Abstract
An epidemiological investigation was carried out on one hundred Salmonella isolates from broiler farms, slaughterhouses, and human patients in the Constantine region of Algeria, in order to explore the contribution of avian strains to human salmonellosis cases in this region over the same period of time. The isolates were characterized by phenotypic as well as genotypic methods. A large variety of antimicrobial resistance profiles was found among human isolates, while only seven profiles were found among avian isolates. Enterobacterial Repetitive Intergenic Consensus-PCR (ERIC-PCR), Insertion Sequence 200-PCR (IS200-PCR), and Pulsed Field Gel Electrophoresis (PFGE) resulted in the allocation of the isolates to 16, 20, and 34 different profiles, respectively. The 3 genotyping methods led to complementary results by underlining the clonality of some serovars with the diffusion and persistence of a single clone in the Constantine area as well as stressing the polymorphism present in isolates belonging to other serovars, indicating the diversity of potential reservoirs of nontyphoidal Salmonella. Altogether, our results seem to indicate that nontyphoidal avian Salmonella may play an important role in human salmonellosis in the Constantine region.
1. Introduction
Salmonella remains a major cause of illness in both humans and animals worldwide [1, 2]. It is estimated that Salmonella spp. are responsible for 93.8 million cases of human gastroenteritis and 155,000 deaths worldwide each year [3]. In the European Union, over 100,000 cases of salmonellosis were reported to EnterNet in 2003 [4] and over 90,000 cases in 2012, even though human salmonellosis cases have decreased regularly since 2005 [5]. It should be stressed that the observed reduction in salmonellosis cases is presumably the result of successful Salmonella control programmes in poultry populations [5]. Salmonella is also a major public health concern in developing countries [6–8].
Salmonellosis due to nontyphoidal Salmonella is mainly associated with eating contaminated eggs, poultry meat, and pork. Contaminated poultry meat is identified as one of the principal sources of Salmonella in humans [2, 9]. Furthermore, one of the most frequent causes of infection by Salmonella reported in humans is the handling of raw poultry carcasses and products, together with the consumption of undercooked poultry meat [10].
The contamination of food products with Salmonella generates serious consequences for public health and the economy. This has motivated numerous studies designed to investigate the survival capacity of this bacterium and its transmission routes in farm-animals and their environment [11].
In the Constantine region (Algeria), a recent study showed that 37% of broiler farms and 53% of slaughterhouses were positive for Salmonella [12], with a predominance of S. Hadar, S. Virchow, S. Infantis, S. Albany, and S. Typhimurium. In a nearby region, 44% of laying hen flocks were reported to be positive for Salmonella [13].
In this study, we report on the epidemiological investigation of a certain number of serovars, isolated from broiler breeding farms, slaughterhouses, and human patients within the Constantine region.
Combined phenotypic and genotypic methods were used to assess the relationships between Salmonella strains isolated from these sources, in order to evaluate the contribution of avian strains to human salmonellosis in the region during the 2-year study. Phenotypic methods consisted of serotyping and antimicrobial susceptibility testing, whereas genotypic techniques were based on polymerase chain reaction (PCR) (i.e., Enterobacterial Repetitive Intergenic Consensus-PCR (ERIC-PCR) and Insertion Sequence 200-PCR (IS200-PCR)) and macrorestriction (i.e., Pulsed Field Gel Electrophoresis (PFGE)).
2. Materials and Methods
2.1. Bacterial Strains
For this study, we analysed 100 isolates recovered over a 2-year period (2006 through 2007) in the Constantine region (Table 1). The 45 human isolates studied (named H1 to H45) were obtained from the Constantine Hospital, whereas the 55 isolates of avian origin (named A1 to A55) were collected from poultry farms and slaughterhouses.
Table 1.
