase-1 and generates secretory IL-1β and IL-18 (206, 207). Hypermorphic missense mutations of NLRP3 and consequently increased IL-1β are linked to rare autoinflammatory disorders (206). Notably, CD-associated NLRP3 polymorphisms appear to be linked to decreased IL-1β secretion from LPS-stimulated peripheral blood cells (208). A primary relationship between IL-1/IL-18 and IBD is also highlighted by a CD- and UC-associated SNP within the IL-18 receptor accessory protein gene (IL18RAP) that identifies a 350-kb haplotype block in strong linkage disequilibrium containing IL1RL1-IL18R1-IL18RAP-SLC9A4 (209). Decreased IL-1β production upon MDP stimulation has similarly been reported in myeloid cells of patients carrying risk-associated NOD2 variants (94, 210, 211). These genetic studies suggest that inadequate innate IL-1β (and possibly IL-18) activity could be a risk pathway for CD and UC, perhaps at the level of the epithelial barrier.
However, these considerations contrast with observations in human IBD tissues and in experimental models. IL-1β expression, relative to IL-1 receptor antagonist (IL-1ra), is increased in IBD intestinal tissues (212). Moreover, administration of anti-IL-1β (213) or IL-1ra (214) ameliorates experimental rabbit immune complex colitis, whereas neutralization of IL-1ra increases its severity (215). Similarly, IL-18 expression is increased in IBD, particularly in IECs (216, 217). Neutralization of IL-18 ameliorates DSS colitis, and, of note, increased IL-18 expression in this model localizes to IECs (218). Administration of the natural IL-18 antagonist, IL-18 binding protein (IL-18BP), also ameliorates DSS colitis (219). A major proinflammatory role of IL-18 in experimental intestinal inflammation was also deduced from experiments in the TNBS (220, 221) and scid transfer (222) models of colitis. Genetic deletion of caspase-1, which impairs the downstream function of the NALP3 inflammasome and hence IL-1β and IL-18 processing, similarly ameliorates DSS colitis (223). These studies suggest that excessive IL-1 and IL-18 may promote chronic intestinal inflammation.
To reconcile these data with the genetic studies, it is reasonable to consider the possibility that an inadequate IL-1 (and potentially IL-18) response during innate management of the luminal interface may predispose to IBD through an uncontrolled adaptive immune response. IL-1 and IL-18 are also both important links to balanced expression of Th17 (224–226) and Th1 (206) pathways, respectively. In addition, IL-1 may intersect with other primary (genetically mediated) risk pathways. For example, IL-1β expression is increased in Atg16l1−/− myeloid cells (136); IL-1β is an important inducer of IL-6, and, interestingly, several α-defensins, including human α-defensin 5, decrease IL-1β secretion, raising the final possibility that elevated IL-1β may result from Paneth cell dysfunction (227).
ADAPTIVE IMMUNE PATHWAYS AND THEIR RELATIONSHIPS WITH INNATE IMMUNE PATHWAYS
As reviewed above, a model for the pathogenesis of IBD emerges that supports an inappropriate relationship between the commensal microbiota—the IEC barrier—and innate immunity that leads to inappropriate release of cytokines and other mediators that are inflammatory in their own right, abnormalities of the physiologic barrier that exists between the intestinal lumen and lamina propria and its consequences as well as cytokines that promote the inflammatory activity of adaptive immune cells. There are many layers in this model, some of which are outlined above and include both an inappropriate drive to innate immune signaling from an abnormal commensal bacterial architecture and an inadequate degree of innate immune regulation through pathways intrinsic to the pattern-recognition receptor (PRR)-associated pathways themselves. In this section, we review the manner in which innate immune signaling links to the adaptive immune system that leads to the chronic inflammation characteristic of IBD as well as the abnormalities that appear to reside within regulatory pathways that typically provide restraint to both innate and adaptive immune pathways. Because these latter topics have been extensively reviewed recently (228), they are only discussed in an abbreviated manner here.
IL-12 and IL-23
IL-12 and IL-23, secreted by DCs secondary to PRR-derived signals, are highly related heterodimeric cytokines sharing the IL-12p40 subunit (12). IL-12 (p35 and p40 heterodimer) supports Th1, while IL-23 (p19 and p40) supports Th17 pathways (229). IL-12 was originally linked with intestinal inflammation in the TNBS colitis model based upon studies that neutralized the p40 chain (230). Anti-IL-12p40 treatment resulted in decreased IFN-γ production from lamina propria CD4+ T cells (230). Further support for the IL-12/Th1 pathway emerged when abrogation of the Th1 effector cytokine IFN-γ in the CD4+ CD45RBhigh/Rag−/− transfer model potently protected from colitis and when T-bet-deficient CD4+CD45RBhigh cells were unable to induce colitis in Rag−/− recipients (178, 231, 232). Moreover, human lamina propria mononuclear cells exhibit increased IFN-γ secretion in CD in contrast to UC and controls (233). In the context of the epidemiologically detrimental relationship of smoking and CD, it is notable that chronic stimulation of the α7 nicotinic acetylcholine receptor on T cells leads to Th1 responses (234, 235).
These studies led to the development of anti-IL-12p40 (ABT864 and ustekinumab) and anti-IFN-γ antibodies as potential therapeutics for CD. Treatment with ABT864 increased response rates compared to placebo, although remission rates were not different (Supplemental Table 2). Neutralization of IL-12p40 decreased Th1- and Th17-associated cytokines within the intestinal tissues. In contrast, IFN-γ neutralization with fontolizumab was unexpectedly not or only partially effective in CD (Supplemental Table 2). Although these studies are consistent with a role for IL-12p40, the effector arm of this pathway in human IBD remains unclear.
The first GWAS in Caucasians detected a strong association of IL23R polymorphisms with CD and UC (13). Specifically, an uncommon coding variant (rs11209026, IL23RG1142A, Arg381Gln) conferred protection from disease, and additional noncoding IL23R variants were independently shown to confer risk (13). Because the IL23R locus is also associated with psoriasis and ankylosing spondylitis, this pathway may be of general importance for auto-inflammatory diseases (15, 236). Similarly, polymorphisms at IL12B (encoding IL-12p40) have been associated with both CD (139) and UC (196, 237).
These observations followed the first report on the then novel cytokine IL-23 in 2000 (238) and the realization that several pathologies, in particular experimental autoimmune encephalitis (EAE) (239) and collagen-induced arthritis (240), that had been ascribed to IL-12 are also linked to IL-23. In EAE and uveitis models, IL-12 and IL-23 are both pathogenic and contribute to distinct patterns of inflammation (241, 242). The relative roles of IL-12 and/or IL-23 have also been reevaluated in experimental colitis. In a model of innate experimental colitis induced in immunodeficient Rag1−/− mice by agonistic anti-CD40 antibodies, IL-23 blockade via anti-IL-23p19 blocked intestinal pathology but did not affect wasting disease or systemic inflammation; in contrast, anti-IL-12p40 blocked all aspects of disease (243). Similar results were obtained in p19−/− and p40−/− mice (243). Notably, in this innate model of colitis lacking T cells (see below), colonic IL-17A mRNA was 65-fold up-regulated upon anti-CD40, which was IL-23-dependent (243). A similar role for IL-23 was reported in Helicobacter hepaticus–induced typhlocolitis in Rag2−/− mice, which was also associated with upregulation of IL-17 (in granulocytes, monocytes, and Gr1−CD11b− cells) and confined to the colon (244). In a variation of the H. hepaticus model in T cell sufficient hosts treated with anti-IL-10R, p35−/− mice developed colitis similar in severity to wild-type mice, but p40−/− mice were protected (245). In the T cell–mediated CD45RBhigh model, colitis was prevented in Rag2−/− recipients deficient in p40 or p19, but not p35 (244), again supporting a role for IL-23. The results with p35−/− mice (244) must be interpreted with caution because p35 is also a component of a third IL-12 family member, IL-35 [heterodimer with Epstein Barr virus–induced gene 3 (EBI3)] (246). IL-35 is mainly produced by FoxP3+ Tregs and exerts significant regulatory functions (246).
Nonetheless, these studies support a proinflammatory role for IL-23 in intestinal inflammation, and, consistent with this, IL-23 administration accelerates colitis development in Rag−/− mice reconstituted with CD45RBhigh naive CD4+ T cells or with memory CD4+ T cells from Il10−/− mice (247). Moreover, blockade of IL-6 and IL-17, but not of either cytokine alone starting prior to reconstitution of Rag−/− mice with CD45RBhigh T cells (derived from Il10−/− mice), significantly ameliorates colitis (247). IL-17 is thought to exert proinflammatory activities by inducing CXC chemokines and other chemoattractants from endothelial and epithelial cells, which broadly express the IL-17 receptor (248). IL-17 also contributes to an inflammatory response syndrome subsequent to systemic TNF administration; Paneth cells appear to be an important source of IL-17 in this model. Taken together, these studies support an important role for not only IL-12 but also IL-23 derived from innate immune cells as a driver of chronic intestinal inflammation in CD and, potentially, UC and have focused attention on Th17 cells because of the potential role of Th-derived IL-17 in mediating these processes.
Th17 Cells
Th17 cells, characterized by IL-17 (also known as IL-17A), IL-17F, and IL-22 production, constitute a distinct T helper cell lineage that differentiates from naive T helper cells (249, 250). Th17 cells are important in host defense against bacterial and fungal infections, in particular at mucosal surfaces (248). TCR ligation, IL-6, and TGF-β are required for Th17 lineage commitment (193, 224). IL-6 via STAT3 and TGF-β induce transcription of RORγt (251), a member of the retinoic acid–related orphan nuclear hormone receptor family. RORγt directs the differentiation of Th17 cells, and Rorc−/− mice lack Th17 cells (251). IL-23R is upregulated on RORγt+ Th17 cells by IL-6, and IL-23 expands committed Th17 cells (251). An intriguing feature of IL-23 and Th17 cells is their selective and constitutive presence in the intestinal lamina propria especially within the small intestine, which is dependent on the microbial flora (251, 252). The germ-free state or treatment with broad-spectrum antibiotics decreases lamina propria Th17 cells (39, 251, 253). Specific components drive the expansion of Th17 cells in the intestine, as a single commensal microbe, segmented filamentous bacterium (SFB), which adheres tightly to the surface of IECs in the terminal ileum, induces Th17 cells upon colonization of germ-free mice (40, 41). Adenosine 5′-triphosphate (ATP), which may derive from commensal bacteria, may activate CD70highCD11clow lamina propria cells leading to colonic Th17 differentiation (253). Germ-free mice contain much lower ATP levels in their intestinal lumen compared to specific pathogen–free mice (253) and exhibit a corollary increase in FoxP3+ Tregs. Moreover, CD70highCD11clow lamina propria cells express IL-6, IL-23p19, and TGF-β-activating integrin αVβ8 after ATP stimulation, which are required for Th17 differentiation and consequently for intestinal inflammation (253, 254). Hence, microbial-derived metabolic factors such as ATP, which act upon purinergic receptors (P2X and P2Y), might be critical factors for Th17 differentiation in the intestine.
A recent study assessed the role of Th17-derived IL-17 in the CD45RBhigh transfer model (255). Transfer of Il17a−/− CD45RBhigh T cells into Rag1−/− hosts unexpectedly increased the severity of colitis compared to wild-type transfer, which was associated with increased IFN-γ in the colon (255). Investigators proposed that IL-17A modulates Th1 polarization, suggesting that excessive IFN-γ is the inflammatory factor in the absence of Th-derived IL-17A. Similarly, transfer of Il17r−/− T cells also induced more severe disease compared to wild-type cells, suggesting T cells as both source and target of IL-17 (255). In this context, it is noteworthy that neutralization of IL-17 (256) or genetic deletion of IL-17 (257) is also associated with exacerbation of DSS-induced colitis, while Il17f−/− mice were protected from DSS colitis (257). These studies suggest that IL-17A is surprisingly anti-inflammatory through an undefined mechanism. However, to the contrary, in the TNBS colitis model IL-17 is proinflammatory (258). These apparently ambiguous functions of IL-17 as an effector cytokine contrast with the clearly demonstrated proinflammatory role of IFN-γ, at least in the experimental models discussed above (178, 231). Given recent studies showing that reversal of IL-17-mediated inflammation requires neutralization of both IL-17A and IL-17F (259), we may consider that (a) IL-17A provides a protective function during mucosal inflammation, perhaps through inhibition of IFN-γ, but that (b) unopposed IL-17F may drive intestinal inflammation through a pathway that involves promotion of IFN-γ production. Clearly, the functional relationships between IL-17-related family members and IFN-γ and their relationship with IL-12p40-related cytokines deserves attention in the normal intestine and in IBD.
IL-17 expression, which maps to T lymphocytes and monocytes/macrophages, is increased in both CD and UC (260). In normal human colonic mucosa, IL-17-producing CD4+ T cells are markedly infrequent compared to IFN-γ-producing cells, but their frequency is substantially increased in CD mucosa (261). How this increase in IL-17-producing cells within the lamina propria relates to the presence of SFB and their metabolic factors is unknown, but it appears to be independent of ATP (40). Notably, human Th17 cells in CD mucosa may also express IFN-γ and IL-4, as well as IL-23R and IL-12Rβ2 (Th17/Th1). Furthermore, IL-12 may downregulate RORγt and IL-17 expression while upregulating T-bet (261), consistent with the inhibition of Th17 cells by IFN-γ and IL-4 (262).
