Abstract
Arterial and venous thrombosis are among the most common causes of death and hospitalization worldwide. Nanotechnology approaches hold great promise for molecular imaging and diagnosis as well as tissue-targeted delivery of therapeutics. In this study, we developed and investigated bioengineered nanoprobes for identifying thrombus formation; the design parameters of nanoparticle shape and surface chemistry, i.e. incorporation of fibrin-binding peptides CREKA and GPRPP, were investigated. Two nanoparticle platforms based on plant viruses were studied — icosahedral cowpea mosaic virus (CPMV) and elongated rod-shaped tobacco mosaic virus (TMV). These particles were loaded to carry contrast agents for dual-modality magnetic resonance (MR) and optical imaging, and both modalities demonstrated specificity of fibrin binding in vitro with the presence of targeting peptides. Preclinical studies in a carotid artery photochemical injury model of thrombosis confirmed thrombus homing of the nanoprobes, with the elongated TMV rods exhibiting significantly greater attachment to thrombi than icosahedral (sphere-like) CPMV. While in vitro studies confirmed fibrin-specificity conferred by the peptide ligands, in vivo studies indicated the nanoparticle shape had the greatest contribution toward thrombus targeting, with no significant contribution from either targeting ligand. These results demonstrate that nanoparticle shape plays a critical role in particle deposition at the site of vascular injury. Shaping nanotechnologies opens the door for the development of novel targeted diagnostic and therapeutic strategies (i.e., theranostics) for arterial and venous thrombosis.
Introduction
Cardiovascular disease continues to be a significant challenge and has remained one of the most common causes of death and hospitalization in the world.1 Arterial and venous thrombus formation are fundamental sources of many acute coronary syndromes such as myocardial infarction, stroke, and sudden ischemic death.1, 2 The obstruction of blood flow during thrombus formation could lead to tissue death and impaired function. To combat and effectively treat thrombotic disorders, methods for sensitive detection are necessary for identifying the source thrombus and for monitoring treatment progression.3 Thrombosis involves two main processes: platelet recruitment and fibrin formation.4 While the ratio of platelets and fibrin vary among patients, they are both major components of thrombi.5 Fibrin formation is a polymerization reaction caused by thrombin-mediated cleavage of fibrinogen such that exposed domains can cause crosslinking of fibrin units.6 Thus, as thrombi spontaneously break loose or are dissolved by a therapeutic, fibrin continues to be present on the surface. In this way, targeting of fibrin is advantageous for monitoring of thrombosis and treatment.
The main strategies for targeted delivery of nanoparticles utilize either antibodies or peptides. Although monoclonal antibodies have been widely used in the past due to their high affinity, they are limited by their large size and random orientation upon conjugation. Peptides have emerged as an attractive alternative to antibodies due to their smaller size, easier production, lower cost, lower immunogenicity, and long-term stability.7 CREKA8-10 and GPRPP11, 12 are two pentapeptide amino acid sequences that have been identified through screening and shown to be effective for molecular imaging of fibrin.
When selecting a nanocarrier, shape is an important consideration that affects a delivery system's in vivo fate.13 For vascular targeting applications, the success of a nanoparticle reaching the site of disease depends on its ability to drift laterally toward the wall of the blood vessel. Studies performed in the setting of cancer nanotechnology indicate that non-spherical materials such as filamentous rods have favorable margination properties and, additionally, the enhanced vascular interaction of filamentous rods increases targeting efficiency.14-17 Many nanoparticle platforms are available, such as polymeric nanoparticles,18 silica nanoparticles,19 liposomes,20 and dendrimers.21 Current synthetic chemistries are still lacking the efficiency and spatial control for synthesizing monodisperse filamentous nanomaterials with high reproducibility. Therefore, we turned toward a bio-inspired approach utilizing plant viral nanoparticles (VNPs) that come naturally in a diverse array of shapes and sizes, including elongated filaments. Aside from the ease of manufacturing large quantities of identical nanofilaments through molecular farming in plants, VNPs are noninfectious in mammals and are advantageous due to their low density, monodispersity, atomic-level controllability, and biocompatibility.22 Additionally, these particles have been shown to be both cytocompatible and hemocompatible, with no induction of cell toxicity, clotting, or hemolysis.23, 24 Naked VNPs are moderately immunogenic and eliminated by antibodies,25-27 but this can be overcome by PEGylation.28-33
In this study, we investigated the combined role of shape and ligand display on targeting efficacy using dual-modality optical and magnetic resonance (MR) imaging. Fibrin-targeted VNP platforms based on rod-shaped tobacco mosaic virus (TMV) were developed and evaluated against formulations based on icosahedral cowpea mosaic virus (CPMV). Their targeting profiles were assessed and outcomes compared for thrombus-specific delivery. Evaluation was performed using phantoms along with an in vivo mouse model of thrombosis and dual optical and MR imaging. Successful thrombus homing of the rod-shaped particles was achieved, which prepares the groundwork for the development of targeted therapeutic formulations.
Experimental
Virus propagation and purification
Viruses were propagated by mechanical inoculation of 5 μg of virus per leaf. CPMV was spread with Vigna unguiculata plants, while wild-type TMV and TMVLys mutants were propagated with Nicotiana benthamiana. Isolation of the viral nanoparticles by established procedures yielded approximately 1 mg of virus per gram of infected leaf material.34-36
Bioconjugation
For CREKA conjugation, N3-(PEG)8-COOH was purchased from Novabiochem (product named N3-(PEG)7-COOH but there are 8 PEG monomers, see Fig. S1), while N3-(PEG)8-CREKA was synthesized by the Molecular Biotechnology Core Laboratory at Cleveland Clinic using standard peptide chemistries. CPMV in 0.1 M potassium phosphate buffer, pH 7.0 was mixed with 6000 molar equivalents of propargyl-NHS ester (Click Chemistry Tools) in 10% (v/v) DMSO at a final concentration of 2 mg/mL virus to attach alkyne ligation handles. Purification of CPMV for this and subsequent reactions was performed using centrifugal spin filters with a cut-off of 10 kDa. Copper(I)-catalyzed azide-alkyne cycloaddition (“click”) was then carried out by mixing CPMV at a 2 mg/mL concentration final concentration in 0.1 M potassium phosphate buffer, pH 7.0 with 1500 molar excess of either N3-(PEG)8-COOH or N3-(PEG)8-CREKA and 900 equivalents of sCy5 azide with the addition of 20 mM aminoguanidine (AMG), 20 mM L-ascorbic acid (Asc), and the combination of 2 mM CuSO4 together with 10 mM tris-(benzyltriazolylmethyl)amine (THPTA). TMV exterior modification was first performed by mixing TMV with 15 equivalents per coat protein of diazonium salt (produced by reaction of 100 mM 3-ethynylaniline with 150 mM sodium nitrite in p-toluenesulfonic acid) in 100 mM borate buffer, pH 8.8 for 30 minutes on ice. Purification of TMV for all reactions was performed by ultracentrifugation at 42,000 rpm for 3 hours over a 40% (w/v) sucrose cushion. N3-(PEG)8-COOH or N3-(PEG)8-CREKA was then attached by click chemistry of 2 mg/mL TMV with 2 equivalents of PEG or peptide in the presence of 2 mM AMG, 2 mM Asc, and 1 mM CuSO4 in 10 mM phosphate buffer, pH 7.4 on ice for 30 minutes. Interior labeling with alkynes was achieved by EDC coupling over 24 hours using 15 equivalents of propargylamine per coat protein with 45 equivalents of EDC (15 equivalents added at 0, 6, and 18 hours), and 90 equivalents of hydroxybenzotriazole (HOBt) in 100 mM HEPES buffer, pH 7.4. Afterwards, 4 equivalents of sCy5 per coat protein were added by click using the same protocol above as for CREKA.
