Abstract
The fungus Sclerotinia sclerotiorum (Lib.) de Bary, one of the most important plant pathogens, causes white mold on a wide range of crops. Crop yield can be dramatically decreased due to this disease, depending on the plant cultivar and environmental conditions. In this study, a suppression subtractive hybridization cDNA library approach was used for the identification of pathogen and plant genes that were differentially expressed during infection of the susceptible cultivar BRS Pérola of Phaseolus vulgaris L. A total of 979 unigenes (430 contigs and 549 singletons) were obtained and classified according to their functional categories. The transcriptional profile of 11 fungal genes related to pathogenicity and virulence were evaluated by reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR). Additionally, the temporal expression profile obtained by RT-qPCR was evaluated for the following categories of plant defense-related genes: pathogenesis-related genes (PvPR1, PvPR2, and PvPR3), phenylpropanoid pathway genes (PvIsof, PvFPS1, and 4CL), and genes involved in defense and stress-related categories (PvLox, PvHiprp, PvGST, PvPod, and PvDox). Data obtained in this study provide a starting point for achieving a better understanding of the pathosystem S. sclerotiorum–P. vulgaris.
Keywords: necrotrophic fungi, common bean, gene expression, Sclerotinia sclerotiorum, suppression subtractive hybridization, stress-induced transcripts
Introduction
Common beans (Phaseolus vulgaris L.) are the world’s most important grain legume for direct human consumption (Broughton et al., 2003). However, bean production is affected by several diseases, such as white mold, which is one of the most important diseases that affect the quality and yield of crops on a global scale. The pathogen associated with white mold, Sclerotinia sclerotiorum (Lib.) de Bary, is a necrotrophic pathogen with worldwide distribution and is known to infect over 400 species of plants (Boland and Hall, 1994). The sclerotia of this pathogen are capable of surviving in the soil for several years and infect the majority of hosts indirectly, i.e., they germinate to produce apothecia, which release ascospores (Bolton et al., 2006). Because of its ability to infect economically important crops causing major losses, S. sclerotiorum has been the focus of many research programs. As a consequence, scientists have successfully annotated its genome (Amselem et al., 2011), investigated the fungus at the level of gene expression, and performed proteome-level studies (Yajima and Kav, 2006).
Previous studies on the pathogenicity of S. sclerotiorum mainly focused on the secretion of oxalic acid (Lumsden, 1979; Godoy et al., 1990) and hydrolytic enzymes (Marciano et al., 1983; Cotton et al., 2003), which act in concert to macerate plant tissues and generate necrosis. Lytic enzymes, such as cellulases, hemicellulases, pectinases, and proteases, sequentially secreted by the fungus facilitate penetration, colonization, and maceration and also generate an important source of nutrients (Hegedus and Rimmer, 2005; Bolton et al., 2006). However, oxalic acid is likely to have more important roles because it suppresses the oxidative burst and resistance of the host plant and triggers mediated apoptotic-like programmed cell death (PCD; Kim et al., 2008). In the compatible interactions between S. sclerotiorum and its host plant, host cells maintain viability and a suppression of the oxidative burst is observed, which is akin to compatible biotrophic pathogens during the early stage of infection (Williams et al., 2011; Kabbage et al., 2013, 2015). Furthermore, direct acidification within the middle lamella enhances the activity of many cell wall-degrading enzymes, including polygalacturonases (PGs; Riou et al., 1991). Secretion of oxalic acid may help inactivate plant PG-inhibiting proteins, thereby allowing the pathogen to overcome this specific host defense response (Favaron et al., 2004). Nonetheless, despite extensive studies on S. sclerotiorum, its pathogenic mechanisms are still incompletely understood because its pathogenesis is very complex (Hegedus and Rimmer, 2005; Amselem et al., 2011; Williams et al., 2011).
Several attempts to genetically study the pathosystem S. sclerotiorum–P. vulgaris have been performed to establish methods to detect physiological resistance (Kolkman and Kelly, 2000; Schwartz and Singh, 2013). The infection process and establishment of compatible interactions have also been well characterized at the cytological level (Lumsden, 1979; Lumsden and Wergin, 1980; Tariq and Jeffries, 1986). Differential accumulation of specific defense-related transcripts, such as mRNA for polygalacturonase-inhibiting protein (PGIP) and pathogen related proteins, during the S. sclerotiorum–P. vulgaris interaction has also been reported (Oliveira et al., 2010, 2013; Kalunke et al., 2011).
Increasing our knowledge of the infection strategies of necrotrophic pathogens, in general, and of S. sclerotiorum, specifically, should be helpful in designing novel rational disease control strategies. This study, in which genes differentially expressed in the S. sclerotiorum–P. vulgaris interaction were identified and analyzed, provides a starting point for achieving a better understanding of the pathosystem S. sclerotiorum–P. vulgaris.
Materials and Methods
Plant Material and Pathogen Inoculation
Dry bean plants (Phaseolus vulgaris L. cv. BRS Pérola), which are susceptible to S. sclerotiorum, were grown in 2-kg plastic pots containing soil fertilized with NPK (4-39-16) (1 g/kg of soil) in a greenhouse at 24 ± 2°C with 60 ± 5% relative humidity and a 16/8 h light/dark period.
Sclerotinia sclerotiorum isolate SPS was collected from a naturally infected dry bean plant and grown on Petri dishes containing potato-dextrose agar (PDA) culture medium for 5 days at 20°C, and 3-mm plugs of this culture were used to inoculate the axillary region of dry bean plants at the flowering stage (R6), which is the main stage of infection in field. The control group was composed of a set of plants mock-inoculated with sterile agar plugs. All of the plants were kept at 20°C and 90% relative humidity to provide adequate conditions for infection. Tissue samples were collected from both the necrotic part and the chlorotic area of each lesion within 10 mm of the lesion edge at 6-, 12-, 24-, 48-, and 72-h post inoculation (hpi) and immediately frozen in liquid nitrogen prior to RNA and protein extraction. The experimental design was completely randomized and consisted of three biological replicates for each of the treatments. For each of the experimental conditions, stem segments (∼10 mm) from 20 plants different plants were pooled together to form one of three biological replicates.
RNA Extraction
Total RNA was extracted from the plant tissues (stem segment) and fungal material using the standard Trizol protocol (Invitrogen Corp., Carlsbad, CA, USA). Total RNA samples were examined quantitatively using the Qubit fluorometry assay (Invitrogen Corp., Carlsbad, CA, USA) and qualitatively using agarose gel electrophoresis.
Two different RNA mixtures were prepared as the source of the tester and driver samples for the suppressive subtractive hybridization (SSH) library construction. For the tester samples, equal amounts of the total RNA isolated from the stem tissues inoculated with S. sclerotiorum at 6, 12, 24, 48, and 72 hpi were mixed. In the case of the driver samples, RNA from non-inoculated stem tissues harvested at the same times (6–72 h) plus RNA extracted from the mycelium of the fungus [grown separately in minimal medium (2 g/L NH4NO3, 1 g/L KH2PO4, 0.1 g/L MgSO4.7H2O, 0.5 g/L yeast extract, 3 g/L DL-malic acid, 1 g/L NAOH) supplemented with 1% glucose] were mixed at a 2:1 ratio (estimated based on qPCR experiments – data not shown).
Suppressive Subtractive Hybridization Library Construction
For subtractive hybridization, 1.0 μg of each total RNA mixture was employed to produce double-stranded cDNA using the Super SMARTTM cDNA Synthesis Kit (Clontech Laboratories Inc., Palo Alto, CA, USA) according to the manufacturer’s instructions. To determine which genes were differentially expressed during the interaction with S. sclerotiorum, a forward subtractive cDNA library was constructed using the PCR-SelectTM cDNA Subtraction Kit (Clontech Laboratories Inc., Palo Alto, CA, USA) according to the standard protocol provided. The subtracted cDNA population was cloned into a PCR 4®-TOPO vector (Invitrogen Corp., Carlsbad, CA, USA) and used to transform One Shot® Top 10 EletrocompTM Escherichia coli cells (Invitrogen Corp., Carlsbad, CA, USA). The plasmid DNA was obtained using alkaline lysis. The cloned products were sequenced using a M13 forward primer and the Big Dye Terminator v 3.1 Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA, USA). The automated capillary electrophoresis sequencing runs were performed on an ABI prism 3130 analyzer (Applied Biosystems, Carlsbad, CA, USA).
Bioinformatic Analyses of Expressed Sequence Tags (ESTs)
The sequences were pre-processed using Phred (Ewing and Green, 1998) and Cross Match software1. Sequences with at least 100 nucleotides and a Phred quality greater than or equal to 20 were considered for further analysis. Cleaned ESTs were assembled into contigs using the CAP3 assembly program and compared against the GenBank non-redundant (nr) database using the BLASTx algorithm available at the National Center for Biotechnology Information2. Functional annotation using Gene Ontology terms3 was analyzed with the Blast2GO program (Conesa and Götz, 2008). KEGG pathway annotation was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database4.