Overall, while IL-23-regulated pathways represent an important new concept for understanding the mechanisms of intestinal inflammation, the relative roles of Th17-derived IL-17 (and Th17 cells themselves) and/or non–T cell–derived IL-17 in IBD remain unclear. Similarly, the limited efficacy of IFN-γ blockade in human IBD (Supplemental Table 2), in comparison with its clear inflammatory role in experimental colitis, remains a conundrum. A small subset of CD patients exhibit granulomas, the pathologic hallmark of IFN-γ expression, suggesting that this subset might benefit from IFN-γ blockade, which could be tested. The limited efficacy of anti-IFN-γ in humans could also be due to the release of Th17 cells, highlighting the complexity of consequences of pharmacological intervention. What seems clear, however, is the importance of IL-23 in promoting chronic mucosal inflammation in animal models (243–245, 247) and likely human disease (13). This draws attention to the role IL-23 plays in blocking Treg function, suggesting that innate immune cell–derived IL-23 may promote colitis not as much by mediating IL-17 production but by inhibiting FoxP3+ Treg cell function and its consequent regulation of adaptive and innate immune cell–driven inflammation (263, 264).
Th17–T Regulatory Cell Balance in IBD
A fine balance exists between Th17 cells and the FoxP3+ subset of regulatory CD4+CD25+ T cells that are induced (iTregs) under the control of many of the factors discussed above. The function of natural FoxP3+CD4+CD25+ Tregs emanating from the thymus and iTregs has been extensively reviewed in relation to IBD (228). They build upon the now classic studies of Powrie (70, 265) and Sakaguchi (266) showing that transfer of CD4+CD45RBhigh or CD4+CD25−/− T cells into scid mice promotes intestinal inflammation or autoimmune gastritis, respectively, with prevention of gastrointestinal inflammation provided by cotransfer of CD4+CD25+ Tregs. Both Treg and Th17 differentiation requires TGF-β, which induces FoxP3 and RORγ, and their differentiation is reciprocally regulated in a highly dynamic manner depending on further signals (195, 267). Such signals include low concentrations of TGF-β together with IL-6 (193, 224, 268) and IL-21 (269, 270), which induce Il23r expression and favor development of Th17 cells, in contrast to high concentrations of TGF-β, which represses Il23r expression (267). Similarly, retinoic acid metabolites, provided by IECs and DCs, tip the balance toward Treg differentiation and inhibit Th17 differentiation (271–273). Mechanistically, FoxP3 directly interacts with RORγt to inhibit its function, resulting in decreased Il17 transcription, while IL-6, IL-21, and IL-23, in turn, decrease the FoxP3-mediated inhibition of RORγt (267). It appears increasingly clear that lineage commitment to either Treg or Th17 is substantially more plastic than previously appreciated and under control of both metabolic and innate factors (195).
Despite the massive evidence in support of a central role for natural FoxP3+CD4+CD25+ Tregs and iTregs in experimental intestinal inflammation, mostly obtained through Treg transfer studies into lymphopenic hosts (228), GWASs in CD or UC have not revealed polymorphisms in FOXP3 or in genes directly related to Treg differentiation (11, 274). CD4+CD25+FoxP3+ Treg numbers are increased in the inflamed lamina propria of patients with CD and UC compared to uninflamed mucosa and healthy controls, whereas their numbers are decreased in peripheral blood (275–277). Moreover, CD4+CD25+ Tregs from peripheral blood and mesenteric lymph nodes retain their suppressive activity toward the CD4+CD25− subset (275, 276). Patients with a rare immunodeficiency due to mutant FOXP3, characterized by immune dysregulation, polyendocrinopathy, enteropathy, and inherited as an X-linked monogenic disorder, IPEX, may exhibit profound intestinal inflammation (278), which is in accordance with the scurfy mouse model secondary to a Foxp3 defect (279, 280). Notably, another rare immunodeficiency, Wiskott-Aldrich Syndrome (WAS)—due to genetic defects in the WAS protein—may also develop an UC-like disease along with abnormalities in natural and likely iTregs (281, 282). The WAS protein is functionally linked to specific cytoskeletal elements, making it interesting that genetic polymorphisms in one such protein, encoded by ARPC2 (Arp2/3), have been specifically linked to UC (283).
There are a number of potential interpretations of these observations. One possibility is that some IBD patients might harbor rare mutations in the genetic programs associated with natural FoxP3+CD4+CD25+ Tregs and iTregs, a possibility worth study by novel methodologies in familial and, especially, early-onset IBD. Another possibility is that in sporadic IBD, which is likely to be based upon polygenic inheritance, more subtle abnormalities exist in the pathways that determine the balance between induced Th17-Treg development due to both primary (e.g., IL23R polymorphisms) and secondary (e.g., composition of the microbiome) factors. Strategies aimed to increase the relative balance of Tregs to Th17 (and other inflammatory T cells), such as histone-deacetylase inhibitors that ameliorate DSS colitis through expansion of Tregs (284), could thus have therapeutic potential in human IBD. Furthermore, development and suppressive function of Tregs depend on the expression of factors such as the IL-2 receptor α chain (CD25), the glucocorticoid-induced TNFR-related protein (GITR; TNFRSF18), and cytotoxic T lymphocyte-associated protein 4 (CTLA-4), predicting that targeting these in the pursuit of limiting inflammatory T cells may unintentionally also target Tregs (Supplemental Table 2). Another model that has been promoted by MacDonald and colleagues is that FoxP3+ Tregs are normal in IBD (275–277) but that the effector T cells are resistant to the effects of inhibitory cytokines such as TGF-β in the context of inflammation (285). This is supported by increased phosphorylated SMAD7, an inhibitor of phosphorylated SMAD2/3—which are directly downstream of the TGF-β-receptor—in lamina propria T cells of inflamed intestines (286). Another anti-inflammatory mediator, IL-10, also important in Treg function, is discussed in the next paragraphs.
IL-10 and IL-10 Receptor in IBD
Spontaneous enterocolitis in Il10−/− mice represents one of the first models of intestinal inflammation and unequivocally supports the involvement of immune mechanisms in IBD (5). Colitis was attenuated in Il10−/− mice held under specific pathogen–free conditions compared to those raised conventionally (5), whereas germ-free Il10−/− mice were completely protected (287), highlighting the critical contribution of the microbiota. Moreover, H. hepaticus causes severe inflammation in Il10−/−, but not in wild-type, mice (67), indicating that this bacterium (and potentially others) has pathogenic potential in this context. The importance of IL-10 in preventing intestinal inflammation is further supported by the spontaneous colitis in Il10rb−/− mice (25) and Blimp1−/− mice, which exhibit a defect in IL-10 production (288).
IL-10 exerts a multitude of anti-inflammatory and immunoregulatory functions and may be produced by a variety of regulatory subsets of T cells, B cells, DCs, parenchymal cells, and IECs (289, 290), making it important to define the sources in intestinal tissues in order to understand the mechanisms of action. In situ hybridization has suggested constitutive expression in human IECs and increased expression in lamina propria mononuclear cells in both CD and UC (290). IL-10 expression in mice (as detected through an IRES GFP knock-in) is low in bone marrow–derived DCs and macrophages but was potently induced in the small intestinal intraepithelial lymphocyte (IEL) and colonic lamina propria lymphocyte compartments upon TCR stimulation, highlighting the intestine as a unique site for induction of IL-10-producing T cells (291). Consistent with this, T cell–specific deletion of floxed Il10 alleles results in spontaneous colitis under specific pathogen–free conditions (292). Similarly, Il10 deletion in FoxP3+ Tregs results in spontaneous colitis, but not in systemic autoimmunity (293), which contrasts with the severe autoimmune disease (including enterocolitis) in FoxP3−/− mice (294, 295) or in patients with FOXP3 mutations causing IPEX (280). These studies highlight the unique importance of IL-10 derived primarily from FoxP3+ Tregs, intestinal T cells, and, possibly, IECs in the prevention of inflammation in the intestines. Importantly, whereas most T cell–derived IL-10 in the colon is from natural and induced FoxP3+ Treg, in the small intestine it derives from FoxP3− T cells, which appear to localize to the epithelium, possibly reflecting other regulatory subsets of T cells such as Tr1 cells (296, 297).
As noted earlier, IL-10 signaling involves STAT3, a genetic risk factor for IBD. IL-10-induced SOCS3 expression is abrogated in Stat3-deficient macrophages, leading to unabated IL-12/23p40 expression upon exposure to microbial products like LPS (205). One important pathway maintaining mucosal homeostasis (or tolerance), which is important in IBD, might therefore be mediated by IL-10 derived from FoxP3+ Tregs that act upon IL-10R on mucosal macrophages to stimulate STAT3-induced SOCS3 expression, which quells TLR-mediated secretion of IL-23 and other inflammatory cytokines. Moreover, STAT3 signaling, perhaps via IL-10, further enables FoxP3 in natural Tregs to specifically suppress Th17 cells (298).
Based upon these insights, subcutaneous recombinant IL-10 was examined as a potential therapeutic in CD and UC (Supplemental Table 2), but it was not effective. As local delivery could offer advantages, Lactococcus lactis engineered to secrete IL-10 ameliorated colitis in the DSS and Il10−/− models (299), but efficacy in humans remains to be shown. Nonetheless, it is extremely notable that a recent GWAS discovered polymorphisms in a SNP flanking IL10 as the most significant association outside the MHC for UC (283). Resequencing of IL10 revealed numerous rare and private variants; several nsSNPs are predicted to affect binding to the IL-10RA receptor chain (283). The identification of such rare, private variants is notable given the recent identification of a monogenic form of CD associated with mutations in IL10RA or IL10RB, as discussed earlier (23). In summary, impairment of IL-10-associated regulatory pathways appears to be critical in both forms of IBD as deduced from functional and genetic studies.
LEUKOCYTE HOMING PATHWAYS AS MODIFIERS OF IBD
The ability of the host to regulate the movement of innate (monocytes and DCs) and adaptive (Tregs) regulatory cells relative to inflammatory populations into the intestines via the sequential processes of tethering (e.g., selectins), rolling (e.g., ICAM), adhesion (e.g., integrins), and transmigration (e.g., chemokines) through the endothelium and consequently localization to their specific destinations within the intestinal microenvironment is of increasingly recognized importance in human IBD and has been extensively reviewed (300, 301). Recent studies indicate that environmental and genetic factors might impact these pathways, which are therapeutic targets as well, as detailed below.
Compartmentalization of Leukocyte Homing and the Role of Environmental and Metabolic Factors
Small intestinal homing of T and B cells is imprinted by DCs in Peyer’s patches and mesenteric lymph nodes by induction of α4β7 integrin and CCR9 (see below), which interact with mucosal vascular addressin cell adhesion molecule-1 (MAdCAM1) and CCL25, respectively, on the surface of lamina propria postcapillary venules (302, 303). Effector T cell entry into the small IEL compartment involves αEβ7 integrin, which interacts with E-cadherin on the basolateral surface of small IECs (301).
The selectins are more broadly expressed, with L-selectin on most leukocytes, P-selectin on inflamed endothelial cells and activated platelets, and E-selectin on inflamed endothelial cells (300). Blockade of the pan-selectin lig- and P-selectin glycoprotein ligand-1 (PSGL1) attenuates spontaneous ileitis in SAMP1/YitFc mice (304, 305) and established DSS colitis (306). In contrast, blockade of L-selectin increases severity of TNBS colitis potentially because of effects on neutrophil recruitment (307).
Chemokine receptor signaling is important in homeostasis and inflammation; two pathways, CCL20-CCR6 and CCL25-CCR9, are particularly relevant for IBD in view of genetic insights, the role of environmental factors (e.g., microbiota and metabolic factors), and their therapeutic manipulation. Inflammatory signals, including IL-17, induce CCL20, which exhibits increased expression in the colonic epithelium of IBD (308–310). Th17 cells, in turn, express CCR6 and CCR4 (311, 312). As a result, Ccr6−/− Th17 cells exhibit an altered homing pattern and migrate to different compartments of the intestine (312). Adoptive transfer of Ccr6−/− Th17 cells into scid mice augments intestinal inflammation along with Th1 deviation of intestinal T cells (312), and Ccr6−/− mice are more susceptible to TNBS colitis (313), implying a potential regulatory function of Th17 cells as noted above (255). However, CCL20 neutralization ameliorates TNBS colitis (314) and reduces severity in DSS colitis (313). Given the pleiotropic activities of chemokine ligands, it is conceivable that CCL20, although the only CCR6 ligand, may interact with other chemokine receptors. Notably, Tregs also express CCR6, and Th17 cells may express CCL20, which could cross-regulate Treg recruitment as another example of the dynamic tension between these two functional populations (315).
CCR6-CCL20 also affects the development of intestinal lymphoid structures, as Ccr6 deletion compromises Peyer’s patch and isolated lymphoid follicle (ILF) development, and some c-kit+ lymphoid precursors in cryptopatches express CCR6 (316). The generation of ILFs requires signaling from the intestinal microbiota via NOD1 in IECs, resulting in regulation of CCL20/CCR6 signaling and β-defensin 3 production (317). Notably, in the absence of ILFs, the microbial community is profoundly altered in a corollary manner (317). Hence, there might be reciprocal regulation of the bacterial flora and ILFs, which could be relevant for the lymphoid aggregations observed within the colon of IBD.
In contrast to CCL20, CCL25 production is confined to small IECs and tethered to the endothelium, where it recruits T effector populations to the IEL and lamina propria compartments (301). Although microbial factors modify CCL20-CCR6 signaling pathways as discussed above, metabolic factors, specifically retinoic acid metabolites, affect CCL25-CCR9 pathways. Retinoic acids enhance TGF-β-mediated signaling (318), increase FoxP3 expression, and promote expression of α4β7 and CCR9 in gut T cells activated by IECs and DCs from gut-associated lymphoid tissue DCs, but not in other tissues, owing to the restricted presence of the retinoic acid biosynthetic pathway (319). These studies underscore the multidirectional interactions between the (microbial and metabolic) environment, the innate immune system, and the adaptive immune system.
