Abstract
Although the thermodynamics of protein denaturation at temperatures over 100 °C is essential for the rational design of highly stable proteins, it is not understood well because of the associated technical difficulties. We designed certain hydrophobic mutant proteins of CutA1 from Escherichia coli, which have denaturation temperatures (Td) ranging from 101 to 113 °C and show a reversible heat denaturation. Using a hydrophobic mutant as a template, we successfully designed a hyperthermostable mutant protein (Td = 137 °C) by substituting six residues with charged ones. Thermodynamic analyses of these mutant proteins indicated that the hydrophobic mutants were stabilized by the accumulation of denaturation enthalpy (ΔH) with no entropic gain from hydrophobic solvation around 100 °C, and that the stabilization due to salt bridges resulted from both the increase in ΔH from ion-ion interactions and the entropic effect of the electrostatic solvation over 113 °C. This is the first experimental evidence that has successfully overcome the typical technical difficulties.
The tertiary structures of proteins, which are vital for their physiological functions, are related to amino-acid sequences and stabilized by thermodynamic rules. To elucidate the mechanisms of protein folding and protein stabilization, it is critically important to obtain the thermodynamic parameters of protein denaturation as a function of temperature. One difficulty for studying the stabilization mechanism of proteins with denaturation temperatures above 100 °C is that the heat denaturation of proteins is usually irreversible at temperatures higher than 80 °C1,2,3,4,5,6,7,8, because under these conditions proteins generally aggregate after heat denaturation. Thus, the thermodynamic features of protein stabilization at temperatures above 100 °C are not well understood. One important question in this field is whether the hydrophobic interactions that make the largest contributions to protein stability9,10,11,12,13 still occur at temperatures above 100 °C14,15. Next, it is necessary to perform thermodynamic analyses of salt bridges in proteins that have denaturation temperatures above 100 °C, because many proteins from hyperthermophiles seem to be stabilized by an abundance of ion pairs formed by charged residues8,16,17,18,19,20,21,22,23,24,25,26. Thermodynamics of protein denaturation at temperatures over 100 °C is also essentially important for the rational design of hyperthermostable proteins that would be highly useful for industrial and bio-technological processes.
The CutA1 protein from the hyperthermophile Pyrococcus horikoshii (PhCutA1) has an unusually high stability, with a denaturation temperature (Td) of nearly 150 °C at pH 7.0 and an unusually high content of charged residues26,27. The stabilities and structures of CutA1 proteins from species with various growth temperatures have been also examined, including those of Thermus thermophilus (TtCutA1)28, Oryza sativa (OsCutA1)28, Homo sapiens (brain) (HsCutA1)29, and Escherichia coli (EcCutA1)30. Their Td values are also unusually high relative to the growth temperatures of each species: 113.9 °C for TtCutA1, 98.9 °C for OsCutA1, 96.2 °C for HsCutA1, and 89.9 °C for EcCutA1. The X-ray crystal structure of PhCutA1 clearly resembles those of other CutA1 proteins. The monomeric structure consists of three α-helices and five β-strands. Three monomers are assembled into a trimer through interactions between the edges of three β-strands (Supplementary Fig. 1). This tightly intertwined interaction contributes to the stabilization of the trimer structures for the CutA1 proteins26.
EcCutA1 and its mutants are not heat-reversible30. However, other CutA1 proteins, such as TtCutA1 and OsCutA1, which have fewer cysteine sulfhydryl (SH) groups, exhibit remarkable heat reversibility28. EcCutA1 has three SH groups per subunit. Therefore, we designed an SH-free EcCutA1 mutant, referred to as EcCutA1_0SH (Cys16 → Ala, Cys39 → Ala, Cys79 → Ala), with the aim of achieving high heat reversibility.
In this study, we used the EcCutA1_0SH protein, which has excellent heat reversibility, as a template to design thermostabilized mutants. First, we constructed hydrophobic mutants from EcCutA1_0SH, which were effective in our previous work30. Then, to achieve a hyperthermostability comparable to that of PhCutA1, we designed ionic mutants in which charged residues were introduced into a hydrophobic mutant (EcCutA1_0SH_S11V/E61V) by substitution. We describe our use of these EcCutA1 mutants to assess the thermodynamic characteristics of proteins at temperatures over 100 °C, in regard to both hydrophobic and ion–ion interactions.
