Abstract
This study aimed to analyse the diversity of the Campylobacter jejuni population in broilers and to evaluate the major source of contamination in poultry meat. Eight rearing cycles over one year provided samples from three different broiler farms processed at the same slaughterhouse. A total of 707 C. jejuni were isolated from cloacal swabs before slaughter and from the breast skin of carcasses after slaughter and after chilling. All suspected Campylobacter colonies were identified with PCR assays and C. jejuni was genotyped by sequence analysis of the flaA short variable region (SVR) and by pulsed-field gel electrophoresis (PFGE) using SmaI enzyme. Phenotypic antibiotic resistance profiles were also assayed using minimal inhibitory concentration (MIC). The flocks carried many major C. jejuni clones possibly carrying over the rearing cycles, but cross contamination between farms may happen. Many isolates were resistant to fluoroquinolones, raising an issue of high public concern. Specific Campylobacter populations could be harboured within each poultry farm, with the ability to contaminate chickens during each new cycle. Thus, although biosecurity measures are applied, with a persistent source of contamination, they cannot be efficient. The role of the environment needs further investigation to better address strategies to control Campylobacter.
1. Introduction
Campylobacter is the most common cause of bacterial gastroenteritis in Europe. The incidence of human campylobacteriosis is increasing worldwide, as well as the number of isolates resistant to fluoroquinolones which are one of the primary classes of antimicrobials used to treat Campylobacter infection in human therapy and thus considered of high public concern [1]. In the European Union, Campylobacter is still the most commonly reported cause of bacterial foodborne illness with a notification rate of 55.49 cases per 100,000 of population in 2012 [2]. Poultry is a natural reservoir of Campylobacter species, constituting the most important source of human infection. The consumption of undercooked poultry meat or the mishandling of raw poultry products is considered to be the main risk factors associated with human campylobacteriosis [3–5].
The prevalence of Campylobacter in broiler chicken flocks ranges from 3 to 90% depending on their location [6, 7] and the isolation rates within positive flocks at slaughter are high (around 80%) [8–10]. Recent studies have reported that the prevalence of Campylobacter in retail chicken products ranges from 90 to 100% across several countries [11, 12]. Campylobacter colonization in chickens takes place at poultry farms, approximately 7 days after hatching [13], while widespread carcass contamination occurs at the slaughterhouse, especially from cross contamination by intestinal contents after the evisceration phase or from dirty surfaces [14]. Nevertheless, there have been few studies on the contamination of poultry carcasses from the farm through the entire production chain up to the retailer [15, 16] so the contamination routes in broiler flocks are still unknown.
The objective of the present study was to perform a comprehensive molecular characterization of C. jejuni isolated from poultry on the farm and during the slaughter process. Different typing methods, such as PFGE and flaA-SVR sequencing, will be used to trace the contamination of chicken products and to investigate the potential of specific isolates to persist or be predominant in the poultry production. PFGE has been successfully applied to track Campylobacter during poultry production [16–18] and, together with flaA-SVR sequencing, it represents a highly discriminatory method for a better understanding of Campylobacter population structures. In addition, antibiotic susceptibility will also be investigated to determine the resistance pattern of Campylobacter that spread from chickens to humans along the poultry food chain, although the correlation between resistant bacteria in people and the use of antibiotics in feed is still a matter of debate [19].
2. Materials and Methods
2.1. Broiler Farms
Three different broiler farms (A, B, and C), randomly selected in the Abruzzo region of central Italy and spaced about 40 kilometres apart in a narrow zone, were enrolled in the study. The farms were managed similarly as part of the same integrated broiler company under good hygiene practices, rearing flocks of 40,000–60,000 birds with an average age at slaughter of 38–42 days.
2.2. Experimental Set-Up
Four different flocks were monitored on farm A, and two flocks each on farms B and C, amounting to a total of eight different rearing cycles under study between July 2011 and July 2012 with detailed dates shown in Table 1. For each flock, one day before slaughter, 50 different chickens, individually identified by leg rings, were randomly chosen and cloacal swabs taken (F), which were transported immediately to the laboratory using Ames transportation medium. The following day, the birds were transported 50 kilometres to the company abattoir, where samples were taken after slaughter (S) and after the chilling process (C). Samples S and C consisted of breast skin sampled under aseptic conditions, which were transported to the laboratory in a portable cooler at 2–4°C for immediate processing. The flocks tested were the first to be slaughtered on these days, using a slaughter line disinfected after the last batch processed on the previous day.
Table 1.
Distribution of flaA and peptide types according to rearing cycle from farms A, B, and C.