Genetic Factors Controlling Leukocyte Homing in IBD
A meta-analysis of GWAS data identified the CCR6 locus as associated with CD (20), which might affect lymphocyte homing, but also ILF induction and, hence, host-microbiota homeostasis. Similarly, NKX2-3, encoding a homeobox transcription factor involved in the localization of T and B lymphocytes to mesenteric lymph nodes, appears to be a risk factor for UC and CD (139, 196, 237).
Therapeutic Evidence for Leukocyte Homing as a Mediator of Human IBD
Importance of integrin blockade in IBD was originally implied from studies of colitis in the cotton-top tamarin (320). Blockade of specific integrins has been extensively studied in several rodent models of intestinal inflammation. As an illustrative example, TnfΔAREβ7−/− mice are protected from the development of colitis owing to the absence of β7-integrin-containing (α4β7 and αEβ7) heterodimers (321). Consistent with these animal studies, a blocking anti-α4β7 antibody (MLN002) exhibited some evidence of therapeutic efficacy in phase II clinical trials in UC and CD (Supplemental Table 2), while an α4-blocking antibody (Natalizumab) was beneficial in a subpopulation of CD patients (Supplemental Table 2). Although no direct comparisons have been performed, the reported remission rates suggest that α4β7-based anti-integrin strategies might be effective in a more confined patient population compared to anti-TNF-based strategies, and this apparently reduced effectiveness could be related to the fact that anti-integrin strategies might affect the homing of effector, but also regulatory populations. Similarly, CCR9 antagonists (Ccx282-B) appear to be efficacious in TnfΔARE-associated enterocolitis (322), albeit mice deficient in either CCL25 or CCR9 were not protected from colitis in the TnfΔARE model (321). Nonetheless, Ccx282-B exhibits some therapeutic activity in CD in preliminary studies (Supplemental Table 2).
LIPID METABOLISM AND ITS ROLE IN THE REGULATION OF INFLAMMATION
Recent evidence from genetic and immunologic studies suggests that metabolism and inflammation are tightly linked processes that cross-regulate each other, with significant implications for IBD (323). Specifically, mammalian and commensal microbial lipids have been demonstrated to play key roles in the regulation of inflammation through their function as (a) ligands of lipid-activated nuclear receptors such as PPAR, retinoic acid receptors, and steroid-hormone receptors; (b) regulators of gene expression through histone deacetylase inhibition, as demonstrated for SCFA; (c) pro- or anti-inflammatory mediators, including those associated with prostaglandins, leukotrienes, lipoxins, resolvins, and protectins; (d) intracellular signaling molecules including sphingolipid- and phosphatidylinositol-derived second messengers; and (e) antigens in CD1-mediated immune responses as discussed previously in this review. In this final section, we discuss the regulation of intestinal inflammation by arachidonic acid metabolites, PPARs, and polyunsaturated fatty acid (PUFA) derivatives in view of recent available information linking these to the immunogenetic basis of IBD and their special importance as potential environmental modifying agents.
Arachidonic Acid Pathways
The two isoforms of the enzyme cyclooxygenase (COX1 and COX2) catalyze the production of prostanoids from arachidonic acid, including prostaglandin D2 (PGD2), PGE2, PGF2α, PGI2, and thromboxane (TX). These act through their respective receptors, PGD receptor (DP), PGE receptor (EP), PGF receptor (FP), PGI receptor (IP), and TX receptor (TP) (324). Nonsteroidal anti-inflammatory drugs (NSAIDs), which share COX inhibition as a common mechanism, have been associated with the relapse of IBD, hence representing an environmental factor intersecting with the immunogenetic biology of IBD. In line with this is the increased susceptibility of mice deficient in COX1 (Ptgs1−/−) and COX2 (Ptgs2−/−) to DSS colitis (325), and pharmacological inhibition of COX1 and COX2 points toward PGE2 as the protective factor in colitis (326). Indeed, genetic deletion of the aforementioned prostanoid receptors has shown that only mice deficient in EP4 (Ptger4−/−), the receptor for PGE2, exhibit increased susceptibility to DSS colitis (326). In human colonic mucosa, EP4 is expressed in the intestinal epithelium and in lamina propria mononuclear cells, including CD3+ and myeloid cells (327, 328). Notably, mucosal barrier function is impaired early after DSS administration upon treatment with an EP4 antagonist (326). It is unclear whether mucosal barrier dysfunction is through a direct effect on IECs or secondary to inflammatory mediators. Consistent with the latter possibility, PGE2-EP4 signaling has been shown to downregulate lamina propria CD4+ T cell proliferation and IFN-γ production (326), which may directly damage IEC tight junctions (329). PGE2 may also suppress chemokine secretion from LPS-stimulated macrophages via EP4 (330), stimulate IL-10 production (331), and suppress the degradation of NF-κB1 p105/p50 (331). Thus, PGE2-EP4 signaling may facilitate mucosal protection through anti-inflammatory IEC, macrophage, and CD4+ T cell function.
However, PGE2-EP4 and -EP2 signaling was recently shown to facilitate IL-1- and IL-23-mediated Th17 differentiation from naive CD4+ T cells (332, 333). Notably, EP4 antagonist treatment ameliorated EAE and contact hypersensitivity (CHS) (332), suggesting disparate roles of PGE2-EP4 signaling in experimental colitis compared to EAE and CHS, two models that can be driven by IL-23 and Th17 cells in a specific immunopathologic pathway.
Recently, PTGER4 has been implicated in the genetic basis of CD (334). Specifically, a GWAS revealed strong evidence for an association of a 250-kb region on chromosome 5p13.1, which is contained within a 1.25 Mb gene desert (334). PTGER4 is the closest gene, 270 kb proximally from the CD-associated block. Quantitative trait locus analysis revealed that individuals harboring two specific 5p13.1 risk alleles exhibited increased PTGER4 mRNA expression (possibly through cis acting elements), suggesting PTGER4 as the causal gene (334). However, increased PTGER4 expression associated with CD is at odds with the increased severity of DSS colitis in Ptger4−/− mice (326). Further studies are required to reconcile these apparent inconsistencies. Nonetheless, a significant body of epidemiologic, immunologic, and genetic information supports a role for arachidonic acid metabolites in IBD.
PPARγ
PPARs are lipid-activated transcription factors that form obligate heterodimers with the retinoid X receptor (RXR) (323). In the absence of ligands, RXR heterodimers are bound to DNA and repress expression of target genes, whereas ligand binding causes release of corepressors leading to the initiation of gene transcription (323). PPARγ is expressed in liver, adipose tissue, IECs, and hematopoietic cells and plays a key role in the regulation of lipid metabolism, inflammation, and cancer (335). The molecular mechanisms of gene regulation by PPARγ are manifold, including direct peroxisome proliferator response element (PPRE)-driven transcriptional activation and PPRE-independent transrepression through interference with AP1, NF-κB, p38, and transcriptional coactivators and repressors (323).
A variety of naturally occurring ligands have been proposed for PPARγ, including unsaturated fatty acids, eicosanoids (e.g., 15dPGJ2), and lysophosphatidic acid (335). In addition, PPARγ is the target of thiazolidinedione drugs and 5-aminosalicylic acid (5-ASA) (336). PPARγ is highly expressed in the intestinal epithelium, and its expression is dependent on PRR signaling by the intestinal microbiota and direct transcriptional regulation by microbial metabolites such as butyrate (337, 338). Consistent with this, both germ-free and TLR4−/− mice exhibit decreased intestinal epithelial expression of PPARγ (337).
Several lines of evidence suggest involvement of PPARγ in IBD. First, thiazolidinediones were shown to be effective in the treatment of human IBD and various mouse models of intestinal inflammation. Thus, both PPARγ agonists block CXCL8 and CCL2 production through IκBα-dependent inhibition of NF-κB and consequently protect from DSS-induced colitis (339). These findings are confirmed in various other models of colitis induced by chemicals, bacteria, ischemia-reperfusion, T cell transfer into immunocompromised hosts, and spontaneous models of IBD including IL-10-deficient mice and SAMP1/YitFc mice (335). The protective effects of thiazolidinediones in these models seem to be mediated mainly via IECs and not via hematopoietic cells because conditional PPARγ deletion in the intestinal epithelium leads to mild spontaneous colitis, increased susceptibility to DSS colitis, and prevention of the therapeutic effect of some but not all PPARγ agonists (340).
Importantly, these observations extend to human IBD because rosiglitazone was shown to be efficacious in the treatment of mild to moderate UC (341). In addition, the therapeutic efficacy of 5-ASA derivatives seems to be at least partially mediated via PPARγ. Thus, 5-ASA was shown to bind to PPARγ leading to activation of PPRE-driven gene transcription (336). In addition, PPARγ heterozygous mice, which exhibit a 70% decrease in colonic PPARγ levels, exhibit pronounced intestinal inflammation upon TNBS challenge and cannot be protected by 5-ASA (336).
These findings demonstrate a crucial role of PPARγ signaling in the prevention of intestinal inflammation and suggest that agonistic targeting of PPARγ might be an effective treatment for IBD. However, these observations also raise the question of whether impaired PPARγ signaling might be a causal factor in IBD. Support for this idea stems from the notion that patients with UC show decreased expression of PPARγ in IECs but not in hematopoietic cells (337). In addition, resistance to ileitis in Samp1/YitFc mice on the AKR background has been linked to increased PPARγ expression in the intestinal crypt epithelium. The reason for impaired PPARγ expression in UC remains enigmatic. However, given the crucial role of commensal-derived signals in the induction of PPARγ, it is possible that changes in the microbiota or its products contribute to impaired PPARγ expression in UC.
Notably, several genes implicated in IBD pathogenesis are functionally linked to PPARs. Thus, mice hypomorphic for the CD-associated gene ATG16L1 showed altered Paneth cell morphology and dramatically increased PPAR signaling (45). These findings might explain why CD patients, in contrast to UC patients, show normal PPARγ expression. In addition, recent evidence also suggests cross-regulation of PPAR signaling and ER stress. Specifically, pioglitazone was shown to reduce ER stress leading to pancreatic islet cell protection and prevention of diabetes (342). In addition to being a regulator of the UPR, PPARs also seem to be targets of ER stress, as demonstrated by increased expression of PPARγ but decreased expression of PPARα in response to ER stress (343). These findings demonstrate the existence of a feedback loop between ER stress and PPARγ signaling and suggest that agonistic targeting of PPARγ might exert its beneficial effects at least in part through alleviation of ER stress.
Lipoxins, Resolvins, and Protectins
It is increasingly recognized that resolution of inflammation is not a passive process but is actively regulated not only by cell surface receptors such as CEACAM1 and cytokines (e.g., TGF-β, IL-10, IL-35) but also by several families of lipid-derived signaling molecules, namely lipoxins, resolvins, and protectins (344). Of these, resolvins and protectins are derived from omega-3 PUFAs, whereas lipoxins are derived from arachidonic acid, the same precursors that give rise to proinflammatory leukotrienes and prostaglandins in a process called lipid class switching (345). Thus, after an acute inflammatory phase driven by leukotrienes and prostaglandins, these same mediators, mostly PGE2 and PGD2, induce key enzymes involved in the biosynthesis of lipoxins leading to resolution of inflammation (345). Importantly, proresolution pathways driven by lipoxins, resolvins, and protectins are distinct from classical anti-inflammatory pathways in that they are not immunosuppressive but actively accelerate resolution of inflammation by inhibition of neutrophil and eosinophil recruitment and stimulation of macrophage phagocytosis with clearance of microorganisms and apoptotic cells (344). Given that the intestinal mucosa is permanently exposed to microorganisms, it is not surprising that some of these lipid mediators, including lipoxins, are constitutively expressed in the healthy mucosa (346). However, in contrast to healthy controls, patients with UC express significantly lower levels of 15-lipoxygenase-2, a key enzyme involved in lipoxin production, and consequently low levels of lipoxin (346). In addition to the putative role of anti-inflammatory lipid mediators in the pathogenesis of IBD, both resolvins and lipoxins are well characterized for their protective therapeutic role in intestinal inflammation. Thus, resolvin E1 was shown to protect from TNBS colitis through regulation of innate and adaptive immune responses (347), and mice overexpressing n3-PUFA are protected from DSS colitis. In addition, omega-3 PUFA may reduce the rates of relapse in CD (348, 349). Similar observations have been made with lipoxin analogs in that they attenuate TNBS colitis concomitant with decreased expression of proinflammatory cytokines (350). In addition to their anti-inflammatory effects on immune cells, lipoxins regulate immune responses of IECs, given that lipoxin A4 analogs reduce NF-κB-mediated transcriptional activation, inhibit degradation of IκBα in response to Salmonella typhimurium in IECs, and protected from DSS colitis (351). Lipoxins, resolvins, and protectins might therefore be of therapeutic efficacy in the prevention and treatment of intestinal inflammation and await testing in human IBD.