Results
Hydrophobic mutants of EcCutA1 with no SH group
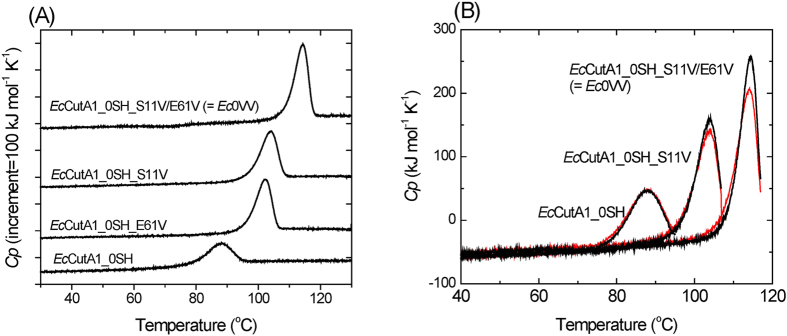
We constructed hydrophobic mutants with no SH groups (EcCutA1_0SH_S11V, EcCutA1_0SH_E61V, EcCutA1_0SH_S11V/E61V), which we expected to increase stability30. Figure 1A shows typical differential scanning calorimetry (DSC) curves of EcCutA1_0SH and its hydrophobic mutant without SH groups at pH 9.0. As shown in Fig. 1B, the reheating curve (second scan) of EcCutA1_0SH agrees completely with the first scan. The other two proteins also exhibited good reproducibility. These results indicate that the removal of SH groups facilitates excellent reversibility of heat denaturation under these conditions and that we can reliably determine the denaturation enthalpies of these proteins. The denaturation temperature (Td) of EcCutA1_0SH decreased by 4.3 °C, relative to that (89.9 °C) of EcCutA1 with SH groups, whereas those of EcCutA1_0SH_S11V and EcCutA1_0SH_E61V were 103 and 101 °C, respectively, which were remarkably improved relative to the template. Furthermore, the Td of a double mutant, EcCutA1_0SH_S11V/E61V, was 113 °C, which is 28 °C higher than that of the template (Table 1). Hereafter, EcCutA1_0SH_S11V/E61V is abbreviated as Ec0VV. These changes in stability due to the hydrophobic mutations were comparable to those observed in mutant proteins with SH groups30.
Figure 1. DSC curves of hydrophobic EcCutA1 mutants with no SH group at pH 9.0.
(A)Typical DSC curves of four mutants: EcCutA1_0SH, EcCutA1_0SH_S11V, EcCutA1_0SH_E61V, and EcCutA1_0SH_S11V/E61V. Scan rates were 60 °C/h. (B)Reversibility of heat denaturation of EcCutA1_0SH, EcCutA1_0SH_S11V, and EcCutA1_0SH_S11V/E61V. The red curves of three proteins are the second runs of DSC, just after the cooling step of the first run (the black curves). Scan rates of both curves were 60 °C/h.
Table 1. Denaturation enthalpy of examined EcCutA1_0SH mutants at denaturation temperatures.
| Td( °C)* | ΔH (kJ mol−1)* | |||||
|---|---|---|---|---|---|---|
| EcCutA1_0SH | 85.6 | ± | 0.3 | 870 | ± | 21 |
| EcCutA1_0SH_S11V | 102.7 | ± | 0.3 | 1302 | ± | 21 |
| EcCutA1_0SH_E61V | 101.0 | ± | 0.2 | 1228 | ± | 15 |
| Ec0VV** | 113.2 | ± | 0.2 | 1569 | ± | 15 |
| Ec0VV_T17K | 107.2 | ± | 0.3 | 1315 | ± | 15 |
| Ec0VV_S48D | 105.4 | ± | 0.2 | 1242 | ± | 14 |
| Ec0VV_T17K/S48D | 112.0 | ± | 0.1 | 1387 | ± | 22 |
| Ec0VV_A39D | 105.4 | ± | 0.5 | 1197 | ± | 31 |
| Ec0VV_S48 K | 112.2 | ± | 0.9 | 1173 | ± | 35 |
| Ec0VV_A39D/S48 K | 118.3 | ± | 0.7 | 1410 | ± | 33 |
| Ec0VV_H72 K | 118.4 | ± | 0.4 | 1521 | ± | 34 |
| Ec0VV_S82 K | 116.9 | ± | 0.5 | 1479 | ± | 22 |
| Ec0VV_S82R | 117.1 | ± | 0.5 | 1446 | ± | 24 |
| Ec0VV_T88R | 117.6 | ± | 0.6 | 1501 | ± | 58 |
| Ec0VV_Q87 K | 116.8 | ± | 0.5 | 1424 | ± | 13 |
| Ec0VV_Q87K/T88R | 122.4 | ± | 0.6 | 1582 | ± | 56 |
| Ec0VV_S110R | 117.3 | ± | 0.4 | 1637 | ± | 35 |
| Ec0VV_6*** | 136.8 | ± | 0.9 | 1739 | ± | 59 |
*Average value and its standard deviation of at least 6 data.
**Ec0VV represents EcCutA1_0SH_S11V/E61V mutant.
***Ec0VV_6 represents Ec0VV_A39D/S48K/H72 K/S82 K/Q87 K/T88R mutant.
Ionic mutants of EcCutA1_0SH_S11V/E61V (Ec0VV)
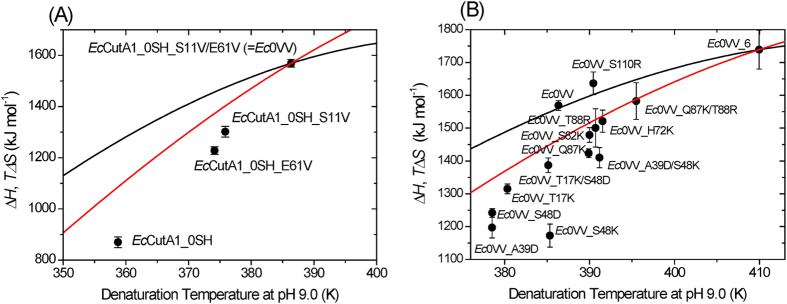
To examine the thermodynamic parameters of stabilization by ion–ion interaction at temperatures over 100 °C, we constructed several mutant proteins containing substitutions with charged residues, using Ec0VV as a template. Ionic mutants, whose denaturation temperatures are improved and whose DSC curves are suitable for thermodynamic analysis, were selected from our pool of stock mutants (Table 1). Typical DSC curves and reversibility curves are shown in Fig. 2 and Supplementary Fig. 2, respectively. Although the Td of a double mutant, Ec0VV_T17K/S48D, was lower than that of the template, it was selected because the Td was over 100 °C and higher than those of the original single mutants (Ec0VV_T17K and Ec0VV_S48D) (Table 1). In the case of Ec0VV_A39D/S48K/H72K/S82K/Q87K/T88R, which is abbreviated as Ec0VV_6, the DSC curve was suitable for analysis (Fig. 2), but the reversibility curve could not be properly obtained due to certain side reactions that occurred at high temperatures. The Td of Ec0VV_6 was 136.8 ± 0.9 °C, improved by 23.6 °C with the introduction of six charged residues.
