Abstract
Cancer of unknown primary (CUP) comprises of 3–5% of new cancer diagnoses in the USA. Diagnostic work up typically includes CT of the chest, abdomen and pelvis, and histopathological review of tissue specimens. These measures are neither sensitive nor specific in determining tissue of origin (ToO) of primary tumours and, therefore, are unable to guide therapy. We present two cases of CUP for which we utilised ultra-deep genomic sequencing to identify the candidate ToO and to propose treatment. Patient 1 presented with metastases involving the lung, lymph nodes and bone. Patient 2 presented with an acute pathological fracture of the T7 vertebral body and metastases involving the bone, lymph nodes and soft tissue. No primary renal mass was found. Sequencing revealed SETD2 and NF2 mutations, and heterozygous loss of the short arm of chromosome 3 (3p). Mutations in conjunction with clinicopathological features strongly support a diagnosis of renal cell carcinoma. Both patients initially responded to mTORC1 inhibition therapy.
Background
Cancers of unknown primary (CUP) comprise of 3–5% of new cancer diagnoses in the USA annually.1 In patients presenting with CUP, work up often includes imaging and immunohistochemical (IHC) staining, and may also involve gene expression profiling to determine a putative tissue of origin (ToO).2 Determining the ToO may improve prognosis in these patients through use of site-directed cancer therapy instead of empiric chemotherapeutic regimens.3 An emerging tool in the clinician's armamentarium is genomic sequencing, which could provide diagnostic and therapeutic information for these rare patients with CUP. We present two young patients with CUP whose cancers, following genomic interrogation, favoured renal cell carcinoma (RCC). Both patients benefited from treatment with an mTORC1 inhibitor, an Food and Drug Administration (FDA)-approved targeted therapy for kidney cancer.
Case presentation
Patient 1