CONCLUDING REMARKS
IBD results from a continuum of complex interactions between a quartet of host-derived and external elements that involve various aspects of the intestinal microbiota, the immune system, the genetic composition of the host, and specific environmental factors. Recent studies into the complexity of these arrangements increasingly support not only the syndromic nature of this disorder, but also the need for systems-based approaches in understanding the biologic pathways involved and the correlation of these arrangements with specific phenotypic outcomes that go beyond the assigned clinical descriptors currently in practice, namely UC and CD. Studies of the microbiota, immune system, and genetics have revealed more similarities than differences between these two extreme phenotypes, suggesting this continuum of interactions is similarly reflected in a continuous lineage of functional pathways and, consequently, phenotypes. Genetic studies, for example, increasingly support the concept of familial and sporadic forms of IBD whose inheritance ranges from monogenic to polygenic and involve a wide range of biologic pathways that affect innate immunity, adaptive immunity, ER stress and autophagy, and metabolic pathways associated with cellular homeostasis and the regulation of inflammation per se. Moreover, these genetic observations, together with immunologic studies, emphasize the particularly important role played by abnormalities of the innate immune functions of hematopoietic and non-hematopoietic cells, especially within the intestinal epithelium and its unique relationship with the commensal microbiota, in influencing and being influenced by the adaptive immune system. Such observations increasingly support a long-held view that the chronic intestinal inflammation associated with IBD may be a secondary consequence of innate immune deficiency (or dysfunction). These immunogenetic observations, together with the “reality-test” provided by biologic therapies in human IBD, also provide a foundation for this model given the evidence to date that supports targets such as TNF, IL-6, and IL-12/23 as important mediators of the major IBD phenotypes. Finally, given these comments, it can be anticipated that environmental factors that modify the risk for development of IBD have the common attribute of affecting the relationship between the commensal microbiota and the immune system in a manner that intersects with the functionally relevant immunogenetic pathway(s) that are uniquely operative within a particular context of IBD.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Stephen B. Hanauer, University of Chicago, and Dr. Andre Franke, Christian-Albrechts University of Kiel, for helpful discussions. The authors acknowledge support by NIH grants DK51362, DK44319, and DK53056, as well as the Harvard Digestive Diseases Center and the Crohn’s and Colitis Foundation of America (to R.S.B.); the Deutsche Forschungsgemeinschaft (Ze 814/1-1) and the Crohn’s and Colitis Foundation of America (to S.Z.); and grants START-Y446 from the Austrian Ministry of Science, P21530 from the Austrian Science Fund, and MFI 2007-407 from Innsbruck Medical University (to A.K.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Halme L, Paavola-Sakki P, Turunen U, Lappalainen M, Farkkila M, Kontula K. Family and twin studies in inflammatory bowel disease. World J. Gastroenterol. 2006;12:3668–72. doi: 10.3748/wjg.v12.i23.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsironi E, Feakins RM, Probert CS, Rampton DS, Phil D. Incidence of inflammatory bowel disease is rising and abdominal tuberculosis is falling in Bangladeshis in East London, United Kingdom. Am. J. Gastroenterol. 2004;99:1749–55. doi: 10.1111/j.1572-0241.2004.30445.x. [DOI] [PubMed] [Google Scholar]
- 3.Thia KT, Loftus EV, Jr, Sandborn WJ, Yang SK. An update on the epidemiology of inflammatory bowel disease in Asia. Am. J. Gastroenterol. 2008;103:3167–82. doi: 10.1111/j.1572-0241.2008.02158.x. [DOI] [PubMed] [Google Scholar]
- 4.Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75:253–61. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 5.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 6.Mombaerts P, Mizoguchi E, Grusby MJ, Glimcher LH, Bhan AK, Tonegawa S. Spontaneous development of inflammatory bowel disease in T cell receptor mutant mice. Cell. 1993;75:274–82. doi: 10.1016/0092-8674(93)80069-q. [DOI] [PubMed] [Google Scholar]
- 7.Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu. Rev. Immunol. 2002;20:495–549. doi: 10.1146/annurev.immunol.20.100301.064816. [DOI] [PubMed] [Google Scholar]
- 8.Baumgart DC, Sandborn WJ. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369:1641–57. doi: 10.1016/S0140-6736(07)60751-X. [DOI] [PubMed] [Google Scholar]
- 9.Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu. Rev. Immunol. 2009;27:363–91. doi: 10.1146/annurev.immunol.021908.132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–27. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Limbergen J, Wilson DC, Satsangi J. The genetics of Crohn’s disease. Annu. Rev. Genomics Hum. Genet. 2009;10:89–116. doi: 10.1146/annurev-genom-082908-150013. [DOI] [PubMed] [Google Scholar]
- 12.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 2008;8:458–66. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 13.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–63. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007;39:207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 15.Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat. Rev. Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- 16.Budarf ML, Labbe C, David G, Rioux JD. GWA studies: rewriting the story of IBD. Trends Genet. 2009;25:137–46. doi: 10.1016/j.tig.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Zielenski J. Genotype and phenotype in cystic fibrosis. Respiration. 2000;67:117–33. doi: 10.1159/000029497. [DOI] [PubMed] [Google Scholar]
- 18.Targan SR, Karp LC. Defects in mucosal immunity leading to ulcerative colitis. Immunol. Rev. 2005;206:296–305. doi: 10.1111/j.0105-2896.2005.00286.x. [DOI] [PubMed] [Google Scholar]
- 19.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–67. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein DB. Common genetic variation and human traits. N. Engl. J. Med. 2009;360:1696–98. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 22.Casanova JL, Abel L. Revisiting Crohn’s disease as a primary immunodeficiency of macrophages. J. Exp. Med. 2009;206:1839–43. doi: 10.1084/jem.20091683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 2009;361:2033–45. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez-Cubero SO, Sullivan KM, McDonald GB. Course of Crohn’s disease after allogeneic marrow transplantation. Gastroenterology. 1998;114:433–40. doi: 10.1016/s0016-5085(98)70525-6. [DOI] [PubMed] [Google Scholar]
- 25.Spencer SD, Di Marco F, Hooley J, Pitts-Meek S, Bauer M, et al. The orphan receptor CRF2–4 is an essential subunit of the interleukin 10 receptor. J. Exp. Med. 1998;187:571–78. doi: 10.1084/jem.187.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 27.Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J. Clin. Invest. 2008;118:534–44. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–94. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 30.Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J. Exp. Med. 1994;180:2359–64. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iqbal N, Oliver JR, Wagner FH, Lazenby AS, Elson CO, Weaver CT. T helper 1 and T helper 2 cells are pathogenic in an antigen-specific model of colitis. J. Exp. Med. 2002;195:71–84. doi: 10.1084/jem.2001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cong Y, Brandwein SL, McCabe RP, Lazenby A, Birkenmeier EH, et al. CD4+ T cells reactive to enteric bacterial antigens in spontaneously colitic C3H/HeJBir mice: increased T helper cell type 1 response and ability to transfer disease. J. Exp. Med. 1998;187:855–64. doi: 10.1084/jem.187.6.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–59. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–84. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 36.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. USA. 2008;105:20858–63. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Umesaki Y, Setoyama H, Matsumoto S, Okada Y. Expansion of αβ T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology. 1993;79:32–37. [PMC free article] [PubMed] [Google Scholar]
- 38.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–25. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 39.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–49. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–89. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 42.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–86. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nieuwenhuis EE, Matsumoto T, Lindenbergh D, Willemsen R, Kaser A, et al. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J. Clin. Invest. 2009;119:1241–50. doi: 10.1172/JCI36509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–56. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–34. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 48.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–97. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–59. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 50.Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Invest. 2007;117:1566–74. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–65. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 52.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA. 2007;104:13780–85. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, et al. Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc. Natl. Acad. Sci. USA. 2005;102:18129–34. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petnicki-Ocwieja T, Hrncir T, Liu Y-J, Biswas A, Hudcovic T, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl. Acad. Sci. USA. 2009;106:15813–18. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, et al. Nod2 mutation in Crohn’s disease potentiates NF-κB activity and IL-1β processing. Science. 2005;307:734–38. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- 56.Pinedo PJ, Buergelt CD, Donovan GA, Melendez P, Morel L, et al. Association between CARD15/NOD2 gene polymorphisms and paratuberculosis infection in cattle. Vet. Microbiol. 2009;134:346–52. doi: 10.1016/j.vetmic.2008.09.052. [DOI] [PubMed] [Google Scholar]
- 57.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–29. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 59.Targan SR, Landers CJ, Yang H, Lodes MJ, Cong Y, et al. Antibodies to CBir1 flagellin define a unique response that is associated independently with complicated Crohn’s disease. Gastroenterology. 2005;128:2020–28. doi: 10.1053/j.gastro.2005.03.046. [DOI] [PubMed] [Google Scholar]
- 60.Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–82. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 61.Neish AS, Gewirtz AT, Zeng H, Young AN, Hobert ME, et al. Prokaryotic regulation of epithelial responses by inhibition of IκB-α ubiquitination. Science. 2000;289:1560–63. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 62.Kelly D, Campbell JI, King TP, Grant G, Jansson EA, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat. Immunol. 2004;5:104–12. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 63.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310:847–50. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 65.Ha EM, Lee KA, Seo YY, Kim SH, Lim JH, et al. Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in drosophila gut. Nat. Immunol. 2009;10:949–57. doi: 10.1038/ni.1765. [DOI] [PubMed] [Google Scholar]
- 66.Bernstein CN, Shanahan F. Disorders of a modern lifestyle: reconciling the epidemiology of inflammatory bowel diseases. Gut. 2008;57:1185–91. doi: 10.1136/gut.2007.122143. [DOI] [PubMed] [Google Scholar]
- 67.Cahill RJ, Foltz CJ, Fox JG, Dangler CA, Powrie F, Schauer DB. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect. Immun. 1997;65:3126–31. doi: 10.1128/iai.65.8.3126-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, et al. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect. Immun. 1998;66:5157–66. doi: 10.1128/iai.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kullberg MC, Andersen JF, Gorelick PL, Caspar P, Suerbaum S, et al. Induction of colitis by a CD4+ T cell clone specific for a bacterial epitope. Proc. Natl. Acad. Sci. USA. 2003;100:15830–35. doi: 10.1073/pnas.2534546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int. Immunol. 1993;5:1461–71. doi: 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- 71.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA. 2008;105:16731–36. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaser A, Blumberg RS. Endoplasmic reticulum stress in the intestinal epithelium and inflammatory bowel disease. Semin. Immunol. 2009;21:156–63. doi: 10.1016/j.smim.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, et al. Bacterial flagellin is a dominant antigen in Crohn disease. J. Clin. Invest. 2004;113:1296–306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brandtzaeg P, Carlsen HS, Halstensen TS. The B-cell system in inflammatory bowel disease. Adv. Exp. Med. Biol. 2006;579:149–67. doi: 10.1007/0-387-33778-4_10. [DOI] [PubMed] [Google Scholar]
- 75.Kobayashi K, Qiao SW, Yoshida M, Baker K, Lencer WI, Blumberg RS. An FcRn-dependent role for anti-flagellin immunoglobulin G in pathogenesis of colitis in mice. Gastroenterology. 2009;137:1746–56. doi: 10.1053/j.gastro.2009.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–20. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc. Natl. Acad. Sci. USA. 2009;106:19256–61. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vijay-Kumar M, Sanders CJ, Taylor RT, Kumar A, Aitken JD, et al. Deletion of TLR5 results in spontaneous colitis in mice. J. Clin. Invest. 2007;117:3909–21. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 80.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 81.Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet. 2001;357:1925–28. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 82.Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, et al. CARD15 mutations in Blau syndrome. Nat. Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 83.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 84.Lesage S, Zouali H, Cezard JP, Colombel JF, Belaiche J, et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am. J. Hum. Genet. 2002;70:845–57. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Coulombe F, Divangahi M, Veyrier F, de Leseleuc L, Gleason JL, et al. Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide. J. Exp. Med. 2009;206:1709–16. doi: 10.1084/jem.20081779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, et al. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 2009;10:1073–80. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gutierrez O, Pipaon C, Inohara N, Fontalba A, Ogura Y, et al. Induction of Nod2 in myelomonocytic and intestinal epithelial cells via nuclear factor-κB activation. J. Biol. Chem. 2002;277:41701–5. doi: 10.1074/jbc.M206473200. [DOI] [PubMed] [Google Scholar]