Primers were designed using Software Primer Express (Applied Biosystems, Carlsbad, CA, USA) for the amplification of gene fragments that were approximately 80–150 bp in length and with an annealing temperature of 60°C (Table 1). The primer specificity was checked in silico against the NCBI database2 through the Primer-BLAST tool. The selected primers were tested with PCR, and their specificity was checked by nucleotide sequencing on an ABI 3130 sequencer (Applied Biosystems, Carlsbad, CA, USA) using DyeTerminator chemistry 4 to confirm their identities.
Table 1.
Oligonucleotide primer pairs used for quantitative real-time polymerase chain reaction (RT-qPCR) analysis.
| Gene | Description/Sequence name | Forward/Reverse |
|---|---|---|
| Fungal Primers | ||
| Sspg1 | Endopolygalacturonase 1 | F: TCTTGCAGCAGTCGAGAAG R: GTGTTGTGTCCGAGGGAGT |
| Sspg3 | Endopolygalacturonase 3 | F: ACCCACCACTTTGGCTACTG R: TGAGACGGTAAGACCCTTG |
| Sspg6 | Endopolygalacturonase 6 | F: AAGCTTATTGGAATGGGTAT R: CTGGAGTTGACGATTTGACTA |
| SsBgsidase | Glucan endo-1,3-β-glucosidase/SS_038 | F: CTGACGGTTCGACCCTGTAT R: GTACTCCCAACGGAGAACCA |
| β-1,4 | β-1,4-glucanase/SS_041 | F: CAAGGCAGCTTAACCGCTAC R: GATCCATCGGAGTCGAGGTA |
| acp1 | Acid protease/SS_011 | F: GCCACCCAAAACGGAGAAT R: GAGGTGAGGACGGAGTTTTGTT |
| aspS | Aspartyl protease/SS_014 | F: TGCTACTGGGTCCAACATCGT R: TGCGCTTGATGCACTTGGT |
| ScXylo | β-xylosidase/SS_024 | F: CGCTCTTTTTCCCCATACAA R: AGCGATGCGTATCTTCGAGT |
| Cellobio | Cellobiohydrolase/SS_091 | F: ACTCTCTGCCCTGATGCAGT R: AGGAGTGTAAGCGGCAGAAA |
| oah | Oxaloacetate acetylhydrolase | F: CCCAATCGTCGAGGACAAGC R: TGCCTGCTCCGGTCATGTAA |
| Perilipin | Perilipin/SS_008 | F: GAAATTCGGCAAGCCATTTA R: TGCCCTTTGAAATCGGATAG |
| gpdh | Glyceraldehyde-3-phosphate dehydrogenase | F: TGGCTCCTACTAAAGTTG R: CAAGCAGTTGGTTGTGCAAG |
| Plant primers | ||
| PvPR1 | Pathogenesis-related PR-1 | F: AAAGCCAAGAGCGATTCTCTTTTCA R: GAACACTCTGATTTGATAACACTTC |
| PvPR2 | B1-3 endoglucanase | F: GAAGATGAGCtCAAAGCTGGTAA R: CAAGGATTGGCCAAAAGGTA |
| PvPR3 | Chitinase class I | F: ATTGTTGTGCCAATCCCTTT R: CACCGCCATACAGTTCAAAA |
| PvPAL | Phenylalanine ammonia-lyase | F: GACACACAAGTTGAAGCACCA R: TGCAGCTTCTTAGCATCCTTC |
| PvLOX | Lipoxygenase | F: AGCACTGTGCCTGTTTTCAGT R: AACACACGAGAAGATTCAACCA |
| PvDOX | α-dioxigenase | F: CACAAATCCTCCCAAAATGG R: AAAGGTTCACACCATTGATTGG |
| PvFPS1 | Farnesyl pyrophosphate syntase 1 | F: CGGAATAGACGTTGAGGAATG R: AACACCTACCCGAATTTCTGC |
| PvISOF | Isoflavonoid glucosyltransferase | F: AGCTGAAACACAGCACCAACT R: AACACCATGCTTGGCAAATAG |
| Pvcallose | Callose synthase-like protein | F: TGGCTTAGTATTCGGATGTACCA R: CGTTGTAATGAAAGCGAAGTGT |
| Pv4CL | 4-coumarate CoA-ligase | F: AGGTTGTTGGTGCTGAGAATG R: CCAACCAAGTCAAAGATTCCA |
| PvHPRP | Hypersensitive-induced response protein | F: ATTGCATGGTTCATAGCCAGT R: CCTCCACACAAGTATCAAAGGA |
| PvGST | Glutatione S-transferase | F: AGCTCTTCAAGGACACTGAGCCAA R: AAAGGCTGTGGATGCTGCACTAGA |
| PvPOD | Peroxidase | F: TCCTTTTCAGCACTTTCACT R: AGAAAGCAGTGTTCTTGTGG |
| Act11 | Actin11 | F: TGCATACGTTGGTGATGAGG R: AGCCTTGGGGTTAAGAGGAG |
Reverse Transcriptase – quantitative PCR (RT-qPCR)
RNA was extracted from plant tissues and fungal material using the standard Trizol protocol (Invitrogen Corp., Carlsbad, CA, USA). After DNase I (Invitrogen Corp., Carlsbad, CA, USA) treatment in the presence of a RNase inhibitor (Invitrogen Corp., Carlsbad, CA, USA), equal amounts of RNA (1 μg) were reverse-transcribed using an oligo(dT)12–18 primer and evaluated with qPCR. RT-qPCR analysis was carried out using the StepOnePlusTM System and Power SYBR® Green PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA) in 10 μL reactions containing 0.4 μM of each oligonucleotide (Table 1), 6 μL of SYBR Green PCR Master mix (2x), and 0.2 μL of template cDNA. After initial denaturation at 95°C for 10 min, amplifications were performed for 40 cycles at 95°C for 15 s and at 60°C for 1 min. To check the specificity of the PCR product, the melting curves were analyzed for each data point. The relative expression levels of the target genes were calculated using the ΔΔCt method (Livak and Schmittgen, 2001) and the reference genes Actin-11 for P. vulgaris (Borges et al., 2012a) and gpdh for S. sclerotiorum. The reference sample was chosen to represent 1x expression of the gene of interest. Three samples were analyzed for each treatment. The values are expressed as the mean ± standard deviation (SD).
Light Microscopy
Resin sections of stems inoculated with fungal samples (12–48 hpi) and without inoculation (control) were prepared for histological observation using light microscopy according to the method described by Mendoza et al. (1993). Briefly, the samples were dehydrated using a graded alcohol series, embedded in a mixture of absolute ethanol and resin (1:1, v/v; Historesin, Leica Microsystems Nußloch GmbH, Heidelberg, Germany) for 4 h, and stored for 24 h in pure Historesin. The resin was polymerized using a hardener, and the samples were sectioned into 8-μm thick slices with a rotary microtome. The sections were stained with 0.05% toluidine blue for 2 min and mounted. Tissue sections were photographed using a Leica DM500 microscope with a Leica ICC50 camera and the Leica Application Suite (LAS) EZ V3.0.0 software (Leica Microsystems, Switzerland).
Results
Disease Development: Time Course Analysis of S. sclerotiorum Infection
To evaluate the S. sclerotiorum behavior in the interaction with P. vulgaris cv BRS Pérola, stem tissues of inoculated plants were sampled at 6-, 12-, 24-, 48-, and 72-hpi. The pathogen showed an infection capacity at 12 hpi, although the tissue from the inoculated plants remained healthy (Figure 1A). Necrotic spots appeared on stem tissues after 24 hpi (Figure 1B), and small necrotic areas were clearly visible at 48 hpi (Figure 1C). In addition, at 72 hpi, late extensive necrosis was observed in the vascular tissues of inoculated stems (Figure 1D).
FIGURE 1.
Plant disease symptoms observed (arrow) in Phaseolus vulgaris stems inoculated with plugs of Sclerotinia sclerotiorum PDA culture. (A) 12 hpi. (B) 24 hpi. (C) 48 hpi. (D) 72 hpi.
Functional Classification of the Significantly Differentially Expressed Genes
Aiming to isolate the pathogen and plant genes that were differentially expressed during the S. sclerotiorum–P. vulgaris interaction, the susceptible common bean cultivar BRS Pérola was used, and suppression subtractive hybridization (SSH) was employed for the generation of a cDNA library. The SSH library was constructed using cDNA from stem tissues inoculated with S. sclerotiorum as the tester and cDNA from non-inoculated stem tissue plus RNA from the fungus grown in minimal medium as the driver.