For GPRPP conjugation, GPRPPC was purchased from Genscript. Synthesis of GPRPP with azide-terminated PEG spacer proved to be challenging, so an alternative method was adopted. Instead of an azide functionality, GPRPP was synthesized with a C-terminal cysteine residue, which provides a handle for conjugation of the peptide to the particles while leaving the pentapeptide essential for targeting free. To carry out the reactions, a TMVLys mutant was utilized, which displays a lysine residue at position 158 (Thr→Lys mutation).34 CPMV and TMVLys were mixed with SM(PEG)8 (3000 and 10000 molar excess, respectively) and Alexa Fluor 647 (A647; 2000 and 8000 molar excess, respectively) at a final concentration of 2 mg/mL virus in 0.1 M potassium phosphate buffer, pH 7.0 with 10% (v/v) DMSO for 1 hour at room temperature. The particles were then quickly purified using 10 kDa centrifugal spin filters before further reaction with GPRPPC overnight (3000 and 10000 excess for CPMV and TMV, respectively) with the same conditions of 2 mg/mL virus and 10% (v/v) DMSO. The GPRPP-labeled particles were then purified by dialysis over a week. For particles for MR imaging, TMVLys was reacted with gadolinium (Gd) before modification with GPRPP and A647. To make Gd(DOTA) azide, GdCl3 was incubated with azido-mono-amide-DOTA (Macrocyclics) at a 1.3:1 ratio and a concentration of 15 mM while maintaining the pH between 6 and 7 with NaOH. After a week, the final concentration was brought down to 10 mM, and the pH was increased to 8 to precipitate out any free Gd. TMVLys was labeled on the interior with alkynes using propargylamine using the same protocol as above for TMV. After purification, the particles were reacted with 6 equivalents per coat protein of Gd(DOTA) azide using the click chemistry procedure for TMV above. CPMV was first functionalized with propargyl-NHS ester then labeled with 2000 excess Gd(DOTA) azide using the procedures described above.
UV/visible spectroscopy
A Thermo Scientific NanoDrop 2000 Spectrophotometer was used to determine the concentration of the particles and the extent of fluorophore labeling. The extinction coefficients for CPMV and TMV at 260 nm are 8.1 mg−1 mL cm−1 and 3 mg−1 mL cm−1, respectively, while the extinction coefficients for sCy5 and A647 are 271,000 M−1 cm−1 at 646 nm and 270,000 M−1 cm−1 at 651 nm, respectively.
Fluorescence measurements
Particles were diluted to a concentration of 0.25 mg/mL with 0.1 M potassium phosphate buffer, pH 7. Samples were measured in triplicate on a black 384 well plate, using a volume of 50 μL per well. Fluorescence intensity was measured using a Tecan Infinite 200 plate reader at excitation and emission wavelengths of 600 and 665 nm, respectively.
SDS gel electrophoresis
Denaturing gel electrophoresis was performed using 4–12% NuPAGE gels (Invitrogen). 10 μg samples were denatured at 100°C for 5 minutes then ran on a polyacrylamide gel in 1× MOPS running buffer at 200 V for 50 minutes. The gels were stained with Coomassie Blue and imaged with an AlphaImager imaging system (Biosciences).
Transmission electron microscopy
Particles were adsorbed to carbon-coated copper grids at a concentration of 0.1 mg/mL, rinsed with deionized water, then negatively stained with 2% (w/v) uranyl acetate for 1 minute. Samples were analyzed with a Zeiss Libra 200FE TEM at 200 kV.
Inductively coupled plasma optical emission spectroscopy
The amount of gadolinium attached to CPMV and TMV was determined by ICP-OES (Agilent 730 Axial ICP-OES) based on the emission spectral line at 342.246 nm.
MALDI-TOF mass spectrometry
Native TMV and TMVLys along with formulations modified with PEG, CREKA, and GPRPP were denatured by adding 3 μL of 6 M guanidine hydrochloride to 12 μL of each sample diluted to 1 mg/mL concentrations. The samples were then spotted on an MTP 384 polished steel target (Bruker) using C18 ZipTips (Millipore). MALDI-TOF MS analysis was performed using an Ultraflex I TOF/TOF mass spectrometer (Bruker).
Fibrin clot assay
4 mg/mL of non-plasminogen deleted fibrinogen (Enzyme Research Labs) was polymerized with 8 U/mL of thrombin in the presence of 7 mM CaCl2 in TBS to form 200 μL fibrin clots in a black 96-well assay plate with a clear bottom (Corning). After 2 hours, either 100 μL of PBS or 50 μg of TMV-PEG, TMV-CREKA, TMV-SMPEG, or TMV-GPRPP in 100 μL of KP buffer were added on top of the clots and allowed to incubate for a further 2 hours. The clots were washed 3 times with 100 μL of TBS, then the plate was analyzed using Maestro imaging (see below).
Carotid artery photochemical injury and thrombosis model
All animal procedures were performed using approved protocols from the Institutional Animal Care and Use Committee at Case Western Reserve University. Particles (200 μg/100 μL) and Rose Bengal dye (10 mg/mL in PBS) at a dose of 50 mg/kg body weight were simultaneously administered to C57BL/6 mice by tail vein injection. A 1.5 mW, 540 nm green laser (Melles Griot) was used to illuminate the right common carotid artery 5 cm from the artery to induce thrombus formation, and blood flow was monitored with a Doppler flow probe (Transonic Systems). After vessel occlusion, the mice were sacrificed, and the injured artery and the contralateral control were excised and fixed in formalin. Particle accumulation was analyzed by Maestro imaging (see below).
