Abstract
Considering the importance of macrophages as the first line of defense against fungal infection and the different roles played by the two M1- and M2-like polarized macrophages, we decided to evaluate the effects of Paracoccidioides brasiliensis infection on GM-CSF- and M-CSF-induced bone marrow-derived macrophages (BMM) from the A/J and B10.A mouse strains, an established model of resistance/susceptibility to PCM, respectively. Upon differentiation, the generated GM- or M-BMMs were characterized by morphological analyses, gene expression profiles, and cytokines production. Our main results demonstrate that GM-BMMs derived from A/J and B.10 produced high levels of pro- and anti-inflammatory cytokines that may contribute to generate an unbalanced early immune response. In accordance with the literature, the B10.A susceptible mice lineage has an innate tendency to polarize into M1-like phenotype, whereas the opposite phenotype occurs in A/J resistance mice. In this context, our data support that susceptibility and resistance are strongly correlated with M1 and M2 polarization, respectively.
1. Introduction
The increased incidence of fungal diseases has been ascribed to the rise in both the number of immunocompromised patients [1–3] and the number of cases in so-called immunocompetent individuals [4–8]. These data indicate that fungal infections are a worldwide health problem with high rates of mortality and morbidity. In Brazil, the situation is not very different, since, between 1996 and 2006, about 3,583 deaths occurred as a result of fungal diseases [9, 10], with Paracoccidioidomycosis (PCM) being the most common systemic mycosis not only in Brazil but also in Latin America [11]. This number may be even higher considering that the notification of patients diagnosed with systemic mycosis is not mandatory [12].
A murine model of resistance/susceptibility to PCM has been established, in which isogenic B10.A mice develop an immune response analogous to a susceptible human host, and the isogenic A/Sn or A/J strain develops a response equivalent to a resistant host [11, 13–16]. The resistance to PCM in both human and murine hosts is associated to a more efficient cell mediated immune response and activation of phagocytes throughout the infection. Although there is no classic Th1/Th2 polarization response, the secretion of IL-12 and IFNγ has been demonstrated as protective [11, 17–19]. The controlled progression of the disease is associated with an initially slower proinflammatory immune response, which further allows the development of a more robust resistance pattern during the course of infection [20]. In contrast, the susceptibility is associated to a decreased immune cellular response due to premature deactivation of T-cell mediated immunity and preferential B-cell activation, in addition to increased levels of IL-10 or TGF-β [11, 17]. Its progression is connected to a more efficient initial proinflammatory immune response, which is later downregulated [20]. In both cases, macrophages play a crucial role in the regulation of fungal growth in the early stages of infection [13]. Recently, it has been shown that the cytokine IL-17 plays a key role in innate antifungal defence, contributing to fungal clearance as observed in the best studied model of mucosal candidiasis (revised in [21]). During P. brasiliensis infection, the production of IL-17 was observed and its level was increased in response to the absence of TLR-2 activation and an uncontrolled inflammatory response with low number of regulatory CD4+CD25+FoxP3+ T-cells was observed. However, the survival time was not affected by the presence or the absence of TLR-2 [22].
Macrophages are known for their role in initiating and directing immune responses in vivo. Although there have been arguments about the spectrum to which a macrophage can be activated, differentiated macrophages are usually divided into two major groups, M1/classically activated macrophages and M2/alternatively activated macrophages [23, 24]. In general, M1 cells have IL-12high, IL-23high, and IL-10low phenotype. On the other hand, the various forms of M2 (M2a, M2b, M2c, and M2d TAM) cells share IL-12low, IL-23low, and IL-10high phenotype [24]. This functional and reversible plasticity is dependent on the activation state, which is primed by particular signals specific to tissues and local microenvironments [23, 25]. In this regard, there are compelling evidences indicating that, according to how, when, and the type of differentiation conditions, the phenotypes M1-M2 will determine the multifactorial outcomes in immune response. In Cryptococcus neoformans pulmonary infection, the polarization status changes over time due to either repolarization of individual macrophages or replacement of M2-polarized (nonprotective) by new M1-polarized (protective) cells [26]. Davis et al. [27] demonstrated in vitro that, independent of any previous stimulation, macrophage polarization is “phenotypically and functionally plastic in response to changing cytokine and fungus-sensing environments,” with the final stimulus determining the fungicidal potential. Macrophage plasticity is probably the mechanism used by Candida albicans to increase pathogenicity/survival, by changing environmental cues that induce M2 to M1 switch [28, 29]. The therapeutic repolarization of macrophages may open the door to interventions that could be useful in the treatment of fungal diseases [27].
In PCM, alveolar macrophages are probably one of the first immune cells to interact with P. brasiliensis. The results of this interaction, associated with the host health, other signals (e.g., damage-associated molecular pattern molecules, DAMPs), and genetic background will determine the infection outcome. It has been shown that this fungus is phagocytized by macrophages both in vivo and in vitro, but only properly activated macrophages manage to become fungicidal [30]. Recently, Feriotti et al. [31] demonstrated that when peritoneal macrophages from resistant (A/J) and susceptible (B10.A) mice were exposed to P. brasiliensis, they exhibit increased expression of “M1-like” (iNOS and SOCS3) and “M2-like” (Arginase-1, FiZZ1, YM1, and SOCS1) differentiation markers, respectively. Indeed, and in accordance with related articles, it seems that the apparent susceptibility to P. brasiliensis is associated with an exacerbated initial innate immune response, mediated by classically activated macrophages (M1-like) and a lack of fungal growth control. On the other hand, resistance is associated with an early moderated proinflammatory response, mediated by alternatively activated macrophages, evolving to a better control of fungal burden in the later stages of infection. However, these M2 macrophages did not show the classical differentiation markers [31].
To better understand the role of M1/M2-like macrophage in the PCM murine model, in this work, we evaluated phagocytic and secretory abilities, as well as expression analyses of some genes related to mice antifungal responses in GM-CSF (M1-like) and M-CSF (M2-like) induced bone marrow macrophages obtained from of A/J and B10.A mouse strains infected in vitro with a virulent strain of P. brasiliensis.
2. Materials and Methods
2.1. Fungus and Culture Conditions
The virulent strain Pb18 of P. brasiliensis was maintained by weekly subcultivation in semisolid Fava Netto's medium at 36.5°C and was used in the experiments after 7 days of growth. Yeast cells were resuspended and adjusted to the desired concentration based on hemocytometer counts using the Janus Green B vital dye to determine viability [32]. The fungal viability used in these tests was always higher than 90%. The virulence of the strain was maintained by in vivo passages in mice every 3 months.
2.2. Mice
P. brasiliensis resistant (A/J) and susceptible (B10.A) strains of 6 to 12 weeks old male mice [14, 16, 20, 33] were obtained from the Immunology Department of the University of São Paulo Biomedical Sciences Institute, Brazil. The animals were housed with food and water ad libitum at the Animal Care Center of the Biological Institute of the University of Brasilia, Brazil. The mice were euthanized in a carbon dioxide chamber, and their bone marrows were collected. All procedures involving animal were performed following the guidelines for the use of animals according to Brazilian laws and were approved by the Committee of Ethical Use of Animals (Proc. UnBDoc 52657/2011).
2.3. Ex Vivo Infection of GM-BMM (GM-CSF-Induced Bone Marrow-Derived Macrophage) and M-BMM (M-CSF-Induced) Cells from P. brasiliensis Resistant and Susceptible Mouse Strains
The two different BMMs populations were obtained using recombinant GM-CSF (20 ng/mL PeproTech) or M-CSF (30% (v/v) of L929 cell-conditioned medium) according to Tadokoro and de Almeida Abrahamsohn [34]. In this work, the nomenclature adopted is GM-BMM or M-BMM, related to GM-CSF- or M-CSF-induced macrophage differentiation condition, respectively. Briefly, the isolated BMMs from each mouse strain were cultivated in RMPI 1640 medium with 10% fetal bovine serum containing GM-CSF and M-CSF for 7 days when the adherent cells were recovered [34]. These cells were infected or not with P. brasiliensis at a cell-to-yeast ratio of 5 : 1 (multiplicity of infection, MOI: 5 : 1) for 6 h (transcription and ELISA assays) or 24 h (ELISA assays) in a humidified atmosphere of 5% CO2 at 37°C. This MOI has been previously shown to be nondeleterious to macrophage cultures [35, 36].