Characteristics of the avian and human studied isolates.
| Strains | Serovars | ERIC-PCR | IS-PCR | Antimicrobial resistance pattern* | PFGE profile | Sources |
|---|---|---|---|---|---|---|
| H17 | S. Agona | I | A | AM, CEF, CTX, SXT, SSS, GEN, STR | SAGOXB0004 | Human |
|
| ||||||
| H45 | S. Albany | II | B | NAL, OFX, STR | SABYXB0003 | Human |
| A81, A88, A89 | NAL, OFX, STR | SABYXB0003 | Farm | |||
| A80 | ENR, NAL, OFX, STR | SABYXB0003 | Farm | |||
| A85, A86 | III | C | NAL, OFX, STR | SABYXB0003 | Slaughter | |
|
| ||||||
| H38 | S. Anatum | IV | D | Susceptible | SANAXB0013 | Human |
|
| ||||||
| H21 | S. Blockley | V | E | NAL | SBLOXB0001 | Human |
| H41 | Susceptible | SBLOXB0001 | Human | |||
|
| ||||||
| A78, A79, A90, A91 | S. Carnac | VI | F | Susceptible | SCARXB0001 | Farm |
|
| ||||||
| H9 | S. Enteritidis | VII | G | AMP, CAZ, SSS, TET | SENTXB0026 | Human |
| H11 | Susceptible | SENTXB0026 | Human | |||
| H3, H14, H31, H32, H47 | NAL, OFX | SENTXB0001 | Human | |||
| H10 | AMP, CTX, SSS, TET | SENTXB0001 | Human | |||
| H25 | NAL | SENTXB0001 | Human | |||
| H26 | NAL | SENTXB0013 | Human | |||
| A82, A87 | Susceptible | SENTXB0016 | Human | |||
| H1 | AMP, NAL | SENTXB0035 | Human | |||
| H24 | NAL, OFX | SENTXB0032 | Slaughter | |||
| H2 | NAL | SENTXB0033 | Human | |||
| H7 | H | NAL, OFX | SENTXB0001 | Human | ||
|
| ||||||
| H4 | S. Hadar | VIII | I | ENR, NAL, OFX, STR, TET | SHADXB0003 | Human |
| H5 | KAN, NAL, OFX, STR, TET | SHADXB0003 | Human | |||
| H18, H44 | AMP, NAL, OFX, STR, TET | SHADXB0003 | Human | |||
| A28, A29, A30, A31, A32, A56 | STR, TET | SHADXB0003 | Slaughter | |||
| A36, A37, A38, A39 | STR, TET | SHADXB0003 | Farm | |||
| A33, A34, A35, A40, | STR, TET | SHADXB0003 | Farm | |||
| A41, A42, A43, A44 | STR, TET | SHADXB0003 | Farm | |||
| A26, A27 | J | STR, TET | SHADXB0003 | Slaughter | ||
|
| ||||||
| H13 | S. Heidelberg | IX | K | NAL, OFX | SHIDXB0002 | Human |
| A60 | NAL, OFX, STR | SHIDXB0002 | Farm | |||
| H17 | NAL | SHIDXB0009 | Human | |||
| H23 | NAL, OFX | SHIDXB0010 | Human | |||
| H33 | NAL, OFX | SHIDXB0001 | Human | |||
|
| ||||||
| H46 | S. Indiana | X | L | Susceptible | SINDXB0005 | Human |
|
| ||||||
| A22, A23, A24, A25 | S. Infantis | XI | M | NAL | SINFXB0001 | Farm |
| A48, A49 | Susceptible | SINFXB0001 | Farm | |||
| H12 | Susceptible | SINFXB0005 | Human | |||
|
| ||||||
| H27, H28 | S. Kentucky | XII | N | AMP, CAZ, CEF, CTX, GEN, KAN, SSS, SXT | SKNTXB0006 | Human |
|
| ||||||
| A67 | S. Montevideo | XIII | O | Susceptible | SMVDXB005 | Slaughter |
|
| ||||||
| A21 | S. Rissen | XI | P | NAL | — | Farm |
|
| ||||||
| H34 | S. Senftenberg | XIV | Q | NAL, STR | SSFTXB0039 | Human |
| H15 | AM, CAZ, CF, CTX, GM, K, NAL, S, SSS | SSFTXB0013 | Human | |||
| H30 | NAL, STR | SSFTXB0038 | Human | |||
| H35, H16 | R | NAL, STR | SSFTXB0037 | Human | ||
| H37 | NAL, STR | SSFTXB0040 | Human | |||
|
| ||||||
| H29 | S. Typhimurium | XV | S | NAL | STYMXB0093 | Human |
| H19, H20, H22, H36 | AMP, CHL, SSS, STR, TET | STYMXB0035 | Human | |||