| Peptide type | flaA type | 12/12/2011 | 12/02/2012 | 10/05/2012 | 17/07/2012 | Proportion | Number of strains | |
|---|---|---|---|---|---|---|---|---|
| Farm A | 1 | 1266 | — | 23 (24.46%) | — | — | 9.23% | 23 |
| 260 | 1 (1.38%) | — | — | — | 0.4% | 1 | ||
| 34 | 1 (1.38%) | — | — | — | 0.4% | 1 | ||
| 49 | — | — | 1 (2.94%) | 27 (55.10%) | 11.24% | 28 | ||
| 368 | 1638 | 1 (1.38%) | 67 (71.27%) | 3 (8.82%) | 19 (38.77%) | 36.14% | 90 | |
| 5 | 259 | — | — | — | 1 (2.04%) | 0.4% | 1 | |
| 8 | 117 | — | — | — | 1 (2.04%) | 0.4% | 1 | |
| 287 | 19 (26.38%) | — | 17 (50.00%) | — | 14.45% | 36 | ||
| 67 | — | 1 (1.03%) | 4 (11.76%) | — | 2% | 5 | ||
| 2 | 612 | 1 (1.38%) | — | — | — | 0.4% | 1 | |
| 10 | 1284 | — | — | 3 (8.82%) | — | 1.21% | 3 | |
| 1429 | 8 (11.11%) | — | — | — | 3.21% | 8 | ||
| 103 | 327 | 1 (1.38%) | — | — | — | 0.4% | 1 | |
| 11 | 14 | 37 (51.38%) | 1 (1.03%) | 3 (8.82%) | — | 16.46% | 41 | |
| 17 | 3 (4.16%) | 2 (2.12%) | 2 (5.82%) | — | 2.81% | 7 | ||
| 30 | 1 (2.94%) | 1 (2.04%) | 0.8% | 2 | ||||
| Total | 72 | 94 | 34 | 49 | 249 | |||
|
| ||||||||
| 19/09/2011 | 17/11/2011 | — | — | |||||
|
| ||||||||
| Farm B | 1 | 36 | 18 (15.65%) | 106 (96.36%) | — | — | 55.11% | 124 |
| 49 | — | 3 (2.72%) | — | — | 1.33% | 3 | ||
| 11 | 11 | 1 (0.86%) | — | — | — | 0.44% | 1 | |
| 3 | 161 | 1 (0.86%) | — | — | — | 0.44% | 1 | |
| 5 | — | 1 (0.90%) | — | — | 0.44% | 1 | ||
| 33 | 222 | 53 (46.08%) | — | — | — | 23.55% | 53 | |
| 8 | 287 | 42 (36.52%) | — | — | — | 18.22% | 42 | |
| Total | 115 | 110 | — | — | 225 | |||
|
| ||||||||
| 31/03/2012 | 12/06/2012 | — | — | |||||
|
| ||||||||
| Farm C | 1 | 260 | — | 9 (10%) | — | — | 4.1% | 9 |
| 23 | — | 1 (1.11%) | — | — | 0.5% | 1 | ||
| 265 | — | 72 (80%) | — | — | 32.9% | 72 | ||
| 8 | 117 | 3 (2.32%) | — | — | — | 1.4% | 3 | |
| 287 | 11 (8.52%) | 2 (2.22%) | — | — | 5.9% | 13 | ||
| 2 | 21 | — | 3 (3.33%) | — | — | 1.4% | 3 | |
| 33 | 222 | 1158 (89.14%) | — | — | — | 52.5% | 115 | |
| 9 | 239 | — | 3 (3.33%) | — | — | 1.4% | 3 | |
| Total | 129 | 90 | — | — | 219 | |||
2.3. Culture Conditions and PCR Assays
A total of 1,720 samples were processed during the whole project and Campylobacter was recovered from carcass samples after the enrichment and the enumeration phases, according to parts 1 and 2, respectively, of the NF EN ISO 10272 standard procedure [20, 21]. The isolates were cultured on Columbia blood agar and incubated at 42°C for 48 h in a microaerophilic atmosphere. After a preliminary phenotypic characterization, suspected colonies were confirmed as thermotolerant Campylobacter and identified to species level using a multiplex PCR, as described previously by Di Giannatale et al. [22]. Genomic DNA was extracted using a Wizard genomic DNA purification kit (Promega, Madison, WI, USA). Isolates were stored in a Microbank (Pro-Lab Diagnostics Canada, Richmond Hill, ON, Canada) at 80°C until further analysis.
2.4. PFGE
Pulsed-field gel electrophoresiswas performed according to the 2009 U.S. PulseNet protocol for Campylobacter [23]. Briefly, bacteria previously identified as Campylobacter by PCR were subcultured on Columbia blood agar at 42°C for 2 days in a microaerophilic atmosphere and embedded in agarose blocks (Seakem Gold Agarose, Lonza, Rockland, ME, USA). The blocks were then lysed, washed, digested with 25 U of SmaI restriction endonuclease (Promega, Milan, Italy) at 25°C overnight, and subjected to pulsed-field electrophoresis in 1% agarose gel (Seakem Gold Agarose). PFGE was performed using a Chef Mapper XA (Bio-rad Laboratories, Hercules, CA, USA) and Salmonella serovar Branderup H9812 was used as standard molecular weight size. After electrophoresis, the gel was stained with Sybr Safe DNA gel stain (Invitrogen, Waltham, MA, USA) and photographed with a transilluminator (Alpha Innotech, San Leandro, CA, USA). For image analysis, Bionumerics v. 6.6 software (Applied Maths, Sint Martens Latem, Belgium) was used to identify the clusters of closely related or identical patterns. Pair comparisons and cluster analyses were carried out using the Dice correlation coefficient (position tolerance, 1.0%) and the unweighted pair group mathematical average (UPGMA) clustering algorithm. PFGE clusters were arbitrarily defined at a similarity level of 60% [24]. Untypeable isolates were not included in the analysis.
2.5. flaA SVR Sequencing
Typing was performed by amplifying the flaA-SVR using primers as described by Nachamkin et al. [25], followed by sequencing of the PCR product. Amplification products were verified by gel electrophoresis. PCR products were purified by using ExoSAP-IT reagent (GE Healthcare, Santa Clara, CA, USA) and sequenced using the BigDye Terminator v.3.1 Cycle sequencing kit (Applied Biosystems, Darmstadt, Germany) according to the manufacturer's instructions. After sequencing, DNA was purified with ethanol precipitation using the Agencourt CleanSEQ kit (Beckman Coulter, Brea, CA, USA). Sequencing products were analysed with a Genetic Analyzer 3500 (Life Technologies, Paisley, UK). The nucleotide sequences were compared with the C. jejuni flaA database (http://pubmlst.org/campylobacter/) and allele numbers were assigned accordingly. Confirmed sequences were aligned using MEGA 4 software [26]. For new flaA-SVR alleles, DNA trace files were submitted to the database administrator for confirmation. The peptide sequences were translated from the DNA sequences and named according to the Oxford database available at http://pubmlst.org/campylobacter/ The genetic diversity and the comparison between the molecular methods were determined using the Simpson's diversity index (SDI) and the adjusted Rand index (aRI) via the online tool available at the Comparing Partitions website (http://darwin.phyloviz.net/ComparingPartitions/index.php?link=Home).