- 88.Hisamatsu T, Suzuki M, Reinecker HC, Nadeau WJ, McCormick BA, Podolsky DK. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology. 2003;124:993–1000. doi: 10.1053/gast.2003.50153. [DOI] [PubMed] [Google Scholar]
- 89.Shaw MH, Reimer T, Sánchez-Valdepeñas C, Warner N, Kim YG, et al. T cell-intrinsic role of Nod2 in promoting type 1 immunity to Toxoplasma gondii. Nat. Immunol. 2009;10:1267–74. doi: 10.1038/ni.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Abbott DW, Wilkins A, Asara JM, Cantley LC. The Crohn’s disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr. Biol. 2004;14:2217–27. doi: 10.1016/j.cub.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 91.Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J. Biol. Chem. 2001;276:4812–18. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 92.Abbott DW, Yang Y, Hutti JE, Madhavarapu S, Kelliher MA, Cantley LC. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol. Cell. Biol. 2007;27:6012–25. doi: 10.1128/MCB.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J. Biol. Chem. 2003;278:5509–12. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 94.van Heel DA, Ghosh S, Butler M, Hunt KA, Lundberg AM, et al. Muramyl dipeptide and Toll-like receptor sensitivity in NOD2-associated Crohn’s disease. Lancet. 2005;365:1794–96. doi: 10.1016/S0140-6736(05)66582-8. [DOI] [PubMed] [Google Scholar]
- 95.Hedl M, Li J, Cho JH, Abraham C. Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc. Natl. Acad. Sci. USA. 2007;104:19440–45. doi: 10.1073/pnas.0706097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Watanabe T, Kitani A, Murray PJ, Strober W. NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat. Immunol. 2004;5:800–8. doi: 10.1038/ni1092. [DOI] [PubMed] [Google Scholar]
- 97.Watanabe T, Kitani A, Murray PJ, Wakatsuki Y, Fuss IJ, Strober W. Nucleotide binding oligomerization domain 2 deficiency leads to dysregulated TLR2 signaling and induction of antigen-specific colitis. Immunity. 2006;25:473–85. doi: 10.1016/j.immuni.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 98.Noguchi E, Homma Y, Kang X, Netea MG, Ma X. A Crohn’s disease-associated NOD2 mutation suppresses transcription of human IL10 by inhibiting activity of the nuclear ribonucleoprotein hnRNP-A1. Nat. Immunol. 2009;10:471–79. doi: 10.1038/ni.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Netea MG, Kullberg BJ, de Jong DJ, Franke B, Sprong T, et al. NOD2 mediates anti-inflammatory signals induced by TLR2 ligands: implications for Crohn’s disease. Eur. J. Immunol. 2004;34:2052–59. doi: 10.1002/eji.200425229. [DOI] [PubMed] [Google Scholar]
- 100.Simms LA, Doecke JD, Walsh MD, Huang N, Fowler EV, Radford-Smith GL. Reduced alpha-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn’s disease. Gut. 2008;57:903–10. doi: 10.1136/gut.2007.142588. [DOI] [PubMed] [Google Scholar]
- 101.Barnich N, Aguirre JE, Reinecker HC, Xavier R, Podolsky DK. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor-κB activation in muramyl dipeptide recognition. J. Cell Biol. 2005;170:21–26. doi: 10.1083/jcb.200502153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 2009 doi: 10.1038/ni.1823. doi:10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 103.Cooney R, Baker J, Brain O, Danis B, Pichulik T, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 2009 doi: 10.1038/nm.2069. doi:10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 104.Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 105.Vallabhapurapu S, Karin M. Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 106.Neurath MF, Pettersson S, Meyer zum Buschenfelde KH, Strober W. Local administration of antisense phosphorothioate oligonucleotides to the p65 subunit of NF-κB abrogates established experimental colitis in mice. Nat. Med. 1996;2:998–1004. doi: 10.1038/nm0996-998. [DOI] [PubMed] [Google Scholar]
- 107.Rogler G, Brand K, Vogl D, Page S, Hofmeister R, et al. Nuclear factor κB is activated in macrophages and epithelial cells of inflamed intestinal mucosa. Gastroenterology. 1998;115:357–69. doi: 10.1016/s0016-5085(98)70202-1. [DOI] [PubMed] [Google Scholar]
- 108.MacMaster JF, Dambach DM, Lee DB, Berry KK, Qiu Y, et al. An inhibitor of IκB kinase, BMS-345541, blocks endothelial cell adhesion molecule expression and reduces the severity of dextran sulfate sodium-induced colitis in mice. Inflamm. Res. 2003;52:508–11. doi: 10.1007/s00011-003-1206-4. [DOI] [PubMed] [Google Scholar]
- 109.Eckmann L, Nebelsiek T, Fingerle AA, Dann SM, Mages J, et al. Opposing functions of IKKβ during acute and chronic intestinal inflammation. Proc. Natl. Acad. Sci. USA. 2008;105:15058–63. doi: 10.1073/pnas.0808216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, et al. IKKβ links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–96. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 111.Tanaka K, Namba T, Arai Y, Fujimoto M, Adachi H, et al. Genetic evidence for a protective role for heat shock factor 1 and heat shock protein 70 against colitis. J. Biol. Chem. 2007;282:23240–52. doi: 10.1074/jbc.M704081200. [DOI] [PubMed] [Google Scholar]
- 112.Ungaro R, Fukata M, Hsu D, Hernandez Y, Breglio K, et al. A novel Toll-like receptor 4 antagonist antibody ameliorates inflammation but impairs mucosal healing in murine colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G1167–79. doi: 10.1152/ajpgi.90496.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 114.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 2008;8:411–20. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 115.Allakhverdi Z, Comeau MR, Jessup HK, Yoon BR, Brewer A, et al. Thymic stromal lymphopoietin is released by human epithelial cells in response to microbes, trauma, or inflammation and potently activates mast cells. J. Exp. Med. 2007;204:253–58. doi: 10.1084/jem.20062211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zaph C, Troy AE, Taylor BC, Berman-Booty LD, Guild KJ, et al. Epithelial-cell-intrinsic IKK-β expression regulates intestinal immune homeostasis. Nature. 2007;446:552–56. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
- 117.Rimoldi M, Chieppa M, Salucci V, Avogadri F, Sonzogni A, et al. Intestinal immune homeostasis is regulated by the crosstalk between epithelial cells and dendritic cells. Nat. Immunol. 2005;6:507–14. doi: 10.1038/ni1192. [DOI] [PubMed] [Google Scholar]
- 118.Taylor BC, Zaph C, Troy AE, Du Y, Guild KJ, et al. TSLP regulates intestinal immunity and inflammation in mouse models of helminth infection and colitis. J. Exp. Med. 2009;206:655–67. doi: 10.1084/jem.20081499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–61. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 120.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signaling. Nature. 2004;430:694–99. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 121.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 2004;5:1052–60. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 122.Turer EE, Tavares RM, Mortier E, Hitotsumatsu O, Advincula R, et al. Homeostatic MyD88-dependent signals cause lethal inflammation in the absence of A20. J. Exp. Med. 2008;205:451–64. doi: 10.1084/jem.20071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, et al. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–90. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee EG, Boone DL, Chai S, Libby SL, Chien M, et al. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science. 2000;289:2350–54. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wellcome Trust Case Control Consort Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 127.Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat. Rev. Immunol. 2008;8:663–74. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 128.Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, et al. Increased sensitivity to dextran sodium sulfate colitis in IRE1β-deficient mice. J. Clin. Invest. 2001;107:585–93. doi: 10.1172/JCI11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brandl K, Rutschmann S, Li X, Du X, Xiao N, et al. Enhanced sensitivity to DSS colitis caused by a hypomorphic Mbtps1 mutation disrupting the ATF6-driven unfolded protein response. Proc. Natl. Acad. Sci. USA. 2009;106:3300–5. doi: 10.1073/pnas.0813036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Turner MJ, Sowders DP, DeLay ML, Mohapatra R, Bai S, et al. HLA-B27 misfolding in transgenic rats is associated with activation of the unfolded protein response. J. Immunol. 2005;175:2438–48. doi: 10.4049/jimmunol.175.4.2438. [DOI] [PubMed] [Google Scholar]
- 132.Shkoda A, Ruiz PA, Daniel H, Kim SC, Rogler G, et al. Interleukin-10 blocked endoplasmic reticulum stress in intestinal epithelial cells: impact on chronic inflammation. Gastroenterology. 2007;132:190–207. doi: 10.1053/j.gastro.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 133.Kaser A, Blumberg RS. Endoplasmic reticulum stress and intestinal inflammation. Mucosal Immunol. 2010;3:11–6. doi: 10.1038/mi.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–68. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 137.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS ONE. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat. Genet. 2008;40:1107–12. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat. Genet. 2007;39:830–32. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Taylor GA, Feng CG, Sher A. p47 GTPases: regulators of immunity to intracellular pathogens. Nat. Rev. Immunol. 2004;4:100–9. doi: 10.1038/nri1270. [DOI] [PubMed] [Google Scholar]
- 141.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–41. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 142.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-γ-inducible LRG-47. Science. 2003;302:654–59. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 143.Garabedian EM, Roberts LJ, McNevin MS, Gordon JI. Examining the role of Paneth cells in the small intestine by lineage ablation in transgenic mice. J. Biol. Chem. 1997;272:23729–40. doi: 10.1074/jbc.272.38.23729. [DOI] [PubMed] [Google Scholar]
- 144.Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, et al. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–17. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 145.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–96. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–35. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ouyang Q, Tandon R, Goh KL, Ooi CJ, Ogata H, Fiocchi C. The emergence of inflammatory bowel disease in the Asian Pacific region. Curr. Opin. Gastroenterol. 2005;21:408–13. [PubMed] [Google Scholar]
- 148.Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat. Genet. 2004;36:471–75. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 149.Koepsell H, Lips K, Volk C. Polyspecific organic cation transporters: structure, function, physiological roles, and biopharmaceutical implications. Pharm. Res. 2007;24:1227–51. doi: 10.1007/s11095-007-9254-z. [DOI] [PubMed] [Google Scholar]
- 150.Rinaldo P, Matern D, Bennett MJ. Fatty acid oxidation disorders. Annu. Rev. Physiol. 2002;64:477–502. doi: 10.1146/annurev.physiol.64.082201.154705. [DOI] [PubMed] [Google Scholar]
- 151.Shekhawat PS, Srinivas SR, Matern D, Bennett MJ, Boriack R, et al. Spontaneous development of intestinal and colonic atrophy and inflammation in the carnitine-deficient jvs (OCTN2−/−) mice. Mol. Genet. Metab. 2007;92:315–24. doi: 10.1016/j.ymgme.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Roediger WE, Nance S. Metabolic induction of experimental ulcerative colitis by inhibition of fatty acid oxidation. Br. J. Exp. Pathol. 1986;67:773–82. [PMC free article] [PubMed] [Google Scholar]
- 153.Tupin E, Kinjo Y, Kronenberg M. The unique role of natural killer T cells in the response to microorganisms. Nat. Rev. Microbiol. 2007;5:405–17. doi: 10.1038/nrmicro1657. [DOI] [PubMed] [Google Scholar]
- 154.Fuss IJ, Heller F, Boirivant M, Leon F, Yoshida M, et al. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J. Clin. Invest. 2004;113:1490–97. doi: 10.1172/JCI19836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Heller F, Fuss IJ, Nieuwenhuis EE, Blumberg RS, Strober W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity. 2002;17:629–38. doi: 10.1016/s1074-7613(02)00453-3. [DOI] [PubMed] [Google Scholar]
- 156.Dougan SK, Kaser A, Blumberg RS. CD1 expression on antigen-presenting cells. Curr. Top. Microbiol. Immunol. 2007;314:113–41. doi: 10.1007/978-3-540-69511-0_5. [DOI] [PubMed] [Google Scholar]
- 157.Spehlmann ME, Eckmann L. Nuclear factor-κ B in intestinal protection and destruction. Curr. Opin. Gastroenterol. 2009;25:92–99. doi: 10.1097/MOG.0b013e328324f857. [DOI] [PubMed] [Google Scholar]
- 158.Brozovic S, Nagaishi T, Yoshida M, Betz S, Salas A, et al. CD1d function is regulated by microsomal triglyceride transfer protein. Nat. Med. 2004;10:535–39. doi: 10.1038/nm1043. [DOI] [PubMed] [Google Scholar]
- 159.Perera L, Shao L, Patel A, Evans K, Meresse B, et al. Expression of nonclassical class I molecules by intestinal epithelial cells. Inflamm. Bowel Dis. 2007;13:298–307. doi: 10.1002/ibd.20026. [DOI] [PubMed] [Google Scholar]
- 160.Page MJ, Poritz LS, Tilberg AF, Zhang WJ, Chorney MJ, Koltun WA. Cd1d-restricted cellular lysis by peripheral blood lymphocytes: relevance to the inflammatory bowel diseases. J. Surg. Res. 2000;92:214–21. doi: 10.1006/jsre.2000.5940. [DOI] [PubMed] [Google Scholar]