Maestro fluorescence imaging
sCy5 and A647 signal from in vitro and in vivo clots were analyzed using Maestro fluorescence imaging with yellow excitation (576 to 621 nm) and emission (635-nm longpass) filters with 800 ms exposure times. The exposure time was changed to 150 ms for the cylindrical clots made for dual-modality imaging.
Immunofluorescence
Excised arteries were embedded in OCT medium after isolating the region of thrombosis. Then, 10 μm sections were cut with a cryostat (Leica) and mounted on ColorFrost Plus microscope slides (Fisher). The sections were blocked with 10% (v/v) goat serum (GS) in PBS for an hour, stained for platelets with integrin αIIbβ3 specific antibodies (Santa Cruz Biotech) diluted 1:100 in PBS with 1% (v/v) GS followed by secondary Alexa Fluor 555 anti-goat antibody staining diluted 1:250 in PBS with 1% (v/v) GS, then mounted with Fluoroshield with DAPI (Sigma). Imaging was performed on a Zeiss Axio Observer Z1 fluorescence microscope.
Pharmacokinetics
Six to eight week old Balb/C mice (Charles River) were placed on an alfalfa free diet (Teklad) for three weeks before tail vein injection of 10 mg/kg A647-TMV-PEG and A647-TMV-GPRPP formulations. Blood samples were collected by orbital sinus bleeds at 3, 5, 10, 15, 20, 30, 60, 100, 140, and 180 min (n = 3) in heparin-coated tubes (Fisher). Serum was separated and collected by centrifugation at 7000 rpm in a tabletop centrifuge for 10 minutes. 50 μL of serum from each time point were added to a black 384-well plate and fluorescence read using a Tecan Infinite 200 microplate reader (λEx = 600 nm, λEm = 665 nm). The results were analyzed using Prism software.
MR imaging
MR imaging was performed on a horizontal 7 T Bruker scanner (Bruker Biospec) using a 35-mm inner diameter volume coil. To determine the T1 relaxivity of the particles, phantom images of increasing concentrations of particles up to a Gd concentration of 32 μM were taken using a multi-slice saturation recovery Look-Locker (MSRLL) sequence. For imaging of clots, the tapered section was cut off from 0.5 mL Eppendorf microcentrifuge tubes, a small hole was pierced into the cap of the tubes, a suture string was threaded through the hole, the hole was sealed with parafilm, and 500 μL cylindrical clots were formed in the tubes around the string after inversion of the tube. The same proportions of fibrinogen, thrombin, and CaCl2 were used as in the fibrin clot assay above. The clots were allowed to form for 3 hours at room temperature, then they were transferred to a 1.5 mL Eppendorf tube and incubated with 400 μL of a 2 mg/mL solution of the TMV particles overnight. The clots were then washed thoroughly in a 1 L beaker of PBS before being transferred and placed in a new 1.5 mL tube completely filled with PBS. A spin echo multi-slice multi-echo (MSME) sequence was applied with the following imaging parameters: TR = 400 ms; TE= 10.6 ms; slice thickness = 1 mm; slice number = 15; FOV = 3.0 × 3.0 cm2; matrix size = 256 × 256; and number of average = 4.
Statistical analysis
Data represent means ± standard deviation. p values were determined by unpaired Student's t test. * p < 0.05 is considered significant, ** p < 0.01 is considered highly significant, and *** p < 0.001 is considered extremely significant.
Results and discussion
Fibrin-targeted nanoparticles and nontargeted controls were produced by modification of TMV nanorods (300×18 nm) with one of two fibrin-specific peptide ligands, either CREKA or GPRPP. These ligands were attached via an intervening PEG spacer, and controls with only the PEG spacer were compared alongside the targeted particles. To investigate the impact of nanoparticle shape, icosahedral CPMV nanoparticle formulations (30 nm diameter) were also created. Conjugation of the PEG and peptides was achieved through modification of exterior tyrosine residues for TMV-CREKA and exterior lysine side chains for CPMV-CREKA, CPMV-GPRPP, and TMV-GPRPP. For dual-modality thrombus imaging, near infrared fluorophores (sulfo-Cy5 for CREKA particles and Alexa Fluor 647 for GPRPP particles) and/or chelated gadolinium-tetraazacyclododecanetetraacetic acid complexes (Gd(DOTA)) were also incorporated into the formulations. All formulations and bioconjugation schemes are outlined in Fig. S1 and described in detail in the Experimental section.
A combination of UV/Vis spectroscopy, fluorescence analysis, inductively coupled plasma optical emission spectrometry (ICP-OES), denaturing gel electrophoresis, matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectroscopy (MS), and transmission electron microscopy (TEM), was performed to determine the degree of particle labeling, the relative fluorescence intensity of the particles, and the structural integrity of the particles after modification (Fig. 1, S2, and S3).
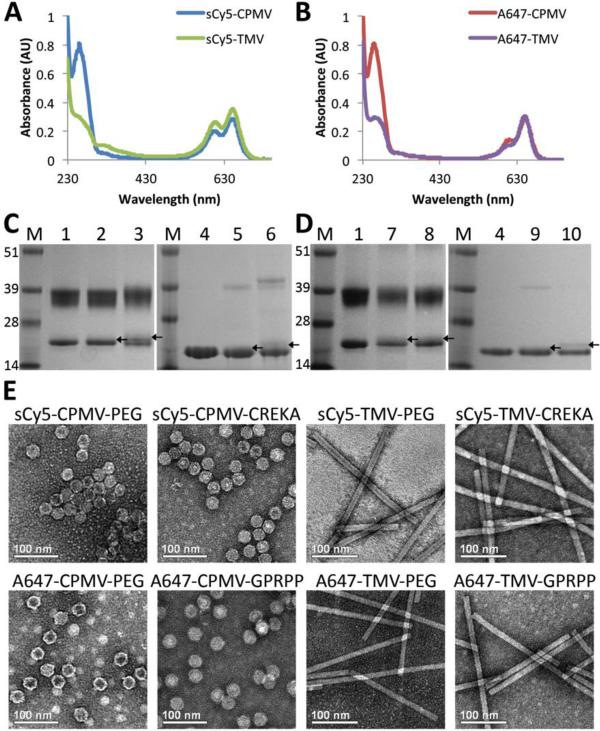
Fig. 1. Characterization of particles.