2.6. Antimicrobial Susceptibility
Campylobacter susceptibility to antibiotics was evaluated using the microdilution method by the “Sensititre” automated system (TREK Diagnostic Systems/Biomedical Service, Venice, Italy). Colonies were harvested in Columbia agar for 24 hours, inoculated in Mueller Hinton Broth supplemented with blood, and dispensed into Eucamp microtiter plates (TREK Diagnostic Systems/Biomedical Service), containing known scalar concentrations of the following antibiotics: gentamicin (0.12–16 μg/mL), streptomycin (1–16 μg/mL), ciprofloxacin (0.06–4 μg/mL), tetracycline (0.25–16 μg/mL), erythromycin (0.5–32 μg/mL), nalidixic acid (2–64 μg/mL), and chloramphenicol (2–32 μg/mL). The plates were then incubated at 42°C in a microaerophilic atmosphere for 24 hours. C. jejuni NCTC 11351 was included for the quality control in the MIC test.
3. Results
3.1. Campylobacter Prevalence
Campylobacter spp. was isolated in 1,081 of the samples. Further differentiation within the Campylobacter genus was obtained by PCR, resulting in 374 C. coli and 707 C. jejuni. The isolates were recovered from the different sources as follows: 281 C. jejuni and 56 C. coli from broiler flocks from the three farms, 366 C. jejuni and 248 C. coli from carcasses processed in the slaughterhouse, and 60 C. jejuni and 70 C. coli after chilling. At farm level, the prevalence of C. jejuni (65.77%) was significantly higher (P < 0.05, χ 2 test) than C. coli isolates (12.62%). All the flocks investigated from the different farms were positive for Campylobacter with high rates of prevalence, ranging from 58 to 90% of positive chicken (data not shown). In contrast, after chilling, the prevalence of C. coli (39.10%) was significantly higher (P < 0.05, χ 2 test) than C. coli groups at farm level (12.62%).
3.2. Typing
From farm A, 249 samples of C. jejuni were isolated during all four sampling periods (Table 1). Molecular investigation of the short variable region of the flagella revealed 16 different nucleotide types that corresponded to eight different peptide types (Table 1). Each flock was characterized by 5 to 9 different flaA types with one type predominant. In 6 instances, the same flaA type was recovered from different samples. The flaA type 1638 was isolated from all four rearing cycles, while flaA types 14 and 17 and flaA types 30, 49, 67, and 287 were isolated from 3 and 2 cycles, respectively (Table 1). From the analysis of isolates from the single flock of farm A, only 6 (4.38%) out of 137 C. jejuni isolated in the slaughterhouse did not belong to flaA types recovered from the farm. These remaining isolates showed the same fla type as those from the farm (Table 2). At a 60% similarity level, the PFGE clustering analysis revealed a high diversity within the isolates, grouping most of the isolates in three major clusters. The first cluster included 33 C. jejuni isolated from three rearing cycles (12.12.2011–10.05.2012–17.07.2012); the second cluster contained 130 C. jejuni isolated from all cycles analysed (12.12.2011–12.02.2012–10.05.2012–17.07.2012); the last cluster included 21 isolates from two cycles (12.12.2011–17.07.2012). All C. jejuni isolates in the three PFGE clusters were detected at farm, slaughter, and postchilling level. At a 100% similarity level, a dendrogram combining the data from farm A resulted in 56 different PFGE pulsotypes. Four pulsotypes comprised 49.57% (117/236) of the C. jejuni isolates from farm A, while 35 of the 56 pulsotypes included only a single C. jejuni isolate (Table 3). The polymorphisms resulting from the PFGE were higher than flaA typing with an SDI of 0.84 against 0.79; nevertheless the agreement between the methods resulted in an aRI of 0.44. A total of 225 strains of C. jejuni were isolated from farm B, 115 in the summer (19.09.2011) and 110 in the autumn (17.11.2011). Molecular investigation of the flagella determined seven different nucleotide types corresponding to five different peptide types (Table 1). Only fla type 36 was recovered in both rearing cycles analysed. All the isolates collected after chilling showed fla types previously detected in the live chicken. Six isolates out of 123 collected from the slaughterhouse featured four fla types (49, 11, 161, and 5) that were different from those collected on the farm (Table 2). At a 60% similarity level, the PFGE clustering showed a high variability with four major clusters. The first cluster included 80 C. jejuni isolated from two rearing cycles (19.09.2011–17.11.2011) obtained at farm, slaughter, and postchilling phases; the second cluster contained 96 C. jejuni from one flock (19.09.2011) but they were present in all the phases analysed; the third cluster included 26 isolates from one flock (17.11.2011) at farm and slaughter level; the last cluster included 16 isolates from two rearing cycles (19.09.2011–17.11.2011) obtained at the farm and slaughter phases. At a 100% similarity level, all the C. jejuni from farm B were clustered in 39 different pulsotypes with four that comprised 53.73% of the isolates and 22 pulsotypes represented by a single isolate (Table 3). The polymorphisms of PFGE showed an SDI of 0.927, higher than the fla typing value of 0.591, but agreed well with an aRI of 0.819. A total of 219 C. jejuni were recovered from farm C, 126 in the winter (31.03.2012) and 93 in the spring (12.06.2012). The fla SVR sequencing identified seven fla SVR sequences, corresponding to five peptide alleles (Table 1). Both samplings from this farm revealed the fla type 287 (Table 1). From the slaughterhouse, 4 out of 219 isolates provided two fla types not present in those C. jejuni from the cloacal swabs (Table 2). At a 60% similarity level, the PFGE clustering showed a high variability with three major clusters. The first cluster included 49 C. jejuni isolated from one rearing cycle (12.06.2012) collected during the farm, slaughter, and postchilling phases; the second cluster contained 11 C. jejuni isolated in two rearing cycles (31.03.2012–12.06.2012) during the farm and slaughter phases; the third cluster included 134 C. jejuni isolated in two rearing cycles (31.03.2012–12.06.2012) during the farm, slaughter, and postchilling phases. At a 100% similarity level, the samples from farm C were divided into 60 different pulsotypes, with three of them comprising 30.85% (58/188) of the isolates and 43 pulsotypes represented by a single isolate (Table 3). For PFGE, the SDI for this cycle was 0.944 while that for the fla type was 0.627 and the aRI displayed a fairly high value of 0.864. The distribution of flaA alleles and peptides isolates in the three farms A, B, and C is summarized in Figure 1. Sixty-nine isolates (9.76%) were untypeable with PFGE, appearing to be a case of DNA smearing rather than restriction.