Figure 2. Typical DSC curves of six ionic EcCutA1 mutants from Ec0VV at pH 9.0.
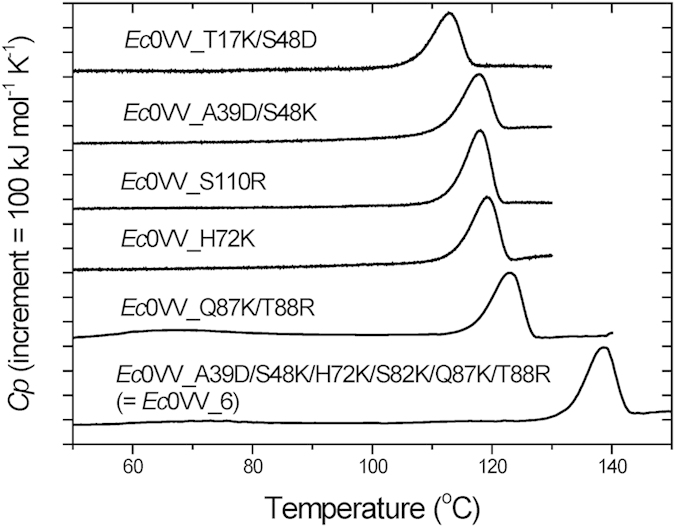
The six mutants are Ec0VV_T17 K/S48D, Ec0VV_A39D/S48 K, Ec0VV_S110R, Ec0VV_H72 K, Ec0VV_Q87 K/T88R, and Ec0VV_6. Scan rates were 60 °C/h.
In acidic pH, negatively charged residues of a protein should be protonated, leading to a decrease in conformational stability. In the case of CutA1 from P. horikoshii, which is stabilized by many ionic interactions, the Td of 148.5 °C at pH 7.0 is drastically reduced to 75.6 °C at pH 2.5, whereas the Td of CutA1 from T. thermophilus changes from 112.8 °C at pH 7.0 to 86.6 °C at pH 2.528. To confirm the stabilization resulted from ionic interactions, the stabilities of ionic mutants were examined under acidic conditions at pH 2–3. The Td values of the ionic mutants monotonically decreased as the pH was lowered, reaching a constant minimum at pH 2–2.5 (Supplementary Table 1). We plotted the Td shift (Td value at pH 9.0 vs. pH 2.0–2.5) versus the Td value at pH 9.0 for several ionic mutants (closed circles in Supplementary Fig. 3). Clearly, the Td shift became greater as Td increased. These results suggest that the electrostatic interactions dominats the thermo-stabilization of the ionic mutant proteins.
Temperature dependence of denaturation enthalpy at higher temperatures
The denaturation heat capacity (ΔCp) is generally assumed to be constant at temperatures below 80 °C31, but it gradually decreases at higher temperatures14. Therefore, it is important to elucidate the temperature function of ΔCp at higher temperatures. To this end, we measured the Cp values of Ec0VV in the native state by DSC at temperatures up to 95 °C (Y2 of Supplementary Fig. 4A). Unfortunately, the temperature dependence of Cp values in the denatured state could not be determined experimentally due to the high reversibility of denatured Ec0VV. Alternatively, assuming that the heat-capacity contribution of amino-acid groups is additive, the heat capacity of proteins in the denatured state can be calculated from their amino-acid composition32. Y1 in Supplementary Fig. 4A also shows the temperature function of the heat capacity of Ec0VV in the denatured state, which was estimated from its amino-acid composition using the parameters in Table II of Makhatadze and Privalov33. Next, we were able to estimate the temperature function of the denaturation heat capacity (ΔCp) for Ec0VV from these native and denatured Cp values (Y3 of Supplementary Fig. 4A). The temperature function obtained of ΔCp can be expanded around Td as a second-order polynomial:
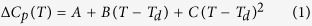 |
Then, the temperature functions of denaturation enthalpy (ΔH) and denaturation entropy (ΔS) can be calculated by the following equations (2) and (3), respectively.
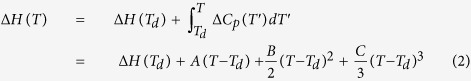 |
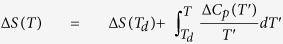 |
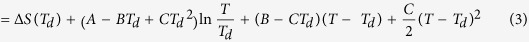 |
Figure 3A shows the temperature function of ΔH for Ec0VV. The ΔH values of Ec0VV were higher than those at each denaturation temperature of other proteins (EcCutA1_0SH, EcCutA1_0SH_S11V, and EcCutA1_0SH_E61V). If we assume that the temperature function of ΔCp is not largely affected by the constitution of the protein, this observation indicates that stabilization of hydrophobic mutants at residue positions 11 and 61 is mainly caused by enthalpic effects.