| H8 | NAL | STYMXB0089 | Human | |||
| H6 | NAL | STYMXB0005 | Human | |||
| A17, A18, A45, A46 | T | STR | STYMXB0021 | Slaughter | ||
| A19 | NAL, STR | STYMXB0021 | Slaughter | |||
|
| ||||||
| A63 | S. Virchow | XVI | U | Susceptible | SVIRXB0017 | Farm |
| A66 | NAL | SVIRXB0017 | Farm | |||
| H40 | Susceptible | SVIRXB0005 | Human | |||
| A20 | NAL | SVIRXB0005 | Slaughter | |||
| A53, A65 | Susceptible | SVIRXB0005 | Slaughter | |||
| A62, A64, A77, A92 | Susceptible | SVIRXB0005 | Farm | |||
*Susceptible: susceptible to all tested antibiotics. AMP: ampicillin; AMC: amoxicillin-clavulanic acid; CAZ: ceftazidime; CEF: cephalothin; CHL: chloramphenicol; CST: colistin; CTX: cefotaxime; ENR: enrofloxacin; GEN: gentamicin; K: kanamycin; NAL: nalidixic acid; OFX: ofloxacin; SSS: sulphonamides; STR: streptomycin; SXT: trimethoprim-sulfamethoxazole; TET: tetracycline.
The isolation of avian strains was performed according to the NF U47-100 and NF U47-101 procedures [14, 15] at the Food Hygiene Laboratory from the Constantine Veterinary Sciences Department. Serotyping was carried out according to the White-Kauffmann-Le Minor scheme [16], as previously described [12].
2.2. Bacterial Susceptibility to Antibiotics
The antimicrobial susceptibility tests were performed using the disk diffusion method and interpreted as recommended by the “Comité de l'Antibiogramme de la Société Française de Microbiologie” [17]. Antimicrobials tested (load, breakpoints (mm)) were ampicillin (10 μg, 19–14), amoxicillin-clavulanic acid (20/10 μg, 21–14), cephalothin (30 μg, 18–12), cefotaxime (30 μg, 21–15), ceftazidime (30 μg, 21–15), streptomycin (10 IU, 15–13), gentamicin (10 IU, 16–14), kanamycin (30 IU, 17–15), chloramphenicol (30 μg, 23–19), tetracycline (30 IU, 19–17), sulphamethoxazole-trimethoprim (23.75 μg + 1.25 μg, 16–10), sulphonamides (200 μg, 17–12), nalidixic acid (30 μg, 20–15), ofloxacin (5 μg, 22–16), enrofloxacin (5 μg, 22–17), and colistin (50 μg, 15). Zone diameters were read using the automated scanner Osiris (Bio-Rad).
2.3. PCR Methods
DNA was extracted by a boiling method as described previously [18]. The intergenic segments were amplified using the primers' sequences described by Millemann et al. [18] and Versalovic et al. [19]. All amplifications were performed on a Perkin Elmer 9700 thermal cycler (Courtaboeuf, France) as previously described [18].
2.4. PFGE Genotyping
PFGE was performed using a CHEF-DR III system (Bio-Rad, Marnes La-Coquette, France) according to the Salm-gene and PulseNet standardized protocol [20–22]. Two endonucleases were used, XbaI for all serovars and BlnI for S. Hadar. The S. enterica Braenderup H9812 strain was used as an internal control and molecular size marker [23]. DNA patterns were analysed with BioNumerics software (V 6.6, Applied Maths, Sint-Martens-Latem, Belgium). Dendrograms were produced using the Dice coefficient and an unweighted pair group method with arithmetic averages (UPGMA) with a 1% tolerance limit and 1% optimization (Pulsenet Europe recommendation [20]).