A 43-year-old man with no family history of cancer presented with malignant pericardial effusion, bilateral pulmonary nodules, and mediastinal and hilar lymphadenopathy. He underwent pericardial window formation and right upper lobe video-assisted thoracoscopic surgery, which revealed carcinoma with clear cell features suggestive of RCC. CT of the chest, abdomen and pelvis revealed additional lytic lesions involving the lumbar spine and left acetabulum, but no primary renal mass (figure 1).
Figure 1.
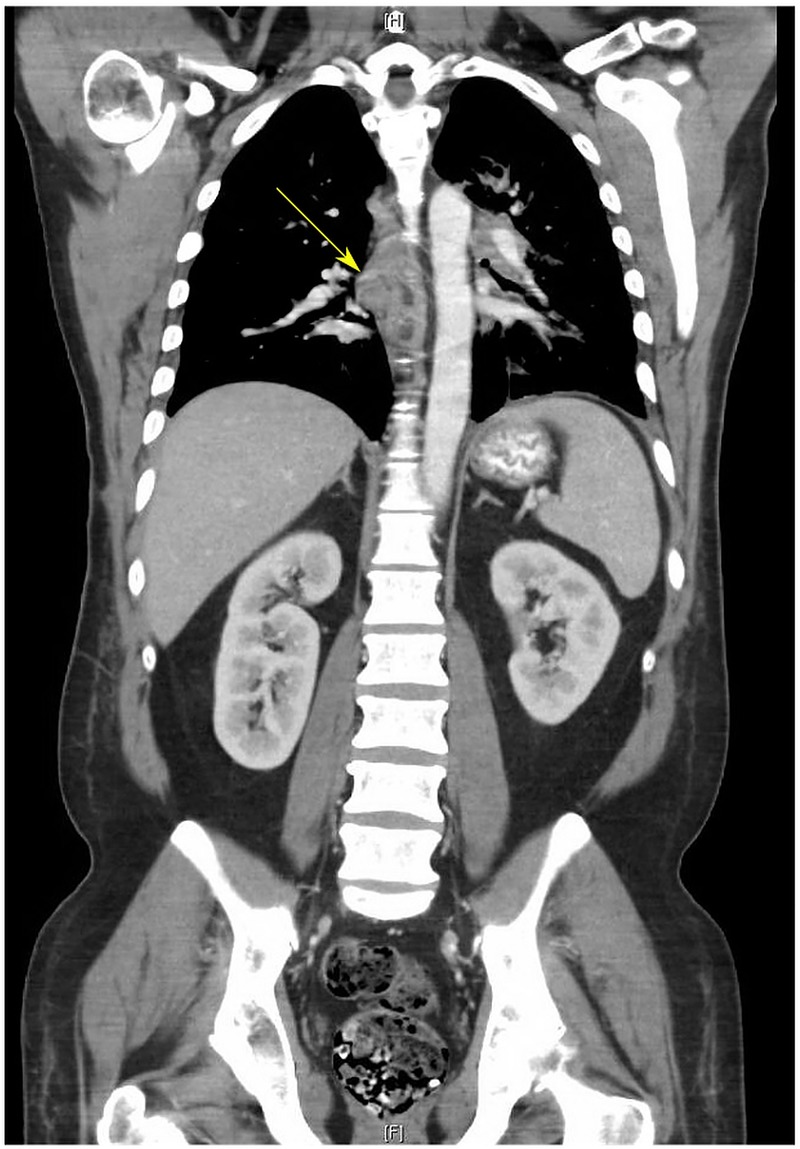
CT of the chest, abdomen and pelvis of patient 1. CT scan demonstrating normal kidneys bilaterally without evidence of renal masses or cysts. The chest reveals hilar lymphadenopathy (arrow). Mediastinal masses and lung nodules were also discovered.
Patient 2
A 55-year-old woman with a remote history of papillary thyroid cancer and an unremarkable family cancer history presented with acute pathological compression fracture of the T7 vertebral body. Follow-up imaging with positron emission tomography/CT and CT of the chest, abdomen and pelvis revealed bilateral pulmonary nodules and metastases involving the lymph nodes (LNs), but no primary renal tumours or suspicious breast masses. While undergoing diagnostic work up, the patient developed an anterior lower neck mass, which was subsequently resected.
Investigations
Patient 1
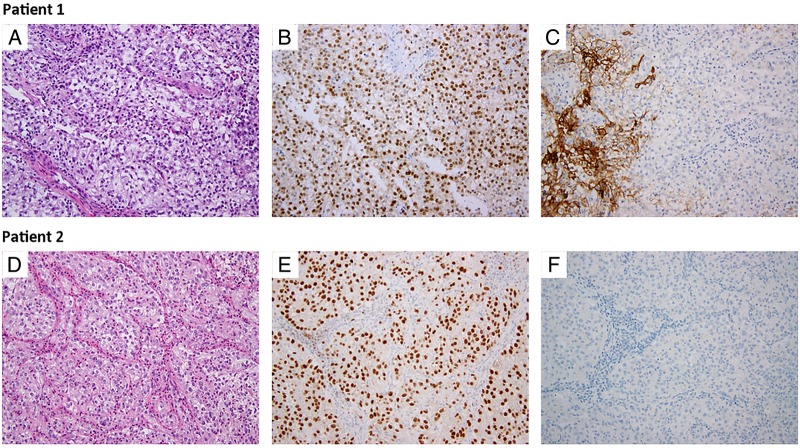
Morphologically, the findings in the right lung biopsy were of interstitial and intravascular clusters of atypical epithelial cells with clear cell features, suggestive of metastatic clear cell RCC (ccRCC) (figure 2A). Immunohistochemical staining revealed weak positivity for pan-cytokeratin, strong positivity for PAX8 (figure 2B) and patchy membranous staining for CA-IX (figure 2C). IHC was negative for TTF-1, thyroglobulin, p40, WT-1 and calretinin.
Figure 2.
H&E and Immunohistochemical staining with PAX8 and CA-IX. H&E staining demonstrating eosinophilic cells with voluminous cytoplasm from the subcarinal lymph node and anterior neck metastasis in patients 1 and 2, respectively (A and D). Positive nuclear PAX8 staining is present in both samples (B and E). CA-IX staining is absent in tumour cells (C and F), but present in the normal stromal cells of patient 1's lymph node sample (C).