Table 2.
Number of C. jejuni grouped by flaA type, rearing cycle, and sampling point from farms A, B, and C.
(a).
| flaA type | 12/12/2011 | 12/02/2012 | 10/05/2012 | 17/07/2012 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F | S | C | F | S | C | F | S | C | F | S | C | ||
| Farm A | 1266 | — | — | — | 19 | 4 | — | — | — | — | — | — | — |
| 260 | — | 1 | — | — | — | — | — | — | — | — | — | — | |
| 34 | — | 1 | — | — | — | — | — | — | — | — | — | — | |
| 49 | — | — | — | — | — | — | — | 1 | — | 7 | 20 | — | |
| 1638 | — | 1 | — | 16 | 48 | 3 | 1 | 2 | — | 2 | 15 | 2 | |
| 259 | — | — | — | — | — | — | — | — | — | — | 1 | — | |
| 117 | — | — | — | — | — | — | — | — | — | 1 | — | — | |
| 287 | 9 | 10 | — | — | — | — | 5 | 11 | 1 | — | — | — | |
| 67 | — | — | — | — | 1 | — | 3 | 1 | — | — | — | — | |
| 612 | — | 1 | — | — | — | — | — | — | — | — | — | — | |
| 1284 | — | — | — | — | — | — | 2 | 1 | — | — | — | — | |
| 1429 | 2 | 6 | — | — | — | — | — | — | — | — | — | — | |
| 327 | — | 1 | — | — | — | — | — | — | — | — | — | — | |
| 14 | 15 | 22 | — | — | 1 | — | 1 | 2 | — | — | — | — | |
| 17 | 2 | 1 | — | — | 1 | 1 | 2 | — | — | — | — | — | |
| 30 | — | — | — | — | — | — | — | — | 1 | — | 1 | — | |
| Total | 28 | 44 | 0 | 35 | 55 | 4 | 14 | 18 | 2 | 10 | 37 | 2 | |
(b).
| flaA type | 19/09/2011 | 17/11/2011 | |||||
|---|---|---|---|---|---|---|---|
| F | S | C | F | S | C | ||
| Farm B | 36 | 3 | 13 | 2 | 45 | 49 | 12 |
| 49 | — | — | — | — | 3 | — | |
| 11 | — | 1 | — | — | — | — | |
| 161 | — | 1 | — | — | — | — | |
| 5 | — | — | — | — | 1 | — | |
| 222 | 21 | 29 | 3 | — | — | — | |
| 287 | 6 | 26 | 10 | — | — | — | |
| Total | 30 | 70 | 15 | 45 | 53 | 12 | |
(c).
| flaA type | 31/03/2012 | 12/06/2012 | |||||
|---|---|---|---|---|---|---|---|
| F | S | C | F | S | C | ||
| Farm C | 260 | — | — | — | 3 | 6 | — |
| 23 | — | — | — | — | 1 | — | |
| 265 | — | — | — | 34 | 38 | — | |
| 117 | — | 3 | — | — | — | — | |
| 287 | 5 | 5 | 1 | 1 | 1 | — | |
| 21 | — | — | — | — | 3 | ||
| 222 | 36 | 60 | 19 | — | — | — | |
| 239 | — | — | — | 2 | 1 | — | |
| Total | 41 | 68 | 20 | 40 | 47 | 3 | |
F = cloacal swabs; S = slaughterhouse line; C = postchilling phase.
Table 3.
Distribution of PFGE pulsotypesat 100% similarity according to sampling point from farms A, B, and C.
| PFGE pulsotypes | Cycles | Major PFGE pulsotype | F (Number of isolates/total number of samples) |
S (Number of isolates/total number of samples) |
C (Number of isolates/total number of samples) |
|
|---|---|---|---|---|---|---|
| Farm A | 56 | 12.12.2011 | A | (10/236) | (9/236) | (7/236) |
| D | (7/236) | (5/236) | (1/236) | |||
| 22.02.2012 | B | (22/236) | (31/236) | (5/236) | ||
| 17.07.2012 | B | (5/236) | (1/236) | (1/236) | ||
| C | (8/236) | (5/236) | (0/236) | |||
|
| ||||||
| Farm B | 39 | 19.09.2011 | E | (16/214) | (18/214) | (2/214) |
| F | (5/214) | (12/214) | (8/214) | |||
| 17.11.2011 | G | (10/214) | (14/214) | (0/214) | ||
| H | (17/214) | (8/214) | (5/214) | |||
|
| ||||||
| Farm C | 60 | 31.03.2012 | I | (0/188) | (2/188) | (0/188) |
| L | (14/188) | (5/188) | (0/188) | |||
| M | (0/188) | (5/188) | (12/188) | |||
| 12.06.2012 | I | (1/188) | (9/188) | (0/188) | ||
Sampling point: F = cloacal swabs; S = slaughterhouse line; C = postchilling phase.