2.4. Ratio of Internalized/Adhered P. brasiliensis Yeast Cells by the Two Different BMMs of A/J and B10.A Mouse Strains
After 6 or 24 h of infection, adherent macrophages were washed with medium at 37°C, fixed, and stained with Panotic Staining kit. The number of macrophages with phagocytized or adhered yeasts was recorded by optical microscopy from a total of 300 cells to determine the percentage of adhered/internalized P. brasiliensis yeast cells. The experiments were performed in triplicate and five to ten microscopic fields were analyzed.
2.5. Cytokines and Chemokine Measurements
The cytokines TNF-α, IL1-β, IL6, and IL10 and the chemokine MCP-1 levels present in cell culture supernatants were measured by a capture enzyme-linked immunosorbent assay (ELISA) using the specific kits from eBioscience, according to the manufacturer's instructions. The absorbance values were measured in spectrophotometer (SpectraMax M5, Molecular Devices) and analyzed with SoftMax 5.2 software. Cytokines and chemokine concentrations were determined using a standard curve, following the kit recommendations. All determinations were performed in triplicate.
2.6. Quantitative Real-Time PCR (qRT-PCR) and PCRarray
The total RNA of the cultured macrophages was obtained employing the RNAeasy Plus Mini Kit (QIAGEN cat. number 74134), in accordance with the manufacturer's protocol. After DNase I treatment (included in the RNeasy Mini Kit Plus), first-strand cDNAs were synthesized from 500 ng of total RNA for each sample following the instructions of SuperScript III (Invitrogen). To confirm the GM- and M-CSF phenotype, the expression of markers genes was tested using specific primers for iNOS (forward-CGAAACGCTTCACTTCCAA, reverse-TGAGCCTATATTGCTGTGGCT) and Arginase-1 (forward-GTTCCCAGATGTACCAGGATTC, reverse-CGATGTCTTTGGCAGATATGC). The internal control used was 40S ribosomal protein S9 (RPS9) gene (forward-CGCCAGAAGCTGGGTTTGT, reverse-CGAGACGCGACTTCTCGAA) [36]. The qRT-PCR was performed using SyBr Green Master Mix (Applied Biosystems) with the standard cycling condition for this dye.
For the PCR array, after quantitative and qualitative analysis of total RNA, 1 μg was reversely transcribed to cDNA using the RT2 First-Strand Kit (SA Biosciences), according to manufacturer's protocol. Subsequently, the cDNA samples were labeled with RT2 Real-Time SYBR Green PCR Master Mix (SA Biosciences) and added to 96-well plates of Mouse Antifungal Response RTC Profiler PCR Array (PAMM 00147Z, SA Biosciences/Quiagen). This array profiles the transcriptional levels of 84 critical genes involved in the innate immune response to fungal pathogens. These genes encompass those related to fungal pattern recognition receptors (PRRs) and their associated signal transduction, inflammation, and phagocytosis. In addition, 5 housekeeping genes for normalization of the PCR data and controls for genomic DNA contamination, reverse transcription efficiency, and PCR performance are included on each array. In our experimental conditions, two housekeeping genes (B2M and GUSB) had constant mRNA levels between control and experimental group and were used for data normalization. Product amplification, data acquisition (obtained as threshold cycle (Ct) values), and melting curve were performed by the ABI 7500 qRT-PCR system (Applied Biosystems, software version 2.0.3). Fold differences in gene expression between control and experimental groups were determined using the comparative threshold method (2−ΔΔCt algorithm) [37]. Genes significantly modulated were identified based on the following two criteria: (i) the fold difference in average 2−ΔΔCt values was greater than 2 or less than −2 (indicative of upregulation or downregulation, resp.) and (ii) the difference of the replicate 2−ΔΔCt values for each gene in the control group and treatment groups was statistically significant (p < 0.05) according to Student's t-test between control and experimental groups. Data were analyzed by RT2 PCR Array Data Analyses profile version 3.5 available at http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php.
2.7. Statistical Analysis
The differences between the groups were analyzed by Student's t-test or by two-way ANOVA with Turkey's multiple comparisons posttest performed using GraphPad Prism Mac 6.0d, GraphPad Software, La Jolla California USA, http://www.graphpad.com/. A p ≤ 0.05 was considered significant.
3. Results and Discussion
3.1. Characterization of GM-BMM and M-BMM of A/J and B10.A Mice Strains
Our group is interested in studying the host innate immune responses to pathogenic fungi, with emphasis on the role of macrophages from A/J e B10.A mouse strains, which is a well-established model of resistance/susceptibility to PCM [14, 16]. Considering the importance of these phagocytic cells as the first line of defense against fungal infection and the different roles played by the two subtypes of polarized macrophages, we decided to evaluate the response of GM- and M-CSF-induced BMMs of A/J and B10.A mouse strains infected with P. brasiliensis. Upon differentiation, the generated GM- or M-BMMs were characterized by morphological analyses and by their gene expression profiles, described as important markers to each polarized macrophage subtype.
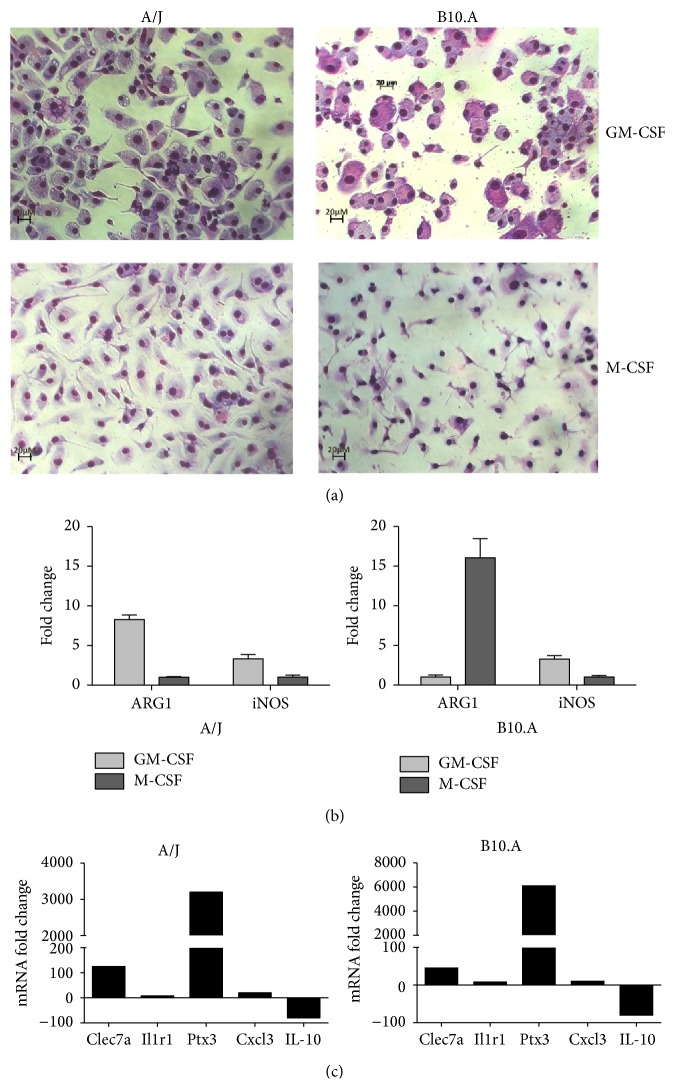
A similar morphology was observed in the GM-BMM from A/J and B10.A mice. Both cell subtypes revealed an abundant and slightly acid granular cytoplasm, filled with vesicles/vacuoles (Figure 1(a)). This feature was also described by McWhorter et al. [38], who observed that M1-polarizing stimuli (LPS+IFNγ) caused cells to flatten into a round, pancake-like shape. Another aspect in common was related to their tendency to form multinucleated giant cells, similar to those found in several pathological conditions, including the foreign body response and infection sites of tuberculosis and cryptococcosis, which were described in vitro with human monocytes [39, 40] (data not shown). Conversely, M-BMM from the two mouse strains showed a reduced, smooth, and basophilic cytoplasm (Figure 1(a)). Again, this elongated cell morphology was similar to that observed with monocytes exposed to M2-polarizing stimuli (IL-4+IL-13), as also described by McWhorter et al. [38]. These macrophages also tend to form cell aggregates around P. brasiliensis yeasts cells (data not shown). Although the GM-BMM and M-BMM revealed morphological resemblances in the two mouse strains, the expression profiles of Arginase-1 (Arg-1) and induced Nitric Oxide synthase (iNOS) marker genes were different.