(A,B) UV/Vis spectroscopy of particles conjugated with dye and either PEG or targeting peptides normalized for 1 mg/mL particle concentration show similar dye concentrations for the particles. (C,D) SDS-PAGE gels of same particles, demonstrating attachment of PEG and targeting peptides to coat proteins. M = SeeBlue Plus2 molecular weight marker; 1 = CPMV; 2 = sCy5-CPMV-PEG; 3 = sCy5-CPMV-CREKA; 4 = TMV; 5 = sCy5-TMV-PEG; 6 = sCy5-TMV CREKA; 7 = A647-CPMV-PEG; 8 = A647-CPMV-GPRPP; 9 = A647-TMV-PEG; 10 = A647-TMV GPRPP (attachment indicated by arrows pointing to bands slightly above unmodified bands). (E) TEM images show particles remain intact after modification. Scale bar = 100 nm.
From the absorbance spectra normalized to 1 mg/mL particle concentration (Fig. 1A,B), it is apparent that the dye concentration (determined by absorbance around 650 nm) is very similar between CPMV and TMV formulations. Quantification of sCy5 and A647 labeling based on the spectra indicated that sCy5-CPMV (PEG and CREKA) were labeled with 60 ± 10% dyes per particle, or 50% of the 120 available subunits (60 each of the small and large coat proteins), while sCy5-TMV-PEG and sCy5-TMV-CREKA were labeled with 460 ± 10% dyes, or approximately 22% of the available coat proteins (TMV is formed by 2130 identical copies of a coat protein). Similarly, A647-CPMV (PEG and GPRPP) were labeled with about 60 ± 10% dyes, while A647-TMV-PEG and A647-TMV-GPRPP contained 400 ± 10% dyes.
Optical properties were also examined, specifically the fluorescence output. The density of dyes around the particles, the method of conjugation, and the local microenvironment could all affect the fluorescence intensity despite similar dye concentration across the formulations.37 Therefore, fluorescence analysis of the particle solutions using a plate reader was employed for determining the relative fluorescence intensities of each formulation (Fig. S2). For data interpretation comparing the various nanocarriers, we performed a normalization of the data by fluorescence intensity. The time period of the experiments (30 min) is unlikely to be sufficient for cellular internalization and degradation to affect the fluorescence properties of the particles, especially in this application where the particles are bound extracellularly to the fibrin. Therefore, the fluorescence of the particles should be similar within the vasculature to bulk measurements performed on the benchtop, and particle concentration was normalized accordingly.
The extent of Gd(DOTA) loading of the particles was quantified by ICP-OES, which revealed ~1,600 Gd conjugated per particle. Additionally, labeling of particles with PEG and fibrin-binding ligands was confirmed with SDS-PAGE denaturing gel electrophoresis of the coat proteins of the particles, with higher molecular weight bands indicating attachment to coat proteins (Fig. 1C,D). Densitometric analysis of the lanes suggests approximately 10-15% coverage of the coat proteins with PEG or peptides. PEG and peptide labeling of TMV was additionally confirmed with MALDI-TOF MS (Fig. S3 and Table S1). Analysis by TEM after complete modification of the particles demonstrated that the integrity of the particles is maintained (Fig. 1E).
The targeting ability of the nanoparticles was then tested in vivo using a mouse model of thrombosis: thrombosis was induced in wild-type C57Bl6 mice by photochemical injury of the right common carotid artery, which is caused by reaction of a 540 nm green laser light with intravenously injected Rose Bengal dye.38 The nanoparticle formulations were administered intravenously and allowed to circulate until occlusion occurred at approximately 30 minutes after initiation of laser injury. Thrombus binding was evaluated by excising the carotid artery after occlusion along with the uninjured contralateral artery then imaging the arteries using Maestro fluorescence imager (Fig. 2A,B). (All animal procedures were performed using approved protocols from the Institutional Animal Care and Use Committee at Case Western Reserve University; detailed procedures are listed in the Experimental section.)
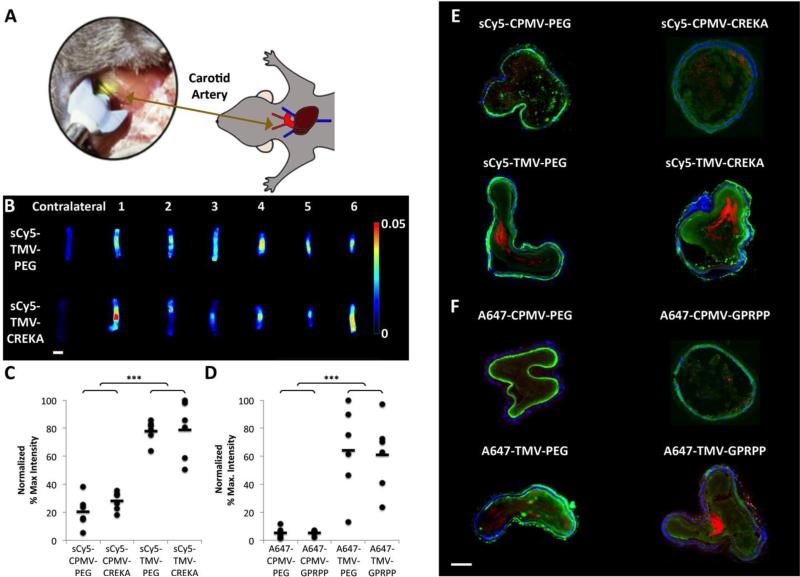
Fig. 2. Evaluation of particle localization in a photochemical mouse model of thrombosis.
(A) Schematic detailing the experimental set-up. (B) Maestro fluorescence imaging of excised carotid arteries with nanoparticles localized at site of thrombosis (example shows PEG- vs. CREKA-modified TMV particles). Contralateral controls of uninjured artery show no particle accumulation. Scale bar = 1 mm. (C,D) Normalized fluorescence intensity based on particle fluorescence values (*** p < 0.001). Asterisks denote significance of TMV samples relative to both nontargeted and targeted CPMV samples. (E,F) Immunofluorescence of artery sections. Red = VNP formulations; blue = nuclei (DAPI) from epithelial cells; green = platelets (αIIbβ3 integrin antibody with Alexa Fluor 555 secondary antibody, pseudocolored green). Scale bar = 100 μm. Multiple tissue sections were imaged and representative images are shown here.
It was apparent that the effect of shape outweighed surface chemistry: the elongated TMV, whether targeted or not, showed the most enhanced thrombus accumulation compared to the icosahedral CPMV (Fig. 2C,D). The shape-mediated differences may be explained by the distinct flow properties of high-aspect versus low-aspect ratio materials. Elongated materials experience lateral torque due to their anisotropy, therefore increasing margination toward the vessel wall; in contrast, spherical particles are more likely to remain in a direct laminar flow path.39 These factors contribute to enhanced thrombus targeting of the TMV formulations and suggest that CPMV has a lower probability to interact with and bind to the thrombus.