A total of 1440 plasmids were sequenced. After removing the vector and adaptor sequences, the cleaned expressed sequence tags (ESTs) were assembled into contigs using the CAP3 assembly program comprising a total of 979 unigenes (representing 430 contigs and 549 singletons). The putative function of the 979 ESTs was assigned using the BLASTX program of the NCBI nr database employing the Blast2GO program with an E-value cutoff of <1e-06 and the Gene Ontology database.
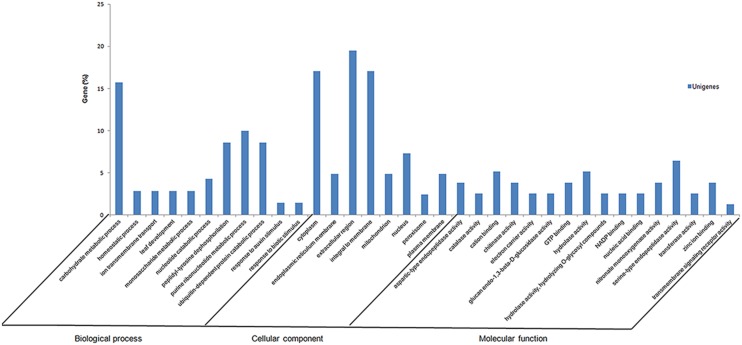
Initially, to identify metabolic processes that are specifically modulated by fungal infection, the 979 unigenes were classified into functional categories (biological processes, cellular components, and molecular functions) according to their putative biological functions reported by the Gene Ontology database (Figure 2). Of 979 unigenes derived from the S. sclerotiorum–P. vulgaris interaction, 29% were categorized as fungal, and 58.3% as plant genes. Of these, 12.7% had no significant match and therefore could not be classified.
FIGURE 2.
Summary of Gene Ontology annotation as assigned by Blast2GO. Functional annotation of the expressed sequence tags (ESTs) in Phaseolus vulgaris infected with Sclerotinia sclerotiorum SSH library using gene ontology terms (http://www.geneontology.org) by Blast2GO program. Most consensus sequences were grouped into major functional categories: biological process, cellular component, and molecular function.
The SSH library was normalized to reduce the redundancy of the most highly expressed genes. Nevertheless, some genes are very highly expressed and thus remain overrepresented in the normalized library. The number of reads/contig and their length distribution are shown in Supplementary Table S1. Most contigs were assembled from two to three reads. As expected, the largest contigs (i.e., those composed of most sequences) are involved in stress response pathways, such as contig 161, which has 28 sequences encoding a binuclear zinc transcription factor (Colletotrichum gloeosporioides).
Gene Expression Profiles in S. sclerotiorum during P. vulgaris Infection
A total of 979 unigenes (representing 430 contigs and 549 singletons) were generated from the S. sclerotiorum–P. vulgaris interaction and aligned to the Sclerotinia genome database (Broad Institute, Cambridge, MA, USA) for the identification and screening of fungal sequences. Preliminary analysis of the fungal unigenes (163 contigs and 120 singletons) revealed a total of 67 unique fungal ESTs in this database. The proteins encoded by this unique EST set were assigned to the following major categories: cell wall degradation, protein degradation, intracellular transport, oxidative stress, among others (Table 2). Global analysis detected a small but significant group of genes related to virulence during the penetration stages of development (6–72 hpi).
Table 2.
Annotation of Sclerotinia sclerotiorum expressed sequence tags (ESTs) (contigs) derived from its interaction with Phaseolus vulgaris.
| Contig | Accession numbera | Name and/or putative function | Organism | E -value |
|---|---|---|---|---|
| Cell wall degradation | ||||
| Contig 27 – SS_024 | SS1G_01493 | β-xylosidaseb | S. sclerotiorum | 3e-15 |
| Contig 45 – SS_041 | SS1G_13255.3 | β-1,4-glucanaseb | S. sclerotiorum | 6e-73 |
| Contig 57 – SS_052 | SS1G_08474 | β-1,3-glucanase precursor | S. sclerotiorum | 6e-05 |
| Contig 98 – SS_091 | SS1G_07146.3 | Cellobiohydrolaseb | S. sclerotiorum | 2e-40 |
| Contig 124 – SS_116 | SS1G_12937 | Glycosyl hydrolase | S. sclerotiorum | 2e-32 |
| Contig 145 – SS_136 | SS1G_07393 | β-1,3-glucanase | S. sclerotiorum | 1e-64 |
| Contig 42 – SS_038 | SS1G_02789 | Glucan endo-1,3-β-glucosidaseb | S. sclerotiorum | 3e-22 |
| Contig 33 – SS_029 | SS1G_01005 | α-glucosidaseputative | S. sclerotiorum | 3e-40 |
| Contig 18 – SS_016 | SS1G_12021 | β-1,6-glucanase putative | S. sclerotiorum | 5e-18 |
| Protein degradation | ||||
| Contig 25 – SS_022 | SS1G_0336 | Serine peptidase putative | S. sclerotiorum | 3e-43 |
| Contig 26 – SS_023 | SS1G_06534 | Serin endopeptidase | S. sclerotiorum | 2e-53 |
| Contig 16 – SS_014 | SS1G_03181 | Aspartyl proteaseb | S. sclerotiorum | 2e-20 |
| Contig 13 – SS_011 | SS1G_00624 | Acid proteaseb | S. sclerotiorum | 8e-15 |
| Contig 62 – SS_057 | SS1G_11818 | Vacuolar protease A | S. sclerotiorum | 4e-53 |
| Contig 74 – SS_068 | SS1G_00477 | Ubiquitin homeostasis protein lub1 (phospholipase) | S. sclerotiorum | 1e-12 |
| Contig 63 – SS_058 | SS1G_0770 | Proteasome component PRE6 | S. sclerotiorum | 3e-47 |
| Contig 61 – SS_056 | SS1G_01331 | Translocation protein Sec62 putative | S. sclerotiorum | 1e-22 |
| Intracellular transport | ||||
| Contig 40 – SS_036 | SS1G_07860 | Protein SEY1 | S. sclerotiorum | 6e-42 |
| Contig 77 – SS_071 | SS1G_09620 | Hypothetical protein (GTPase) | S. sclerotiorum | 2e-04 |
| Contig 103 – SS_096 | SS1G_10229 | GTP-binding protein EsdC | S. sclerotiorum | 1e-51 |
| Contig 160 – SS_150 | SS1G_08784 | GTP-binding protein | S. sclerotiorum | 6e-33 |
| Contig 148 – SS_139 | SS1G_11149 | SEC24 related gene family (GTPase) | S. sclerotiorum | 3e-31 |
| Contig 9 – SS_007 | SS1G_02490 | DENN domain-containing protein | S. sclerotiorum | 2e-32 |
| Contig 86 – SS_080 | SS1G_09635 | Geranylgeranyl pyrophosphate synthetase (biosynthesis of secondary metabolites) | S. sclerotiorum | 5e-25 |
| Oxidative stress | ||||
| Contig 53 – SS_048 | SS1G_14466 | Oxidoreductase, 2-nitropropane dioxygenase family | S. sclerotiorum | 9e-19 |
| Contig 44 – SS_040 | SS1G_12928 | Peroxidase/catalase | S. sclerotiorum | 2e-38 |
| Contig 88 – SS_082 | SS1G_10037 | Cytochrome P450 | S. sclerotiorum | 4e-29 |