Figure 3.
(A) Temperature dependence of ΔH for hydrophobic EcCutA1 mutants at pH 9.0. ΔH values (closed circles) of denaturation for EcCutA1_0SH, EcCutA1_0SH_S11V, EcCutA1_0SH_E61V, and Ec0VV come from Table 1. The black curves represent the temperature function of ΔH upon denaturation using the temperature function of ΔCp for Ec0VV obtained from Y3 of Supplementary Fig. 4A. The red curve represents TΔS of Ec0VV. In the case of Ec0VV, the parameters A, B, and C of ΔCp (in kJ mol−1 K−1) in equation (1) were calculated to be 7.61029, −0.26614, and −8.4434 × 10−4, respectively. (B) Temperature dependence of ΔH for ionic Ec0VV mutants at pH 9.0. ΔH values (closed circles) of denaturation for EcCutA1 mutants come from Table 1. Black curves represent the temperature function of ΔH upon denaturation, using the temperature function of ΔCp for Ec0VV_6 obtained from Y3 of Supplementary Fig. 4B. The red curve represents TΔS of Ec0VV_6. In the case of Ec0VV_6, the parameters A, B, and C of ΔCp (in kJ mol−1 K−1) in equation (1) were calculated to be 4.22658, −0.29835, and −10.0757 × 10−4, respectively.
The temperature function of ΔCp for Ec0VV_6 was also determined using the native Cp values (Supplementary Fig. 4B), which were directly measured up to 110 °C. Figure 3B shows the temperature function of ΔH for Ec0VV_6 and the denaturation enthalpy values at the denaturation temperatures of several ionic Ec0VV mutants. This Figure indicates that the ΔH value of Ec0VV is similar to those of Ec0VV_S110R and Ec0VV_6 at each denaturation temperature, but remarkably higher than those of other mutants derived from Ec0VV by the further addition of charged residues.
The thermodynamic parameters of denaturation for EcCutA1_0SH mutants at the denaturation temperature (113.2 °C) of Ec0VV are listed in Table 2. The ΔG values, estimated using the ΔCp temperature function obtained from Ec0VV, agreed well with those from Ec0VV_6, around the denaturation temperature of Ec0VV. Figure 4 also shows the temperature functions of ΔG, ΔH, and TΔS for Ec0VV and Ec0VV_6 over a larger temperature range (between 280 and 420 K), indicating that ΔG values of Ec0VV_6 are positive over a broad range of temperatures.
Table 2. Thermodynamic parameters of denaturation for EcCutA1_0SH mutants at the denaturation temperature (113.2 oC) of Ec0VV.
| ΔH | TΔS | ΔG (=ΔΔG) | ΔΔH | TΔΔS | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| a* | b* | a | b | a | b | a | b | a | b | |
| EcCutA1_0SH | 1175 | 1254 | 1255 | 1336 | −80 | −82 | −394 | −315 | −314 | −233 |
| EcCutA1_0SH_S11V | 1396 | 1440 | 1434 | 1478 | −38 | −38 | −173 | −129 | −135 | −91 |
| EcCutA1_0SH_E61V | 1340 | 1377 | 1382 | 1419 | −42 | −42 | −229 | −192 | −187 | −150 |
| Ec0VV | 1569 | 1569 | 1569 | 1569 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ec0VV_A39D | 1265 | 1289 | 1290 | 1315 | −26 | −26 | −304 | −280 | −279 | −254 |
| Ec0VV_S48 K | 1181 | 1184 | 1184 | 1187 | −3 | −3 | −388 | −385 | −385 | −382 |
| Ec0VV_A39D/S48 K | 1375 | 1358 | 1356 | 1340 | 18 | 18 | −194 | −211 | −213 | −229 |
| Ec0VV_H72 K | 1485 | 1469 | 1465 | 1449 | 20 | 20 | −84 | −100 | −104 | −120 |
| Ec0VV_S82 K | 1453 | 1442 | 1439 | 1428 | 14 | 14 | −116 | −127 | −130 | −141 |
| Ec0VV_T88R | 1470 | 1456 | 1453 | 1440 | 17 | 17 | −99 | −113 | −116 | −129 |
| Ec0VV_Q87 K | 1398 | 1387 | 1385 | 1374 | 13 | 13 | −171 | −182 | −184 | −195 |
| Ec0VV_Q87 K/T88R | 1524 | 1495 | 1488 | 1459 | 36 | 36 | −45 | −74 | −81 | −110 |
| Ec0VV_S110R | 1608 | 1595 | 1590 | 1578 | 17 | 17 | 39 | 26 | 21 | 9 |
| Ec0VV_6 | 1638 | 1560 | 1540 | 1465 | 98 | 96 | 69 | −9 | −29 | −104 |
The unit is kJ mol−1.
*a and b represent the calculated results using the temperature function of ΔCp obtained from Ec0VV and Ec0VV_6, respectively.
Figure 4. Temperature functions of ΔG, ΔH, and TΔS for Ec0VV, and Ec0VV_6 between 280 and 420 K.