3. Results
Salmonella isolates were grouped into 16 different serovars (Table 1). Six serovars, namely, Agona, Anatum, Blockley, Indiana, Kentucky, and Senftenberg, were only recovered from humans during the two-year study, whereas 3 serovars, namely, Carnac, Montevideo, and Rissen, were only isolated from poultry. Isolates belonging to the 7 remaining serovars, that is, Albany, Enteritidis, Hadar, Heidelberg, Infantis, Typhimurium, and Virchow, were recovered from both poultry and humans.
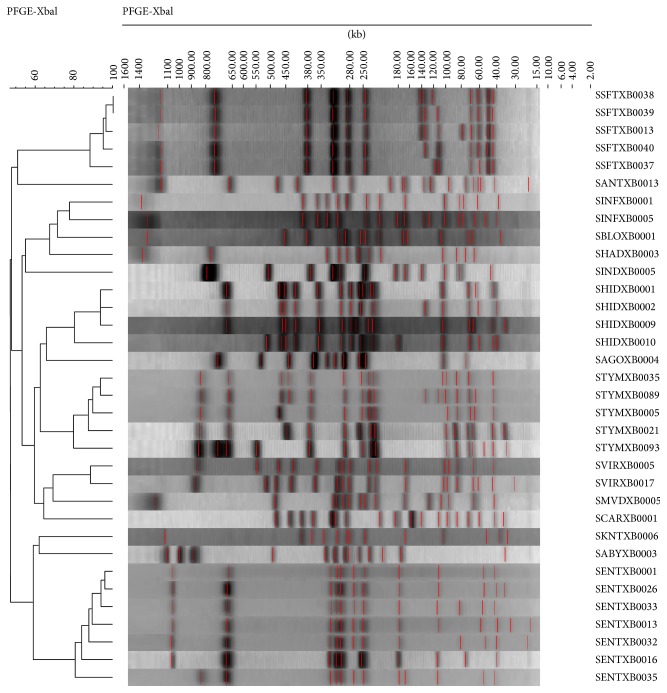
A total of 16 ERIC-PCR, 20 IS200-PCR, 30 antimicrobial resistance, and 34 PFGE profiles were generated from the 100 isolates. For all isolates studied, those belonging to the same serovar clustered together (Table 1 and Figure 1).
Figure 1.
Dendrogram based on XbaI Pulsed Field Gel Electrophoresis (PFGE) profiles of major Salmonella serovars identified in Constantine (Algeria). Similarity percentages are figured on the left; names of the profiles are listed on the right. SABY: Salmonella Albany; SANT: Salmonella Anatum; SAGO: Salmonella Agona; SBLO: Salmonella Blockley; SCAR: Salmonella Carnac; SENT: Salmonella Enteritidis; SHAD: Salmonella Hadar; SHID: Salmonella Heidelberg; SIND: Salmonella Indiana; SINF: Salmonella Infantis; SKNT: Salmonella Kentucky; SMVD: Salmonella Montevideo; SSFT: Salmonella Senftenberg; STYM: Salmonella Typhimurium; SVIR: Salmonella Virchow.
The different ERIC-PCR profiles obtained were numbered from I to XVI and IS-PCR profiles were identified by the letters A through T (Table 1). Rissen and Infantis isolates shared the same ERIC- and IS200-PCR profiles.
The 34 different PFGE profiles obtained were numbered according to the preexisting database. Based on PFGE patterns, different situations were established among the analysed isolates, which led to various hypotheses. All PFGE results are shown in Table 1 and Figure 1.
4. Discussion
Evaluating the contribution of various animal sources to the burden of human salmonellosis is very difficult and requires microbial subtyping approaches [24] that depend on the comparison of the phenotypic and genotypic characteristics of the isolates studied. This consists of comparing serovars isolated from animals and humans to normal findings in both national and international serovar-based surveillance databases. Finally, the use of molecular markers for which there is a database may be also useful.