| Contig 161 – SS_151 | SS1G_06754 | Zinc transcription factor | S. sclerotiorum | 4e-42 |
| Contig 153 – SS_143 | SS1G_11235 | 2-nitropropane dioxygenase | S. sclerotiorum | 2e-44 |
| Others | ||||
| Contig 10 – SS_008 | SS1G_06319 | Perilipin MPL1- like protein CAP 20b | S. sclerotiorum | 1e-07 |
| Contig 97 – SS_090 | SS1G_03197 | pH-response regulator protein | S. sclerotiorum | 2e-09 |
| Contig 75 – SS_069 | SS1G_10538 | Ceramidase | S. sclerotiorum | 6e-20 |
| Contig 142 – SS_133 | SS1G_00477 | Phospholipase | S. sclerotiorum | 1e-12 |
a GenBank database (http://www.ncbi.nlm.nih.gov)
b Validated by RT-qPCR
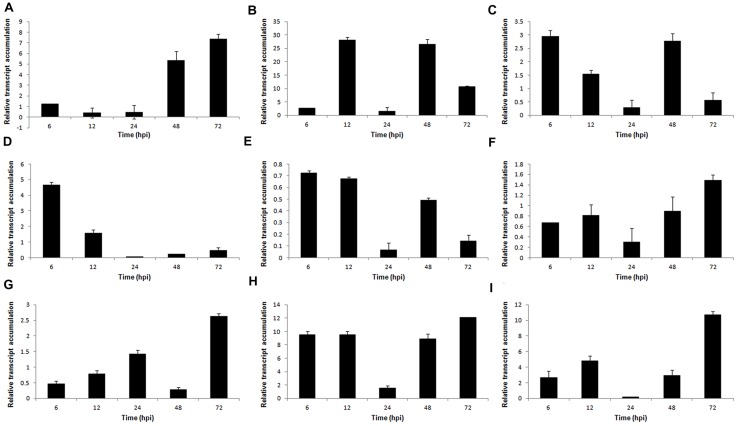
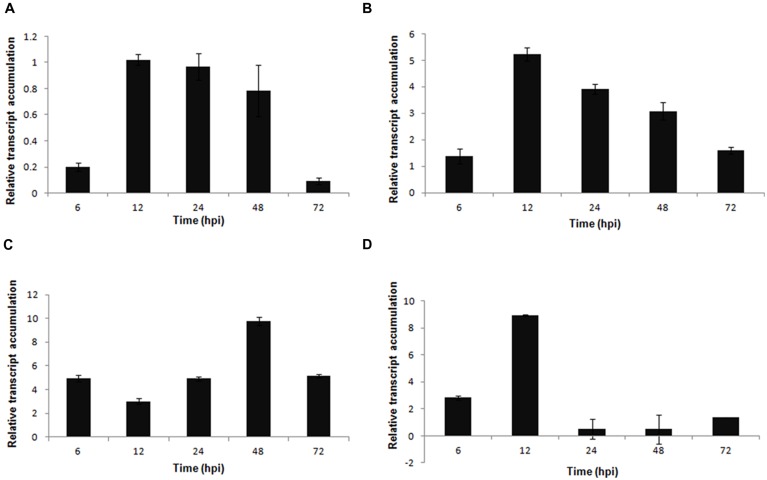
To gain a better understanding of the mechanisms of S. sclerotiorum pathogenesis, 9 genes from the S. sclerotiorum–P. vulgaris SSH library related to plant cell wall degradation were selected for expression patterns analysis: PGs (Sspg1, Sspg3, and Sspg6); cellulases (β-1,3-glucosidase, β-1,4-glucanase, and cellobiohydrolase); hemicellulases (β-xylosidase); and proteases (aspartyl protease – aspS and acid protease – acp1). These genes were evaluated at 6, 12, 24, 48, and 72 hpi by reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR). The relative expression levels of all genes tested were normalized to the S. sclerotiorum gpdh gene.
The PG Sspg1 gene showed higher transcript levels in the late stage of the interaction. Sspg3 expression was induced at 12 hpi and was reduced at 72 hpi. The Ssp6 gene showed higher transcript levels at the early stages (6–12 hpi) after inoculation (Figures 3A–C). Previously, we showed that the Sspg1 and Sspg5 genes are highly expressed during S. sclerotiorum infection of P. vulgaris plants and also that the Sspg3 gene is expressed during the initial phase of colonization, whereas Sspg6 is weakly expressed during this process (Oliveira et al., 2010).
FIGURE 3.
Gene expression profiles of S. sclerotiorum during infection to P. vulgaris. Expression analyses using RT-qPCR were performed and transcript levels were calculated in triplicate using a comparative method. GPDH gene was used as the reference gene in S. sclerotiorum and a health plant as the reference sample. Tissue samples were collected at 6, 12, 24, 48, and 72 hpi. Results are reported as means ± standard deviation (SD) of three samples for each treatment. (A) Sspg1. (B) Sspg3. (C) Sspg6. (D) β-1,3-glucosidase. (E) β-xylosidase. (F) β-1,4-glucanase. (G) cellobiohydrolase. (H) acp1 (acid protease). (I) aspS (aspartyl protease).
The genes encoding β-1,3-glucosidase and β-xylosidase were also upregulated during the early stages of the S. sclerotiorum–P. vulgaris interaction (Figures 3D,E). Conversely, the genes encoding β-1,4-glucanase and cellobiohydrolase showed increased expression levels in the later stages of infection (Figures 3F,G). The gene acp1, which encodes an acid protease, was highly expressed from the beginning of infection, and presented the highest level of expression at 72 hpi. A similar profile was also observed for the aspS gene, which encodes an aspartyl protease (Figures 3H,I).
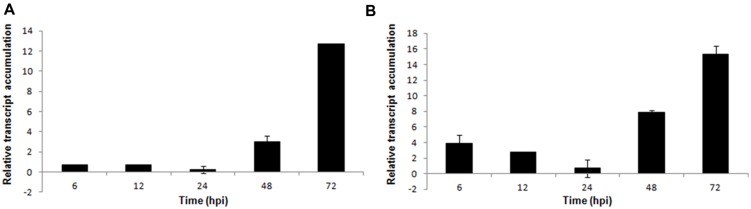
In S. sclerotiorum, as well as in other fungi, oxalic acid is produced from oxaloacetate in a reaction catalyzed by oxaloacetate acetylhydrolase (OAH). In this work, oah transcripts were detected in the early stage of infection (6 hpi), and its expression was higher in the late stage (72 hpi; Figure 4A).
FIGURE 4.
Gene expression profiles of S. sclerotiorum during infection to P. vulgaris. Expression analyses using RT-qPCR were performed and transcript levels were calculated in triplicate using a comparative method. GPDH gene was used as the reference gene in S. sclerotiorum and a health plant as the reference sample. Tissue samples were collected at 6, 12, 24, 48, and 72 hpi. Results are reported as means ± standard deviation (SD) of three samples for each treatment. (A) oah (oxaloacetate acetylhydrolase). (B) perilipin.
Additionally, 15 unigenes (5 contigs and 10 singlets) that encode perilipin-like protein were detected in the S. sclerotiorum–P. vulgaris SSH library. Perilipin-like protein is largely responsible for the regulation of lipid storage and metabolism in filamentous fungi (Duan et al., 2013). Our analysis showed that although this gene was upregulated starting at the early stage of infection, the highest expression level was observed in the final stage (72 hpi; Figure 4B).
Defense Genes Expressed and Regulated Over Time in the Common Bean Stem During Interaction with S. sclerotiorum
As a result, the majority of the cDNAs from the SSH library were derived from P. vulgaris. In total 570 unigenes (267 contigs and 303 singletons) showed significant similarities to plant sequences, that indicates that 58.3% (979 unigenes) of our database represent common bean genes. The list of P. vulgaris the most common contigs/reads and their length distribution are shown in Supplementary Table S1. The proteins encoded by the unique EST set were assigned to five major categories: metabolism, energy, protein degradation, response to biotic and abiotic stimulus, and others. An important group of the identified genes are related to biotic and abiotic stress responses, among these are PR1 (pathogenesis related protein 1), and peroxidase genes. Although most of the defense-related genes were not detected in our EST collection (979 unigenes), primers were designed based on the bean transcriptome sequence (Melotto et al., 2005) to investigate the plant response during the interaction with S. sclerotiorum. We searched for ESTs that may play an important role in the defense response to S. sclerotiorum disease and can be used as reference for comparison among different pathosystems.