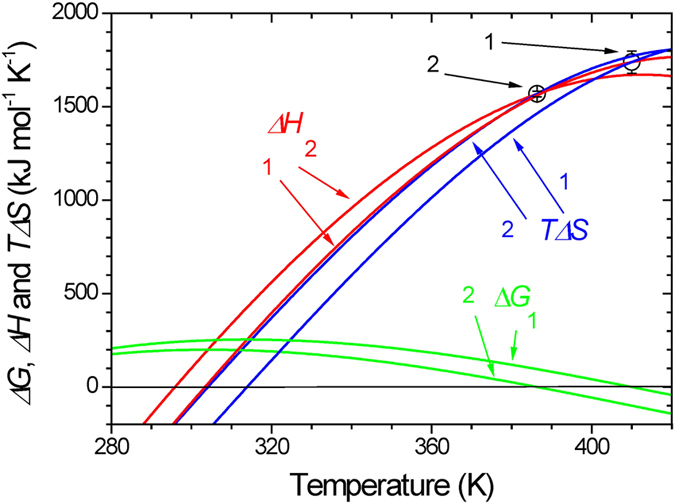
Temperature functions of ΔH and ΔS were obtained using equations (2) and (3), respectively, in which each temperature function of ΔCp was used for the calculation of Ec0VV and Ec0VV_6. The green, red, and blue curves represent values of ΔG(=ΔH–TΔS), ΔH, and TΔS, respectively. Numbers 1 and 2 represent Ec0VV_6 and Ec0VV, respectively. Open circles with error bars show the ΔH value of each protein at the denaturation temperature, as indicated in the figure.
Discussion
Forty years ago, Privalov et al.14,31 developed highly qualified adiabatic differential micro-calorimeters for determining the thermodynamic parameters of protein denaturation. They reported that specific characters of amino-acid residues disappear during protein unfolding near 110 °C, and that all the observed entropy originates from the increase in conformational freedom of the polypeptide upon unfolding, because the specific ΔH and ΔS of unfolding for several proteins intersect at a single point near 110 °C. On the other hand, the thermodynamics of transfer of hydrocarbons to water provides a model for the temperature dependence of the hydrophobic interaction in protein folding. Baldwin15 examined the solution thermodynamics of several liquid hydrocarbons in water. He found that the extrapolated temperatures at which the transfer ΔS reaches zero, around 112.8 °C, were similar for six hydrocarbons. The extrapolated temperature of the transfer ΔH is 22.0 °C. This means that at 113 °C, the hydrophobic interaction changes from being entropy-driven at 22 °C to being enthalpy-driven at 113 °C, and the contribution of water to the entropy of protein unfolding (hydrophobic hydration) is removed15. Regarding these estimations, the heat capacity change (ΔCp) is assumed to be constant against temperature. Later, Makhatadze and Privalov33,34 reported that the temperature at which ΔS is zero approaches 145 °C when the decreasing nature of ΔCp against temperature is taken into account, because the ΔCp value of hydrocarbon hydration decreases with increasing temperature.
In this study, the heat capacities of the native states of Ec0VV and Ec0VV_6 could be directly measured by DSC up to 95 and 110 °C, respectively, and the temperature functions of their ΔCp values were estimated as shown in Supplementary Fig. 4A and Supplementary Fig. 4B, respectively.
Hydrophobic effects strongly contribute to ΔH at temperatures around 100 °C
The temperature function of ΔH of Ec0VV is depicted in Fig. 3A. Using the same temperature function of ΔCp, the ΔH values of Ec0VV, EcCutA1_0SH_S11V, EcCutA1_0SH_E61V, and EcCutA1_0SH were estimated to be 1569, 1396, 1340, and 1175 kJ/mol at 113.2 °C, respectively (Table 2). The increase in ΔH (394 kJ/mol) of Ec0VV (EcCutA1_0SH_S11V/E61V) agrees well with the sum (386 kJ/mol) of the increases in ΔH of EcCutA1_0SH_S11V (221 kJ/mol) and EcCutA1_0SH_E61V (165 kJ/mol). On the other hand, the TΔS values of Ec0VV (the red curve in Fig. 3A) were larger than the ΔH value (=TΔS at Td) at each Td of EcCutA1_0SH_S11V, EcCutA1_0SH_E61V, and EcCutA1_0SH, indicating that Ec0VV is entropically unfavorable compared with other proteins. That is, Ec0VV, which contains hydrophobic substitutions for hydrophilic residues in the interior of a molecule, is mainly stabilized by the enthalpic gain upon substitutions, but is partly destabilized by the entropic loss, probably due to the disruption of the hydrophilic solvation in the denatured state upon substitutions. These results at high temperatures around 100 °C are contrary to the well-accepted belief that the entropic gain from hydrophobic solvation can account for the stabilization effect of hydrophobic substitutions at lower temperatures15. Whereas, the estimations from the hydration of amino acids34 are generally consistent with our results. This is the first experimental evidence pertaining to the hydrophobic effects on protein stability, which is obtained by direct measurement at temperatures around 100 °C.