- 161.Rachitskaya AV, Hansen AM, Horai R, Li Z, Villasmil R, et al. Cutting edge: NKT cells constitutively express IL-23 receptor and RORγt and rapidly produce IL-17 upon receptor ligation in an IL-6-independent fashion. J. Immunol. 2008;180:5167–71. doi: 10.4049/jimmunol.180.8.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Akbari O, Stock P, Meyer E, Kronenberg M, Sidobre S, et al. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat. Med. 2003;9:582–88. doi: 10.1038/nm851. [DOI] [PubMed] [Google Scholar]
- 163.Fernando MM, Stevens CR, Walsh EC, De Jager PL, Goyette P, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4:e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ware CF. Network communications: lymphotoxins, LIGHT, and TNF. Annu. Rev. Immunol. 2005;23:787–819. doi: 10.1146/annurev.immunol.23.021704.115719. [DOI] [PubMed] [Google Scholar]
- 165.Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–98. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 166.Armaka M, Apostolaki M, Jacques P, Kontoyiannis DL, Elewaut D, Kollias G. Mesenchymal cell targeting by TNF as a common pathogenic principle in chronic inflammatory joint and intestinal diseases. J. Exp. Med. 2008;205:331–37. doi: 10.1084/jem.20070906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kontoyiannis D, Boulougouris G, Manoloukos M, Armaka M, Apostolaki M, et al. Genetic dissection of the cellular pathways and signaling mechanisms in modeled tumor necrosis factor-induced Crohn’s-like inflammatory bowel disease. J. Exp. Med. 2002;196:1563–74. doi: 10.1084/jem.20020281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pender SL, MacDonald TT. Matrix metalloproteinases and the gut—new roles for old enzymes. Curr. Opin. Pharmacol. 2004;4:546–50. doi: 10.1016/j.coph.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 169.Jung HC, Eckmann L, Yang SK, Panja A, Fierer J, et al. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J. Clin. Invest. 1995;95:55–65. doi: 10.1172/JCI117676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zeissig S, Bojarski C, Buergel N, Mankertz J, Zeitz M, et al. Downregulation of epithelial apoptosis and barrier repair in active Crohn’s disease by tumor necrosis factor α antibody treatment. Gut. 2004;53:1295–302. doi: 10.1136/gut.2003.036632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Marini M, Bamias G, Rivera-Nieves J, Moskaluk CA, Hoang SB, et al. TNF-α neutralization ameliorates the severity of murine Crohn’s-like ileitis by abrogation of intestinal epithelial cell apoptosis. Proc. Natl. Acad. Sci. USA. 2003;100:8366–71. doi: 10.1073/pnas.1432897100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nancey S, Holvoet S, Graber I, Joubert G, Philippe D, et al. CD8+ cytotoxic T cells induce relapsing colitis in normal mice. Gastroenterology. 2006;131:485–96. doi: 10.1053/j.gastro.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 173.Desreumaux P, Ernst O, Geboes K, Gambiez L, Berrebi D, et al. Inflammatory alterations in mesenteric adipose tissue in Crohn’s disease. Gastroenterology. 1999;117:73–81. doi: 10.1016/s0016-5085(99)70552-4. [DOI] [PubMed] [Google Scholar]
- 174.Neurath MF, Fuss I, Pasparakis M, Alexopoulou L, Haralambous S, et al. Predominant pathogenic role of tumor necrosis factor in experimental colitis in mice. Eur. J. Immunol. 1997;27:1743–50. doi: 10.1002/eji.1830270722. [DOI] [PubMed] [Google Scholar]
- 175.Gratz R, Becker S, Sokolowski N, Schumann M, Bass D, Malnick SD. Murine monoclonal anti-TNF antibody administration has a beneficial effect on inflammatory bowel disease that develops in IL-10 knockout mice. Dig. Dis. Sci. 2002;47:1723–27. doi: 10.1023/a:1016476024293. [DOI] [PubMed] [Google Scholar]
- 176.Watkins PE, Warren BF, Stephens S, Ward P, Foulkes R. Treatment of ulcerative colitis in the cottontop tamarin using antibody to tumor necrosis factor α. Gut. 1997;40:628–33. doi: 10.1136/gut.40.5.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Corazza N, Eichenberger S, Eugster HP, Mueller C. Nonlymphocyte-derived tumor necrosis factor is required for induction of colitis in recombination activating gene (RAG)2−/− mice upon transfer of CD4+CD45RBhi T cells. J. Exp. Med. 1999;190:1479–92. doi: 10.1084/jem.190.10.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Powrie F, Leach MW, Mauze S, Menon S, Caddle LB, Coffman RL. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity. 1994;1:553–62. doi: 10.1016/1074-7613(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 179.Corazza N, Brunner T, Buri C, Rihs S, Imboden MA, et al. Transmembrane tumor necrosis factor is a potent inducer of colitis even in the absence of its secreted form. Gastroenterology. 2004;127:816–25. doi: 10.1053/j.gastro.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 180.Murch SH, Braegger CP, Walker-Smith JA, MacDonald TT. Location of tumor necrosis factor α by immunohistochemistry in chronic inflammatory bowel disease. Gut. 1993;34:1705–9. doi: 10.1136/gut.34.12.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lugering A, Lebiedz P, Koch S, Kucharzik T. Apoptosis as a therapeutic tool in IBD? Ann. NY Acad. Sci. 2006;1072:62–77. doi: 10.1196/annals.1326.013. [DOI] [PubMed] [Google Scholar]
- 182.Iijima H, Neurath MF, Nagaishi T, Glickman JN, Nieuwenhuis EE, et al. Specific regulation of T helper cell 1-mediated murine colitis by CEACAM1. J. Exp. Med. 2004;199:471–82. doi: 10.1084/jem.20030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fichtner-Feigl S, Strober W, Kawakami K, Puri RK, Kitani A. IL-13 signaling through the IL-13α2 receptor is involved in induction of TGF-β1 production and fibrosis. Nat. Med. 2006;12:99–106. doi: 10.1038/nm1332. [DOI] [PubMed] [Google Scholar]
- 184.Boirivant M, Fuss IJ, Chu A, Strober W. Oxazolone colitis: A murine model of T helper cell type 2 colitis treatable with antibodies to interleukin 4. J. Exp. Med. 1998;188:1929–39. doi: 10.1084/jem.188.10.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yamazaki K, McGovern D, Ragoussis J, Paolucci M, Butler H, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum. Mol. Genet. 2005;14:3499–506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- 186.Yang SK, Lim J, Chang HS, Lee I, Li Y, et al. Association of TNFSF15 with Crohn’s disease in Koreans. Am. J. Gastroenterol. 2008;103:1437–42. doi: 10.1111/j.1572-0241.2007.01752.x. [DOI] [PubMed] [Google Scholar]
- 187.Papadakis KA, Prehn JL, Landers C, Han Q, Luo X, et al. TL1A synergizes with IL-12 and IL-18 to enhance IFN-γ production in human T cells and NK cells. J. Immunol. 2004;172:7002–7. doi: 10.4049/jimmunol.172.11.7002. [DOI] [PubMed] [Google Scholar]
- 188.Bamias G, Mishina M, Nyce M, Ross WG, Kollias G, et al. Role of TL1A and its receptor DR3 in two models of chronic murine ileitis. Proc. Natl. Acad. Sci. USA. 2006;103:8441–46. doi: 10.1073/pnas.0510903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Takedatsu H, Michelsen KS, Wei B, Landers CJ, Thomas LS, et al. TL1A (TNFSF15) regulates the development of chronic colitis by modulating both T-helper 1 and T-helper 17 activation. Gastroenterology. 2008;135:552–67. doi: 10.1053/j.gastro.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kishimoto T. Interleukin-6: from basic science to medicine—40 years in immunology. Annu. Rev. Immunol. 2005;23:1–21. doi: 10.1146/annurev.immunol.23.021704.115806. [DOI] [PubMed] [Google Scholar]
- 191.Atreya R, Mudter J, Finotto S, Mullberg J, Jostock T, et al. Blockade of interleukin 6 trans signaling suppresses T-cell resistance against apoptosis in chronic intestinal inflammation: evidence in Crohn disease and experimental colitis in vivo. Nat. Med. 2000;6:583–88. doi: 10.1038/75068. [DOI] [PubMed] [Google Scholar]
- 192.Yamamoto M, Yoshizaki K, Kishimoto T, Ito H. IL-6 is required for the development of Th1 cell-mediated murine colitis. J. Immunol. 2000;164:4878–82. doi: 10.4049/jimmunol.164.9.4878. [DOI] [PubMed] [Google Scholar]
- 193.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–38. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 194.Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, et al. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–55. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 196.Franke A, Balschun T, Karlsen TH, Hedderich J, May S, et al. Replication of signals from recent studies of Crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat. Genet. 2008;40:713–15. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 197.Anderson CA, Massey DC, Barrett JC, Prescott NJ, Tremelling M, et al. Investigation of Crohn’s disease risk loci in ulcerative colitis further defines their molecular relationship. Gastroenterology. 2009;136:523–29. doi: 10.1053/j.gastro.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ernst M, Inglese M, Waring P, Campbell IK, Bao S, et al. Defective gp130-mediated signal transducer and activator of transcription (STAT) signaling results in degenerative joint disease, gastrointestinal ulceration, and failure of uterine implantation. J. Exp. Med. 2001;194:189–203. doi: 10.1084/jem.194.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tebbutt NC, Giraud AS, Inglese M, Jenkins B, Waring P, et al. Reciprocal regulation of gastrointestinal homeostasis by SHP2 and STAT-mediated trefoil gene activation in gp130 mutant mice. Nat. Med. 2002;8:1089–97. doi: 10.1038/nm763. [DOI] [PubMed] [Google Scholar]
- 200.Wang L, Walia B, Evans J, Gewirtz AT, Merlin D, Sitaraman SV. IL-6 induces NF-κB activation in the intestinal epithelia. J. Immunol. 2003;171:3194–201. doi: 10.4049/jimmunol.171.6.3194. [DOI] [PubMed] [Google Scholar]
- 201.Becker C, Fantini MC, Schramm C, Lehr HA, Wirtz S, et al. TGF-β suppresses tumor progression in colon cancer by inhibition of IL-6 trans-signaling. Immunity. 2004;21:491–501. doi: 10.1016/j.immuni.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 202.Bollrath J, Phesse TJ, von Burstin VA, Putoczki T, Bennecke M, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 203.Mashimo H, Wu DC, Podolsky DK, Fishman MC. Impaired defense of intestinal mucosa in mice lacking intestinal trefoil factor. Science. 1996;274:262–65. doi: 10.1126/science.274.5285.262. [DOI] [PubMed] [Google Scholar]
- 204.Welte T, Zhang SS, Wang T, Zhang Z, Hesslein DG, et al. STAT3 deletion during hematopoiesis causes Crohn’s disease-like pathogenesis and lethality: a critical role of STAT3 in innate immunity. Proc. Natl. Acad. Sci. USA. 2003;100:1879–84. doi: 10.1073/pnas.0237137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kobayashi M, Kweon MN, Kuwata H, Schreiber RD, Kiyono H, et al. Toll-like receptor-dependent production of IL-12p40 causes chronic enterocolitis in myeloid cell-specific Stat3-deficient mice. J. Clin. Invest. 2003;111:1297–308. doi: 10.1172/JCI17085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 2009;27:519–50. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 207.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu. Rev. Immunol. 2009;27:229–65. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 208.Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, et al. Common variants in the NLRP3 region contribute to Crohn’s disease susceptibility. Nat. Genet. 2009;41:71–76. doi: 10.1038/ng285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zhernakova A, Festen EM, Franke L, Trynka G, van Diemen CC, et al. Genetic analysis of innate immunity in Crohn’s disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am. J. Hum. Genet. 2008;82:1202–10. doi: 10.1016/j.ajhg.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Li J, Moran T, Swanson E, Julian C, Harris J, et al. Regulation of IL-8 and IL-1β expression in Crohn’s disease associated NOD2/CARD15 mutations. Hum. Mol. Genet. 2004;13:1715–25. doi: 10.1093/hmg/ddh182. [DOI] [PubMed] [Google Scholar]
- 211.van Beelen AJ, Zelinkova Z, Taanman-Kueter EW, Muller FJ, Hommes DW, et al. Stimulation of the intracellular bacterial sensor NOD2 programs dendritic cells to promote interleukin-17 production in human memory T cells. Immunity. 2007;27:660–69. doi: 10.1016/j.immuni.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 212.Casini-Raggi V, Kam L, Chong YJ, Fiocchi C, Pizarro TT, Cominelli F. Mucosal imbalance of IL-1 and IL-1 receptor antagonist in inflammatory bowel disease. A novel mechanism of chronic intestinal inflammation. J. Immunol. 1995;154:2434–40. [PubMed] [Google Scholar]
- 213.Cominelli F, Nast CC, Llerena R, Dinarello CA, Zipser RD. Interleukin 1 suppresses inflammation in rabbit colitis. Mediation by endogenous prostaglandins. J. Clin. Invest. 1990;85:582–86. doi: 10.1172/JCI114476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cominelli F, Nast CC, Clark BD, Schindler R, Lierena R, et al. Interleukin 1 (IL-1) gene expression, synthesis, and effect of specific IL-1 receptor blockade in rabbit immune complex colitis. J. Clin. Invest. 1990;86:972–80. doi: 10.1172/JCI114799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ferretti M, Casini-Raggi V, Pizarro TT, Eisenberg SP, Nast CC, Cominelli F. Neutralization of endogenous IL-1 receptor antagonist exacerbates and prolongs inflammation in rabbit immune colitis. J. Clin. Invest. 1994;94:449–53. doi: 10.1172/JCI117345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Monteleone G, Trapasso F, Parrello T, Biancone L, Stella A, et al. Bioactive IL-18 expression is up-regulated in Crohn’s disease. J. Immunol. 1999;163:143–47. [PubMed] [Google Scholar]
- 217.Pizarro TT, Michie MH, Bentz M, Woraratanadharm J, Smith MF, Jr, et al. IL-18, a novel immunoregulatory cytokine, is up-regulated in Crohn’s disease: expression and localization in intestinal mucosal cells. J. Immunol. 1999;162:6829–35. [PubMed] [Google Scholar]
- 218.Siegmund B, Fantuzzi G, Rieder F, Gamboni-Robertson F, Lehr HA, et al. Neutralization of interleukin-18 reduces severity in murine colitis and intestinal IFN-γ and TNF-α production. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001;281:R1264–73. doi: 10.1152/ajpregu.2001.281.4.R1264. [DOI] [PubMed] [Google Scholar]
- 219.Sivakumar PV, Westrich GM, Kanaly S, Garka K, Born TL, et al. Interleukin 18 is a primary mediator of the inflammation associated with dextran sulphate sodium induced colitis: blocking interleukin 18 attenuates intestinal damage. Gut. 2002;50:812–20. doi: 10.1136/gut.50.6.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kanai T, Watanabe M, Okazawa A, Sato T, Yamazaki M, et al. Macrophage-derived IL-18-mediated intestinal inflammation in the murine model of Crohn’s disease. Gastroenterology. 2001;121:875–88. doi: 10.1053/gast.2001.28021. [DOI] [PubMed] [Google Scholar]