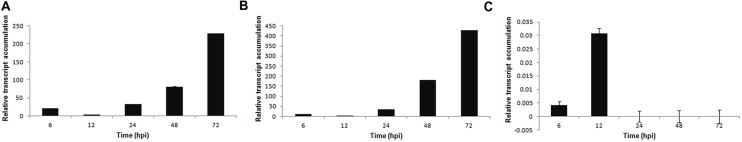
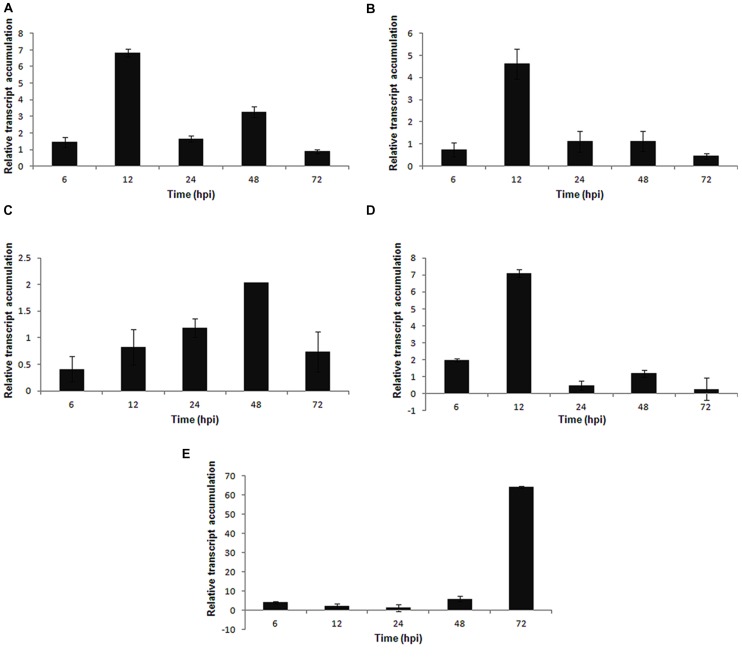
We evaluated the temporal expression profile of the three main categories of defense-related genes, namely: (1) pathogenesis-related (PR) genes PvPR1 (function unknown), PvPR2 (β-1,3-endoglucanase), and PvPR3 (chitinase); (2) phenylpropanoid pathway genes, such as PvISOF (isoflavonoid glucosyltransferase), PvFPS1 (farnesyl pyrophosphate synthetase 1), PvPAL (phenylalanine ammonia-lyase), and Pv4CL (4-coumarate CoA-ligase), which are involved in phytoalexin biosynthesis; and (3) genes involved in defense and stress-related categories, such as PvLOX (lipoxygenase), PvHIPRP (hypersensitive-induced response protein), PvGST (glutathione S-transferase), PvPOD (peroxidase), and PvDOX (alpha-dioxygenase). The time-course expression patterns were determined using control and infected samples at 6, 12, 24, 48, and 72 hpi. The relative expression levels of all of the genes were normalized using the P. vulgaris Actin 11 gene (Act11) as the reference gene (Borges et al., 2012a).
Significant fold-change differences in expression levels were observed for all of the selected genes in the infected plant (fungus-inoculated) compared with the mock-inoculated plant. The temporal expression of the PR genes PvPR1 (function unknown) and PvPR2 (β-1,3-endoglucanase) showed a similar expression profile. Both genes presented a basal expression during the early stages of infection (6–12 hpi) and a high expression level at 72 hpi; however, at this time, the expression of PvPR2 was two-fold higher than that of PvPR1 (Figures 5A,B). The expression of the PvPR3 gene was observed only in the early stages of infection (6–12 hpi), but at lower levels compared with the other PvPR genes analyzed (Figure 5C).
FIGURE 5.
Expression pattern of pathogenesis-related genes in P. vulgaris in response to S. sclerotiorum infection. Expression analyses using RT-qPCR were performed and transcript levels were calculated in triplicate using a comparative method. Actin-11 gene was used as the reference gene in P. vulgaris and a health plant as the reference sample. Tissue samples were collected at 6, 12, 24, 48, and 72 hpi. Results are reported as means ± standard deviation of three samples for each treatment. (A) PvPR1 (pathogenesis related protein 1). (B) PvPR2 (pathogenesis related protein 2). (C) PvPR3 (pathogenesis related protein 3).
The genes involved in the phenylpropanoid pathway, PvISOF (isoflavonoid glucosyltransferase), PvFPS1 (farnesyl pyrophosphate synthetase 1), PvPAL (phenylalanine ammonia-lyase), and Pv4CL (4-coumarate CoA-ligase), were upregulated in dry bean plants during the interaction with S. sclerotiorum. The highest levels of expression were registered for PvPAL (a two-fold increase compared with PvFPS1 and a 10-fold increase compared with PvISOF at 48 hpi (Figures 6A–C) and Pv4CL at 12 hpi (Figure 6D).
FIGURE 6.
Expression pattern of phenylpropanoid pathway genes in P. vulgaris in response to S. sclerotiorum infection. Expression analyses using RT-qPCR were performed and transcript levels were calculated in triplicate using a comparative method. Actin-11 gene was used as the reference gene in P. vulgaris and a health plant as the reference sample. Tissue samples were collected at 6, 12, 24, 48, and 72 hpi. Results are reported as means ± standard deviation of three samples for each treatment. (A) PvISOF (isoflavonoid glucosyltransferase). (B) PvFPS1 (farnesyl pyrophosphate synthetase 1). (C) PvPAL (phenylalanine ammonia-lyase). (D) Pv4CL (4-coumarate CoA-ligase).
Among the genes involved in the defense and stress-related categories, PvLOX was activated in the early stages of infection and reached a maximum expression at 12 hpi (Figure 7A). The PvHIPRP gene, which is related to hypersensitive responses (HR), was induced at the early stages of infection and peaked in expression at 12 hpi (Figure 7B). The PvGST (glutathione S-transferase) gene presented a progressive increase in expression up to 48 hpi, followed by a decrease at 72 hpi (Figure 7C), whereas the PvPOD (peroxidase) gene had a maximum expression at 12 hpi (Figure 7D). Finally, the PvDOX gene, which encodes α-dioxygenase, showed a high level of expression at 72 hpi (Figure 7E) in the S. sclerotiorum–P. vulgaris pathosystem.
FIGURE 7.
Expression pattern of defense and stress related genes in P. vulgaris plant in response to S. sclerotiorum infection. Expression analyses by RT-qPCR were performed and transcript levels were calculated in triplicate using a comparative method. Actin-11 gene was used as the reference gene in P. vulgaris and a health plant as the reference sample. Tissue samples were collected at 6, 12, 24, 48, and 72 hpi. Results are reported as means ± standard deviation of three samples for each treatment. (A) PvLOX (lipoxygenase). (B) PvHIPRP (hypersensitive-induced response protein). (C) PvGST (glutathione S-transferase). (D) PvPOD (peroxidase). (E) PvDOX (alpha-dioxygenase).
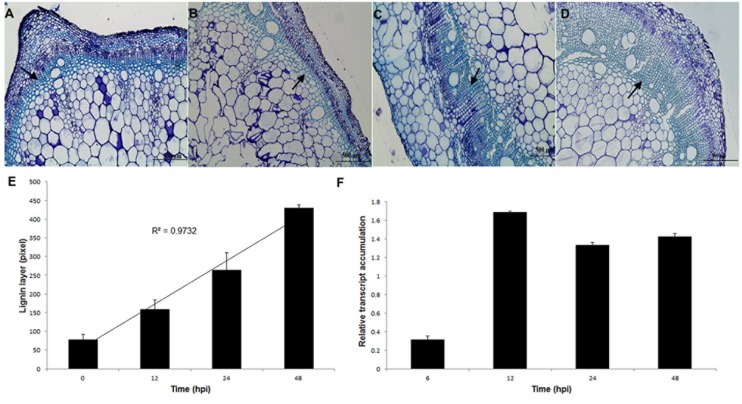
Distinct vascular changes in the stem were noticed in response to S. sclerotiorum infection. Histological sections of bean stem showed that under S. sclerotiorum infection (12–72 hpi; Figures 8B–D), bean plants had evident lignification, which was not observed in the (Figure 8A) control (mock-inoculated). The uppermost 3–4 cell layers of the epidermis were stained a greenish–blue color with Aniline blue, indicating an accumulation of phenolic compounds. Thickness of lignin layer (Figure 8E) was measured using Leica LAS EZ V3.0.0 software (Leica Application Suite, Leica Microsystems, Switzerland).
FIGURE 8.
Thickening layers of lignin in P. vulgaris stems in response to S. sclerotiorum infection. (A) Without infection. (B) 12 hpi. (C) 24 hpi. (D) 48 hpi. Bar = 500 μm. (E) Thickness of lignin layer (arrows) measured using Leica LAS EZ V3.0.0 software (Leica Application Suite, Leica Microsystems, Switzerland). (F) Relative transcript accumulation of Pvcallose gene. Expression analyses using RT-qPCR were performed and transcript levels were calculated in triplicate using a comparative method. Actin-11 gene was used as the reference gene in P. vulgaris and a health plant as the reference sample. Results are reported as means ± standard deviation of three experiments.
To assess whether the callose gene (Pvcallose) was modulated during the S. sclerotiorum invasion of P. vulgaris stem tissues, the level of its transcription was evaluated using RT-qPCR. The results indicate that this gene was activated in the early stages of infection (6 hpi) and showed an increased accumulation of transcripts from 12 hpi until 48 hpi (Figure 8F).