Immunofluorescence of artery sections further confirmed presence of the nanoparticles within the thrombi (Fig. 2E,F). 10 μm cross-sections of the arteries were taken and stained for cell nuclei with DAPI and for platelets with an αIIbβ3 integrin antibody followed by an Alexa Fluor 555 secondary antibody. The αIIbβ3 staining demonstrates the presence of the thrombus within the arteries, with some elastin-derived autofluorescence around the perimeter of the arteries. Interestingly, the difference in accumulation of CPMV compared to TMV can be more easily observed from fluorescence signal of artery sections, where a low level of fluorescence is seen for arteries with CPMV while brighter areas of particle localization are observed for TMV (see Fig. 2E,F). This is likely attributable to a greater propensity for TMV to interact with the side of the vasculature as the vessel narrows during thrombus formation due to its oblong shape allowing for easier margination.
To verify that the peptides preserved their fibrin-binding functionality after conjugation, the ability of the particles to bind to thrombi was evaluated in vitro. Because only the TMV formulation exhibited significant thrombus targeting in vivo, we only considered TMV for the in vitro studies. Fibrin clots were created in a 96-well plate by incubating 4 mg/mL fibrinogen with 8 U/mL of thrombin and 7 mM CaCl2 in 200 μL TBS for 2 hours at room temperature. After thrombin-catalyzed fibrin polymerization had occurred, the clots were incubated with PBS, sCy5-TMV-PEG, sCy5-TMV-CREKA, A647-TMV-PEG, or A647-TMV-GPRPP, washed thoroughly with PBS, then imaged using Maestro fluorescence imager for binding affinity (Fig. 3). Binding of both sCy5-TMV-CREKA and A647-TMV-GPRPP targeted formulations to fibrin was significantly increased compared to the nontargeted controls. CREKA-targeted TMV formulations showed 5.5 times greater binding compared to their PEG-only counterparts, and a 9.5-fold increase in signal was measured for GPRPP-targeted TMV particles vs. TMV-PEG. The higher binding rate of the TMV-GPRPP may be explained with the higher affinity of the peptide: CREKA has a Kd of 6 μM, while GPRPP has a Kd of around 8 nM.9, 40
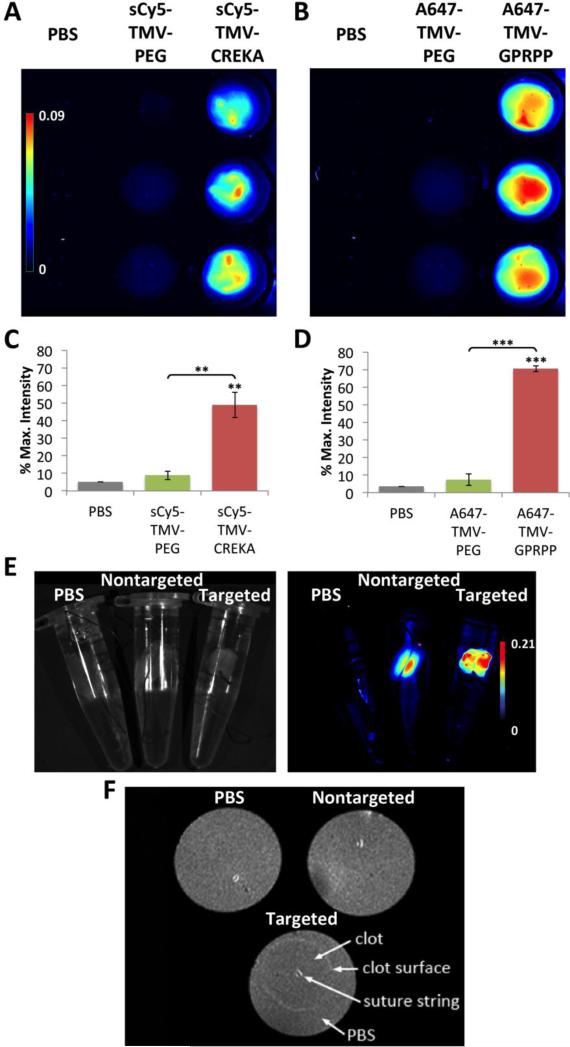
Fig. 3. Fibrin targeting as studied by in vitro assay.
(A,B) Fluorescence imaging of particles binding to fibrin clots formed from fibrinogen and thrombin in a 96-well plate (n = 3). (C,D) Quantification of average fluorescence intensity observed (** p < 0.01, *** p < 0.001). Unpaired asterisks denote significance vs. PBS control. (E) Fluorescent imaging of clots in Eppendorf tubes under white light (left) and heat map (right). (F) MR imaging of the same clots using a preclinical 7 T MRI, with binding of TMV-GPRPP seen on the exterior surface.
The stark differences between the in vitro and in vivo findings may be explained by the fact that the blood stream is not a dilute solution, and the nanoparticles are most likely subject to formation of a so-called protein corona, which may interfere with target recognition.41 Further, the in vitro conditions do not realistically mimic the in vivo conditions where the nanoparticle formulations are subjected flow; at the injury site in particular, turbulent flow and shear stress will affect particle accumulation. The inclusion of flow within in vitro assays has shown contradicting results, with some literature supporting enhanced margination and deposition of rod-shaped nanoparticles,16 while others indicate only particles in the micrometer size range exhibit this enhanced interaction of rods over spheres.42 The variation may arise due to the different flow chamber set-ups and differences in the properties of the particles, such as porosity and density. Our results show that, at least for viral nanoparticles, nanoparticle shape can indeed influence the accumulation of particles at areas of thrombosis.
To carry these studies a step further and demonstrate the translational potential, we investigated the possibility of performing dual-modality imaging (optical-MR) with the TMV formulations, considering GPRPP-targeted and PEG-TMV controls. TMV was loaded with ~1,600 Gd(DOTA) per particle, with conjugation of the MR contrast agents carried out using the 4 nm-wide interior channel of TMV (see Fig. S1). The relaxivity of the particles was determined by imaging phantoms of the particles in solution with Gd concentrations ranging up to 32 μM using a 7 T preclinical MRI and a multi-slice multi-echo sequence, specifically a multi-slice saturation recovery Look-Locker (MSRLL) sequence. The ionic relaxivity was determined to be 5.1 mM−1 s−1 based on linear recursion of the R1 relaxivity (1/T1) versus the gadolinium concentration (Fig. S4A). This corresponds to an ionic relaxivity of approximately 8,160 mM−1 s−1 per particle.