Figure 1.
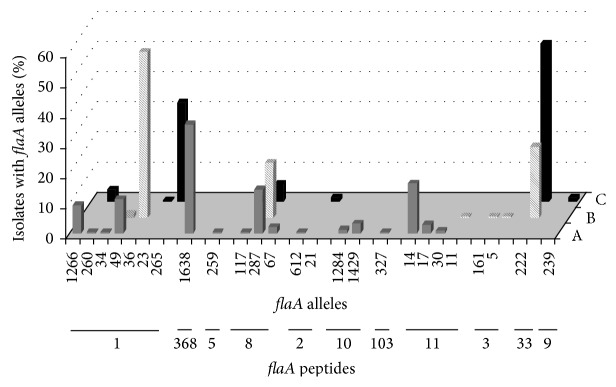
Distribution of flaA alleles and peptides (shown on the x-axis) from farms A, B, and C.
3.3. Antimicrobial Susceptibility Tests
MIC and antimicrobial resistance of all Campylobacter isolates tested in this study are presented in Table 4. The MIC test revealed that 90% of the isolates were resistant to quinolones (NAL and CIP), but 98% were susceptible to chloramphenicol and streptomycin and 99% susceptible to gentamicin. Notably, 64% of the Campylobacter showed resistance to tetracycline, 18% showed resistance to erythromycin, and a few isolates were resistant to other antimicrobials such as chloramphenicol (1.2%), streptomycin (1%), and gentamicin (0.3%). Furthermore, resistance to erythromycin and tetracycline antimicrobials was significantly more frequent in C. coli compared with C. jejuni (P < 0.05, χ 2 test), whereas no differences were observed for the remaining antibiotics.
Table 4.
Antimicrobials, dilution ranges, and cut-off values used for MIC determination of Campylobacter.
| Antimicrobials | MIC breakpoints (µg/mL) | Distribution % of MIC (µg/mL) | MIC50 (µg/mL) | MIC90 (µg/mL) | Number of resistant isolates (%) | Number of resistant C. jejuni (%) | Number of resistant C. coli (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | R | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | ||||||
| Chloramphenicol | ≤8 | ≥32 | 76.5 | 21 | 1.2 | 0 | 1.3 | 2 | 4 | 9 (1.2%) | 8 (1.71%) | 1 (0.34%) | ||||||
|
| ||||||||||||||||||
| Ciprofloxacin | ≤1 | ≥4 | 7.2 | 0.4 | 0 | 0.1 | 0 | 1.9 | 90.4 | 4 | 4 | 696 (90.4%) | 424 (90.79%) | 272 (93.15%) | ||||
|
| ||||||||||||||||||
| Erythromycin | ≤0.5 | ≥8 | 72 | 8.5 | 1.2 | 0.4 | 0.3 | 0.5 | 17.1 | 0.5 | 32 | 139 (18%) | 29 (6.20%) | 110 (41.98%)* | ||||
|
| ||||||||||||||||||
| Gentamicin | ≤4 | ≥16 | 39 | 14.3 | 31 | 13.3 | 1.6 | 0.2 | 0.3 | 0.3 | 0.25 | 1 | 2 (0.3%) | 2 (0.42%) | 0 | |||
|
| ||||||||||||||||||
| Nalidixic acid | ≤16 | ≥32 | 6.6 | 0.7 | 0.7 | 1.6 | 8.4 | 82 | 64 | 64 | 697 (90%) | 428 (91.64%) | 269 (92.12%) | |||||
|
| ||||||||||||||||||
| Streptomycin | ≤2 | ≥8 | 66.5 | 18.2 | 14.3 | 0.4 | 0.6 | 1 | 4 | 8 (1%) | 7 (0.14%) | 1 (0.34%) | ||||||
|
| ||||||||||||||||||
| Tetracycline | ≤4 | ≥16 | 33 | 1.3 | 0.2 | 0.5 | 0.2 | 0.8 | 64 | 16 | 16 | 492 (64%) | 272 (58.24%) | 220 (75.34%)** | ||||
S = sensible, R = resistant.
*Statistically significant versus C. jejuni group (P < 0.05, χ 2 test), **statistically significant versus C. jejuni group (P ≤ 0.01, χ 2 test).