4.1. Serovars Isolated in Poultry and Humans
The serovars isolated from broilers in our study represent those usually present in broilers worldwide, especially in the USA and Europe [5, 25]. In our study, we recovered 6 serovars from broilers (i.e., on farms or in slaughterhouses) that are among the top 10 serovars encountered in Europe, including Enteritidis, Hadar, Indiana, Infantis, Typhimurium, and Virchow [5]. However, our study did not reflect this order as Hadar was isolated most frequently followed by Virchow, Infantis, and Albany.
Enteritidis and Typhimurium were the serovars most often isolated from human clinical cases in this study. This is generally consistent with other worldwide studies, for instance, in the USA and in Europe, as well as in Africa [5, 6, 25]. Senftenberg was ranked third, followed by Heidelberg, Blockley, and Kentucky. The high occurrence of Senftenberg is somewhat surprising and may be related to extensive commercial links with France. On the other hand one would have expected a slightly higher number of S. Kentucky isolates due to the recent emergence and distribution of this serovar in Africa [26, 27].
Interestingly, although it is rarely isolated from broiler and laying hen flocks, Albany was frequently isolated from broilers in our study [13, 26, 28]. Carnac is an extremely rare serovar in both poultry and humans. For instance, only one Carnac isolate was recovered from poultry in the European base line studies in 2008 [27] and the 2013 USA atlas for Salmonella did not report Carnac isolates for humans [29].
Some serovars (i.e., Agona, Anatum, Blockley, Indiana, Kentucky, and especially Senftenberg) were only isolated from humans in our study. However, those serovars are frequently isolated from various poultry species and are associated with chicken consumption when isolated in humans [28]. Senftenberg is mainly isolated in hatcheries and laying hen farms, and, in 2012, it ranked fourth among laboratory-confirmed Salmonella isolates from nonclinical nonhuman sources submitted to the National Veterinary Services Laboratories (NVSL) for typing in the USA [24]. This is one of the most commonly isolated serovars in France. For instance, in 2008, S. Senftenberg ranked first in total isolates collected from nonhuman sources as well as from poultry farm environments [28]. Kentucky is an emerging serovar in poultry and human and, recently, a particular multidrug resistant (MDR) phenotype has emerged in Africa and spread throughout poultry plants [30]. This MDR phenotype has also been isolated from laying hen flocks in Algeria [13]. Nevertheless, the Kentucky isolates from this study, although they were multidrug resistant, could not be linked to the global epidemic described by le Hello et al. [30] as these isolates are fully susceptible to fluoroquinolones.
Thus, considering the 7 serovars isolated in this study from both humans and poultry as well as the 6 serovars usually linked to human infection by poultry, isolates belonging to 13 of the 16 identified serovars suggest the potential link between poultry contamination and human salmonellosis.
4.2. Contribution of Epidemiological Markers to the Comparison of Avian Isolates and Human Isolates
Among the 7 serovars isolated from both humans and poultry in this study, 4 serovars (i.e., Albany, Hadar, Heidelberg, and Virchow) included human and avian isolates with indistinguishable patterns. In contrast, human and avian strain patterns did not match for serovars Enteritidis, Infantis, or Typhimurium.
4.2.1. Matching Avian and Human Patterns
Serovar Albany strains were isolated from 3 different sources (i.e., humans, breeding farms, and slaughterhouses) but could not be differentiated by PFGE after digestion by restriction enzyme XbaI. There were only two strains of this serovar in the ANSES database and the identified profile SABYXB0003 was new. Therefore, it remains difficult to determine any genetic heterogeneity among these isolates. However, the two isolates from the slaughterhouses shared distinct ERIC-PCR and IS-PCR profiles. As a whole, our results suggest an epidemiological link between strains isolated from breeding farms, humans and, to a lesser extent, slaughterhouses. This conclusion is supported by the very similar antimicrobial resistance patterns observed, especially since fluoroquinolones were targeted.