To further characterise this patient's malignancy, tumour and matched adjacent normal DNA samples from two subsequent procedures, a resection of the left iliac lesion and a subcarinal LN biopsy, were profiled for genomic alterations with the Illumina Hi-Seq 2000 platform using an ultra-deep sequencing MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets) assay.4 The assay utilises target-specific probes to hybridise and capture all exons and select introns of 341 key cancer-associated genes. Creation of barcoded libraries from matched samples and subsequent custom analysis allows for the interrogation of somatic single-nucleotide variants, insertions/deletions and copy number alterations.
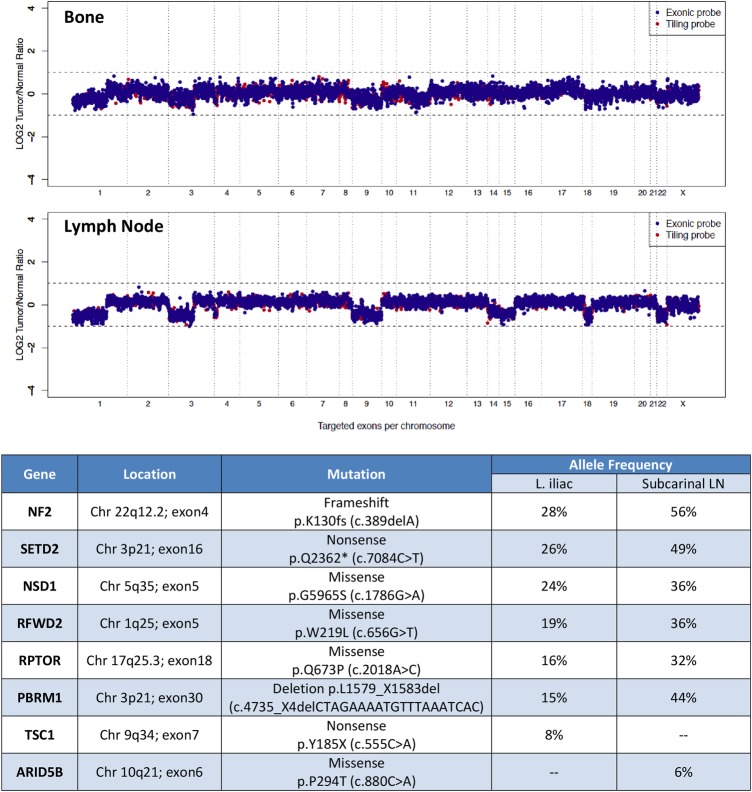
IMPACT sequencing had mean sequencing depths of 420× and 417×, respectively, for the bone and LN lesions. It also revealed somatic mutations in PBRM1 and SETD2, as well as heterozygous loss of 3p, which, taken together, strongly suggested ccRCC despite wild-type VHL (figure 3). Both lesions also contained an NF2 frameshift mutation, and private mutations in TSC1 and ARID5B in the bone and LN, respectively. Other mutations affected RFWD2, NSD1 and RPTOR.
Figure 3.
Copy number alterations and mutation profile from two metastatic sites from patient 1. Copy number analysis (CNA) revealed multiple heterozygous losses involving chromosomes 1p, 3p and 9. Heterozygous losses of chromosomes 14, 15, 18 and 22 were also detected from the lymph node. Mutations were similar across the two metastatic sites with the exception of TSC1 and ARID5B, which were only demonstrated in the left iliac and subcarinal lymph node samples, respectively.
Patient 2
Overall histological appearance of the bone metastasis initially suggested a primary breast adenocarcinoma. Tumour cells showed weak positive staining for cytokeratin 7 and gross cystic disease fluid protein (GCDFP), which is most consistent with a breast carcinoma primary. However, re-evaluation with additional IHC stains suggested a renal or Müllerian origin given the strong positive staining with PAX8 (figure 2E) and negative TTF-1, thyroglobulin and mammaglobin. Histopathological evaluation of an anterior neck metastatic lesion revealed sheets of clear cells compartmentalised by a rich capillary network morphologically suggestive of kidney cancer origin (figure 2D). IHC further demonstrated positive staining for CAM 5.2, vimentin, and CD10, which supported the diagnosis of RCC. However, other RCC markers, such as PAX2, CA-IX (figure 2F) and RCC antigen, were negative.