- 221.Ten Hove T, Corbaz A, Amitai H, Aloni S, Belzer I, et al. Blockade of endogenous IL-18 ameliorates TNBS-induced colitis by decreasing local TNF-α production in mice. Gastroenterology. 2001;121:1372–79. doi: 10.1053/gast.2001.29579. [DOI] [PubMed] [Google Scholar]
- 222.Wirtz S, Becker C, Blumberg R, Galle PR, Neurath MF. Treatment of T cell-dependent experimental colitis in SCID mice by local administration of an adenovirus expressing IL-18 antisense mRNA. J. Immunol. 2002;168:411–20. doi: 10.4049/jimmunol.168.1.411. [DOI] [PubMed] [Google Scholar]
- 223.Siegmund B, Lehr HA, Fantuzzi G, Dinarello CA. IL-1β-converting enzyme (caspase-1) in intestinal inflammation. Proc. Natl. Acad. Sci. USA. 2001;98:13249–54. doi: 10.1073/pnas.231473998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–89. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 225.Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30:576–87. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat. Immunol. 2007;8:950–57. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 227.Shi J, Aono S, Lu W, Ouellette AJ, Hu X, et al. A novel role for defensins in intestinal homeostasis: regulation of IL-1β secretion. J. Immunol. 2007;179:1245–53. doi: 10.4049/jimmunol.179.2.1245. [DOI] [PubMed] [Google Scholar]
- 228.Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annu. Rev. Immunol. 2009;27:313–38. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- 229.Langrish CL, McKenzie BS, Wilson NJ, de Waal Malefyt R, Kastelein RA, Cua DJ. IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunol. Rev. 2004;202:96–105. doi: 10.1111/j.0105-2896.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 230.Neurath MF, Fuss I, Kelsall BL, Stuber E, Strober W. Antibodies to interleukin 12 abrogate established experimental colitis in mice. J. Exp. Med. 1995;182:1281–90. doi: 10.1084/jem.182.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ito H, Fathman CG. CD45RBhigh CD.4+ T cells from IFN-γ knockout mice do not induce wasting disease. J. Autoimmun. 1997;10:455–59. doi: 10.1016/s0896-8411(97)90152-9. [DOI] [PubMed] [Google Scholar]
- 232.Neurath MF, Weigmann B, Finotto S, Glickman J, Nieuwenhuis E, et al. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn’s disease. J. Exp. Med. 2002;195:1129–43. doi: 10.1084/jem.20011956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Fuss IJ, Neurath M, Boirivant M, Klein JS, de la Motte C, et al. Disparate CD4+ lamina propria (LP) lymphokine secretion profiles in inflammatory bowel disease. Crohn’s disease LP cells manifest increased secretion of IFN-γ, whereas ulcerative colitis LP cells manifest increased secretion of IL-5. J. Immunol. 1996;157:1261–70. [PubMed] [Google Scholar]
- 234.Loftus EV., Jr Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–17. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 235.Kikuchi H, Itoh J, Fukuda S. Chronic nicotine stimulation modulates the immune response of mucosal T cells to Th1-dominant pattern via nAChR by upregulation of Th1-specific transcriptional factor. Neurosci. Lett. 2008;432:217–21. doi: 10.1016/j.neulet.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 236.Xavier RJ, Rioux JD. Genome-wide association studies: a new window into immune-mediated diseases. Nat. Rev. Immunol. 2008;8:631–43. doi: 10.1038/nri2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn’s disease. Nat. Genet. 2008;40:710–12. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–25. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 239.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–48. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 240.Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, et al. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J. Exp. Med. 2003;198:1951–57. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kroenke MA, Carlson TJ, Andjelkovic AV, Segal BM. IL-12- and IL-23-modulated T cells induce distinct types of EAE based on histology, CNS chemokine profile, and response to cytokine inhibition. J. Exp. Med. 2008;205:1535–41. doi: 10.1084/jem.20080159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Luger D, Silver PB, Tang J, Cua D, Chen Z, et al. Either a Th17 or a Th1 effector response can drive autoimmunity: conditions of disease induction affect dominant effector category. J. Exp. Med. 2008;205:799–810. doi: 10.1084/jem.20071258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Uhlig HH, McKenzie BS, Hue S, Thompson C, Joyce-Shaikh B, et al. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–18. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 244.Hue S, Ahern P, Buonocore S, Kullberg MC, Cua DJ, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J. Exp. Med. 2006;203:2473–83. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kullberg MC, Jankovic D, Feng CG, Hue S, Gorelick PL, et al. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J. Exp. Med. 2006;203:2485–94. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Collison LW, Workman CJ, Kuo TT, Boyd K, Wang Y, et al. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450:566–69. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 247.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J. Clin. Invest. 2006;116:1310–16. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008;28:454–67. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 2005;6:1123–32. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 250.Park H, Li Z, Yang XO, Chang SH, Nurieva R, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 2005;6:1133–41. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 252.Becker C, Wirtz S, Blessing M, Pirhonen J, Strand D, et al. Constitutive p40 promoter activation and IL-23 production in the terminal ileum mediated by dendritic cells. J. Clin. Invest. 2003;112:693–706. doi: 10.1172/JCI17464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–12. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 254.Manocha M, Svend R, Laouar A, Liao G, Bhan A, et al. Blocking CD27-CD70 costimulatory pathway suppresses experimental colitis. J. Immunol. 2009;183:270–76. doi: 10.4049/jimmunol.0802424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.O’Connor W, Jr, Kamanaka M, Booth CJ, Town T, Nakae S, et al. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat. Immunol. 2009;10:603–9. doi: 10.1038/ni.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ogawa A, Andoh A, Araki Y, Bamba T, Fujiyama Y. Neutralization of interleukin-17 aggravates dextran sulfate sodium-induced colitis in mice. Clin. Immunol. 2004;110:55–62. doi: 10.1016/j.clim.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 257.Yang XO, Chang SH, Park H, Nurieva R, Shah B, et al. Regulation of inflammatory responses by IL-17F. J. Exp. Med. 2008;205:1063–75. doi: 10.1084/jem.20071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zhang Z, Zheng M, Bindas J, Schwarzenberger P, Kolls JK. Critical role of IL-17 receptor signaling in acute TNBS-induced colitis. Inflamm. Bowel Dis. 2006;12:382–88. doi: 10.1097/01.MIB.0000218764.06959.91. [DOI] [PubMed] [Google Scholar]
- 259.Leppkes M, Becker C, Ivanov II, Hirth S, Wirtz S, et al. RORγ-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009;136:257–67. doi: 10.1053/j.gastro.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 260.Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52:65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, et al. Phenotypic and functional features of human Th17 cells. J. Exp. Med. 2007;204:1849–61. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 2007;25:821–52. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 263.Izcue A, Hue S, Buonocore S, Arancibia-Carcamo CV, Ahern PP, et al. Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity. 2008;28:559–70. doi: 10.1016/j.immuni.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Maloy KJ, Salaun L, Cahill R, Dougan G, Saunders NJ, Powrie F. CD4+CD25+ T(R) cells suppress innate immune pathology through cytokine-dependent mechanisms. J. Exp. Med. 2003;197:111–19. doi: 10.1084/jem.20021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Powrie F, Mason D. OX-22high CD4+ T cells induce wasting disease with multiple organ pathology: prevention by the OX-22low subset. J. Exp. Med. 1990;172:1701–8. doi: 10.1084/jem.172.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor α-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 1995;155:1151–64. [PubMed] [Google Scholar]
- 267.Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, et al. TGF-β-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORγt function. Nature. 2008;453:236–40. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, et al. Transforming growth factor-β induces development of the T(H)17 lineage. Nature. 2006;441:231–34. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 269.Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–83. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 270.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 2007;8:967–74. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 271.Mucida D, Park Y, Kim G, Turovskaya O, Scott I, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–60. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 272.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 2007;204:1775–85. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat. Immunol. 2007;8:1086–94. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 274.Park O, Grishina I, Leung PS, Gershwin ME, Prindiville T. Analysis of the Foxp3/scurfin gene in Crohn’s disease. Ann. NY Acad. Sci. 2005;1051:218–28. doi: 10.1196/annals.1361.125. [DOI] [PubMed] [Google Scholar]
- 275.Maul J, Loddenkemper C, Mundt P, Berg E, Giese T, et al. Peripheral and intestinal regulatory CD4+ CD25high T cells in inflammatory bowel disease. Gastroenterology. 2005;128:1868–78. doi: 10.1053/j.gastro.2005.03.043. [DOI] [PubMed] [Google Scholar]
- 276.Saruta M, Yu QT, Fleshner PR, Mantel PY, Schmidt-Weber CB, et al. Characterization of FOXP3+CD4+ regulatory T cells in Crohn’s disease. Clin. Immunol. 2007;125:281–90. doi: 10.1016/j.clim.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 277.Makita S, Kanai T, Oshima S, Uraushihara K, Totsuka T, et al. CD4+CD25bright T cells in human intestinal lamina propria as regulatory cells. J. Immunol. 2004;173:3119–30. doi: 10.4049/jimmunol.173.5.3119. [DOI] [PubMed] [Google Scholar]
- 278.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 279.Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 280.Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 281.Maillard MH, Cotta-de-Almeida V, Takeshima F, Nguyen DD, Michetti P, et al. The Wiskott-Aldrich syndrome protein is required for the function of CD4+CD25+Foxp3+ regulatory T cells. J. Exp. Med. 2007;204:381–91. doi: 10.1084/jem.20061338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Marangoni F, Trifari S, Scaramuzza S, Panaroni C, Martino S, et al. WASP regulates suppressor activity of human and murine CD4+CD25+FOXP3+ natural regulatory T cells. J. Exp. Med. 2007;204:369–80. doi: 10.1084/jem.20061334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat. Genet. 2008;40:1319–23. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 284.Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 2007;13:1299–307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 285.Monteleone G, Boirivant M, Pallone F, MacDonald TT. TGF-β1 and Smad7 in the regulation of IBD. Mucosal Immunol. 2008;1(Suppl. 1):S50–53. doi: 10.1038/mi.2008.55. [DOI] [PubMed] [Google Scholar]
- 286.Fantini MC, Rizzo A, Fina D, Caruso R, Sarra M, et al. Smad7 controls resistance of colitogenic T cells to regulatory T cell-mediated suppression. Gastroenterology. 2009;136:1308–16. doi: 10.1053/j.gastro.2008.12.053. [DOI] [PubMed] [Google Scholar]
- 287.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 1998;66:5224–31. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, et al. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat. Immunol. 2006;7:457–65. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 289.Maynard CL, Weaver CT. Diversity in the contribution of interleukin-10 to T-cell-mediated immune regulation. Immunol. Rev. 2008;226:219–33. doi: 10.1111/j.1600-065X.2008.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Autschbach F, Braunstein J, Helmke B, Zuna I, Schurmann G, et al. In situ expression of interleukin-10 in noninflamed human gut and in inflammatory bowel disease. Am. J. Pathol. 1998;153:121–30. doi: 10.1016/S0002-9440(10)65552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Kamanaka M, Kim ST, Wan YY, Sutterwala FS, Lara-Tejero M, et al. Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity. 2006;25:941–52. doi: 10.1016/j.immuni.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 292.Roers A, Siewe L, Strittmatter E, Deckert M, Schluter D, et al. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J. Exp. Med. 2004;200:1289–97. doi: 10.1084/jem.20041789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–58. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 294.Fontenot JD, Rudensky AY. A well adapted regulatory contrivance: regulatory T cell development and the forkhead family transcription factor Foxp3. Nat. Immunol. 2005;6:331–37. doi: 10.1038/ni1179. [DOI] [PubMed] [Google Scholar]
- 295.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 2007;8:191–97. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 296.Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, et al. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3− precursor cells in the absence of interleukin 10. Nat. Immunol. 2007;8:931–41. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- 297.Groux H, O’Garra A, Bigler M, Rouleau M, Antonenko S, et al. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737–42. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 298.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, et al. CD4+ regulatory T cells control Th17 responses in a Stat3-dependent manner. Science. 2009;326:986–91. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, et al. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289:1352–55. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 300.Luster AD, Alon R, von Andrian UH. Immune cell migration in inflammation: present and future therapeutic targets. Nat. Immunol. 2005;6:1182–90. doi: 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- 301.Agace WW. T-cell recruitment to the intestinal mucosa. Trends Immunol. 2008;29:514–22. doi: 10.1016/j.it.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 302.Mora JR, Bono MR, Manjunath N, Weninger W, Cavanagh LL, et al. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature. 2003;424:88–93. doi: 10.1038/nature01726. [DOI] [PubMed] [Google Scholar]