Discussion
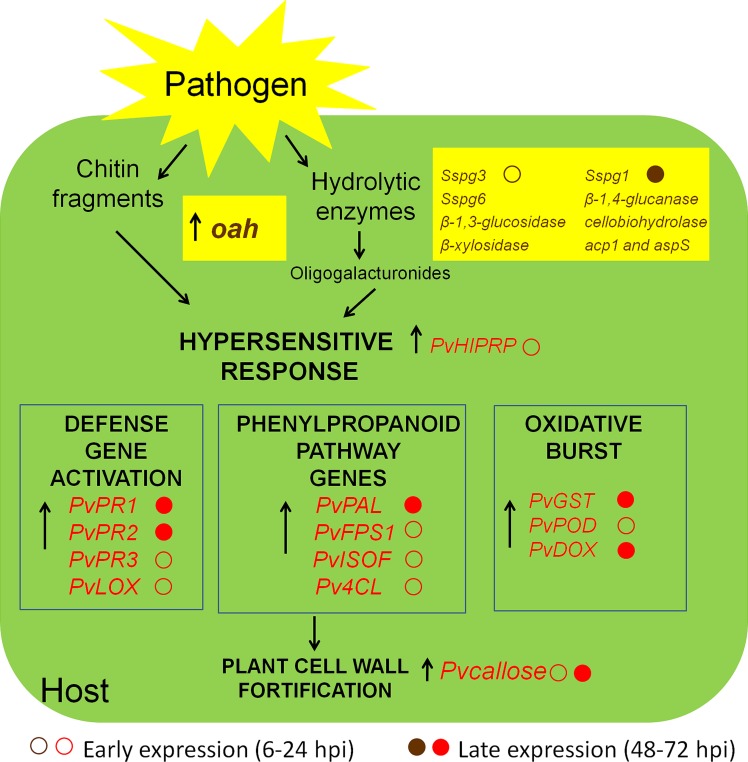
Although S. sclerotiorum is considered to be a typical necrotroph, there is evidence that it colonizes plant tissues through multiple phases involving important transcriptional and physiological reprogramming (Hegedus and Rimmer, 2005; Kabbage et al., 2013, 2015; Guyon et al., 2014). In this study, we evaluated the infective process of S. sclerotiorum in P. vulgaris plants. The gene expression profiles of stem tissues in the inoculation site were examined over 3 days. The steady-state levels of all of the identified genes were quantified at the time of inoculation (0 hpi), at 2 early time points following inoculation (12 and 24 hpi) and at somewhat later time points (48 and 72 hpi), at which point necrotic lesions develop on the stems and intensive mycelial colonization occurs. From our transcript profile results, we propose the model illustrated in Figure 9, which depicts a plausible interpretation of the observed expression patterns of the S. sclerotiorum–P. vulgaris interaction.
FIGURE 9.
Schematic representation of the interaction between S. sclerotiorum and P. vulgaris. In the beginning of the pathogenic process, the fungus secretes hydrolytic enzymes (polygalacturonases, PGs) and oxalic acid. PGs degrade plant cell walls and release oligogalacturonides (OGs), which are elicitors of defense response. The hypersensitive response (HR) of the plant is used by the fungus as a strategy to promote virulence. Chitin fragments of fungus and OGs released from the plant itself allow the recognition of the presence of the pathogen and activate transcription factors which lead to the expression of genes belonging to different pathways of defense. The arrows indicate genes up-regulated in RTqPCR analysis. In brown, S. sclerotiorum genes, and in red, plant response genes.
The recent description of the genome sequences of two fungal necrotrophs, S. sclerotiorum and Botrytis cinerea, revealed 346 and 367 genes, respectively, encoding putative carbohydrate-active enzymes (CAZymes), with over 100 genes potentially associated with plant cell wall degradation (Amselem et al., 2011). In this work, we observed that many genes encoding cell wall-degrading enzymes: PGs (Sspg1, Sspg3, and Sspg6), cellulases (β-1,3-glucosidase, β-1,4-glucanase, and cellobiohydrolase) and hemicellulases (β-xylosidase), were found to be differentially expressed and showed accumulated transcripts in the inoculated sample.
A >2-fold induction in expression was measured at 12 hpi for Sspg3 (28-fold), Sspg6-encoding endo-PGs (3-fold) and β-1,3-glucosidase genes (4.8-fold), even though no necrotic symptoms in the stem are visible at this time. PGs enable a pathogen to invade plant tissues; however, on the other hand, their activity may also trigger plant defense responses. Namely, the degradation of homogalacturonan, the main component of pectin, by PGs results in the release of oligogalacturonides (OGs), which are capable of inducing plant immune reactions. The inactivation of PG activity by plant PGIPs has been shown to reduce disease development (Powell et al., 2000). More recent attempts to delay disease by expressing PGIPs, which are plant proteins that inhibit fungal endopolygalacturonases, have been successful against neurotropic fungi (Borras-Hidalgo et al., 2012).
The increase in β-1,4-glucanase (1.5-fold) and cellobiohydrolase (2.8-fold) transcript accumulation coincided with the phase of symptom development (48 – 72 hpi) in which intensive mycelial colonization of the stem occurred and soaked lesions were observed.
Proteases secreted by this fungus can degrade host plant proteins, some of which are involved in defense responses to fungal inoculation, and also seem to be correlated with symptom development (Bolton et al., 2006). During the S. sclerotiorum–P. vulgaris interaction the expression of the gene (acp1), encoding an acid protease, and the gene aspS, encoding an aspartyl protease, was low at the beginning of infection but increased at the stage of spreading necrosis. Proteases are also regarded as antagonists of the antifungal proteins secreted as part of the defense response by the host. In previous reports, Poussereau et al. (2001) showed that acp1 and aspS were expressed in planta during sunflower cotyledon infection.
In this work, oxaloacetate acetylhydrolase (oah) transcripts were already detected in the early stages of infection (6 hpi), and its expression was increased at the late stage of infection (72 hpi) (Figure 4A). Corroborating this, Liang et al. (2014) recently demonstrated that the Ss-oah1 gene, which encodes an oxaloacetate acetylhydrolase, is required for oxalic acid accumulation and affects the pH-responsive growth, morphogenesis and virulence of S. sclerotiorum on a variety of plant hosts. Recently, oxalic acid was found to create reducing conditions in plant cells ahead of advancing hyphae. It was speculated that reductive conditions dampen the oxidative burst, allowing for precious time for fungal establishment prior to plant recognition (Kim et al., 2008). Also Kabbage et al. (2013, 2015) suggested that S. sclerotiorum, a prototypical necrotroph, has a biotrophic phase that occurs during the initial stages of disease establishment. During the early stages, oxalate dampens the host oxidative burst, an early response associated with plant defense, as part of a potential biotrophic interaction, before triggering generation of reactive oxygen species (ROS) at the later stages of infection, culminating in PCD, the advanced necrotrophic phase of the interaction and disease.
The results presented in this study show that dry bean plants respond to inoculation with S. sclerotiorum in the early stages of the process. The PvHIPRP gene, related to HR, was induced at the early stages of infection in infected tissue and had its expression peak at 12 hpi (4.8-fold). The importance of the HR for successful infection by necrotrophic pathogens is exemplified by studies showing that plants unable to undergo PCD (Dickman et al., 2001) or incapable of generating a HR (Govrin and Levine, 2000) are more resistant to such pathogens.
Among the genes involved in the defense and stress-related categories, LOX was activated in the early stages of infection (12 hpi – 6.8-fold). Lipoxygenase (LOX) catalyzes the oxidation of unsaturated fatty acids, producing oxylipins, which play a pivotal role in plant defense by acting as signaling molecules and/or protective compounds (Blée, 2002).
Plants that have to cope with oxidative stress can improve their ROS scavenging capacity via the upregulation of related enzymatic activities, such as glutathione S-transferase (Mhamdi et al., 2010), catalase, superoxide dismutase, and peroxidase (Nanda et al., 2010). In the present study, PvPOD (peroxidase) was induced at 6 hpi and peaked in expression at 12 hpi (7.5-fold). The PvGST (glutathione S-transferase) and PvDOX (α-dioxygenase) genes presented a progressive increase in expression following infectious development. The plants lacking a dioxigenase, DOX2 (α-DOX2-deficient mutant), which catalyzes oxylipin production from fatty acids, were more susceptible to Botrytis cinerea (Angulo et al., 2015). During the interaction with Rhizoctonia solani–P. vulgaris, this gene was highly expressed, and it was suggested that this enzyme protects plant tissues undergoing excessive necrosis associated with oxidative stress during pathogenesis (Guerrero-González et al., 2011).
Once the plant has identified the attack, it can respond by activating the synthesis of a diverse number of proteins. These proteins include antibiotic proteins, such as PR proteins. The genes encoding the PvPR1 and PvPR2 protein families increased in expression 250-fold and 450-fold at 72 hpi, respectively, following fungal infection. Similarly, Borges et al. (2012b) reported the upregulation of PvPR1 and a β-1,3-glucanase (PvPR2) during the incompatible interaction between the common bean and Colletotrichum lindemuthianum. Although this interaction is not the same, we found a similar expression profile in our experiment in which PvPR1 and PvPR2 were activated in the later stages of infection.
