ABSTRACT
Two full-scale slow sand filters (SSFs) were sampled periodically from April until November 2011 to study the spatial and temporal structures of the bacterial communities found in the filters. To monitor global changes in the microbial communities, DNA from sand samples taken at different depths and locations within the SSFs and at different filters ages was used for Illumina 16S rRNA gene sequencing. Additionally, 15 water quality parameters were monitored to assess filter performance, with functionally relevant microbial members being identified by using multivariate statistics. The bacterial diversity in the SSFs was found to be much larger than previously documented, with community composition being shaped by the characteristics of the SSFs (filter age and depth) and sampling characteristics (month, side, and distance from the influent and effluent pipes). We found that several key genera (Acidovorax, Halomonas, Sphingobium, and Sphingomonas) were associated with filter performance. In addition, at the whole-community level, a strong positive correlation was found between species evenness and filter performance. This study is the first to comprehensively characterize the microbial community of SSFs and link specific microbes to water quality parameters. In doing so, we reveal key patterns in microbial community structure that relate to overall community function.
IMPORTANCE
The supply of sustainable, energy-efficient, and safe drinking water to an increasing world population is a huge challenge faced by the water industry. SSFs have been used for hundreds of years to provide a safe and reliable source of potable drinking water, with minimal energy requirements. However, a lack of knowledge pertaining to the treatment mechanisms, particularly the biological processes, underpinning SSF operation has meant that SSFs are still operated as “black boxes.” Understanding these dynamics alongside performance-induced effects associated with operational differences will promote optimized SSF design, maintenance, and operation, creating more efficient and environmentally sustainable filters. Through a spatial-temporal survey of full-scale SSFs at various points of operation, we present the most detailed characterization to date of the functional microbial communities found in SSFs, linking various taxa and community metrics to optimal water quality production.
INTRODUCTION
The supply of clean and safe drinking water free from any substances or organisms that pose a danger to human health is a major objective of the European Union Drinking Water Directive (1) and the World Health Organization (WHO). For over 200 years, slow sand filtration has been an effective means of treating water for the control of microbiological and chemical contaminants in both small and large community water supplies (2, 3). This ability to remove various contaminants efficiently has underpinned slow sand filter (SSF) deployment in various areas outside drinking water purification, including aquaculture (4), horticulture (5), storm water purification (6), and food and drink waste management (7). However, despite their adoption and use in the energy-efficient production of high-quality water, little is understood about the functional ecology of SSFs, i.e., the biological mechanisms and organisms responsible for producing the diverse and efficient functional capacity of SSFs (3). This lack of knowledge has hindered the optimization of the design, management, and operation of these systems and will continue to do so.
Recently, a number of studies have attempted to characterize the purification mechanisms and the microbes responsible for them (5, 7–10). However, such studies have focused on specific aspects of SSFs (e.g., the Schmutzdecke [11]) or specific purification mechanisms (e.g., nitrate removal [10]) and, with the exception of those reported in references 12 to 13, have been performed in nonverified laboratory scale SSF microcosms, which may not accurately reflect the true microbial community found in real SSFs. Although all of those studies have provided great insights into the biological processes occurring within SSFs, a deeper analysis of the structure and dynamics of the microbial community underpinning SSFs as a function of performance and operational conditions is needed. Such a study has the potential to reveal important and underappreciated structure-function relationships, which could greatly improve the operation, management, and design of these systems but also reveal patterns and processes in microbial communities with more general ecological relevance. Previous microbial ecology papers on engineered systems with a biological component have shown that functional stability and robustness are correlated with several components of biodiversity, such as species richness and evenness (14–18); however, no such study of SSFs has ever been performed. These provide ideal systems for functional ecology research, allowing easily quantifiable measurement of overall community performance that depends on a complex microbial ecosystem.
Here we present results from the periodic sampling of two full-scale SSFs. We determined the spatial and temporal structures of the bacterial communities found in the filters. This enabled us to quantify how specific microbial groups and overall community structure are related to overall filter performance. This study comprises the entire life cycle of the filters (drained, scraped, juvenile, mature, and clogged; Fig. 1) (2). This provides a detailed SSF microbiome blueprint that can be placed into a functional context.
FIG 1 .