- 303.Johansson-Lindbom B, Svensson M, Wurbel MA, Malissen B, Marquez G, Agace W. Selective generation of gut tropic T cells in gut-associated lymphoid tissue (GALT): requirement for GALT dendritic cells and adjuvant. J. Exp. Med. 2003;198:963–69. doi: 10.1084/jem.20031244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Rivera-Nieves J, Burcin TL, Olson TS, Morris MA, McDuffie M, et al. Critical role of endothelial P-selectin glycoprotein ligand 1 in chronic murine ileitis. J. Exp. Med. 2006;203:907–17. doi: 10.1084/jem.20052530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kosiewicz MM, Nast CC, Krishnan A, Rivera-Nieves J, Moskaluk CA, et al. Th1-type responses mediate spontaneous ileitis in a novel murine model of Crohn’s disease. J. Clin. Invest. 2001;107:695–702. doi: 10.1172/JCI10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Rijcken EM, Laukoetter MG, Anthoni C, Meier S, Mennigen R, et al. Immunoblockade of PSGL-1 attenuates established experimental murine colitis by reduction of leukocyte rolling. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;287:G115–24. doi: 10.1152/ajpgi.00207.2003. [DOI] [PubMed] [Google Scholar]
- 307.Kuhl AA, Kakirman H, Janotta M, Dreher S, Cremer P, et al. Aggravation of different types of experimental colitis by depletion or adhesion blockade of neutrophils. Gastroenterology. 2007;133:1882–92. doi: 10.1053/j.gastro.2007.08.073. [DOI] [PubMed] [Google Scholar]
- 308.Kwon JH, Keates S, Bassani L, Mayer LF, Keates AC. Colonic epithelial cells are a major site of macrophage inflammatory protein 3α (MIP-3α) production in normal colon and inflammatory bowel disease. Gut. 2002;51:818–26. doi: 10.1136/gut.51.6.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Lee JW, Wang P, Kattah MG, Youssef S, Steinman L, et al. Differential regulation of chemokines by IL-17 in colonic epithelial cells. J. Immunol. 2008;181:6536–45. doi: 10.4049/jimmunol.181.9.6536. [DOI] [PubMed] [Google Scholar]
- 310.Kaser A, Ludwiczek O, Holzmann S, Moschen AR, Weiss G, et al. Increased expression of CCL20 in human inflammatory bowel disease. J. Clin. Immunol. 2004;24:74–85. doi: 10.1023/B:JOCI.0000018066.46279.6b. [DOI] [PubMed] [Google Scholar]
- 311.Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 2007;8:639–46. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 312.Wang C, Kang SG, Lee J, Sun Z, Kim CH. The roles of CCR6 in migration of Th17 cells and regulation of effector T-cell balance in the gut. Mucosal Immunol. 2009;2:173–83. doi: 10.1038/mi.2008.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Varona R, Cadenas V, Flores J, Martinez-A C, Márquez G. CCR6 has a nonredundant role in the development of inflammatory bowel disease. Eur. J. Immunol. 2003;33:2937–46. doi: 10.1002/eji.200324347. [DOI] [PubMed] [Google Scholar]
- 314.Katchar K, Kelly CP, Keates S, O’Brien MJ, Keates AC. MIP-3α neutralizing monoclonal antibody protects against TNBS-induced colonic injury and inflammation in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;292:G1263–71. doi: 10.1152/ajpgi.00409.2006. [DOI] [PubMed] [Google Scholar]
- 315.Yamazaki T, Yang XO, Chung Y, Fukunaga A, Nurieva R, et al. CCR6 regulates the migration of inflammatory and regulatory T cells. J. Immunol. 2008;181:8391–401. doi: 10.4049/jimmunol.181.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Williams IR. CCR6 and CCL20: partners in intestinal immunity and lymphorganogenesis. Ann. NY Acad. Sci. 2006;1072:52–61. doi: 10.1196/annals.1326.036. [DOI] [PubMed] [Google Scholar]
- 317.Bouskra D, Brezillon C, Berard M, Werts C, Varona R, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–10. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 318.Xiao S, Jin H, Korn T, Liu SM, Oukka M, et al. Retinoic acid increases Foxp3+ regulatory T cells and inhibits development of Th17 cells by enhancing TGF-β-driven Smad3 signaling and inhibiting IL-6 and IL-23 receptor expression. J. Immunol. 2008;181:2277–84. doi: 10.4049/jimmunol.181.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Mora JR, Iwata M, von Andrian UH. Vitamin effects on the immune system: vitamins A and D take center stage. Nat. Rev. Immunol. 2008;8:685–98. doi: 10.1038/nri2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Podolsky DK, Lobb R, King N, Benjamin CD, Pepinsky B, et al. Attenuation of colitis in the cotton-top tamarin by antialpha 4 integrin monoclonal antibody. J. Clin. Invest. 1993;92:372–80. doi: 10.1172/JCI116575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Apostolaki M, Manoloukos M, Roulis M, Wurbel MA, Muller W, et al. Role of β7 integrin and the chemokine/chemokine receptor pair CCL25/CCR9 in modeled TNF-dependent Crohn’s disease. Gastroenterology. 2008;134:2025–35. doi: 10.1053/j.gastro.2008.02.085. [DOI] [PubMed] [Google Scholar]
- 322.Wei Z, Ertl L, Baumgart T, Rubas W, Hor S-Y, et al. CC chemokine receptor 9 (CCR9) antagonist ameliorates experimental ileitis and colitis. Gastroenterology. 2005;128:A204. Abstr. [Google Scholar]
- 323.Bensinger SJ, Tontonoz P. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature. 2008;454:470–77. doi: 10.1038/nature07202. [DOI] [PubMed] [Google Scholar]
- 324.Narumiya S, Sugimoto Y, Ushikubi F. Prostanoid receptors: structures, properties, and functions. Physiol. Rev. 1999;79:1193–226. doi: 10.1152/physrev.1999.79.4.1193. [DOI] [PubMed] [Google Scholar]
- 325.Morteau O, Morham SG, Sellon R, Dieleman LA, Langenbach R, et al. Impaired mucosal defense to acute colonic injury in mice lacking cyclooxygenase-1 or cyclooxygenase-2. J. Clin. Invest. 2000;105:469–78. doi: 10.1172/JCI6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Kabashima K, Saji T, Murata T, Nagamachi M, Matsuoka T, et al. The prostaglandin receptor EP4 suppresses colitis, mucosal damage and CD4 cell activation in the gut. J. Clin. Invest. 2002;109:883–93. doi: 10.1172/JCI14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Cosme R, Lublin D, Takafuji V, Lynch K, Roche JK. Prostanoids in human colonic mucosa: effects of inflammation on PGE(2) receptor expression. Hum. Immunol. 2000;61:684–96. doi: 10.1016/s0198-8859(00)00131-2. [DOI] [PubMed] [Google Scholar]
- 328.Takafuji V, Cosme R, Lublin D, Lynch K, Roche JK. Prostanoid receptors in intestinal epithelium: selective expression, function, and change with inflammation. Prostaglandins Leukot. Essent. Fatty Acids. 2000;63:223–35. doi: 10.1054/plef.2000.0144. [DOI] [PubMed] [Google Scholar]
- 329.Colgan SP, Parkos CA, Delp C, Arnaout MA, Madara JL. Neutrophil migration across cultured intestinal epithelial monolayers is modulated by epithelial exposure to IFN-γ in a highly polarized fashion. J. Cell Biol. 1993;120:785–98. doi: 10.1083/jcb.120.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Takayama K, Garcia-Cardena G, Sukhova GK, Comander J, Gimbrone MA, Jr, Libby P. Prostaglandin E2 suppresses chemokine production in human macrophages through the EP4 receptor. J. Biol. Chem. 2002;277:44147–54. doi: 10.1074/jbc.M204810200. [DOI] [PubMed] [Google Scholar]
- 331.Minami M, Shimizu K, Okamoto Y, Folco E, Ilasaca ML, et al. Prostaglandin E receptor type 4-associated protein interacts directly with NF-κB1 and attenuates macrophage activation. J. Biol. Chem. 2008;283:9692–703. doi: 10.1074/jbc.M709663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Yao C, Sakata D, Esaki Y, Li Y, Matsuoka T, et al. Prostaglandin E2-EP4 signaling promotes immune inflammation through Th1 cell differentiation and Th17 cell expansion. Nat. Med. 2009;15:633–40. doi: 10.1038/nm.1968. [DOI] [PubMed] [Google Scholar]
- 333.Boniface K, Bak-Jensen KS, Li Y, Blumenschein WM, McGeachy MJ, et al. Prostaglandin E2 regulates Th17 cell differentiation and function through cyclic AMP and EP2/EP4 receptor signaling. J. Exp. Med. 2009;206:535–48. doi: 10.1084/jem.20082293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Dubuquoy L, Rousseaux C, Thuru X, Peyrin-Biroulet L, Romano O, et al. PPARγ as a new therapeutic target in inflammatory bowel diseases. Gut. 2006;55:1341–49. doi: 10.1136/gut.2006.093484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Rousseaux C, Lefebvre B, Dubuquoy L, Lefebvre P, Romano O, et al. Intestinal antiinflammatory effect of 5-aminosalicylic acid is dependent on peroxisome proliferator-activated receptor-γ. J. Exp. Med. 2005;201:1205–15. doi: 10.1084/jem.20041948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Dubuquoy L, Jansson EA, Deeb S, Rakotobe S, Karoui M, et al. Impaired expression of peroxisome proliferator-activated receptor gamma in ulcerative colitis. Gastroenterology. 2003;124:1265–76. doi: 10.1016/s0016-5085(03)00271-3. [DOI] [PubMed] [Google Scholar]
- 338.Wachtershauser A, Loitsch SM, Stein J. PPAR-γ is selectively upregulated in Caco-2 cells by butyrate. Biochem. Biophys. Res. Commun. 2000;272:380–85. doi: 10.1006/bbrc.2000.2793. [DOI] [PubMed] [Google Scholar]
- 339.Su CG, Wen X, Bailey ST, Jiang W, Rangwala SM, et al. A novel therapy for colitis utilizing PPAR-γ ligands to inhibit the epithelial inflammatory response. J. Clin. Invest. 1999;104:383–89. doi: 10.1172/JCI7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Bassaganya-Riera J, Reynolds K, Martino-Catt S, Cui Y, Hennighausen L, et al. Activation of PPAR γ and δ by conjugated linoleic acid mediates protection from experimental inflammatory bowel disease. Gastroenterology. 2004;127:777–91. doi: 10.1053/j.gastro.2004.06.049. [DOI] [PubMed] [Google Scholar]
- 341.Lewis JD, Lichtenstein GR, Deren JJ, Sands BE, Hanauer SB, et al. Rosiglitazone for active ulcerative colitis: a randomized placebo-controlled trial. Gastroenterology. 2008;134:688–95. doi: 10.1053/j.gastro.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Evans-Molina C, Robbins RD, Kono T, Tersey SA, Vestermark GL, et al. Peroxisome proliferator-activated receptor gamma activation restores islet function in diabetic mice through reduction of endoplasmic reticulum stress and maintenance of euchromatin structure. Mol. Cell. Biol. 2009;29:2053–67. doi: 10.1128/MCB.01179-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev. Cell. 2008;15:829–40. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: dual anti-inflammatory and proresolution lipid mediators. Nat. Rev. Immunol. 2008;8:349–61. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Levy BD, Clish CB, Schmidt B, Gronert K, Serhan CN. Lipid mediator class switching during acute inflammation: signals in resolution. Nat. Immunol. 2001;2:612–19. doi: 10.1038/89759. [DOI] [PubMed] [Google Scholar]
- 346.Mangino MJ, Brounts L, Harms B, Heise C. Lipoxin biosynthesis in inflammatory bowel disease. Prostaglandins Other Lipid Mediat. 2006;79:84–92. doi: 10.1016/j.prostaglandins.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 347.Arita M, Yoshida M, Hong S, Tjonahen E, Glickman JN, et al. Resolvin E1, an endogenous lipid mediator derived from omega-3 eicosapentaenoic acid, protects against 2,4,6-trinitrobenzene sulfonic acid-induced colitis. Proc. Natl. Acad. Sci. USA. 2005;102:7671–76. doi: 10.1073/pnas.0409271102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Hudert CA, Weylandt KH, Lu Y, Wang J, Hong S, et al. Transgenic mice rich in endogenous omega-3 fatty acids are protected from colitis. Proc. Natl. Acad. Sci. USA. 2006;103:11276–81. doi: 10.1073/pnas.0601280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Belluzzi A, Brignola C, Campieri M, Pera A, Boschi S, Miglioli M. Effect of an enteric-coated fish-oil preparation on relapses in Crohn’s disease. N. Engl. J. Med. 1996;334:1557–60. doi: 10.1056/NEJM199606133342401. [DOI] [PubMed] [Google Scholar]
- 350.Fiorucci S, Wallace JL, Mencarelli A, Distrutti E, Rizzo G, et al. A beta-oxidation-resistant lipoxin A4 analog treats hapten-induced colitis by attenuating inflammation and immune dysfunction. Proc. Natl. Acad. Sci. USA. 2004;101:15736–41. doi: 10.1073/pnas.0404722101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Gewirtz AT, Collier-Hyams LS, Young AN, Kucharzik T, Guilford WJ, et al. Lipoxin a4 analogs attenuate induction of intestinal epithelial proinflammatory gene expression and reduce the severity of dextran sodium sulfate-induced colitis. J. Immunol. 2002;168:5260–67. doi: 10.4049/jimmunol.168.10.5260. [DOI] [PubMed] [Google Scholar]
- 352.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. Isme J. 2007;1:403–18. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 353.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J. Clin. Microbiol. 2006;44:4136–41. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–93. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, Acero D, Garcia-Gil LJ. Abnormal microbiota composition in the ileocolonic mucosa of Crohn’s disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm. Bowel Dis. 2006;12:1136–45. doi: 10.1097/01.mib.0000235828.09305.0c. [DOI] [PubMed] [Google Scholar]
- 357.Bibiloni R, Mangold M, Madsen KL, Fedorak RN, Tannock GW. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn’s disease and ulcerative colitis patients. J. Med. Microbiol. 2006;55:1141–49. doi: 10.1099/jmm.0.46498-0. [DOI] [PubMed] [Google Scholar]
- 358.Conte MP, Schippa S, Zamboni I, Penta M, Chiarini F, et al. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut. 2006;55:1760–67. doi: 10.1136/gut.2005.078824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–23. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 360.Tsang J, Brown R, Anderson G, Schmidt T, Tannock G, et al. Selective expansion of colitogenic commensal bacterial species in SPF IL-10−/− mice. Gastroenterology. 2004;126:A291. Abstr. [Google Scholar]
- 361.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA. 2005;102:11070–75. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 363.Hildebrandt MA, Hoffman C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, et al. High fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–24. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.