Ec0VV_6 substituted with six charged residues is stabilized by both enthalpic and entropic effects around 137 °C
Below 100 °C, the ion–ion interaction (salt bridge) is driven entirely by entropic effects due to the release of water strongly bound to the ions of charged residues35,36,37. The proteins substituted with single charged residues, Ec0VV_H72K, Ec0VV_S82K, Ec0VV_Q87K, and Ec0VV_T88R, are stabilized by electrostatic interactions (Supplementary Fig. 3). The ΔH values of all of these proteins were drastically decreased relative to ΔH at each corresponding temperature on the temperature function of Ec0VV_6 (Fig. 3B). Because the changes in ΔH upon these mutations are unfavorable for folding, the observed improvements in stability were caused by entropic effects due to the release of water at the charged residues that we introduced (electrostatic solvation). The thermodynamic analyses also clearly confirmed stabilization resulted from entropic effects (Table 2). The other single mutants, Ec0VV_A39D and Ec0VV_S48 K, whose mutation sites were located in the interior of the molecule (Table 3), demonstrated drastically decreased Td and ΔH values. However, a double mutant containing both of these single mutants, Ec0VV_A39D/S48 K, in which Asp39 forms a strong salt bridge with Lys48, was stabilized by an increase in ΔH relative to the single mutants (Fig. 3B), probably due to Coulomb’s force resulting from salt bridge formation. A similar result was obtained in the other double mutant, Ec0VV_T17 K/S48D (Fig. 3B). The double mutant Ec0VV_Q87 K/T88R was also stabilized by an increase in ΔH relative to Ec0VV_Q87 K and Ec0VV_T88R. Because this double mutant does not have additional salt bridges, two individual thermo-stabilizations might work synergistically at around 120 °C to promote the desolvation of the ionic residues introduced, thereby reducing both the enthalpic loss and the entropic gain that are mutually attributed to the electrostatic solvation. In addition, the increase in ΔH in this double-ion mutant might have been caused by a hydrophobic interaction due to the alkyl groups of Lys87 and Arg88.
Table 3. Burial rates of target residues of Ec 0VV mutants*.
| Mutants | Residues | All atoms | Charged atoms | location | ||||
|---|---|---|---|---|---|---|---|---|
| Ec0VV_6 | Asp39 | 100 | ± | 1 | 99 | ± | 2 | beta 2 |
| Lys48 | 96 | ± | 3 | 93 | ± | 6 | beta 2 | |
| Lys72 | 80 | ± | 5 | 55 | ± | 18 | alpha 2, N terminal | |
| Lys82 | 10 | ± | 6 | 13 | ± | 8 | alpha 2, C terminal | |
| Lys87 | 10 | ± | 14 | −2 | ± | 21 | loop | |
| Arg88 | 55 | ± | 20 | 40 | ± | 34 | loop | |
| Val11 | 98 | ± | 1 | beta 1 | ||||
| Val61 | 97 | ± | 3 | beta 3 | ||||
| Ec0VV_S110R | Arg110 | 84 | ± | 10 | 66 | ± | 20 | alpah 3, C-terminal |
*The burial rates (%) were estimated from the average of ASA values for nine structures during 40 ns MD at 300 K (in preparation).
Ec0VV_6, with six additional charged residues, was stabilized by an increase in the ΔH value relative to each of the six original ionic mutants (Fig. 3B). The increase in ΔH might indicate that hydrophobic effects due to the alkyl groups of Lys or Arg and Coulomb’s force still function effectively at these high temperatures. According to Coulomb’s law, the strength of an electrical interaction is inversely proportional to the dielectric constant. The dielectric constant of water drops from 80 at 0 °C to 55 at 100 °C38. Furthermore, Elcock39 found that increasing temperature decreases the electrostatic desolvation penalty incurred in forming a salt bridge, leading to an increase in salt bridge stabilization from his continuum solvation model40. The increase in ΔH of Ec0VV_6 might result mainly from the high degree of desolvation of the ionic residues in the denatured state at around 137 °C, in addition to the other effects described above.
The temperature dependence of ΔH for Ec0VV_6 has an intersection near Td (113 °C) of Ec0VV, as shown in Fig. 4, suggesting that the contribution of ΔH to the stability of Ec0VV_6 with additional 6 charged residues becomes favorable in the temperature region above 113 °C, compared with those of Ec0VV. Furthermore, Fig. 4 shows that the increase in ΔG of Ec0VV_6 results largely from the decrease in ΔS of Ec0VV_6 when compared with the template Ec0VV at temperatures below 113 °C. That is, the stabilization due to ionic mutations results mainly from both the enthalpic gain from ion-ion interactions in the native state and the entropic gain from the water release of ionic residues in the denatured state at temperatures over 113 °C.
A mutant, Ec0VV_S110R, whose substitution position is located in the C-terminus of α-helix and almost buried (Table 3), was stabilized by an increase in ΔH (Fig. 3B). In this case, due to local conformations, the decrease in ΔH due to water release might be suppressed by the effects of other stabilizing factors.
Conclusion
The rational design of hyper-thermostable proteins may be possible through the introduction of multiple salt bridges at high temperatures over 113 °C that can be reached through a preceding stabilization by hydrophobic substitutions.
Matrials and Methods
Mutagenesis, expression, and purification of CutA1 mutants from E. coli
The mutagenesis, expression, and purification of CutA1 mutants from E. coli were performed as described30 with minor modifications. The homogeneity and identity of the purified samples were assessed using SDS–PAGE. The protein concentration was estimated from the absorbance at 280 nm, assuming E1cm1% = 14.96, based on the number of aromatic amino acids41.
Differential scanning calorimetry (DSC) experiments
To measure the changes in stability due to mutations, DSC was performed using a scan rate of 60 °C/h on a VP-capillary DSC platform (Microcal, USA) for temperatures up to 130 °C at pressures below 60 psi, or a Nano-DSC 6300Y microcalorimeter (TA Instruments, USA) for higher temperatures up to 160 °C at a pressure of 88 psi. Protein concentrations were around 0.6 mg/ml in a 50 mM glycine buffer at pH 9.0 containing 2 mM EDTA or a 50 mM glycine buffer at pH 2.0–3.5. All samples were dialyzed against the buffers overnight at 4 °C and then filtered through a membrane with 0.22-μm pores. The denaturation temperature (Td) is the temperature at which the area of the denaturation enthalpy (ΔH) is 0.5. The Td and ΔH values in this study represent the averages for at least six experiments.