Metastatic tissue was sent for evaluation with a FoundationOne assay, an ultra-deep sequencing platform that interrogates 315 cancer-related genes and selected introns to a median exon sequencing depth of >500×. The assay identified genomic alterations involving NF2 (p.I448_splice), SETD2 (p.L760fs*10) and TSC1 (p.A883T). Additional mutations were also detected in ARID1A (p.V2263fs*15), BRCA2 (p.D2712N), ABL2 (p.S31C), MET (p.V1294fs*22), PDGFRB (p.V316M), AXIN1 (p.V383M), FANCF (p.L162fs*103) and MAP2K2 (p.I97F). This genomic profile is consistent with ccRCC with 96% probability despite wild-type VHL.
Outcome and follow-up
Patient 1
Given the patient's presentation, the differential diagnosis included primary lung adenocarcinoma as well as metastatic adenocarcinoma of an unknown primary. However, based on initial IHC results, the patient was diagnosed with putative stage IV RCC and began treatment with the mTORC1 inhibitor, temsirolimus, to which the patient demonstrated a beneficial response. Follow-up CT scans revealed reduction of his primary lung metastasis from 4.0 to 2.2 cm after just eight doses of temsirolimus.
Following review of IMPACT results that further suggested ccRCC, the patient was then switched to axitinib, one of the vascular endothelial growth factor receptor (VEGFR) inhibitors recommended for ccRCC therapy. However, the patient progressed in all remaining metastatic sites, including the bone, thus prompting a return to mTOR inhibitor therapy. Furthermore, somatic mutation in TSC1 in the patient's left iliac lesion also hinted at potential benefits from mTOR therapy as opposed to VEGFR therapy.
Concurrent with his treatment with everolimus, the patient also underwent palliative radiation to osseous lesions involving the lumbar spine and left hip. Despite these efforts, the patient died 9 months following initial presentation.
Patient 2
On the basis of the divergence in preliminary histopathological diagnosis and pending a FoundationOne assay, the patient began empiric therapy for adenocarcinoma of unknown primary with carboplatin and gemcitabine. Following receipt of the patient's Cancer TYPE ID report from the FoundationOne assay, the patient was begun on sunitinib, but this was discontinued after one dose due to adverse reactions. Temsirolimus was thus initiated. At the time of report, the patient has been treated for 7 months with stable response.
Discussion
Genetic sequencing continues to make significant contributions to our understanding of cancer. In ccRCC, for example, sequencing has not only identified mutations associated with poor prognosis, survival and recurrence,5 but has also begun to elucidate mutations that contribute to therapeutic response, such as those in TSC1 and MTOR.6 Sequencing has furthermore revealed patterns of genetic alterations and chromosomal losses that are characteristic of certain malignancies, including heterozygous loss of 3p and mutations of VHL, PBRM1, SETD2, or BAP1 in ccRCC.5
The application of sequencing technologies to CUP is in its infancy, but its potential utility is immense as CUP presents as an ideal candidate for personalised therapy. Furthermore, as the cases suggest, histopathological review and IHC staining, particularly for RCC, are fraught with difficulty. Not only can clear cell carcinomas arise from nearly any organ, but IHC can also be non-specific, especially in cases such as these, for which classic ccRCC morphology was lacking. Therefore, genomic sequencing may play an increasingly important role in diagnosing putative RCC lacking a renal primary, a phenomenon of spontaneous regression previously reported in this malignancy.
The genomic profiles of these two patients lack VHL mutations, which are found in nearly 90% of patients with ccRCC.5 Despite this, concurrent mutations in PBRM1, SETD2 and TSC1 are found in over 40% of patients with ccRCC, lending credence to the diagnosis for patient 1.7 8 Patient 2 lacked mutation in PBRM1, but had mutations in SETD2 and TSC1. This duo of gene alterations is found most frequently in ccRCC, followed by bladder cancer, which had been ruled out during this patient's initial assessment.7 8 When the NF2 mutation is considered for both patients, again the most likely malignancies are ccRCC and bladder cancer.7 8
Recent comprehensive genomic profiling of 200 CUP specimens revealed targetable genetic alterations presented in 96% of patients.9 The authors furthermore described two patients for whom targeted therapies selected on the basis of the tumour genomic profile—a MET amplification and EML4-ALK fusion, respectively—led to sustained complete responses on crizotinib.