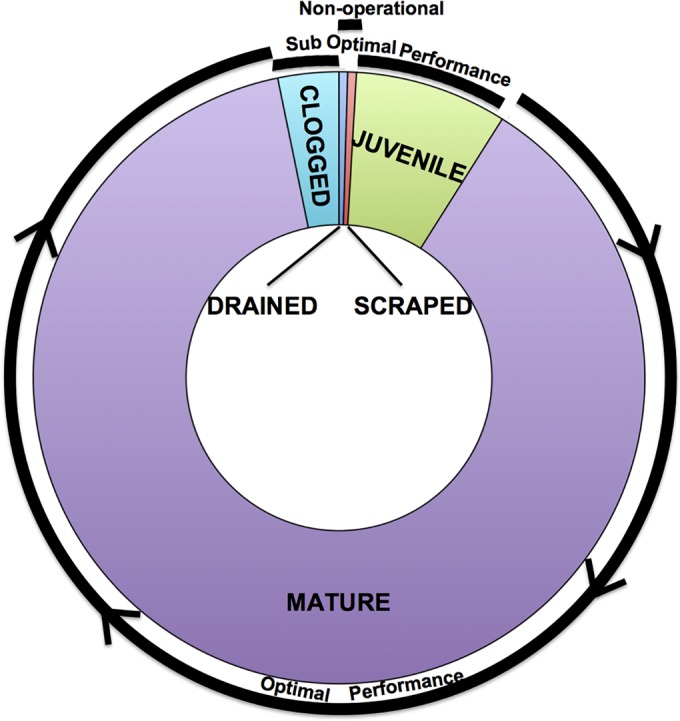
Life cycle of SSFs. The size of each ring segment corresponds to the proportion of time SSFs are at that stage. The black outer lines provide performance-related information.
RESULTS
Water quality.
Two full-scale SSFs were sampled routinely from April 2011 until they were decommissioned in November 2011 (see Fig. S1 in the supplemental material). Both full-scale SSFs performed extremely well in terms of the Water Supply (Water Quality) (Scotland) Regulations 2001 part of the Water (Scotland) Act 1980. For influent, effluent, and percent removal results, see Fig. S2 to S5 in the supplemental material. Overall, the filters failed to meet only one drinking water requirement, the coliform levels. However, it should be noted that these filters are not a single point of purification, with effluent from the filters being chlorinated before being distributed, a process that would remove the low levels of coliforms present in the effluent. In terms of performance, there was no statistically significant difference (Wilcoxon test P value, 0.08) between filters A and B.
There were substantial correlations between the water quality parameters, all of them significantly correlated with at least six other parameters, with the dissolved organic carbon (DOC) correlating the least and NH4 and dissolved oxygen (DO) correlating with every parameter (see Fig. S5 in the supplemental material). Additionally, several parameters (Table 1) showed various strengths of correlation with the age of the filters, with coliform removal showing the strongest positive correlation and optimum coliform removal occurring after 7 weeks. This is consistent with operators’ verbal reports of SSF performance increasing with filter age or maturity.
TABLE 1 .
Significant correlations of filter age with percent removal of water quality parameters, based on 470 samples
| Water quality parameter | Correlation | P value |
|---|---|---|
| Ammonia | −0.283 | 3.89 × 10−10 |
| Coliforms | 0.537 | 1.89 × 10−4 |
| DOC | −0.285 | 3.11 × 10−10 |
| Nitrate | −0.606 | 2.20 × 10−16 |
| Nitrite | −0.171 | 1.90 × 10−4 |
| Orthophosphate | −0.411 | 2.20 × 10−16 |
| Performance metric ▽ | 0.475 | 4.00 × 10−4 |
| Total viable bacteria at: | ||
| 30°C | 0.243 | 1.90 × 10−3 |
| 13°C | 0.117 | 1.09 × 10−3 |
Distinct community compositions of sand and water samples.
A total of 26,163,232 sequences were generated by Illumina sequencing, with an average number of 38,566 ± 503 reads for each sample. In order to account for differences in read number and therefore diversity, samples were rarefied to the lowest read number within the data set (5,909). Rarefied samples were classified below the domain level, being affiliated with 36 phyla, 82 classes, 126 orders, 239 families, 688 genera, and 11,026 operational taxonomic units (OTUs). Proteobacteria was the dominant phylum in all of the samples, accounting for, on average, 51% of the community. Overall, sand from operational SSFs contained the greatest number of OTUs (8,319, of which 2,312 were unique to sand), which was almost double that found in drained SSF sand (4,482, with three unique OTUs) and both influent (4,504) and effluent (3,947) samples. This coincided with significant differences in species diversity and evenness, with operational SSF sand having the greater species diversity and evenness and drained sand samples having the lowest (Wilcoxon test P values, 0.0021 and 0.0004, respectively). Influent water samples possessed more OTUs than effluent samples did, along with a significantly higher species diversity index (P value, 0.001); however, there was no difference between Pielou’s evenness values (0.65 and 0.63, respectively).