To measure the heat capacity of mutant proteins in their native states, the protein concentrations were adjusted to around 2.0 mg/ml in a 50 mM glycine buffer at pH 9.0. Two different scan rates, 60 and 200 °C/h, were used. Each experiment comprised six cycles of reheating to the pre-denaturation temperatures: 95 °C and 110 °C for Ec0VV and Ec0VV_6, respectively. The partial specific volumes for the calculation of heat capacity were estimated from the amino-acid composition of each mutant protein42.
Additional Information
How to cite this article: Matsuura, Y. et al. Thermodynamics of protein denaturation at temperatures over 100 °C: CutA1 mutant proteins substituted with hydrophobic and charged residues. Sci. Rep. 5, 15545; doi: 10.1038/srep15545 (2015).
Supplementary Material
Acknowledgments
This work was partly supported by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science), from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) and Japan Agency for Medical Research and Development (AMED).
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.M., K.O. and K.Y. designed the study; Y.M., M.T., T.T. and N.O. performed the experiments; Y.M., M.T., Y.J., N.K. and K.Y. analyzed the data; and Y.M., Y.J., K.O., N.K. and K.Y. wrote the paper.
References
- Cavagnero S. et al. Kinetic role of electrostatic interactions in the unfolding of hyperthermophilic and mesophilic rubredoxins. Biochemistry 37, 3369–3376 (1998). [DOI] [PubMed] [Google Scholar]
- Pothekin S. A., Ogasahara K. & Yutani K. Transition state of heat denaturation of methionine aminopeptidase from a hyperthermophile. J. Therm. Anal. Calorim. 62, 111–122 (2000). [Google Scholar]
- Wrba A. et al. Extremely thermostable D-glyceraldehyde-3-phosphate dehydrogenase from the eubacterium Thermotoga maritima. Biochemistry 29, 7584–7592 (1990). [DOI] [PubMed] [Google Scholar]
- Klump H. et al. Glutamate dehydrogenase from the hyperthermophile Pyrococcus furiosus. Thermal denaturation and activation. J. Biol. Chem. 267, 22681–22685 (1992). [PubMed] [Google Scholar]
- McAfee J. G., Edmondson S. P., Zegar I. & Shriver J. W. Equilibrium DNA binding of Sac7d protein from the hyperthermophile Sulfolobus acidocaldarius: fuorescence and circular dichroism studies. Biochemistry 35, 4034–4045 (1996). [DOI] [PubMed] [Google Scholar]
- Pfeil W., Gesierich U., Kleemann G. R. & Sterner R. Ferredoxin from the hyperthermophile Thermotoga maritima is stable beyond the boiling point of water. J. Mol. Biol. 272, 591–596 (1997). [DOI] [PubMed] [Google Scholar]
- Pappenberger G., Schurig H. & Jaenicke R. Disruption of an ionic network leads to accelerated thermal denaturation of D-glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic bacterium Thermotoga maritima. J. Mol. Biol. 274, 676–683 (1997). [DOI] [PubMed] [Google Scholar]
- Ogasahara K. et al. Electrostatic stabilization in methionine aminopeptidase from hyperthermophile Pyrococcus furiosus. Biochemistry 37, 5939–5946 (1998). [DOI] [PubMed] [Google Scholar]
- Yutani K., Ogasahara K., Sugino Y. & Matsushiro A. Effect of a single amino acid substitution on stability of conformation of a protein. Nature 267, 274–275 (1977). [DOI] [PubMed] [Google Scholar]
- Yutani K., Ogasahara K., Tsujita T. & Sugino Y. Dependence of conformational stability on hydrophobicity of the amino acid residue in a series of variant proteins substituted at a unique position of tryptophan synthase α subunit. Proc. Natl. Acad. Sci. USA 84, 4441–4444 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalov P. L. & Gill S. J. Stability of protein structure and hydrophobic interaction. Adv. Protein. Chem. 39, 191–234 (1988). [DOI] [PubMed] [Google Scholar]
- Mukaiyama A. & Takano K. Slow unfolding of monomeric proteins from hyperthermophiles with reversible unfolding. Int. J. Mol. Sci. 10, 1369–1385 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace C. N., Scholtz J. M. & Grimsley G. R. Forces stabilizing proteins. FEBS Lett. 588, 2177–2184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalov P. L. Stability of proteins: small globular proteins. Adv. Protein. Chem. 33, 167–241 (1979). [DOI] [PubMed] [Google Scholar]
- Baldwin R. L. Temperature dependence of the hydrophobic interaction in protein folding. Proc. Natl. Acad. Sci. USA 83, 8069–8072 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korndörfer I. et al. The crystal structure of holo-glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic bacterium Thermotoga maritima at 2.5 Å resolution. J. Mol. Biol. 246, 511–521 (1995). [DOI] [PubMed] [Google Scholar]
- Hennig M. et al. 2.0 Å structure of indole-3-glycerol phosphate synthase from the hyperthermophile Sulfolobus solfataricus: possible determinants of protein stability. Structure 3, 1295–1306 (1995). [DOI] [PubMed] [Google Scholar]