Figure 1.
Characterization of GM-BMM and M-BMM of A/J and B10.A mice. Murine bone marrow cells were differentiated into macrophages (BMM) in the presence of GM-CSF (GM-BMM) or M-CSF (M-BMM) as described in Section 2. (a) Photomicrography of GM-BMM and M-BMM from A/J and B10.A mouse strains stained with Panotic kit (×200). (b) Quantitative PCR analysis (qRT-PCR) of induced nitric oxide synthase (iNOS) or Arginase-1 (ARG-1) mRNA expression from GM-BMM or M-BMM. Bars with SD represent the mean of fold change of the gene expression and are shown as n-fold difference of GM-BMM to the M-BMM cells. Fold change values were determined after each gene was normalized to the constitutively expressed rps9 gene. Data is representative of three separate experiments. (c) qRT-PCR analysis of Clec7a, Ilr1, Ptx3, Cxcl3, or Il10 mRNA expression from GM-BMM or M-BMM. Bars represent the mean of fold gene expression and are shown as n-fold difference of GM-BMM to the M-BMM cells. Fold change values were determined after each gene was normalized to the constitutively expressed Gusb (A/J) and B2m (B10.A) genes using the comparative threshold method.
The in vitro expression profile of these marker genes is well known from studies with typical polarized M1 and M2 macrophages stimulated with IFN-γ and IL-4, respectively, which resulted in iNOShigh and Arg-1low in M1 and iNOSlow and Arg-1high in M2 [23, 41]. In the present work, the culture conditions used to obtain the different types of BMMs employed only GM-CSF or M-CSF, without any other postdifferentiation mediator. The analysis of the expression profiles revealed that only the BMMs of B10.A strain developed the expected pattern (iNOShigh and Arg-1low in response to GM-CSF and iNOSlow and Arg-1high to M-CSF stimulus), as shown in Figure 1(b). A different expression pattern was observed with GM-BMM of A/J strain, revealing a discrete induction of iNOS, a slight upregulation of Arg-1, and a lack of induction of both genes in M-BMM (Figure 1(b)). The paucity of data related to marker genes expression profiles of in vitro polarized BMMs from A/J and B10.A mouse strains causes some difficulty in exploring our data. Altogether, the culture conditions and the different mouse strains could explain the absence of a clear expression profile of the marker genes when compared to in vitro IFN-γ and IL-4 poststimulated M1 and M2 macrophages, respectively. Of note, several other genes besides iNOS and Arg-1 are differently expressed in GM-BMM and M-BMM cells. In fact, Lacey et al. [42] using a whole murine genome microarray showed 4206 genes differentially regulated between the murine C57BL/6 GM-BMM and M-BMM, with no poststimulation, including Clec7a, Il1r1, Ptx3, Cxcl3 (upregulated), and IL-10 (downregulated). As shown in Figure 1(c), these genes showed the same transcriptional pattern regardless of the mice strain, when we compared GM-BMM versus M-BMM. In sum, these results suggest a polarized differentiation in our cells.
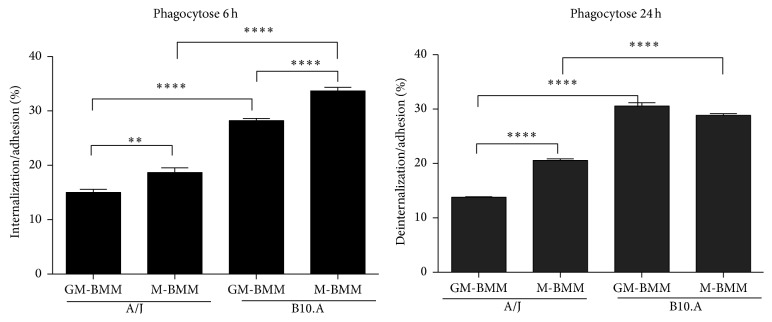
To comparatively evaluate the initial interaction between the two types of BMMs, from both susceptible/resistant mouse strains and P. brasiliensis yeast cells, we determined the number of adhered/internalized fungal cells by the different BMMs. Since the formation of multinucleated giant cells observed in GM-BMMs and cells aggregates in M-BMM made counting individual macrophages difficult, the percentage of yeast cells internalized/adhered only took into consideration individual infected/noninfected macrophages. After 6 h of infection, regardless of the differentiated macrophage type, BMMs derived from B10.A strain showed higher percentage of internalization/adhesion than A/J strain (Figure 2). This agrees with the hypothesis that the susceptibility to P. brasiliensis is associated with an exacerbated initial innate immune response, while resistance is associated with an initially moderate response [11, 20]. Furthermore, as shown in Figure 2, a significantly higher percentage of internalized/adhered yeast cells was observed in M-BMMs when compared to GM-BMMs in both mouse strains (to both AJ and B10.A, M-BMM > GM-BMM). After 24 h of coculture, the same pattern was observed, with significantly higher percentage of internalization/adhesion in B10.A than A/J BMMs. In addition, despite being higher than A/J BMMs, both BMMs from B10.A presented relatively similar results (GM-BMM similar to M-BMM), while in A/J there was a significantly higher percentage of internalization/adhesion in M-BMMs than GM-BMMs (M-BMM > GM-BMM).
Figure 2.
Ratio of internalized/adhered P. brasiliensis yeast cell by GM-BMMs and M-BMMs of A/J and B10.A mouse strains. Phagocytosis assays were performed employing a MOI (multiplicity of infection) of 5 : 1 macrophage to P. brasiliensis (Pb18) yeast cells, for incubation times of 6 h and 24 h. After the coculture, the cells were stained with Panotic kit. An average of 300 macrophages was counted and the number of ingested and/or adherent yeasts was determined. (∗∗∗∗ p ≤ 0.0001, ∗∗∗ p ≤ 0.001, ∗∗ p ≤ 0.01, and ∗ p ≤ 0.05).
4. Gene Expression Profiling of GM- and M-BMMs from A/J and B10.A Mice Infected with Paracoccidioides brasiliensis and Cytokines Production
The pattern of gene expression in GM- and M-BMMs from A/J and B10.A mice infected with P. brasiliensis yeast cells was assessed using the Antifungal Response RT2 Profiler PCR Array (Quiagen). As described in the methodology, this array profiles the transcriptional levels of 84 critical genes, classified in important functional categories of the innate immune response to fungal pathogens. Figure 3(a) shows the gene expression heat map of all 84 genes in GM- and M-BMMs from both mice strains infected with P. brasiliensis. Specifically, in GM- and M-BMMs from A/J mice, fungal infection results in a significant transcriptional modulation (up- or downregulation) of 55 genes. Among them, 10 genes were similarly induced in both BMMs, 30 exclusively in GM-BMM, and 15 in M-BMM. Similarly, in GM- and M-BMMs from B10.A mice, 7 genes were commonly modulated, 24 were exclusively in GM-BMM, and 24 were in M-BMM (Figure 3(b)). Based on the findings of previous fungal-phagocyte interaction studies, we selected modulated genes and clustered them into different functional categories as shown in Table 1.