To evaluate fibrin binding and dual-modality imaging, in vitro cylindrical clots were formed around a suture string in a tube by incubating fibrinogen with thrombin and CaCl2 using a similar procedure as described above. The clots were then transferred to an Eppendorf tube containing a buffered solution of the nanoparticles with binding allowed to proceed overnight. After washing vigorously with water, the clots were placed in a tube filled with fresh buffer and imaged with both Maestro fluorescence and MR imaging (Fig. 3E,F). Consistent with in vitro results from the plate reader assay (Fig. 3A,B), fluorescence imaging indicated greater binding of the GPRPP-targeted particles compared to nontargeted particles (Fig. 3E). It appeared that some PEG-modified TMV particles were present in the center of the clots that were not removed during the washing steps. Without the targeting ligand and lack of driving force to accumulate at the fibrin surface, it is likely that TMV-PEG accumulated non-specifically in the opening in the center where the suture string is present. The high relaxivity in combination with molecular specificity also enabled detection of the thrombus by MR imaging (Fig. 3F). Matching the fluorescence data, the targeted nanoparticles accumulated around the exterior surface of the dense fibrin network formed in vitro,43 while the nontargeted particles are likely only present in the central pore.
These results illustrate an additional advantage of the TMV platform. Along with favorable in vivo partitioning properties, TMV provides a greater surface area to carry medical payloads. For example, the TMV nanorod provides a large surface area for modification with Gd(DOTA), allowing a significantly higher payload compared to CPMV, which is directly correlated to greater contrast enhancement (as measured by the T1 relaxivity). Whereas TMV loaded with ~1,600 Gd(DOTA) per particle, with an ionic relaxivity of 5.1 mM−1 s−1, corresponded to a per particle relaxivity of approximately 8,160 mM−1 s−1, CPMV loaded with ~80 Gd(DOTA) per particle (as determined by ICP-OES), with a similar ionic relaxivity per gadolinium ion of 5.2 mM−1 s−1 (Fig. S4B), corresponded to an ionic relaxivity 20 times lower than that of TMV, at 420 mM−1 s−1 per particle. Toward translational efforts, it is expected that a significantly higher sensitivity of the TMV-based contrast agent would be achieved in clinical studies. Based on previous work, Gd-TMV has a T1 ionic relaxivity of 11.0 mM−1 s−1 per Gd in a clinical 1.5 T MRI, compared to relaxivities of around 5 mM−1 s−1 in the preclinical 7 T scanner used in this study.44 This increase in ionic relaxivity upon nanoparticle conjugation is explained by reduced tumbling rates of the Gd probe, a phenomenon that is lost at higher field strength. From the degree of labeling here, we extrapolate relaxivities as high as 17,600 mM−1 s−1 per particle, which would provide even greater contrast enhancement. (It should be noted that in vivo MRI studies were not feasible using the photochemical injury mouse model of thrombosis due to the requirement of surgery and exposure of the carotid artery during laser illumination; future studies will investigate other small animal models to further drive the development of the TMV-based contrast agent.)
An additional consideration for the difference in behavior observed between CPMV and TMV may be pharmacokinetic properties. From the literature, short clearance times in mice were found for both nanoparticles, with CPMV having a plasma half-life of 4 to 7 minutes,25 while TMV displays a two-phase decay with a fast initial clearance having a half-life of 3.5 minutes followed by a phase II half-life of 94.9 minutes.24 The clearance times of the TMV particles used in this study were not drastically affected by change in surface chemistry, as pharmacokinetic studies performed for A647-TMV-PEG and A647-TMV-GPRPP indicated short half-lives of 7-8 minutes (Fig. S5). Thus, both CPMV and TMV particles are quickly cleared and clearance time is unlikely to account for the stark differences in thrombus targeting.
Despite the similarity in clearance time, shape-based features would result in differences in the partitioning of the nanoparticles between organs of the mononuclear phagocyte system (MPS) and the target tissue. While both CPMV and TMV are cleared through the liver and spleen,24, 25 we and others have previously shown that phagocytosis is reduced for rod-shaped particles,45 including TMV,46 therefore reducing accumulation in non-target organs and increasing the partitioning toward the target site. Additionally, upon contact with the thrombus site, a greater surface area and lower curvature may allow TMV to have stronger binding interactions that grant it the ability to remain more tightly bound to the thrombus under flow (in particular turbulent flow at the disease site). In other words, the spatial orientation of the peptides along the length of TMV allows greater multivalent interactions and, consequently, higher affinity. Some non-specific interactions appear to contribute to the accumulation of particles as well, which may be attributed to a number of factors, including the “stickiness” of activated platelets within the thrombus and hemodynamic drag forces from the porous nature and irregularity of the thrombus.47
In the present study we chose to compare TMV and CPMV, as opposed to spherical aggregates that can be obtained from denatured TMV rods.24, 44, 48 The primary advantage for comparing CPMV, instead of TMV-derived spherical nanoparticles, with TMV is their natural self-assembly into monodisperse particles with knowledge of their precise structure at atomic scale resolution. These factors provide a high degree of reproducibility and spatio-structural control over peptide display and geometrical parameters. Considering their volumes, CPMV and TMV are quite similar in size, with an equivalent sphere to TMV having a diameter of around 50 nm. It should be noted, that while spherical TMV-based nanoparticles can be obtained, their structural confirmation and protein organization differs from rod-shaped TMV counterparts.24, 44, 48
To overcome these technological challenges, we recently developed two independent approaches to yield protein-based nanoparticle assemblies of distinct aspect ratio but composed of the same protein make-up. In one approach, we used an RNA-templated self-assembly approach to synthesize TMV rods of distinct aspect ratios, specifically 60 nm, 130 nm in 300 nm in length;46 and in another approach CPMV nanoparticle chains were obtained through symmetry-breaking and polymer-assisted assembly.49 While beyond the current study, these materials would offer attractive building blocks for follow-up studies to further delineate the effect of aspect ratio on vascular targeting.
Conclusion
In summary, we developed nanoparticle-based probes for imaging of thrombi in vivo, and thrombosis targeting and dual modality optical-MR imaging was demonstrated. Thrombosis targeting was achieved in vivo based on the choice of nanoparticle shape. The effect of nanoparticle shape has been extensively studied in the setting of oncology; our data indicate that nanocarrier shape also has implications in cardiovascular nanomedicine. While in vitro studies confirmed fibrin-specificity conferred by the peptide ligands, in vivo studies indicated the nanoparticle shape had the greatest contribution toward thrombus targeting, with no significant contribution from either targeting ligand.