- Ermler U., Merckel M. C., Thauer R. & Shima S. Formylmethanofuran: tetrahydromethanopterin formyltransferase from Methanopyrus kandleri - new insights into salt-dependence and thermostability. Structure 5, 635–646 (1997). [DOI] [PubMed] [Google Scholar]
- Aguilar C. F. et al. Crystal structure of the β-glycosidase from the hyperthermophilic archeon Sulfolobus solfataricus: resilience as a key factor in thermostability. J. Mol. Biol. 271, 789–802 (1997). [DOI] [PubMed] [Google Scholar]
- Vetriani C. et al. Protein thermostability above 100 oC: a key role for ionic interactions. Proc. Natl. Acad. Sci. USA 95, 12300–12305 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov T. H. et al. Crystal structure of methionine aminopeptidase from hyperthermophile, Pyrococcus furiosus. J. Mol. Biol. 284, 101–124 (1998). [DOI] [PubMed] [Google Scholar]
- de Bakker P. I., Hünenberger P. H. & McCammon J. A. Molecular dynamics simulations of the hyperthermophilic protein Sac7d from Sulfolobus acidocaldarius: contribution of salt bridges to thermostability. J. Mol. Biol. 285, 1811–1830 (1999). [DOI] [PubMed] [Google Scholar]
- Karshikoff A. & Ladenstein R. Ion pairs and the thermotolerance of proteins from hyperthermophiles: a “traffic rule” for hot roads. Trends Biochem. Sci. 26, 550–556 (2001). [DOI] [PubMed] [Google Scholar]
- Yamagata Y. et al. Entropic stabilization of the tryptophan synthase α-subunit from a hyperthermophile, Pyrococcus furiosus: X-ray analysis and calorimetry. J. Biol. Chem. 276, 11062–11071 (2001). [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M., Alibés A. & Godzik A. Contribution of electrostatic interactions, compactness and quaternary structure to protein thermostability: lessons from structural genomics of Thermotoga maritima. J. Mol. Biol. 356, 547–557 (2006). [DOI] [PubMed] [Google Scholar]
- Tanaka T. et al. Hyper-thermostability of CutA1 protein, with a denaturation temperature of nearly 150 oC. FEBS Lett. 580, 4224–4230 (2006). [DOI] [PubMed] [Google Scholar]
- Matsuura Y. et al. Role of charged residues in stabilization of Pyrococcus horikoshii CutA1, which has a denaturation temperature of nearly 150 oC. FEBS J. 279, 78–90 (2012). [DOI] [PubMed] [Google Scholar]
- Sawano M. et al. Thermodynamic basis for the stabilities of three CutA1s from Pyrococcus horikoshii, Thermus thermophilus, and Oryza sativa, with unusually high denaturation temperatures. Biochemistry 47, 721–730 (2008). [DOI] [PubMed] [Google Scholar]
- Bagautdinov B. et al. Thermodynamic analysis of unusually thermostable CutA1 protein from the human brain and its protease susceptibility. J. Biochem. 157, 169–176 (2015). [DOI] [PubMed] [Google Scholar]
- Matsuura Y. et al. Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design. J. Biochem. 148, 449–458 (2010). [DOI] [PubMed] [Google Scholar]
- Privalov P. L. & Khechinashvili N. N. A thermodynamic approach to the problem of stabilization of globular protein structure: a calorimetric study. J. Mol. Biol. 86, 665–684 (1974). [DOI] [PubMed] [Google Scholar]
- Privalov P. L. & Makhatadze G. I. Heat capacity of proteins II. Partial molar heat capacity of the unfolded polypeptide chain of proteins: protein unfolding effects. J. Mol. Biol. 213, 385–391 (1990). [DOI] [PubMed] [Google Scholar]
- Makhatadze G. I. & Privalov P. L. Energetics of protein structure. Adv. Protein Chem. 47, 307–425 (1995). [DOI] [PubMed] [Google Scholar]
- Makhatadze G. & Privalov P. L. Hydration effects in protein unfolding. Biophys. Chem. 51, 291–309 (1994). [DOI] [PubMed] [Google Scholar]
- Kauzmann W. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 14, 1–63 (1959). [DOI] [PubMed] [Google Scholar]
- Fersht A. R. Conformational equilibria in α and δ chymotrypsin. The energetics and importance of the salt bridge. J. Mol. Biol. 64, 497–509 (1972). [DOI] [PubMed] [Google Scholar]
- Lam S. Y. et al. A rigidifying salt-bridge favors the activity of thermophilic enzyme at high temperatures at the expense of low-temperature activity. PLoS Biol. 9, e1001027 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace C. N. Single surface stabilizer. Nat. Struct. Biol. 7, 345–346 (2000). [DOI] [PubMed] [Google Scholar]
- Elcock A. H. The stability of salt bridges at high temperatures: Implications for hyperthermophilic proteins. J. Mol. Biol. 284, 489–502 (1998). [DOI] [PubMed] [Google Scholar]
- Elcock A. H. & McCammon J. A. Continuum solvation model for studying protein hydration thermodynamics at high temperatures. J. Phys. Chem., 101, 9624–9634 (1997). [Google Scholar]
- Pace C. N. et al. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4, 2411–2423 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durchschlag H. Specific volumes of biological macromolecules and some other molecules of biological interest. Thermodynamic Data for Biochemistry and Biotechnology, ed Hinz H.-J. (Springer-Verlag, Berlin, Germany), pp. 45–128 (1986).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.