PR1 proteins have frequently been used as markers for systemic-acquired resistance (SAR) and have been associated with antifungal properties, such as the hydrolysis of fungal cell walls. These signaling events occur normally by hormones such as salicylic acid (SA), ethylene and jasmonic acid (JA), which can activate PR genes (Edreva, 2005; Ferreira et al., 2007; Leon-Reyes et al., 2009).
The expression of the PvPR3 gene (chitinases) was observed only in the early stages of infection (6–12 hpi). PvPR3, like PvPR2 (β-1,3-endoglucanase), may act directly by inhibiting the growth of the pathogen or indirectly by aiding in the generation of signaling molecules that may function as elicitors of further defensive mechanisms (Edreva, 2005; Van Loon et al., 2006; Ferreira et al., 2007).
Apart from inducible proteins, active plant defenses against pathogens also include phenylpropanoid pathway enzymes, which are known to be involved in plant disease resistance. The comparative analysis of the bean plant inoculated with S. sclerotiorum and the mock-inoculated plant (without infection) proved that the PvISOF (isoflavonoid glucosyltransferase), PvFPS1 (farnesyl pyrophosphate synthetase 1), PvPAL (phenylalanine ammonia-lyase), and Pv4CL (4-coumarate CoA-ligase) genes, which are involved in the phenylpropanoid pathway, were upregulated during infectious development.
PAL is the primary enzyme involved in the phenylpropanoid biosynthetic pathway and is responsible for the production of phenolic compounds, such as flavonoids, phytoalexins, lignins, and benzoic acid derivatives (Zhao et al., 2007). Several studies have demonstrated that the levels of PAL activity have important functions in the plant defense response to pathogen infection. PvPAL gene expression was induced by anthracnose fungal attack in P. vulgaris (Borges et al., 2012b), and PAL-suppressed plants were more susceptible to fungal infection (Maher et al., 1994).
In this study, the expression level of the Pv4CL gene was transient during the analyzed time period, with the greatest upregulation (9.5-fold) in the early stages (12 hpi) of infection. The 4-coumarate CoA-ligase is the third enzyme of the phenylpropanoid pathway in plants, and leads to the synthesis of lignin, pigments, and many defense molecules (Zabala et al., 2006).
In accordance with these findings, the results obtained here with aniline blue staining showed a progressive thickening of the lignin layers in dry bean plants during S. sclerotiorum infection. Certain phenylpropanoid compounds are polymerized to form defensive barriers, such as lignin, which is a crucial component of the plant defense repertoire against abiotic and biotic stress factors (Dixon et al., 2002). It has been strongly demonstrated that lignification is a common phenomenon in the expression of disease resistance in plants. Lignin synthesis is induced in response to mechanical damage or wounding, and many plants respond to invading pathogens with the deposition of lignin and lignin-like material (Vance et al., 1980; Nicholson and Hammerschmidt, 1992; Boudet et al., 1995; Dixon and Paiva, 1995). Additionally, callose is an effective barrier induced at the site of attack during the early stages of pathogen invasion and is an established marker associated with incompatible responses (Williams et al., 2011).
Conclusion
This study contributes to a better understanding of the kinetics of induced defenses against a fungal pathogen of the common bean, as well fungal genes related to pathogenicity and virulence, and provides a valuable first step toward the understanding and analysis of the Phaseolus vulgaris – S. sclerotiorum interaction.
Author Contributions
MO carried out the experiments and drafted the manuscript. RA participated in the transcription analysis experiments. MG participated in the design of the study and drafted the manuscript. SP conceived the study and participated in its design and coordination. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação de Amparo à Pesquisa do Estado de Goiás (FAPEG). The authors are thankful to: Dr Murillo Lobo Junior for experimental support and Embrapa Arroz e Feijão, for providing the seeds and preparing the plant material; the authors gratefully acknowledge the support of CNPq for the grants provided (project number 56.4675/2010-5).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01162
References
- Amselem J., Cuomo C. A., van Kan J. A., Viaud M., Benito E. P., Couloux A., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230 10.137/journal.pgen.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo C., de la O Leyva M., Finiti I., López-Cruz J., Fernández-Crespo E., García-Agustín P., et al. (2015). Role of dioxygenase α-DOX2 and SA in basal response and in hexanoic acid-induced resistance of tomato (Solanum lycopersicum) plants against Botrytis cinerea. J. Plant Physiol. 175 163–173. 10.1016/j.jplph.2014.11.004 [DOI] [PubMed] [Google Scholar]
- Blée E. (2002). Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 7 315–322. 10.1016/S1360-1385(02)02290-2 [DOI] [PubMed] [Google Scholar]
- Boland G., Hall R. (1994). Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 16 93–108. 10.1080/07060669409500766 [DOI] [Google Scholar]
- Bolton M. D., Thomma B. P., Nelson B. D. (2006). Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7 1–16. 10.1111/j.1364-3703.2005.00316.x [DOI] [PubMed] [Google Scholar]
- Borges A., Tsai S. M., Caldas D. G. G. (2012a). Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep. 31 827–838. 10.1007/s00299-011-1204-x [DOI] [PubMed] [Google Scholar]
- Borges A., Melotto M., Tsai S. M., Caldas D. G. G. (2012b). Changes in spatial and temporal gene expression during incompatible interaction between common bean and anthracnose pathogen. J. Plant Physiol. 169 1216–1220. 10.1016/j.jplph.2012.04.003 [DOI] [PubMed] [Google Scholar]
- Borras-Hidalgo O., Caprari C., Hernandez-Estevez I., De Lorenzo G., Cervone F. (2012). A gene for plant protection: expression of a bean polygalacturonase inhibitor in tobacco confers a strong resistance against Rhizoctonia solani and two oomycetes. Front. Plant Sci. 3:268 10.3389/fpls.2012.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet A. M., Lapierre C., Grima-Pettenati J. (1995). Biochemistry and molecular biology of lignification. New Phytol. 129 203–236. 10.1111/j.1469-8137.1995.tb04292.x [DOI] [PubMed] [Google Scholar]
- Broughton W. J., Hernandez G., Blair M., Beebe S., Gepts P., Vanderleyden J. (2003). Beans (Phaseolus spp.)–model food legumes. Plant Soil 252 55–128. 10.1023/A:1024146710611 [DOI] [Google Scholar]
- Conesa A., Götz S. (2008). Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008 619832 10.1155/2008/619832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton P., Kasza Z., Bruel C., Rascle C., Fèvre M. (2003). Ambient pH controls the expression of endopolygalacturonase genes in the necrotrophic fungus Sclerotinia sclerotiorum. FEMS Microbiol. Lett. 227 163–169. 10.1016/S0378-1097(03)00582-2 [DOI] [PubMed] [Google Scholar]
- Dickman M. B., Park Y. K., Oltersdorf T., Li W., Clemente T., French R. (2001). Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc. Natl. Acad. Sci. U.S.A. 98 6957–6962. 10.1073/pnas.091108998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. A., Achnine L., Kota P., Liu C. J., Reddy M. S. S., Wang L. (2002). The phenylpropanoid pathway and plant defence-a genomics perspective. Mol. Plant Pathol. 3 371–390. 10.1046/j.1364-3703.2002.00131.x [DOI] [PubMed] [Google Scholar]
- Dixon R. A., Paiva N. L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7 1085–1097. 10.1105/tpc.7.7.1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z., Chen Y., Huang W., Shang Y., Chen P., Wang C. (2013). Linkage of autophagy to fungal development, lipid storage and virulence in Metarhizium robertsii. Autophagy 9 538–549. 10.4161/auto.23575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edreva A. (2005). Geration and scavenging of reactive oxygen species in chloroplasts: a submolecular approach. Agric. Ecosyst. Environ. 106 119–133. 10.1016/j.agee.2004.10.022 [DOI] [Google Scholar]
- Ewing B., Green P. (1998). Base-calling of automated sequencer traces using Phred. II. error probabilities. Genome Res. 8 186–194. 10.1101/gr.8.3.186 [DOI] [PubMed] [Google Scholar]