Despite these encouraging results and our patients’ beneficial responses to mTORC1 inhibitor therapy, large prospective randomised clinical trials are necessary to confirm these observations and to determine whether targeted therapy based on genomic profiles affects patient prognosis in this rare malignancy.10 Currently, genetic sequencing is used to find similarities between CUP and metastases from typical solid tumours. The reversal of this model—sequencing to elucidate differences between CUP and other malignancies—may also provide key insights into carcinogenesis and metastatic potential.
The correlation between genotype and phenotype also remains unclear. Mutations affecting the same gene do not necessarily result in the same phenotype. Determining which genes become drivers and which genes become passengers warrants further investigation and may benefit from continued sequencing of responders to therapy.11 Analysis of responders can aid in identification of shared pathways involved in the unique presentation of CUP.
Other limitations to genetic sequencing include intratumour heterogeneity, a characteristic feature of ccRCC and many other solid tumours. Our first patient, for instance, demonstrated two distinct mutations in TSC1 and ARID5B in his bone and LN metastases, respectively. While TSC1 is associated with response to mTOR inhibitor therapy, particularly in conjunction with NF2 mutation,12 the lack of TSC1 in all metastatic sites might have contributed to the progression of disease on everolimus.
Learning points.
Cancer of unknown primary (CUP) is rare and portends a poor prognosis.
Tissue of origin determination is a main focus in patient work up, as determining a putative primary site and using disease-specific therapeutic regimens may improve survival.
Genetic sequencing of patients with CUP may reveal the tissue of origin, but, more importantly, can indicate the most appropriate targeted therapies.
Footnotes
Contributors: The authors gratefully thank the patients and their families. They also thank the research staff members for their valuable assistance.
Competing interests: None declared.
Patient consent: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Varadhachary GR, Raber MN. Cancer of unknown primary site. N Engl J Med 2014;371:757–65. 10.1056/NEJMra1303917 [DOI] [PubMed] [Google Scholar]
- 2.Varadhachary GR, Greco FA. Overview of patient management and future directions in unknown primary carcinoma. Semin Oncol 2009;36:75–80. 10.1053/j.seminoncol.2008.10.008 [DOI] [PubMed] [Google Scholar]
- 3.Handorf CR, Kulkarni A, Grenert JP et al. . A multicenter study directly comparing the diagnostic accuracy of gene expression profiling and immunohistochemistry for primary site identification in metastatic tumors. Am J Surg Pathol 2013;37:1067–75. 10.1097/PAS.0b013e31828309c4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng DT, Mitchell TN, Zehir A et al. . Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–64. 10.1016/j.jmoldx.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hakimi AA, Pham CG, Hsieh JJ. A clear picture of renal cell carcinoma. Nat Genet 2013;45:849–50. 10.1038/ng.2708 [DOI] [PubMed] [Google Scholar]
- 6.Voss MH, Hakimi AA, Pham CG et al. . Tumor genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin Cancer Res 2014;20:1955–64. 10.1158/1078-0432.CCR-13-2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao JJ, Aksoy BA, Dogrusoz U et al. . Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerami E, Gao JJ, Dogrusoz U et al. . The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross JS, Wang K, Gay L et al. . Comprehensive genomic profiling of carcinoma of unknown primary site: new routes to targeted therapies. JAMA Oncol 2015;1:40–9. 10.1001/jamaoncol.2014.216 [DOI] [PubMed] [Google Scholar]
- 10.Ettinger DS, Handorf CR, Agulnik M et al. . Occult primary, version 3.2014. J Natl Compr Canc Netw 2014;12:969–74. [DOI] [PubMed] [Google Scholar]
- 11.Wei EY, Hsieh JJ. A river model to map convergent cancer evolution and guide therapy in RCC. Nat Rev Urol 2015; doi:10.1038/nrurol.2015.260. [DOI] [PubMed] [Google Scholar]
- 12.Iyer G, Hanrahan AJ, Milowsky MI et al. . Genome sequencing identifies a basis for everolimus sensitivity. Science 2012;338:221–2. 10.1126/science.1226344 [DOI] [PMC free article] [PubMed] [Google Scholar]