Sand samples from operational filters had only 55 and 73% of their OTUs in common with influent and effluent water samples, respectively. This may suggest that some of the microbiota is being acquired from other sources, for example, through aerosol deposition or transmission by wildlife. However, this does not account for sampling effects; some OTUs may be present in the influent but undetected at these sampling levels. It also does not account for errors; some OTUs in the SSFs may be sequencing or PCR artifacts. To determine the accuracy of OTU diversity prediction, we also sequenced two positive controls found in mock communities of known diversity (12); this revealed a substantial overestimation of diversity, so absolute OTU richness values must be treated with caution (see Fig. S6 in the supplemental material). The reasons for this overestimation are unknown, but one recent study (19) has shown that library preparation, bar code choice, and sample complexity all impact sequencing results. However, it is reassuring that we correctly determined that one of the mock communities was more diverse than the other; therefore, even if we cannot be confident of the absolute diversities determined, the relative differences in diversity obtained should be correct.
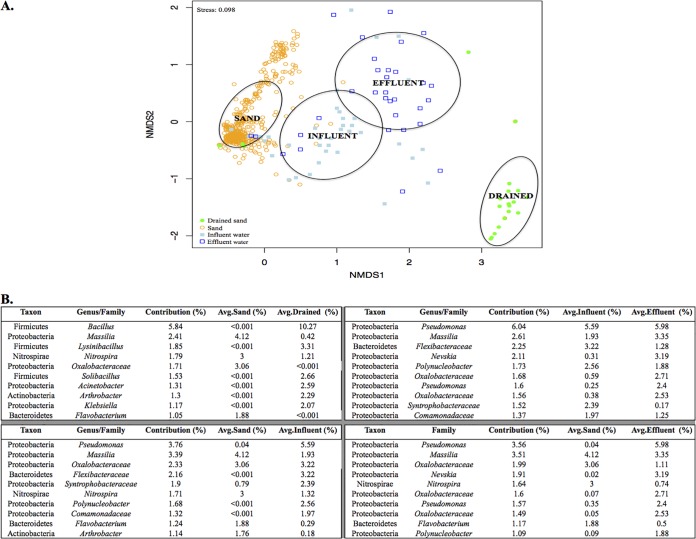
Visualization in two dimensions of the 97% similarity OTU community structures by nonmetric multidimensional scaling (NMDS) with Bray-Curtis dissimilarities (Fig. 2A) revealed that the samples clustered into the following four groups: influent water, effluent water, sand from operational filters, and drained sand from two SSFs. Multivariate analysis of variance (MANOVA) confirmed that the four groups were significantly different (P value = 0.001). The similarity percentage (SIMPER) procedure was used to identify the top 10 OTUs responsible for the dissimilarities between water and sand samples (Fig. 2B). As shown in Fig. 2, a difference in the community compositions of drained SSFs and operational SSFs is apparent; members of the family Bacillaceae of the Firmicutes phylum, appear to be responsible for the greatest proportion of the difference, being 16,000 times as abundant in drained samples as in operational SSFs.
FIG 2 .
(A) NMDS ordination of the microbial community structures of all of the samples at the 97% OTU level. Ellipses designate the 95% confidence intervals of the four groups. (B) SIMPER analysis identified the top 10 taxa (at the 97% OTU level) that account for most of the dissimilarities among the four water groups.
Spatial and temporal community diversity in sand samples.
To resolve the factors shaping the microbial community in the SSFs, we used canonical correspondence analysis (CCA). The environmental and sampling parameters used in the CCA were month, filter age (time in weeks since filter scraping), side, distances from both the influent and effluent pipes, depth, and filter identity (Table 2). All of these factors except the filter identity significantly impacted the community structure. This suggests that, with these environmental factors accounted for, there were no further differences between the filters. Of the characteristics evaluated, filter age, the side of the filter, and the month when the sample was collected were the major drivers of the bacterial community structure, with filter age being the most significant factor. To determine how filter age impacts the community structure, we divided the samples into three filter age categories (early, 0 to 4 weeks; mid, 5 to 8 weeks; late, ≥9 weeks). We observed differences in the abundance of the top 18 families between these categories; most notably, there were differences among Flavobacteriaceae, Micrococcaceae, Nitrospiraceae, and Oxalobacteraceae (see Fig. S7 in the supplemental material). Further analysis revealed that there is a strong positive correlation with the total number of OTUs and the age of the filters (early, 4,790 OTUs; mid, 5,234 OTUs; late, 6,798 OTUs). As the filters age, the number and diversity of OTUs increase, which is consistent with previous studies (7). SIMPER analysis confirmed that significant differences in the community compositions of the various filter age categories were due mainly to members of the Flavobacteriaceae, Micrococcaceae, Nitrospiraceae, and Oxalobacteraceae families. However, Ruminococcaceae, a less abundant family, was also found to explain a significant amount of the community variation, with percent abundances of this family being significantly higher in older filters (early, <0.001%; mid, 0.0013%; late, 0.268%). Furthermore, there were nine families (see Table S1 in the supplemental material), ranging in relative percent abundance from <0.0001 to 0.027%, that were present only in the oldest filters.