- Favaron F., Sella L., D’Ovidio R. (2004). Relationships among endo-polygalacturonase, oxalate, pH, and plant polygalacturonase-inhibiting protein (PGIP) in the interaction between Sclerotinia sclerotiorum and soybean. Mol. Plant Microbe Interact. 17 1402–1409. 10.1094/MPMI.2004.17.12.1402 [DOI] [PubMed] [Google Scholar]
- Ferreira R. B., Monteiro S., Freitas R., Santos C. N., Chen Z., Batista L. M., et al. (2007). The role of plant defence proteins in fungal pathogenesis. Mol. Plant Pathol. 8 677–700. 10.1111/j.1364-3703.2007.00419.x [DOI] [PubMed] [Google Scholar]
- Godoy G., Steadman J. R., Dickman M. B., Dam R. (1990). Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol. Mol. Plant Pathol. 37 179–191. 10.1016/0885-5765(90)90010-U [DOI] [Google Scholar]
- Govrin E. M., Levine A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10 751–757. 10.1016/S0960-9822(00)00560-1 [DOI] [PubMed] [Google Scholar]
- Guerrero-González M. L., Rodríguez-Kessler M., Rodríguez-Guerra R., González-Chavira M., Simpson J., Sanchez F., et al. (2011). Differential expression of Phaseolus vulgaris genes induced during the interaction with Rhizoctonia solani. Plant Cell Rep. 30 1465–1473. 10.1007/s00299-011-1055-5 [DOI] [PubMed] [Google Scholar]
- Guyon K., Balagué C., Roby D., Raffaele S. (2014). Secretome analysis reveals effector candidates associated with broad host rang necrotrophy in fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics 15:336 10.1186/1471-2164-15-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus D. D., Rimmer S. R. (2005). Sclerotinia sclerotiorum: when “to be or not to be” a pathogen? FEMS Microbiol. Lett. 251 177–184. 10.1016/j.femsle.2005.07.040 [DOI] [PubMed] [Google Scholar]
- Kabbage M., Williams B., Dickman M. B. (2013). Cell Death Control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 9:e1003287 10.1371/journal.ppat.1003287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage M., Yarden O., Dickman M. B. (2015). Pathogenic attributes of Sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 233 53–60. 10.1016/j.plantsci.2014.12.018 [DOI] [PubMed] [Google Scholar]
- Kalunke R. M., Janni M., Sella L., David P., Geffroy V., Favaron F., et al. (2011). Transcript analysis of the bean polygalacturonase-inhibiting protein gene family reveals that Pvpgip2 is expressed in the whole plant and is strongly induced by pathogen infection. J. Plant Pathol. 93 141–148. 10.4454/jpp.v93i1.284 [DOI] [Google Scholar]
- Kim K. S., Min J. Y., Dickman M. B. (2008). Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant Microbe Interact. 21 605–612. 10.1094/MPMI-21-5-0605 [DOI] [PubMed] [Google Scholar]
- Kolkman J. M., Kelly J. D. (2000). An indirect test using oxalate to determine physiological resistance to white mold in common bean. Crop Sci. 40 281–285. 10.2135/cropsci2000.401281x [DOI] [Google Scholar]
- Leon-Reyes A., Spoel S. H., De Lange E. S., Abe H., Kobayashi M., Tsuda S., et al. (2009). Ethylene modulates the role of nonexpressor of pathogenesis-related genes1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 149 1797–1809. 10.1104/pp.108.133926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Liberti D., Li M., Kim Y. T., Hutchens A., Wilson R., et al. (2014). Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 16 559–571. 10.1111/mpp.12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lumsden R. (1979). Histology and physiology of pathogenesis in plant diseases caused by Sclerotinia species. Phytopathology 69 890–895. 10.1094/Phyto-69-890 [DOI] [Google Scholar]
- Lumsden R., Wergin W. P. (1980). Scanning-electron microscopy of infection of bean by species of Sclerotinia. Mycologia 72 1200–1209. 10.2307/3759575 [DOI] [Google Scholar]
- Maher E. A., Bate N. J., Ni W., Elkind Y., Dixon R. A., Lamb C. J. (1994). Increased disease susceptibility of transgenic tobacco plants with suppressed levels of preformed phenylpropanoid products. Proc. Natl. Acad. Sci. U.S.A. 91 7802–7806. 10.1073/pnas.91.16.7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciano P., Di Lenna P., Magro P. (1983). Oxalic acid, cell wall-degrading enzymes and pH in pathogenesis and their significance in the virulence of two Sclerotinia sclerotiorum isolates on sunflower. Physiol. Plant Pathol. 22 339–345. 10.1016/S0048-4059(83)81021-2 [DOI] [Google Scholar]
- Melotto M., Monteiro-Vitorello C. B., Bruschi A. G., Camargo L. E. (2005). Comparative bioinformatic analysis of genes expressed in common bean (Phaseolus vulgaris L.) seedlings. Genome 48 562–570. 10.1139/g05-010 [DOI] [PubMed] [Google Scholar]
- Mendoza A. B., Hattori K., Nishimura T., Futsuhara Y. (1993). Histological and scanning electron microscopic observations on plant regeneration in mungbean cotyledon (Vigna radiata (L.) Wilczek) cultured in vitro. Plant Cell Tissue Organ Cult. 32 137–143. 10.1007/BF00029835 [DOI] [Google Scholar]
- Mhamdi A., Hager J., Chaouch S., Queval G., Han Y., Taconnat L., et al. (2010). Arabidopsis glutathione reductase1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 153 1144–1160. 10.1104/pp.110.153767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda A. K., Andrio E., Marino D., Pauly N., Dunand C. (2010). Reactive oxygen species during plant-microorganism early interactions. J. Integr. Plant Biol. 52 195–204. 10.1111/j.1744-7909.2010.00933.x [DOI] [PubMed] [Google Scholar]
- Nicholson R. L., Hammerschmidt R. (1992). Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 30 369–389. 10.1146/annurev.py.30.090192.002101 [DOI] [Google Scholar]
- Oliveira M. B., Barbosa S. C., Petrofeza S. (2013). Comparative in vitro and in planta analyses of extracellular enzymes secreted by the pathogenic fungus Sclerotinia sclerotiorum. Genet. Mol. Res. 12 1796–1880. 10.4238/2013 [DOI] [PubMed] [Google Scholar]
- Oliveira M. B., Nascimento L. B., Junior M. L., Petrofeza S. (2010). Characterization of the dry bean polygaiacturonase-inhibiting protein (PGIP) gene family during Sclerotinia sclerotiorum (Sclerotiniaceae) infection. Genet. Mol. Res. 9 994–1004. 10.4238/2013.June.6.3 [DOI] [PubMed] [Google Scholar]
- Poussereau N., Gente S., Rascle C., Billon-Grand G., Fèvre M. (2001). aspS encoding an unusual aspartyl protease from Sclerotinia sclerotiorum is expressed during phytopathogenesis. FEMS Microbiol. Lett. 194 27–32. 10.1111/j.1574-6968.2001.tb09441.x [DOI] [PubMed] [Google Scholar]
- Powell A. L., van Kan J., ten Have A., Visser J., Greve L. C., Bennett A. B., et al. (2000). Transgenic expression of pear PGIP in tomato limits fungal colonization. Mol. Plant Microbe Interact. 13 942–950. 10.1094/MPMI.2000.13.9.942 [DOI] [PubMed] [Google Scholar]
- Riou C., Freyssinet G., Fevre M. (1991). Production of cell wall-degrading enzymes by the phytopathogenic fungus Sclerotinia sclerotiorum. Appl. Environ. Microbiol. 57 1478–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz H. F., Singh S. P. (2013). Breeding common bean for resistance to white mold: a review. Crop Sci. 53 1832–1844. 10.2135/cropsci2013.02.0081 [DOI] [Google Scholar]
- Tariq V. N., Jeffries P. (1986). Ultrastructure of penetration of Phaseolus spp. by Sclerotinia sclerotiorum. Can. J. Bot. 64 2909–2915. 10.1139/b86-384 [DOI] [Google Scholar]
- Vance C. P., Kirk T. K., Sherwood R. T. (1980). Lignification as a mechanism of disease resistance. Annu. Rev. Phytopathol. 18 259–288. 10.1146/annurev.py.18.090180.001355 [DOI] [Google Scholar]
- Van Loon L. C., Rep M., Pieterse C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44 135–162. 10.1146/annurev.phyto.44.070505.143425 [DOI] [PubMed] [Google Scholar]
- Williams B., Kabbage M., Kim H. J., Britt R., Dickman M. B. (2011). Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7:e1002107 10.1371/journal.ppat.1002107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima W., Kav N. N. V. (2006). The proteome of the phytopathogenic fungus Sclerotinia sclerotiorum. Proteomics 6 5995–6007. 10.1002/pmic.200600424 [DOI] [PubMed] [Google Scholar]
- Zabala G., Zou J., Tuteja J., Gonzalez D. O., Clough S. J., Vodkin L. O. (2006). Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection. BMC Plant Biol. 6:26 10.1186/1471-2229-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., She X., Du Y., Liang X. (2007). Induction of antiviral resistance and stimulary effect by oligochitosan in tobacco. Pestic. Biochem. Physiol. 87 78–84. 10.1016/j.pestbp.2006.06.006 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.