Twenty-four Hadar isolates isolated from slaughterhouses, farms, and humans were characterized. All isolates merged with a single PFGE profile, with digestion by either XbaI or BlnI restriction enzymes, which seems to demonstrate the clonal character of the strains isolated from broiler chickens and humans. Nevertheless, we must be cautious since Hadar is considered to be a genetically homogeneous serovar (DI = 0.70 [20]). The comparison with the ANSES database showed that, with XbaI, 24 profiles had been identified out of the 153 strains of this previously studied serovar and the DI was only 0.48. This possible epidemiological link also seems to be supported by the single profile found by ERIC-PCR and the IS-PCR profile, with the exception of 2 strains isolated from slaughterhouses. The 2 dissimilar Hadar isolates were associated with turkeys slaughtered in the same slaughterhouse. Antibiotyping also gave a different reading in that human isolates were multiresistant and therefore differentiated, whereas all the other isolates shared a single resistance pattern.
For Heidelberg and Virchow, we identified at least one common pulsotype in avian and human isolates, which may indicate an avian source for human infection. Additionally, the SHIDXB0001 profile, identified in a human Heidelberg strain, had previously been found in the poultry chain.
Two different PFGE profiles were identified for the Virchow isolates. It is possible that isolates exhibiting a SVIRXB0005 profile may have spread from broiler chickens to consumers. This hypothesis is supported by our results where all strains isolated from slaughterhouses shared this profile. To date, 93 strains of this serovar have been recorded in the database and 24 different profiles have been identified.
4.2.2. Nonmatching Human and Avian Patterns
Although human illnesses due to Enteritidis, Infantis, Senftenberg, and Typhimurium are commonly linked to avian sources, we did not find any matching pulsotypes between the avian and human isolates of these serovars. This must be emphasized particularly for Enteritidis and Senftenberg, even though they tend to originate in laying hens rather than broilers [28, 31, 32]. However, Cardinale et al. [33] highlighted the genetic similarity of S. Enteritidis PFGE profiles from human and broiler sources in Senegal. We may add that the SENTXB0001 profile has already been encountered in isolates of human origin, as well as from poultry, pastries, cooked meals, sea products, and so forth.
5. Conclusion
Our study did not confirm an association between the main serotypes detected in humans and those recovered in poultry production. However, collectively, our results bring to light a probable significant contribution of nontyphoidal Salmonella by avian species to human salmonellosis in the Constantine region. Since the majority of isolates belonged to serovars usually associated with poultry, and despite the very low number of isolates studied, we were able to confirm identical profiles among avian and human isolates. The development of a large monitoring programme is crucial for the surveillance of Salmonella in poultry and the improvement of public health in Algeria.
Acknowledgments
The authors are grateful to V. Carlier, F. Smati, J. C. Augustin, and all the staff of the ENV Alfort former MASQ Laboratory and to the C.H.U Constantine Microbiology Laboratory for their technical training. They also thank Mr. R. Chaouaou from the Veterinary Department of Constantine University for his technical support and Pr. Andrew Ponter and Joshua Ison for their wise edition of this paper.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Rabsch W., Tschäpe H., Bäumler A. J. Non-typhoidal salmonellosis: emerging problems. Microbes and Infection. 2001;3(3):237–247. doi: 10.1016/s1286-4579(01)01375-2. [DOI] [PubMed] [Google Scholar]