Figure 3.
Expression profiling based on Mouse Antifungal Response RTC Profiler PCR Array. (a) A heat map was generated with 84 genes associated with antifungal immune response and five housekeeping genes, using RT2 Profiler Data Analysis Software version 3.5 with the PAMM 00147Z array panel (SABiosciences). Fold change values were determined after each gene was normalized to the constitutively expressed Gusb (A/J) and B2m (B10.A) genes using the comparative threshold method. AIGs 1, 2, and 3 and AIMs 1, 2, and 3 stand for A/J GM-BMM and M-BMM, respectively, infected with P. brasiliensis in relation to its respective control cells. BIGs 1, 2, and 3 and BIMs 1, 2, and 3 stand for B10.A GM-BMM and M-BMM, respectively, infected with P. brasiliensis in relation to its respective control cells. (b) Venn diagram summarizing the results of differentially expressed genes (p ≤ 0.05; fold change ≥2) between the noninfected and P. brasiliensis infected group of GM- or M-BMM of A/J and B10.A mice strains.
Table 1.
Comparison of differentially expressed genes in GM-BMM versus M-BMM of Paracoccidioides brasiliensis infected A/J and B10.A mice.
| Gene | Name | Fold changes (FC)∗ | |||
|---|---|---|---|---|---|
| A/J | B10.A | ||||
| M-BMM + Pb18 versus M-BMM | GM-BMM + Pb18 versus GM-BMM | M-BMM + Pb18 versus M-BMM | GM-BMM + Pb18 versus GM-BMM | ||
| Pattern of recognition receptor (PRR) | |||||
| TLR2 | Toll-like receptor 2 | −1.7 | 1.4 | 3.7 | −4.3 |
| TLR4 | Toll-like receptor 4 | 1.2 | 1.0 | 1.4 | −6.3 |
| TLR9 | Toll-like receptor 9 | −1.7 | 1.3 | 1.1 | −9.1 |
| Clec4n (Dectin-2) | C-type lectin domain family 4, member n | 6.4 | 2.1 | 6.1 | 1.6 |
| Clec7a (Dectin-1) | C-type lectin domain family 7, member a | 5.3 | 2.1 | 4.4 | 1.1 |
| Mrc1 (MR) | Mannose receptor, C-type 1 | 3.0 | 2.0 | −1.7 | −2.7 |
| Nlrp3 | NLR family, pyrin domain containing 3 | 1.3 | 2.9 | 4.8 | −1.9 |
| Scarf1 | Scavenger receptor class F, member 1 | 3.8 | −1.2 | −1.7 | −2.1 |
| Itgb2 | Integrin beta 2 | −1.4 | 1.2 | 2.7 | −3.0 |
| Mbl2 | Mannose-binding protein (protein C) 2 | 1.8 | 1.2 | −20.2 | 5.5 |
| CD14 | CD14 antigen | 2.0 | 3.0 | −1.3 | 8.8 |
| Itgam | Integrin alpha M | −1.4 | 1.6 | 1.3 | −3.6 |
| Colec12 | Collectin subfamily member 12 | 4.6 | 1.8 | 3.0 | −1.1 |
| PRR signal Transduction | |||||
| Casp-8 | Caspase-8 | 1.5 | 3.9 | 1.3 | 1.6 |
| Irak4 | Interleukin-1 receptor-associated kinase 4 | 1.3 | 2.1 | 1.5 | −2.6 |
| Mapk14 | Mitogen-activated protein kinase 14 | 1.1 | 1.3 | 2.3 | −3.6 |
| Mapk8 | Mitogen-activated protein kinase 8 | −1.7 | 1.4 | 2.0 | −1.6 |
| MyD88 | Myeloid differentiation primary response gene 88 | 1.5 | 4.6 | 2.6 | −3.0 |
| Bcl10 | B-cell leukemia/lymphoma 10 | 3.3 | 4.9 | 1.6 | 2.0 |
| Malt1 | Mucosa associated lymphoid tissue lymphoma translocation gene 1 | 2.4 | 1.7 | 3.1 | −1.7 |
| Pycard | PYD and CARD domain containing | −3.1 | 1.3 | 1.7 | −5.5 |
| Plcg2 | Phospholipase C gamma 2 | −2.2 | −1.1 | 2.0 | −4.9 |
| Tirap (Mal) | Toll-interleukin-1 receptor (TIR) domain-containing adaptor protein | −1.1 | 1.3 | 1.4 | −7.1 |
| Raf1 | V-raf-leukemia viral oncogene 1 | 1.1 | 1.1 | 3.2 | −5.4 |
| Syk | Spleen tyrosine kinase | 1.5 | 1.3 | 2.9 | −6.1 |
| CD40 | CD40 antigen | −1.4 | 10.2 | 2.0 | 10.9 |
| Transcription factor and other proteins | |||||
| Map2k4 (MKK4) | Mitogen-activated protein kinase 4 | −1.3 | 1.8 | 1.9 | −4.5 |
| Mapk8 | Mitogen-activated protein kinase 8 | −1.7 | 1.4 | 2.0 | −1.6 |
| Nfkbia | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | −1.7 | 1.7 | 2.0 | 1.1 |
| Cytokines | |||||
| Csf2 (GM-CSF) | Colony stimulating factor 2 (granulocyte-macrophage) | −3.3 | 3.1 | −2.0 | −8.2 |
| Il1a | Interleukin-1 alpha | 5.4 | 15.0 | 4.5 | 5.8 |
| Il1b | Interleukin-1 beta | 1.8 | 31.4 | 3.4 | 7.0 |
| Il2 | Interleukin-2 | 1.3 | 124.7 | −6.5 | 5.0 |
| Il6 | Interleukin-6 | 1.0 | 61.7 | −1.4 | 4.4 |
| Il10 | Interleukin-10 | −1.8 | 101.1 | 1.5 | 26.9 |
| Il12a | Interleukin-12A | 1.3 | 4.4 | 1.8 | 3.5 |
| Il12b | Interleukin-12B | 1.5 | 21.7 | 1.5 | 9.0 |
| Il18 | Interleukin-18 | −1.2 | 2.4 | 1.0 | −1.3 |
| Il23a | Interleukin-23, alpha subunit p19 | 1.2 | 40.4 | 2.0 | 1.9 |
| Tnf | Tumor necrosis factor alpha | −1.0 | 18.7 | −1.1 | 18.6 |
| Chemokines | |||||
| Ccl5 | Chemokine (C-C motif) ligand 5 | 1.1 | 6.2 | 3.5 | 21.1 |
| Ccl12 | Chemokine (C-C motif) ligand 12 | −4.5 | 6.9 | 1.1 | 7.7 |
| Ccl20 | Chemokine (C-C motif) ligand 20 | −1.1 | 6.0 | 2.0 | 1.0 |
| Cxcl1 (KC) | Chemokine (C-X-C motif) ligand 1 | 8.8 | 53.9 | 6.5 | 19.4 |
| Cxcl3 | Chemokine (C-X-C motif) ligand 3 | 3.5 | 3.4 | 15.2 | 64.4 |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 | 2.1 | 3.9 | 1.5 | 3.7 |
| Cxcl10 (IP-10) | Chemokine (C-X-C motif) ligand 10 | −1.9 | 6.8 | 2.5 | 8.9 |
| Cxcl11 | Chemokine (C-X-C motif) ligand 11 | 2.1 | −1.2 | −2.7 | −1.3 |
| Other proteins | |||||
| Ptgs2 (Cox-2) | Prostaglandin-endoperoxide synthase 2 | 8.3 | 28.0 | 18.8 | 11.3 |
| Chia1 | Chitinase, acidic | −1.7 | −2.0 | −3.4 | −4.9 |
| Stat1 | Signal transducer and activator of transcription 1 | −2.5 | 2.2 | −1.1 | −2.6 |
| C3 | Complement component 3 | 2.6 | 6.0 | 2.2 | 1.1 |
| C5ar1 | Complement component 5a receptor 1 | 3.4 | 2.7 | 2.8 | 1.5 |
| Fcgr3 | Fc receptor, IgG, low affinity III | 4.9 | 1.4 | 1.5 | −1.3 |
∗Genes In bold lettering had their transcript levels significantly modulated (FC ≥ 2 or ≤ −2 and p value < 0.05 as described in Section 2). Positive and negative values represent genes with expression induced and repressed, respectively.