Although we investigated molecular imaging of fibrin in this study due to its essential role in formation of thrombi, another critical component that could be targeted is platelets. Evidence suggests that thrombi contain large numbers of both platelets and fibrin, having a greater abundance of fibrin in cases of ruptured plaque and being more platelet-rich for thrombi on eroded plaques.5 For effective targeting regardless of the underlying nature of the components exposed to circulating nanoparticles, heteromultivalent ligand display that can simultaneously bind to both factors is a possible approach. Dual-targeting of disease-specific markers can be advantageous compared to single targets by being more site-selective and having superior binding affinity. 50
Another direction currently under investigation in our lab is the addition of therapeutics for theranostics. The site-specific delivery of TMV using molecular targeting can be utilized for detection of thrombosis with conjugation of contrast agents, as shown in this investigation, in conjunction with thrombus dissolution with incorporation of thrombolytic drugs such as tissue plasminogen activator, streptokinase, and urokinase.51 The combination of diagnosis and treatment would allow quick action in the management of the disease before, if necessary, additional follow-up personalized therapy based on the results from imaging.
In conclusion, our data support the successful development of shape-optimized, targeted probes for dual MR and optical imaging of thrombi. We have shown that shape plays a crucial role in the delivery of nanoparticles to the site of disease, with rod-shaped nanoparticles faring considerably better than their spherical counterparts. Overall, this study lays the groundwork for designing a new platform for the ultimate objective of targeted treatment of thrombi.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Science Foundation (NSF): CMMI NM 1333651 (for contribution on CPMV and TMV nanomanufacturing), NIH NHLBI R21 HL121130 (for contribution on preclinical evaluation and imaging), NIH NCI R25 CA148052 Cancer Pharmacology training grant (to K.L.L.), NIH T32 HL105338 Cardiovascular Research training grant (to A.M.W), and a pilot grant from Case-Coulter Translational Research Partnership and the Harrington Heart & Vascular Institute. Prof. Christina Wege and team from the University of Stuttgart, Germany are thanked for providing the TMV lysine mutant. CWRU Farm is thanked for help with scaled-up production and growth of N. benthamiana plants.
Footnotes
Supplementary Materials
Electronic supplementary information (ESI) available: Bioconjugation reaction schemes, fluorescence intensity measurements, MALDI-TOF MS spectra, relaxivity measurements, and pharmacokinetics.
References
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Judd SE, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Mackey RH, Magid DJ, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, Moy CS, Mussolino ME, Neumar RW, Nichol G, Pandey DK, Paynter NP, Reeves MJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuster V, Badimon L, Badimon JJ, Chesebro JH. N. Engl. J. Med. 1992;326:242–250. doi: 10.1056/NEJM199201233260406. [DOI] [PubMed] [Google Scholar]
- 3.Ay I, Blasi F, Rietz TA, Rotile NJ, Kura S, Brownell AL, Day H, Oliveira BL, Looby RJ, Caravan P. Circ. Cardiovasc. Imaging. 2014;7:697–705. doi: 10.1161/CIRCIMAGING.113.001806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falk E, Fernandez-Ortiz A. Am. J. Cardiol. 1995;75:3B–11B. doi: 10.1016/0002-9149(95)80003-b. [DOI] [PubMed] [Google Scholar]
- 5.Sato Y, Hatakeyama K, Yamashita A, Marutsuka K, Sumiyoshi A, Asada Y. Heart. 2005;91:526–530. doi: 10.1136/hrt.2004.034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Undas A, Ariens RA. Arterioscler. Thromb. Vasc. Biol. 2011;31:e88–99. doi: 10.1161/ATVBAHA.111.230631. [DOI] [PubMed] [Google Scholar]
- 7.Yu MK, Park J, Jon S. Theranostics. 2012;2:3–44. doi: 10.7150/thno.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters D, Kastantin M, Kotamraju VR, Karmali PP, Gujraty K, Tirrell M, Ruoslahti E. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9815–9819. doi: 10.1073/pnas.0903369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agemy L, Sugahara KN, Kotamraju VR, Gujraty K, Girard OM, Kono Y, Mattrey RF, Park JH, Sailor MJ, Jimenez AI, Cativiela C, Zanuy D, Sayago FJ, Aleman C, Nussinov R, Ruoslahti E. Blood. 2010;116:2847–2856. doi: 10.1182/blood-2010-03-274258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simberg D, Duza T, Park JH, Essler M, Pilch J, Zhang L, Derfus AM, Yang M, Hoffman RM, Bhatia S, Sailor MJ, Ruoslahti E. Proc. Natl. Acad. Sci. U. S. A. 2007;104:932–936. doi: 10.1073/pnas.0610298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarthy JR, Patel P, Botnaru I, Haghayeghi P, Weissleder R, Jaffer FA. Bioconjug. Chem. 2009;20:1251–1255. doi: 10.1021/bc9001163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawasaki K, Miyano M, Hirase K, Iwamoto M. Chem. Pharm. Bull. (Tokyo) 1993;41:975–977. doi: 10.1248/cpb.41.975. [DOI] [PubMed] [Google Scholar]
- 13.Wen AM, Rambhia PH, French RH, Steinmetz NF. J. Biol. Phys. 2013 doi: 10.1007/s10867-013-9314-z. DOI: 101007/s10867-013-9314-z, DOI:10.1007/s10867-10013-19314-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S-Y, Ferrari M, Decuzzi P. Nanotechnology. 2009;20:495101. doi: 10.1088/0957-4484/20/49/495101. [DOI] [PubMed] [Google Scholar]
- 15.Gentile F, Chiappini C, Fine D, Bhavane RC, Peluccio MS, Cheng MM-C, Liu X, Ferrari M, Decuzzi P. J. Biomech. 2008;41:2312–2318. doi: 10.1016/j.jbiomech.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Toy R, Hayden E, Shoup C, Baskaran H, Karathanasis E. Nanotechnology. 2011;22:115101. doi: 10.1088/0957-4484/22/11/115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doshi N, Prabhakarpandian B, Rea-Ramsey A, Pant K, Sundaram S, Mitragotri S. J. Control. Release. 2010;146:196–200. doi: 10.1016/j.jconrel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Chem. Soc. Rev. 2012;41:2971–3010. doi: 10.1039/c2cs15344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Barnes JC, Bosoy A, Stoddart JF, Zink JI. Chem. Soc. Rev. 2012;41:2590–2605. doi: 10.1039/c1cs15246g. [DOI] [PubMed] [Google Scholar]