- 2.Whyte P., Mc Gill K., Collins J. D., Gormley E. The prevalence and PCR detection of Salmonella contamination in raw poultry. Veterinary Microbiology. 2002;89(1):53–60. doi: 10.1016/s0378-1135(02)00160-8. [DOI] [PubMed] [Google Scholar]
- 3.Majowicz S. E., Musto J., Scallan E., et al. The global burden of nontyphoidal Salmonella gastroenteritis . Clinical Infectious Diseases. 2010;50(6):882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 4.Gill N., Reilly B., Threlfall E. J. Surveillance of Enteric Pathogens in Europe and Beyond. Enternet Annual Report. European Commission; 2004. [Google Scholar]
- 5.European Food Safety Authority. The European union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA Journal. 2014;12(2):p. 3547. doi: 10.2903/j.efsa.2014.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendriksen R. S., Vieira A. R., Karlsmose S., et al. Global monitoring of salmonella serovar distribution from the world health organization global foodborne infections network country data bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathogens and Disease. 2011;8(8):887–900. doi: 10.1089/fpd.2010.0787. [DOI] [PubMed] [Google Scholar]
- 7.Sánchez-Vargas F. M., Abu-El-Haija M. A., Gómez-Duarte O. G. Salmonella infections: an update on epidemiology, management, and prevention. Travel Medicine and Infectious Disease. 2011;9(6):263–277. doi: 10.1016/j.tmaid.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Sow A. I., Seydi M., Thiaw M., et al. Les salmonelloses au centre hospitalier universitaire de Fann à Dakar: aspects bactériologiques. Médecine et Maladies Infectieuses. 2000;30:657–660. [Google Scholar]
- 9.Butaye P., Michael G. B., Schwarz S., Barrett T. J., Brisabois A., White D. G. The clonal spread of multidrug-resistant non-typhi Salmonella serotypes. Microbes and Infection. 2006;8(7):1891–1897. doi: 10.1016/j.micinf.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 10.Panisello P. J., Rooney R., Quantick P. C., Stanwell-Smith R. Application of foodborne disease outbreak data in the development and maintenance of HACCP systems. International Journal of Food Microbiology. 2000;59(3):221–234. doi: 10.1016/S0168-1605(00)00376-7. [DOI] [PubMed] [Google Scholar]
- 11.Winfield M. D., Groisman E. A. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli . Applied and Environmental Microbiology. 2003;69(7):3687–3694. doi: 10.1128/aem.69.7.3687-3694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elgroud R., Zerdoumi F., Benazzouz M., et al. Characteristics of Salmonella contamination of broilers and slaughterhouses in the region of Constantine (Algeria) Zoonoses and Public Health. 2009;56(2):84–93. doi: 10.1111/j.1863-2378.2008.01164.x. [DOI] [PubMed] [Google Scholar]
- 13.Bouzidi N., Aoun L., Zeghdoudi M., et al. Salmonella contamination of laying-hen flocks in two regions of Algeria. Food Research International. 2012;45(2):897–904. doi: 10.1016/j.foodres.2011.05.027. [DOI] [Google Scholar]
- 14.Norme française. NF U47-100. Isolement et identification de tout sérovar ou de sérovar(s) spécifié(s) de salmonelles dans l’environnement des productions animales. Editions AFNOR, 2005.
- 15.Norme Française. NF. U47-101. AFNOR; 2005. Isolement et identification de tout sérovar ou de sérovar(s) spécifié(s) de salmonelles chez les oiseaux. [Google Scholar]
- 16.Grimont P. A. D., Weill F.-X. WHO Collaborating Centre for Reference and Research on Salmonella. 9th. Paris, France: Institut Pasteur; 2007. Antigenic formulae of the Salmonella serovars; p. 166. [Google Scholar]
- 17.CA-SFM. Comité de l’antibiogramme de la Société Française de Microbiologie, 2011, Recommandations 2011, http://www.sfm-microbiologie.org/UserFiles/file/CASFM/casfm_2011.pdf.