Activation of macrophages is one of the first events in the innate immune response to fungal infections. This activation occurs upon recognition of conserved components of fungal cells by germline-encoded pattern recognition receptors (PRRs) of the phagocytic innate immune cells. Accordingly, several PRRs and PRRs signal transduction-encoding genes were modulated in BMMs infected with P. brasiliensis. Toll-like receptors (TLR) and C-lectin type receptors (CLR) are considered as being the main PRRs families involved in fungal recognition [21].
Our PCRarray data shows that the infection of GM- and M-BMMs from A/J mice with P. brasiliensis leads to no modulation of TLR2, TLR4, and TLR9 coding genes. Conversely, GM-BMM from B10.A had significant lower transcript levels of these TLRs, whereas TLR2 and MYD88 (protein adaptors that are critically important to TLR signaling pathway) were induced in M-BMM. Interestingly, expression of the gene coding TLR2 increased in dendritic cells after infection of susceptible mice with P. brasiliensis, but not in the resistant ones [43]. Furthermore, [22] demonstrated in vitro and in vivo that the lack of TLR2 usage and signaling resulted in a less-severe fungal infection, despite the fact that TLR2 deficient and normal mice showed equivalent survival times. Thus, TLR2 engagement could be employed as an evasion mechanism of P. brasiliensis and other PRRs may play an important role in the host immune response against P. brasiliensis infection.
Another important PRR to P. brasiliensis recognition and cellular activation is Dectin-1 [44]. According to these authors, the absence of Dectin-1 receptor drives macrophages to M2 phenotype with an anti-inflammatory activity that results in a lower nitric oxide production and an increased fungal growth [44]. Our results showed that Dectin-1 and Dectin-2 genes were upregulated in both BMMs of A/J (Table 1). The Dectin-1 gene was also upregulated in M-BMM of B10.A but was nonmodulated in GM-BMM, while Dectin-2 gene was nonmodulated in BMMs of B10.A. The importance of Dectin-1 in antifungal defense is well established in several fungi as Candida sp., Aspergillus sp., capsule deficient C. neoformans, and Histoplasma capsulatum [45, 46]. Recently, it has been shown that Dectin-1 and Dectin-2 receptors were involved in the interaction with Fonsecaea pedrosoi spores, while spore recognition by Dectin-2 was responsible for the development of antigen-specific Th17 response [47].
The Mannose receptor (MR) gene was upregulated in GM-BMM of A/J; however, it was downregulated in both macrophages from B10.A. Feriotti et al. [31] showed that P. brasiliensis infection of a murine strain where MR was blocked by monoclonal antibodies resulted in a decrease of macrophage killing abilities as well as nitric oxide production. Moreover, the authors using flow cytometry assay have shown that P. brasiliensis induced a lower expression of this receptor on A/J macrophages, different to what we observed here, by transcript level analysis. The MR engagement was associated with classical macrophage activation and susceptibility of B10.A strain [31]. However, this apparent conflicting result may be probably due to a very poor correlation observed for comparisons between mRNA and protein levels [48, 49].
The downstream CLR signaling is important for the generation of an efficient cellular activation and fungal elimination. Dectin-1 transduces downstream signaling via Src and Syk family kinases and/or by a second pathway via Raf. These pathways lead to cytokine genes transcription and protein secretion [45, 46]. The Syk and Raf-1 genes were nonmodulated in GM- and M-BMM from A/J (Table 1), while they were upregulated in M-BMM and downregulated in GM-BMM from B10.A. Following Dectin-1 activation, the intracellular Raf-1 and Syk-dependent signaling pathway was demonstrated crucial for fine-tuning cytokine gene expression [50].
According to Feriotti et al. [31], A/J mice experimentally infected with P. brasiliensis showed M2-like differentiation macrophages, while B10.A mice showed M1-like differentiation macrophages. Analyzing the gene expression profile of GM-BMM from B10.A and M-BMM from A/J, the B10.A macrophages showed a decreased gene expression of CLR signaling molecules that could impair the efficient cell activation and, despite their M1-like phenotype, the control of antifungal defense seems not to be correctly activated.
Another crucial aspect of the innate immunity against fungal pathogen is the activation of the inflammasome, a cytoplasmic protein complex composed of PRRs such as NLRP3 (NOD-like receptor family, pyrin domain-containing 3), an adaptor protein ASC (apoptosis-associated speck-like protein containing a CARD), and procaspase-1. Upon formation of the complex, procaspase-1 is cleaved into an active cysteine protease, which further leads to the maturation of the proinflammatory cytokines IL-1β and IL-18. Recently, we have shown that P. brasiliensis activates NLRP3 inflammasome [51]. Feriotti et al. [31] found that macrophages from the susceptible B10.A mice infected with P. brasiliensis exhibited the typical markers of a proinflammatory “M1-like” (GM-BMM) differentiation. In contrast, A/J macrophages exhibited an alternatively activated “M2-like” (M-BMM) differentiation. In this context, here is shown that B10.A macrophages, regardless of M- or GM-CSF-induced differentiation, showed high IL-1β transcript accumulation, suggesting an “M1-like” differentiation pattern, whereas in M-BMM of A/J, neither NLRP3 nor IL-1β genes were modulated by P. brasiliensis infection. Thus, infected M-BMM from A/J preserves the profile associated with resistance (i.e., alternatively activated macrophages). Of note, GM-BMM from A/J showed similar pattern of NLRP3 and IL-1β expression when compared to M- and GM-BMM from susceptible B10.A mice (i.e., classically activated macrophages).