- 20.Al-Jamal WT, Kostarelos K. Acc. Chem. Res. 2011;44:1094–1104. doi: 10.1021/ar200105p. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y, Zhao L, Li Y, Xu T. Chem. Soc. Rev. 2011;40:2673–2703. doi: 10.1039/c0cs00097c. [DOI] [PubMed] [Google Scholar]
- 22.Pokorski JK, Steinmetz NF. Mol. Pharm. 2011;8:29–43. doi: 10.1021/mp100225y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aljabali AA, Shukla S, Lomonossoff GP, Steinmetz NF, Evans DJ. Mol. Pharm. 2013;10:3–10. doi: 10.1021/mp3002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruckman MA, Randolph LN, VanMeter A, Hern S, Shoffstall AJ, Taurog RE, Steinmetz NF. Virology. 2014;449:163–173. doi: 10.1016/j.virol.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh P, Prasuhn D, Yeh RM, Destito G, Rae CS, Osborn K, Finn MG, Manchester M. J. Control. Release. 2007;120:41–50. doi: 10.1016/j.jconrel.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava AS, Kaido T, Carrier E. J. Virol. Methods. 2004;115:99–104. doi: 10.1016/j.jviromet.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Gatto D, Ruedl C, Odermatt B, Bachmann MF. J. Immunol. 2004;173:4308–4316. doi: 10.4049/jimmunol.173.7.4308. [DOI] [PubMed] [Google Scholar]
- 28.Zhou JG, Chen YM. Bioorg. Med. Chem. Lett. 2011;21:3268–3272. doi: 10.1016/j.bmcl.2011.04.037. [DOI] [PubMed] [Google Scholar]
- 29.Park JB, Kwon YM, Lee TY, Brim R, Ko MC, Sunahara RK, Woods JH, Yang VC. J. Control. Release. 2010;142:174–179. doi: 10.1016/j.jconrel.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basu A, Yang K, Wang M, Liu S, Chintala R, Palm T, Zhao H, Peng P, Wu D, Zhang Z, Hua J, Hsieh MC, Zhou J, Petti G, Li X, Janjua A, Mendez M, Liu J, Longley C, Mehlig M, Borowski V, Viswanathan M, Filpula D. Bioconjug. Chem. 2006;17:618–630. doi: 10.1021/bc050322y. [DOI] [PubMed] [Google Scholar]
- 31.Yang Z, Wang J, Lu Q, Xu J, Kobayashi Y, Takakura T, Takimoto A, Yoshioka T, Lian C, Chen C, Zhang D, Zhang Y, Li S, Sun X, Tan Y, Yagi S, Frenkel EP, Hoffman RM. Cancer Res. 2004;64:6673–6678. doi: 10.1158/0008-5472.CAN-04-1822. [DOI] [PubMed] [Google Scholar]
- 32.O'Riordan CR, Lachapelle A, Delgado C, Parkes V, Wadsworth SC, Smith AE, Francis GE. Hum. Gene Ther. 1999;10:1349–1358. doi: 10.1089/10430349950018021. [DOI] [PubMed] [Google Scholar]
- 33.Cao G, Beyer TP, Zhang Y, Schmidt RJ, Chen YQ, Cockerham SL, Zimmerman KM, Karathanasis SK, Cannady EA, Fields T, Mantlo NB. J. Lipid. Res. 2011;52:2169–2176. doi: 10.1194/jlr.M018069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geiger FC, Eber FJ, Eiben S, Mueller A, Jeske H, Spatz JP, Wege C. Nanoscale. 2013;5:3808–3816. doi: 10.1039/c3nr33724c. [DOI] [PubMed] [Google Scholar]
- 35.Bruckman MA, Steinmetz NF. Methods Mol. Biol. 2014;1108:173–185. doi: 10.1007/978-1-62703-751-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho CF, Shukla S, Simpson EJ, Steinmetz NF, Luyt LG, Lewis JD. Methods Mol. Biol. 2014;1108:211–230. doi: 10.1007/978-1-62703-751-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen AM, Infusino M, De Luca A, Kernan DL, Czapar AE, Strangi G, Steinmetz NF. Bioconjug. Chem. 2015 doi: 10.1021/bc500524f. DOI: 10.1021/bc500524f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furie B, Furie BC. J. Clin. Invest. 2005;115:3355–3362. doi: 10.1172/JCI26987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SY, Ferrari M, Decuzzi P. Nanotechnology. 2009;20:495101. doi: 10.1088/0957-4484/20/49/495101. [DOI] [PubMed] [Google Scholar]
- 40.Aruva MR, Daviau J, Sharma SS, Thakur ML. J. Nucl. Med. 2006;47:155–162. [PMC free article] [PubMed] [Google Scholar]
- 41.Salvati A, Pitek AS, Monopoli MP, Prapainop K, Bombelli FB, Hristov DR, Kelly PM, Aberg C, Mahon E, Dawson KA. Nat. Nanotechnol. 2013;8:137–143. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 42.Thompson AJ, Mastria EM, Eniola-Adefeso O. Biomaterials. 2013;34:5863–5871. doi: 10.1016/j.biomaterials.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 43.Yu X, Song S-K, Chen J, Scott MJ, Fuhrhop RJ, Hall CS, Gaffney PJ, Wickline SA, Lanza GM. Magnet. Reson. Med. 2000;44:867–872. doi: 10.1002/1522-2594(200012)44:6<867::aid-mrm7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 44.Bruckman MA, Hern S, Jiang K, Flask CA, Yu X, Steinmetz NF. J. Mater. Chem. B Mater. Biol. Med. 2013;1:1482–1490. doi: 10.1039/C3TB00461A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Nat. Nanotechnol. 2007;2:249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shukla S, Eber FJ, Nagarajan AS, DiFranco NA, Schmidt N, Wen AM, Eiben S, Twyman RM, Wege C, Steinmetz NF. Adv. Healthc. Mater. 2015 doi: 10.1002/adhm.201400641. DOI: 10.1002/adhm.201400641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson SP, Nesbitt WS, Westein E. J. Thromb. Haemost. 2009;7(Suppl 1):17–20. doi: 10.1111/j.1538-7836.2009.03401.x. [DOI] [PubMed] [Google Scholar]
- 48.Atabekov J, Nikitin N, Arkhipenko M, Chirkov S, Karpova O. J. Gen. Virol. 2011;92:453–456. doi: 10.1099/vir.0.024356-0. [DOI] [PubMed] [Google Scholar]
- 49.Wen AM, Steinmetz NF. Adv. Healthc. Mater. 2014;3:1739–1744. doi: 10.1002/adhm.201400141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Modery-Pawlowski CL, Gupta AS. Biomaterials. 2014;35:2568–2579. doi: 10.1016/j.biomaterials.2013.12.047. [DOI] [PubMed] [Google Scholar]
- 51.Kunamneni A, Abdelghani TT, Ellaiah P. J. Thromb. Thrombolysis. 2007;23:9–23. doi: 10.1007/s11239-006-9011-x. [DOI] [PubMed] [Google Scholar]
- 52.Weidner S, Kühn G, Just U. Rapid Commun. Mass Spectrom. 1995;9:697–702. doi: 10.1002/rcm.1290090813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.