4. Discussion
Over the last five years, campylobacteriosis has become more prevalent in Europe. Campylobacter is found mostly in chicken meat with poultry and poultry farms playing a key role in the epidemiology of human infection [27, 28]. In Italy, a European survey showed a prevalence of Campylobacter-colonized broiler batches of 63.3% [9]. Similar prevalence levels in Italy (60%) have recently been obtained by other studies [29, 30]. The present study aimed to analyse the diversity of the C. jejuni population in poultry and to monitor the contamination process throughout the farm, slaughter, and postchilling phases. The results have shown a very diverse C. jejuni population, even though only three broiler farms from a narrow area were evaluated. A total of 25 flaA-SVR types and 11 flaA peptides were identified among the numerous isolates that were analysed, demonstrating the presence of a heterogeneous population. This is also supported by previous studies where isolates from different continents were assessed by flaA SVR typing revealing a similar degree of diversity [31, 32]. Interestingly, we found a high individual prevalence of Campylobacter, in common with other studies [10]. Tracing back the Campylobacter for each flock showed that the major source of chicken meat contamination remains the flock itself. In the present study, only 10% of the isolates from the abattoir were distinguishable from the live chicken isolates, showing that there were few cases of contamination during slaughtering. Frequently, the most common fla types in live chickens were also the most common genotypes in the processed carcasses and this confirms results reported in previous studies [6, 10, 33, 34]. In a context where all flocks are contaminated, it seems that the slaughterhouse does not play an important role in carcass contamination. However, the situation completely changes when Campylobacter-free flocks meet contaminated flocks at the abattoir. It is therefore sound practice for contaminated poultry flocks to be slaughtered at the end of the working day to contain the cross contamination among the flocks. So diagnostic systems must be able to detect Campylobacter and distinguish uncontaminated from contaminated flocks. The potential of Campylobacter to carry over to succeeding rearing cycles would indirectly suggest its ability to survive within the broiler farm. A comparison of isolates from different samplings for farm A showed that seven fla alleles (83.94%) recurred over a period of almost 8 months. The overlaps of fla genotypes were minor for farms B and C, probably because of the short length of monitoring undertaken, although communities were also demonstrated by the carryover of alleles 36 and 287. To strengthen these findings, we also found that PFGE clustering at 60% of similarity grouped isolates from different rearing cycles. These isolates fell into the same PFGE cluster and featured the same fla allele suspected to be stable over time thus indicating that some isolates were successful in the broilers. The fla SVR analysis also showed that 38.57% of the isolates shared the same fla alleles among the three farms (Figure 1), although a limited correlation between the farms could be argued. fla allele 287, in particular, revealed a PFGE clustering, supporting the hypothesis that all the isolates were strongly related, independently of the farms (Figure 2). This could be explained by cross contamination, probably caused by objects that might transport Campylobacter within the broiler houses most likely during the thinning process. In the present study, the farms were managed as part of a vertically integrated supply chain. Generally, feed mills, breeding farms, hatcheries, and slaughterhouses are owned by the same company and it is probable that the same catching crew could cross contaminate the farms by using unclean crates. Monitoring these practices very thoroughly is required to better address these types of problem. Antibiotic resistance has been a long-standing problem in the field of human and veterinary medicine [31, 35–40] generally related to the indiscriminate use of antibiotics in prophylaxis and therapy or as a growth promoter [39]. Comparative studies of isolates from different geographical areas show a steady and alarming increase in resistance, even to the next generation molecules [31, 32, 37, 40–42]. Particularly worrying is the increase in the frequency of resistance against fluoroquinolones, particularly ciprofloxacin [10], which was confirmed in our study (90%). Moreover our results on resistance against nalidixic acid (90%) and tetracycline (64%) agreed with those in the EFSA Report [42] and other studies [31, 43], confirming this increasing trend. The susceptibility against chloramphenicol (1.2%), streptomycin (1%), and gentamicin (0.3%) could be probably attributable to the lack of extensive use of these drugs in Italy.
Figure 2.
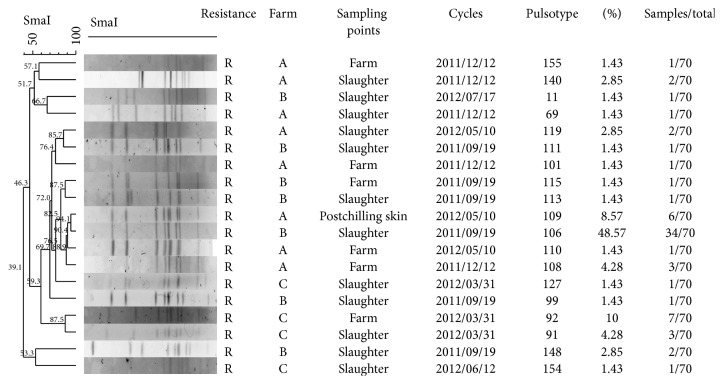
Dendrogram of C. jejuni SmaI PFGE patterns isolated in the three farms characterized by flaA allele 287 and antimicrobial resistance to fluoroquinolones.
5. Conclusions
This study has revealed the usefulness of molecular methods for tracing Campylobacter contamination in the poultry supply chain. These data have provided more information on the presence of Campylobacter clones that have adapted well to poultry and can survive on the farms. The question arises whether Campylobacter has an ecological niche that permits its survival. Several hypotheses have been debated but no data are available to evaluate water supplies and vectors such as flying insects or rodents as potential risk factors involved in the mechanism of contamination [6, 44]. Our results showed a highly diverse C. jejuni population in poultry, suggesting that its introduction or reintroduction on the farm may originate from different sources. Since the main source of poultry meat contamination was confirmed to be the flock, it is reasonable to suggest that Campylobacter-free meat could be achieved by reducing its prevalence at farm level.
Conflict of Interests
The authors declare no conflict of interests relating to this study.