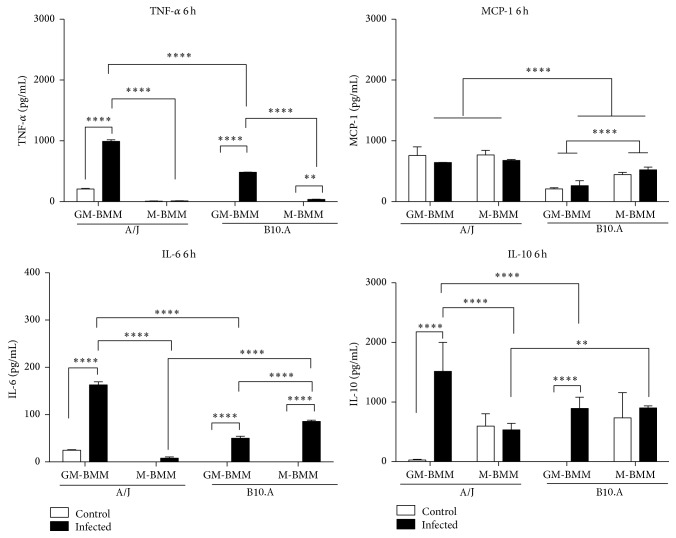
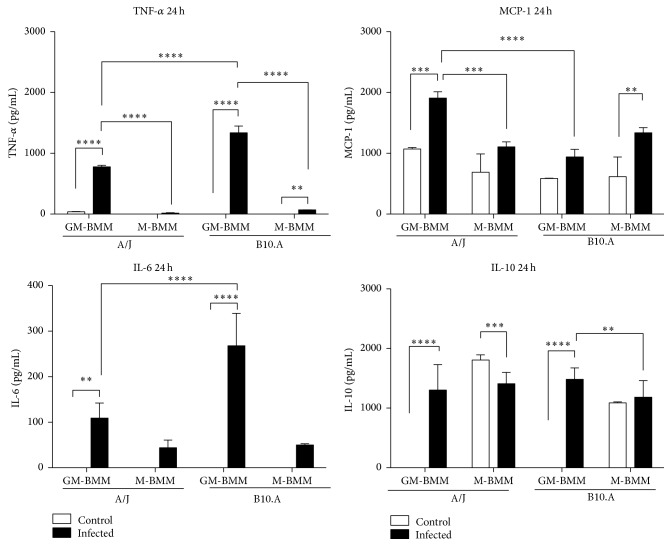
Regarding the transcript levels of the cytokines IL-2, IL-6, IL-10, IL-12, and TNF-α, they were increased in GM-BMMs of both mice lineage when compared to M-BMMs. In fact, the protein levels of the proinflammatory cytokines IL-6 and TNF-α and the anti-inflammatory IL-10 were significantly increased, as assayed by ELISA at 6 and 24 h of infection (Figures 4 and 5), corroborating the transcript levels shown by the PCRarray. Interestingly, monocytes infected with the P. brasiliensis highly virulent isolate Pb18 produced higher levels of IL-6 and IL-10 than Pb265 isolate (low virulence) [52]. Thus, regardless of the mice strain, the isolate 18 of P. brasiliensis is induced in GM-BMMs to release into the environment context large amounts of both pro- and anti-inflammatory cytokines that may contribute to disturbances in immunity, which possibly leads to fungal survival. For instance, IL-6 production was associated to a significant increase in P. brasiliensis (Pb18 isolate) growth in monocytes [53]. Concerning IL-10, a large body of data exists, revealing the role of this regulatory cytokine with a compromised response to P. brasiliensis infection. In this sense, the simultaneous incubation of IL-10 with either IFN-γ or TNF-α inhibits murine macrophage fungicidal activity of these cells when cocultured with P. brasiliensis. The suppression of this activity by IL-10 was associated with the inhibition of nitric oxide production [54]. Furthermore, IL-10-knockout mice develop early T-cell responses, controlling the fungal growth in the lungs, resulting in an increased survival when compared to IL-10-sufficient mice [55].
Figure 4.
Cytokines profile after 6 h of Paracoccidioides brasiliensis infection of GM- and M-BMMs from A/J and B10.A mouse strains. Cytokines levels produced by the two different BMMs of both mouse strains after infection with Pb18, by ELISA assay. Data are means ± SD of triplicate samples representative of three separate experiments (∗∗∗∗ p ≤ 0.0001, ∗∗∗ p ≤ 0.001, and ∗∗ p ≤ 0.01).
Figure 5.
Cytokines profile after 24 h of Paracoccidioides brasiliensis infection of GM- and M-BMMs from A/J and B10.A mouse strains. Cytokines levels produced by the two different BMMs of both mouse strains after infection with Pb18, by ELISA assay. Data are means ± SD of triplicate samples representative of three separate experiments (∗∗∗∗ p ≤ 0.0001, ∗∗∗ p ≤ 0.001, and ∗∗ p ≤ 0.01).
Altogether, our results as shown in Figure 6 demonstrate that GM-BMMs derived from A/J and B.10 produced higher levels of pro- and anti-inflammatory cytokines that may contribute to generate an unbalanced early immune response. It has been shown that the susceptible mouse lineage B10.A has an innate tendency to polarize into M1-like phenotype, whereas the opposite phenotype occurs in resistant A/J mice [31]. In this context, our data support that susceptibility and resistance are strongly associated with M1 and M2 polarization, respectively.
Figure 6.
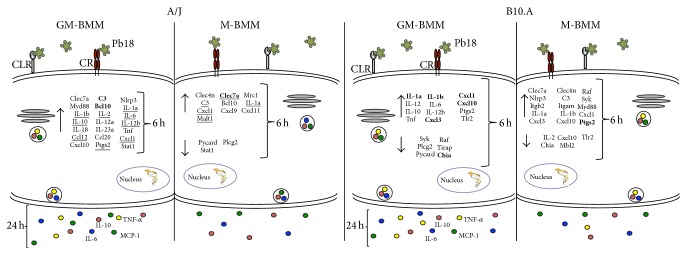
Schematic representation of the major results of gene expression and cytokine production by GM- and M-BMM from A/J and B10.A mouse strains in response to in vitro Pb18 infection. The arrows indicate up- and downregulated genes. The yellow, red, green, and blue circles are representative of TNF-α, IL-10, MCP-1, and IL-6, respectively. The genes indicated in bold demonstrated a higher expression compared between GM- and M-BMM from the same mouse strain. The underlined genes indicated the same expression pattern between GM- and M-BMM from different mice strain.
As final considerations, below we present some relevant and interesting data recently published in the field of innate immune regulation.
Since the development of high-throughput DNA/RNA sequencing methodologies, our understanding of transcriptional and posttranscriptional gene regulation increased at a never thought level [56, 57]. Furthermore, the advances in proteomic methods and the development of a myriad of innovative cell analysis techniques have also contributed to the better understanding of several highly complex biological processes in a more holistic view.
These approaches have been applied to detail the molecular basis involved in the innate immune response, tracing the path from the first molecular event, as PAMP-PRR interaction, to signalling cascades and transcriptional regulation, which in concert defines the control of pathogen-induced gene expression program [58–61]. In view of the importance of innate immune response to properly activate an inflammatory response to fight infection, its precise regulation is crucial to avoid host damage, as observed in several diseases.
Besides the huge amount of data revealing the importance of transcriptional regulation of the inflammatory genes expression, another equally important step of regulation, much less considered, operates at the posttranscriptional level [62, 63]. These authors stressed the role of alternative splicing, mRNA stability, and translational regulation, directly associated with components of the innate immunity. They presented several examples of differential splicing of TLR and signaling protein genes, resulting in functionally different isoforms, as well as the control at the level of mRNA stability and translation, providing a rapid and finely tuned response in its magnitude and extent. Altogether, these recent developments highlight the irrefutable importance of the coordination of regulatory mechanisms operating at the multiple layers of inflammatory genes expression, as a keystone in the control and modulation of host innate immune responses.
Acknowledgments
This study was supported by research funding from Project PRONEX no. 193.000.571/2009 from FAP-DF/CNPq. CSS had a fellowship from CNPq/CAPES in the context of the PhD-Program of Molecular Biology/CEL/IB/UnB. The authors thank Professor Dr. Cynthia Maria Kyaw for her helpful reviewing and proofreading of this paper.
Disclosure
Anamélia Lorenzetti Bocca and Ildinete Silva-Pereira share senior authorship of this paper.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Calliandra de Souza Silva and Aldo Henrique Tavares contributed equally to this work.