- 18.Millemann Y., Gaubert S., Remy D., Colmin C. Evaluation of IS200-PCR and comparison with other molecular markers to trace Salmonella enterica subsp. enterica serovar Typhimurium bovine isolates from farm to meat. Journal of Clinical Microbiology. 2000;38(6):2204–2209. doi: 10.1128/jcm.38.6.2204-2209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Versalovic J., Koeuth T., Lupski J. R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Research. 1991;19(24):6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kérouanton A., Marault M., Lailler R., et al. Pulsed-field gel electrophoresis subtyping database for foodborne Salmonella enterica serotype discrimination. Foodborne Pathogens and Disease. 2007;4(3):293–303. doi: 10.1089/fpd.2007.0090. [DOI] [PubMed] [Google Scholar]
- 21.Ribot E. M., Fair M. A., Gautom R., et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathogens and Disease. 2006;3(1):59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 22.Peters T. M., Maguire C., Threlfall E. J., Fisher I. S., Gill N., Gatto A. J. The Salm-gene project—a European collaboration for DNA fingerprinting for Salm-gene project. Eurosurveillance Monthly Release. 2003;8(2):46–50. doi: 10.2807/esm.08.02.00401-en. [DOI] [PubMed] [Google Scholar]
- 23.Hunter S. B., Vauterin P., Lambert-Fair M. A., et al. Establishment of a universal size standard strain for use with the pulsenet standardized pulsed-field gel electrophoresis protocols: Converting the national databases to the new size standard. Journal of Clinical Microbiology. 2005;43(3):1045–1050. doi: 10.1128/JCM.43.3.1045-1050.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pires S. M., Evers E. G., van Pelt W., et al. Attributing the human disease burden of foodborne infections to specific sources. Foodborne Pathogens and Disease. 2009;6(4):417–424. doi: 10.1089/fpd.2008.0208. [DOI] [PubMed] [Google Scholar]
- 25.Centers for Disease Control and Prevention (CDC) National Salmonella Surveillance Annual Report, 2012. Atlanta, Ga, USA: Department of Health and Human Services, CDC; 2014. http://www.cdc.gov/ncezid/dfwed/pdfs/salmonella-annual-report-2012-508c.pdf. [Google Scholar]
- 26.Picherot M., Guillon F., Pinson M., Danan C., le Hello S., Francart S. Bilan de la surveillance obligatoire des salmonelles dans les troupeaux de l’espèce Gallus gallus en 2009. Bulletin épidémiologique, santé animale et alimentation. 2010;42:11–13. [Google Scholar]
- 27.European Food Safety Authority. Analysis of the baseline survey on the prevalence of Campylobacter in broiler batches and of Campylobacter and Salmonella on broiler carcasses in the EU, 2008—part A: Campylobacter and Salmonella prevalence estimates. EFSA Journal. 2010;8(3):p. 1503. doi: 10.2903/j.efsa.2010.1503. [DOI] [Google Scholar]
- 28.Moury F., Danan C., Frémy S., et al. Inventaire des Salmonella d'origine non humaine. Réseau Salmonella 2008. Maisons-Alfort, France: ANSES; 2011. [Google Scholar]
- 29.Centers for Disease Control and Prevention (CDC) An Atlas of Salmonella in the United States, 1968–2011. Washington, DC, USA: Laboratory-based Enteric Disease Surveillance, Atlanta, Georgia; US Department of Health and Human Services, CDC; 2013. [Google Scholar]
- 30.le Hello S., Hendriksen R. S., Doublet B., et al. International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. Journal of Infectious Diseases. 2011;204(5):675–684. doi: 10.1093/infdis/jir409. [DOI] [PubMed] [Google Scholar]
- 31.Chemaly M., Huneau A., Rouxel S., et al. Enquêtes communautaires sur la prévalence de Salmonella en filières avicoles. Proceedings of the Communication, 10eme Réunion Annuelle du Réseau Salmonella; 2006; Paris, France. Afssa; [Google Scholar]
- 32.EFSA (European Food Safety Authority) Report of Task Force on Zoonoses Data Collection on the analysis of the baseline survey on the prevalence of Campylobacter in broiler batches and of Salmonella on broiler carcasses in EU, 2008, Part A: Campylobacter and Salmonella prevalence estimates. EFSA Journal. 2010;8(3):p. 1503. [Google Scholar]
- 33.Cardinale E., Gros-Claude J. D. P., Rivoal K., et al. Epidemiological analysis of Salmonella enterica ssp. enterica serovars Hadar, Brancaster and Enteritidis from humans and broiler chickens in Senegal using pulsed-field gel electrophoresis and antibiotic susceptibility. Journal of Applied Microbiology. 2005;99(4):968–977. doi: 10.1111/j.1365-2672.2005.02618.x. [DOI] [PubMed] [Google Scholar]