References
- 1.Moore J. E., Barton M. D., Blair I. S., et al. The epidemiology of antibiotic resistance in Campylobacter . Microbes and Infection. 2006;8(7):1955–1966. doi: 10.1016/j.micinf.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 2.FSA (European Food Safety Authority) The European union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA Journal. 2012;12(2, article 3547):p. 312. doi: 10.2903/j.efsa.2014.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buettner S., Wieland B., Staerk K. D. C., Regula G. Risk attribution of Campylobacter infection by age group using exposure modelling. Epidemiology and Infection. 2010;138(12):1748–1761. doi: 10.1017/s095026881000155x. [DOI] [PubMed] [Google Scholar]
- 4.Wingstrand A., Neimann J., Engberg J., et al. Fresh chicken as main risk factor for campylobacteriosis, Denmark. Emerging Infectious Diseases. 2006;12(2):280–284. doi: 10.3201/eid1202.050936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kittl S., Korczak B. M., Niederer L., et al. Comparison of genotypes and antibiotic resistances of Campylobacter jejuni and Campylobacter coli on chicken retail meat and at slaughter. Applied and Environmental Microbiology. 2013;79(12):3875–3878. doi: 10.1128/aem.00493-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newell D. G., Fearnley C. Sources of Campylobacter colonization in broiler chickens. Applied and Environmental Microbiology. 2003;69(8):4343–4351. doi: 10.1128/aem.69.8.4343-4351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofshagen M., Tarpai A., Bruheim T. Annual Report 2010. Oslo, Norway: Norwegian Veterinary Institute; 2011. The surveillance and control programme for Campylobacter spp. in broiler flocks in Norway, Surveillance and control programmes for terrestrial and aquatic animals in Norway. [Google Scholar]
- 8.Jacobs-Reitsma W. F., Bolder N. M., Mulder R. W. Cecal carriage of Campylobacter and Salmonella in Dutch broiler flocks at slaughter: a one-year study. Poultry Science. 1994;73(8):1260–1266. doi: 10.3382/ps.0731260. [DOI] [PubMed] [Google Scholar]
- 9.EFSA. Analysis of the baseline survey on the prevalence of Campylobacter in broiler batches and of Campylobacter and Salmonella on broiler carcasses in the EU, 2008. EFSA Journal. 2010;8:1503–1602. [Google Scholar]
- 10.Colles F. M., McCarthy N. D., Sheppard S. K., Layton R., Maiden M. C. J. Comparison of Campylobacter populations isolated from a free-range broiler flock before and after slaughter. International Journal of Food Microbiology. 2010;137(2-3):259–264. doi: 10.1016/j.ijfoodmicro.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moran L., Scates P. A. M., Madden R. H. Prevalence of Campylobacter spp. in raw retail poultry on sale in Northern Ireland. Journal of Food Protection. 2009;72(9):1830–1835. doi: 10.4315/0362-028x-72.9.1830. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki H., Yamamoto S. Campylobacter contamination in retail poultry meats and by-products in the world: a literature survey: a literature survey. Journal of Veterinary Medical Science. 2009;71(3):255–261. doi: 10.1292/jvms.71.255. [DOI] [PubMed] [Google Scholar]
- 13.Gibbens J. C., Pascoe S. J. S., Evans S. J., Davies R. H., Sayers A. R. A trial of biosecurity as a means to control Campylobacter infection of broiler chickens. Preventive Veterinary Medicine. 2001;48(2):85–99. doi: 10.1016/S0167-5877(00)00189-6. [DOI] [PubMed] [Google Scholar]
- 14.Corry J. E. L., Atabay H. I. Poultry as a source of Campylobacter and related organisms. Journal of Applied Microbiology. 2001;90(30):96S–114S. doi: 10.1046/j.1365-2672.2001.01358.x. [DOI] [PubMed] [Google Scholar]
- 15.Damjanova I., Jakab M., Farkas T., et al. From farm to fork follow-up of thermotolerant campylobacters throughout the broiler production chain and in human cases in a Hungarian county during a ten-months period. International Journal of Food Microbiology. 2011;150(2-3):95–102. doi: 10.1016/j.ijfoodmicro.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Melero B., Juntunen P., Hänninen M.-L., Jaime I., Rovira J. Tracing Campylobacter jejuni strains along the poultry meat production chain from farm to retail by pulsed-field gel electrophoresis, and the antimicrobial resistance of isolates. Food Microbiology. 2012;32(1):124–128. doi: 10.1016/j.fm.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 17.Miller R. S., Miller W. G., Behringer M., Hariharan H., Matthew V., Oyarzabal O. A. DNA identification and characterization of Campylobacter jejuni and Campylobacter coli isolated from caecal samples of chickens in Grenada. Journal of Applied Microbiology. 2010;108(3):1041–1049. doi: 10.1111/j.1365-2672.2009.04507.x. [DOI] [PubMed] [Google Scholar]
- 18.Oyarzabal O. A., Williams A., Zhou P., Samadpour M. Improved protocol for isolation of Campylobacter spp. from retail broiler meat and use of pulsed field gel electrophoresis for the typing of isolates. Journal of Microbiological Methods. 2013;95(1):76–83. doi: 10.1016/j.mimet.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Marshall B. M., Levy S. B. Food animals and antimicrobials: impacts on human health. Clinical Microbiology Reviews. 2011;24(4):718–733. doi: 10.1128/cmr.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ISO FDIS 10272-1. Microbiology of Food and Animal Feeding Stuff—Horizontal for Detection and Enumeration of Campylobacter spp. Part 1: Detection Method. International Organisation for Standardisation (ISO); 2006. [Google Scholar]
- 21.ISO FDIS 10272-2. Microbiology of Food and Animal Feeding Stuff—Horizontal for Detection and Enumeration of Campylobacter spp. Part 2 Enumeration Method. International Organisation for Standardisation (ISO); 2006. [Google Scholar]
- 22.Di Giannatale E., Di Serafino G., Zilli K., et al. Characterization of antimicrobial resistance patterns and detection of virulence genes in Campylobacter isolates in Italy. Sensors (Switzerland) 2014;14(2):3308–3322. doi: 10.3390/s140203308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. http://www.pulsenetinternational.org/protocols.