References
- 1.Chakrabarti A. Microbiology of systemic fungal infections. Journal of Postgraduate Medicine. 2005;51(supplement 1):S16–S20. [PubMed] [Google Scholar]
- 2.Rivera A. Protective immune responses to fungal infections. Parasite Immunology. 2014;36(9):453–462. doi: 10.1111/pim.12098. [DOI] [PubMed] [Google Scholar]
- 3.Menzin J., Meyers J. L., Friedman M., et al. Mortality, length of hospitalization, and costs associated with invasive fungal infections in high-risk patients. American Journal of Health-System Pharmacy. 2009;66(19):1711–1717. doi: 10.2146/ajhp080325. [DOI] [PubMed] [Google Scholar]
- 4.Ali Z. A., Ali A. A., Tempest M. E., Wiselka M. J. Invasive pulmonary aspergillosis complicating chronic obstructive pulmonary disease in an immunocompetent patient. Journal of Postgraduate Medicine. 2003;49(1):78–80. doi: 10.4103/0022-3859.922. [DOI] [PubMed] [Google Scholar]
- 5.Bigliazzi C., Poletti V., Dell'Amore D., Saragoni L., Colby T. V. Disseminated basidiobolomycosis in an immunocompetent woman. Journal of Clinical Microbiology. 2004;42(3):1367–1369. doi: 10.1128/jcm.42.3.1367-1369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J., Varma A., Diaz M. R., Litvintseva A. P., Wollenberg K. K., Kwon-Chung K. J. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerging Infectious Diseases. 2008;14(5):755–762. doi: 10.3201/eid1405.071312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saini J., Gupta A. K., Jolapara M. B., et al. Imaging findings in intracranial aspergillus infection in immunocompetent patients. World Neurosurgery. 2010;74(6):661–670. doi: 10.1016/j.wneu.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 8.Thorpe J., Ahmed I., Hind R. E. Intestinal perforation with invasive candidiasis in an immunocompetent adult. Journal of the College of Physicians and Surgeons—Pakistan. 2004;14(3):187–188. [PubMed] [Google Scholar]
- 9.Prado M., da Silva M. B., Laurenti R., Travassos L. R., Taborda C. P. Mortality due to systemic mycoses as a primary cause of death or in association with AIDS in Brazil: a review from 1996 to 2006. Memorias do Instituto Oswaldo Cruz. 2009;104(3):513–521. doi: 10.1590/s0074-02762009000300019. [DOI] [PubMed] [Google Scholar]
- 10.Coutinho Z. F., Wanke B., Travassos C., Oliveira R. M., Xavier D. R., Coimbra C. E., Jr. Hospital morbidity due to paracoccidioidomycosis in Brazil (1998–2006) Tropical Medicine & International Health. 2015;20(5):673–680. doi: 10.1111/tmi.12472. [DOI] [PubMed] [Google Scholar]
- 11.Calich V. L. G., da Costa T. A., Felonato M., et al. Innate immunity to Paracoccidioides brasiliensis infection. Mycopathologia. 2008;165(4-5):223–236. doi: 10.1007/s11046-007-9048-1. [DOI] [PubMed] [Google Scholar]
- 12.Dignani M. C. Epidemiology of invasive fungal diseases on the basis of autopsy reports. F1000Prime Reports. 2014;6, article 81 doi: 10.12703/p6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calich V. L. G., Kashino S. S. Cytokines produced by susceptible and resistant mice in the course of Paracoccidioides brasiliensis infection. Brazilian Journal of Medical and Biological Research. 1998;31(5):615–623. doi: 10.1590/s0100-879x1998000500003. [DOI] [PubMed] [Google Scholar]
- 14.Garcia Calich V. L., Singer-Vermes L. M., Siqueira A. M., Burger E. Susceptibility and resistance of inbred mice to Paracoccidioides brasiliensis . British Journal of Experimental Pathology. 1985;66(5):585–594. [PMC free article] [PubMed] [Google Scholar]
- 15.Calich V. L. G., Singer-Vermes L. M., Russo M., et al. Immunogenetics in Paracoccidioidomycosis. Boca Raton, Fla, USA: CRC Press; 1994. [Google Scholar]
- 16.Cano L. E., Singer-Vermes L. M., Vaz C. A. C., Russo M., Calich V. L. G. Pulmonary paracoccidioidomycosis in resistant and susceptible mice: relationship among progression of infection, bronchoalveolar cell activation, cellular immune response, and specific isotype patterns. Infection and Immunity. 1995;63(5):1777–1783. doi: 10.1128/iai.63.5.1777-1783.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calich V. L. G., Vaz C. A. C., Burger E. Immunity to Paracoccidioides brasiliensis infection. Research in Immunology. 1998;149(4-5):407–500. doi: 10.1016/s0923-2494(98)80764-5. [DOI] [PubMed] [Google Scholar]
- 18.Ferreira K. S., Lopes J. D., Almeida S. R. Down-regulation of dendritic cell activation induced by Paracoccidioides brasiliensis . Immunology Letters. 2004;94(1-2):107–114. doi: 10.1016/j.imlet.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira K. S., Bastos K. R., Russo M., Almeida S. R. Interaction between Paracoccidioides brasiliensis and pulmonary dendritic cells induces interleukin-10 production and toll-like receptor-2 expression: possible mechanisms of susceptibility. Journal of Infectious Diseases. 2007;196(7):1108–1115. doi: 10.1086/521369. [DOI] [PubMed] [Google Scholar]
- 20.Pina A., Bernardino S., Calich V. L. G. Alveolar macrophages from susceptible mice are more competent than those of resistant mice to control initial Paracoccidioides brasiliensis infection. Journal of Leukocyte Biology. 2008;83(5):1088–1099. doi: 10.1189/jlb.1107738. [DOI] [PubMed] [Google Scholar]
- 21.Drummond R. A., Gaffen S. L., Hise A. G., Brown G. D. Innate defense against fungal pathogens. Cold Spring Harbor Perspectives in Medicine. 2014;5(6) doi: 10.1101/cshperspect.a019620.a019620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loures F. V., Pina A., Felonato M., Calich V. L. G. TLR2 is a negative regulator of Th17 cells and tissue pathology in a pulmonary model of fungal infection. The Journal of Immunology. 2009;183(2):1279–1290. doi: 10.4049/jimmunol.0801599. [DOI] [PubMed] [Google Scholar]
- 23.Murray P. J., Allen J. E., Biswas S. K., et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantovani A., Sica A., Locati M. Macrophage polarization comes of age. Immunity. 2005;23(4):344–346. doi: 10.1016/j.immuni.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Stout R. D., Suttles J. Functional plasticity of macrophages: reversible adaptation to changing microenvironments. Journal of Leukocyte Biology. 2004;76(3):509–513. doi: 10.1189/jlb.0504272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arora S., Olszewski M. A., Tsang T. M., McDonald R. A., Toews G. B., Huffnagle G. B. Effect of cytokine interplay on macrophage polarization during chronic pulmonary infection with Cryptococcus neoformans . Infection and Immunity. 2011;79(5):1915–1926. doi: 10.1128/iai.01270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis M. J., Tsang T. M., Qiu Y., et al. Macrophage M1/M2 polarization dynamically adapts to changes in cytokine microenvironments in Cryptococcus neoformans infection. mBio. 2013;4(3) doi: 10.1128/mbio.00264-13.e00264-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reales-Calderón J. A., Aguilera-Montilla N., Corbí Á. L., Molero G., Gil C. Proteomic characterization of human proinflammatory M1 and anti-inflammatory M2 macrophages and their response to Candida albicans . Proteomics. 2014;14(12):1503–1518. doi: 10.1002/pmic.201300508. [DOI] [PubMed] [Google Scholar]
- 29.Zheng X.-F., Hong Y.-X., Feng G.-J., et al. Lipopolysaccharide-induced M2 to M1 macrophage transformation for IL-12p70 production is blocked by Candida albicans mediated up-regulation of EBI3 expression. PLoS ONE. 2013;8(5) doi: 10.1371/journal.pone.0063967.e63967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brummer E., Hanson L. H., Restrepo A., Stevens D. A. Intracellular multiplication of Paracoccidioides brasiliensis in macrophages: killing and restriction of multiplication by activated macrophages. Infection and Immunity. 1989;57(8):2289–2294. doi: 10.1128/iai.57.8.2289-2294.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feriotti C., Loures F. V., Frank de Araújo E., da Costa T. A., Calich V. L. G. Mannosyl-recognizing receptors induce an M1-like phenotype in macrophages of susceptible mice but an M2-like phenotype in mice resistant to a fungal infection. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0054845.e54845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goihman-Yahr M., Pine L., Albornoz M. C., et al. Studies on plating efficiency and estimation of viability of suspensions of Paracoccidioides brasiliensis yeast cells. Mycopathologia. 1980;71(2):73–83. doi: 10.1007/bf00440612. [DOI] [PubMed] [Google Scholar]
- 33.Singer-Vermes L. M., Sakamoto T. N., Vaz C. A. C., Calich V. L. G. Influence of the genetic pattern and sex of mice in experimental paracoccidioidomycosis. Clinical and Experimental Immunology. 1995;101(1):114–120. doi: 10.1111/j.1365-2249.1995.tb02286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tadokoro C. E., de Almeida Abrahamsohn I. Bone marrow-derived macrophages grown in GM-CSF or M-CSF differ in their ability to produce IL-12 and to induce IFN-gamma production after stimulation with Trypanosoma cruzi antigens. Immunology Letters. 2001;77(1):31–38. doi: 10.1016/s0165-2478(01)00197-3. [DOI] [PubMed] [Google Scholar]
- 35.Tavares A. H. F. P., Silva S. S., Dantas A., et al. Early transcriptional response of Paracoccidioides brasiliensis upon internalization by murine macrophages. Microbes and Infection. 2007;9(5):583–590. doi: 10.1016/j.micinf.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 36.Silva S. S., Tavares A. H. F. P., Passos-Silva D. G., et al. Transcriptional response of murine macrophages upon infection with opsonized Paracoccidioides brasiliensis yeast cells. Microbes and Infection. 2008;10(1):12–20. doi: 10.1016/j.micinf.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 37.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.McWhorter F. Y., Wang T., Nguyen P., Chung T., Liu W. F. Modulation of macrophage phenotype by cell shape. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(43):17253–17258. doi: 10.1073/pnas.1308887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Möst J., Spötl L., Mayr G., Gasser A., Sarti A., Dierich M. P. Formation of multinucleated giant cells in vitro is dependent on the stage of monocyte to macrophage maturation. Blood. 1997;89(2):662–671. [PubMed] [Google Scholar]
- 40.Gasser A., Möst J. Generation of multinucleated giant cells in vitro by culture of human monocytes with Mycobacterium bovis BCG in combination with cytokine-containing supernatants. Infection and Immunity. 1999;67(1):395–402. doi: 10.1128/iai.67.1.395-402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho V. W. H., Sly L. M. Derivation and characterization of murine alternatively activated (M2) macrophages. Methods in Molecular Biology. 2009;531:173–185. doi: 10.1007/978-1-59745-396-7_12. [DOI] [PubMed] [Google Scholar]
- 42.Lacey D. C., Achuthan A., Fleetwood A. J., et al. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. The Journal of Immunology. 2012;188(11):5752–5765. doi: 10.4049/jimmunol.1103426. [DOI] [PubMed] [Google Scholar]
- 43.Ferreira K. S., Bastos K. R., Russo M., Almeida S. R. Interaction between Paracoccidioides brasiliensis and pulmonary dendritic cells induces interleukin-10 production and toll-like receptor-2 expression: possible mechanisms of susceptibility. The Journal of Infectious Diseases. 2007;196(7):1108–1115. doi: 10.1086/521369. [DOI] [PubMed] [Google Scholar]
- 44.Loures F. V., Araújo E. F., Feriotti C., et al. Dectin-1 induces M1 macrophages and prominent expansion of CD8+IL-17+ cells in pulmonary paracoccidioidomycosis. Journal of Infectious Diseases. 2014;210(5):762–773. doi: 10.1093/infdis/jiu136. [DOI] [PubMed] [Google Scholar]
- 45.Wevers B. A., Geijtenbeek T. B., Gringhuis S. I. C-type lectin receptors orchestrate antifungal immunity. Future Microbiology. 2013;8(7):839–854. doi: 10.2217/fmb.13.56. [DOI] [PubMed] [Google Scholar]
- 46.Dambuza I. M., Brown G. D. C-type lectins in immunity: recent developments. Current Opinion in Immunology. 2015;32:21–27. doi: 10.1016/j.coi.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wüthrich M., Wang H., Li M., et al. Fonsecaea pedrosoi-induced Th17-cell differentiation in mice is fostered by Dectin-2 and suppressed by Mincle recognition. European Journal of Immunology. 2015 doi: 10.1002/eji.201545591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Sousa Abreu R., Penalva L. O., Marcotte E. M., Vogel C. Global signatures of protein and mRNA expression levels. Molecular BioSystems. 2009;5(12):1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maier T., Güell M., Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Letters. 2009;583(24):3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 50.Gringhuis S. I., den Dunnen J., Litjens M., et al. Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-κB activation through Raf-1 and Syk. Nature Immunology. 2009;10(2):203–213. doi: 10.1038/ni.1692. [DOI] [PubMed] [Google Scholar]
- 51.Tavares A. H., Magalhães K. G., Almeida R. D. N., Correa R., Burgel P. H., Bocca A. L. NLRP3 inflammasome activation by Paracoccidioides brasiliensis . PLoS Neglected Tropical Diseases. 2013;7(12) doi: 10.1371/journal.pntd.0002595.e2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kurokawa C. S., Araujo J. P., Jr., Soares A. M. V. C., Sugizaki M. F., Peraçoli M. T. S. Pro- and anti-inflammatory cytokines produced by human monocytes challenged in vitro with Paracoccidioides brasiliensis . Microbiology and Immunology. 2007;51(4):421–428. doi: 10.1111/j.1348-0421.2007.tb03929.x. [DOI] [PubMed] [Google Scholar]
- 53.Siqueira K. Z., de Campos Soares Â. M. V., Dias-Melicio L. A., Calvi S. A., Peraçoli M. T. S. Interleukin-6 treatment enhances human monocyte permissiveness for Paracoccidioides brasiliensis growth by modulating cytokine production. Medical Mycology. 2009;47(3):259–267. doi: 10.1080/13693780802244204. [DOI] [PubMed] [Google Scholar]
- 54.Moreira A. P., Dias-Melicio L. A., Soares A. M. V. C. Interleukin-10 but not transforming growth factor beta inhibits murine activated macrophages Paracoccidioides brasiliensis killing: effect on H2O2 and NO production. Cellular Immunology. 2010;263(2):196–203. doi: 10.1016/j.cellimm.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 55.Costa T. A., Bazan S. B., Feriotti C., et al. In pulmonary paracoccidioidomycosis IL-10 deficiency leads to increased immunity and regressive infection without enhancing tissue pathology. PLoS Neglected Tropical Diseases. 2013;7(10) doi: 10.1371/journal.pntd.0002512.e2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shendure J., Ji H. Next-generation DNA sequencing. Nature Biotechnology. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 57.Sultan M., Schulz M. H., Richard H., et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321(5891):956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 58.Medzhitov R., Horng T. Transcriptional control of the inflammatory response. Nature Reviews Immunology. 2009;9(10):692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 59.Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 60.Costa V., Angelini C., De Feis I., Ciccodicola A. Uncovering the complexity of transcriptomes with RNA-Seq. Journal of Biomedicine and Biotechnology. 2010;2010:19. doi: 10.1155/2010/853916.853916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carpenter S., Fitzgerald K. A. Transcription of inflammatory genes: long noncoding RNA and beyond. Journal of Interferon & Cytokine Research. 2015;35(2):79–88. doi: 10.1089/jir.2014.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carpenter S., Ricci E. P., Mercier B. C., Moore M. J., Fitzgerald K. A. Post-transcriptional regulation of gene expression in innate immunity. Nature Reviews Immunology. 2014;14(6):361–376. doi: 10.1038/nri3682. [DOI] [PubMed] [Google Scholar]
- 63.O’Connor B. P., Danhorn T., De Arras L., et al. Regulation of toll-like receptor signaling by the SF3a mRNA splicing complex. PLoS Genetics. 2015;11(2) doi: 10.1371/journal.pgen.1004932.e1004932 [DOI] [PMC free article] [PubMed] [Google Scholar]