- 24.de Boer P., Duim B., Rigter A., van der Plas J., Jacobs-Reitsma W. F., Wagenaar J. A. Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli . Journal of Clinical Microbiology. 2000;38(5):1940–1946. doi: 10.1128/jcm.38.5.1940-1946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nachamkin I., Bohachick K., Patton C. M. Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. Journal of Clinical Microbiology. 1993;31(6):1531–1536. doi: 10.1128/jcm.31.6.1531-1536.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 27.Doorduyn Y., van den Brandhof W. E., van Duynhoven Y. T. H. P., Breukink B. J., Wagenaar J. A., van Pelt W. Risk factors for indigenous Campylobacter jejuni and Campylobacter coli infections in the Netherlands: a case-control study. Epidemiology and Infection. 2010;138(10):1391–1404. doi: 10.1017/s095026881000052x. [DOI] [PubMed] [Google Scholar]
- 28.Schielke A., Rosner B. M., Stark K. Epidemiology of campylobacteriosis in Germany—insights from 10 years of surveillance. BMC Infectious Diseases. 2014;14, article 30 doi: 10.1186/1471-2334-14-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Giannatale E., Prencipe V., Colangeli P., et al. Prevalence of thermotolerant Campylobacter in broiler flocks and broiler carcasses in Italy. Veterinaria Italiana. 2010;46(4):405–423. [PubMed] [Google Scholar]
- 30.Comin D., Valero A., Manfreda G., et al. Microbiological criteria for Campylobacter in broiler carcasses in Italy: a possible approach to derive them. International Journal of Food Microbiology. 2014;184:64–68. doi: 10.1016/j.ijfoodmicro.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 31.Wieczorek K., Osek J. Characteristics and antimicrobial resistance of Campylobacter isolated from pig and cattle carcasses in Poland. Polish Journal of Veterinary Sciences. 2013;16(3):501–508. doi: 10.2478/pjvs-2013-0070. [DOI] [PubMed] [Google Scholar]
- 32.Wassenaar T. M., Fernández-Astorga A., Alonso R., et al. Comparison of Campylobacter fla-SVR genotypes isolated from humans and poultry in three European regions. Letters in Applied Microbiology. 2009;49(3):388–395. doi: 10.1111/j.1472-765x.2009.02678.x. [DOI] [PubMed] [Google Scholar]
- 33.Hue O., Allain V., Laisney M.-J., et al. Campylobacter contamination of broiler caeca and carcasses at the slaughterhouse and correlation with Salmonella contamination. Food Microbiology. 2011;28(5):862–868. doi: 10.1016/j.fm.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Ugarte-Ruiz M., Wassenaar T. M., Gómez-Barrero S., Porrero M. C., Navarro-Gonzalez N., Domínguez L. The effect of different isolation protocols on detection and molecular characterization of Campylobacter from poultry. Letters in Applied Microbiology. 2013;57(5):427–435. doi: 10.1111/lam.12130. [DOI] [PubMed] [Google Scholar]
- 35.Rozynek E., Maćkiw E., Kamińska W., et al. Emergence of macrolide-resistant Campylobacter strains in chicken meat in Poland and the resistance mechanisms involved. Foodborne Pathogens and Disease. 2013;10(7):655–660. doi: 10.1089/fpd.2012.1333. [DOI] [PubMed] [Google Scholar]
- 36.Možina S. S., Kurinčič M., Klančnik A., Mavri A. Campylobacter and its multi-resistance in the food chain. Trends in Food Science & Technology. 2011;22(2-3):91–98. doi: 10.1016/j.tifs.2010.09.003. [DOI] [Google Scholar]
- 37.Chen X., Naren G.-W., Wu C.-M., et al. Prevalence and antimicrobial resistance of Campylobacter isolates in broilers from China. Veterinary Microbiology. 2010;144(1-2):133–139. doi: 10.1016/j.vetmic.2009.12.035. [DOI] [PubMed] [Google Scholar]
- 38.Hanning I., Biswas D., Herrera P., Roesler M., Ricke S. C. Prevalence and characterization of Campylobacter jejuni isolated from pasture flock poultry. Journal of Food Science. 2010;75(7):M496–M502. doi: 10.1111/j.1750-3841.2010.01747.x. [DOI] [PubMed] [Google Scholar]
- 39.Usha M. R., Fauziah M., Tunung R., et al. Occurrence and antibiotic resistance of Campylobacter jejuni and Campylobacter coli in retail broiler chicken. International Food Research Journal. 2010;17(2):247–255. [Google Scholar]
- 40.Zhao S., Young S. R., Tong E., et al. Antimicrobial resistance of campylobacter isolates from retail meat in the united states between 2002 and 2007. Applied and Environmental Microbiology. 2010;76(24):7949–7956. doi: 10.1128/aem.01297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.EFSA (European Food Safety Authority) The European Union summary report on trends and sources of zoonoses , zoonotic agents and food-born outbreaks in 2010. EFSA Journal. 2012;10(3):p. 2597. [Google Scholar]
- 42.EFSA. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animal and food in the European Union in 2010. EFSA Journal. 2012;10(3, article 2598) [Google Scholar]
- 43.EFSA. Community summary report on antimicrobial resistance in zoonotic and indicator bacteria from animals and food in the European Union in 2008. EFSA Journal. 2010;8(7):p. 1658. [Google Scholar]
- 44.Bull S. A., Allen V. M., Domingue G., et al. Sources of Campylobacter spp. colonizing housed broiler flocks during rearing. Applied and Environmental Microbiology. 2006;72(1):645–652. doi: 10.1128/aem.72.1.645-652.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]