TABLE 2 .
CCA of the relative abundances of bacterial OTUs and filter parameters and characteristics in 406 sand samples from two SSFs
| Parameter | Degree(s) of freedom | χ2 | F value | No. of permutations | Pr(>F) |
|---|---|---|---|---|---|
| Month | 3 | 0.1575 | 6.4583 | 99 | 0.01a |
| Filter age | 2 | 0.1936 | 11.3858 | 99 | 0.01a |
| Side | 1 | 0.0691 | 8.5006 | 99 | 0.01a |
| Distance from: | |||||
| Effluent pipe | 1 | 0.0246 | 3.0248 | 99 | 0.04a |
| Influent pipe | 1 | 0.0243 | 2.8756 | 99 | 0.01a |
| Depth | 1 | 0.0131 | 1.6080 | 99 | 0.05a |
| Filter identity | 1 | 0.0093 | 1.1394 | 99 | 0.64 |
| Residual | 190 | 1.5352 |
Significant variable.
Surprisingly, depth was only a marginally significant parameter (P value, 0.05) in explaining differences between sand samples. We might have expected that significant gradients would exist across depths within SSFs and that this would drive depth-dependent community differences. In contrast, the side of the filter did significantly impact community structure (explaining 3.5% of the variance; P value, 0.01), suggesting the existence of lateral gradients within the SSFs. SIMPER analysis revealed that side 1 was most similar to side 2 (47% similarity) but less similar to side 3 (41% similarity) and that side 2 was 43% similar to side 3 (see Fig. S1 in the supplemental material). The majority of the differences between the microbial community compositions at the different sides was due to Acidobacteria and various orders of Proteobacteria. Furthermore, ANOVA showed that the microbial communities in both filters A and B were statistically equivalent (P value = 0.093); the microbial communities in the two filters were indistinguishable.
Impact of draining and chlorination on the microbial community.
In addition to the increase in diversity, we found that as the filters matured, their microbial communities became more even (P value, 1.245 × 10−7), and this was observed consistently throughout all depths of the sand bed. In contrast, perturbation of the filters typically reduced evenness, as seen during the decommissioning of the site when chlorine was added, resulting in significantly lower species evenness (P value, 2.2 × 10−16) in both filters (filter A, 0.558 ± 0.090 before chlorination and 0.502 ± 0.077 after chlorination; filter B, 0.556 ± 0.070 before chlorination and 0.448 ± 0.090 after chlorination) (see Fig. S8 in the supplemental material); an equivalent effect of chlorine was observed in a study by Wang et al. (20). Interestingly, this change in evenness was due to the large increase in the abundance of Deltaproteobacteria (average abundance change from 24.87 to 63.50%; P value, 1.076 × 10−5). However, the size of the impact was dependent on the side of the filter, with side 1 (the side of the filter where the influent pipe is located) being the first and most severely affected. This is not surprising, as it is the closest location to where chlorine delivery occurs. Similarly, when the filters were drained and scraped, a reduction in evenness was observed (P value, 1.276 × 10−11), with a larger effect on the top depths than on the lower depths (P value, 0.0197). This change in evenness was due to a large increase in the proportion of Chloroflexi, Planctomycetes, and Verrucomicrobia, which coincides with a decrease in Acidobacteria, Bacteroidetes, and Deltaproteobacteria (see Fig. S9 in the supplemental material).
Mesoscale spatial variation.
We discuss above the importance of the side and distances from the influent and effluent pipes in explaining the differences seen in microbial communities. In order to resolve these patterns at a higher resolution, six cores at each side of both full-scale SSFs were taken on a single sampling occasion, 21 June 2011 (see Fig. S1 in the supplemental material). Using CCA to perform constrained ordination (Fig. 3), we observed that the sand samples from the mesoscale experiment formed three distinct clusters: distance from the influent pipe (side 1), distance from effluent pipe (side 3), and distance from effluent corner (side 2). The depth and distance from the influent pipe correlated with CA1 and explained 33.97% of the variation, and the distances from the effluent pipe and effluent corner correlated with CA2 and explained 16.89% of the variation. Adonis analysis confirmed that there were significant differences in the microbial community within and between groups (between groups, P value = 0.009; within distance from the influent pipe, P value = 0.034; within distance from the effluent pipe, P value = 0.018; within distance from the effluent corner, P value = 0.053). Such differences among communities can be attributed to the chemical gradients that likely exist within the filters. SIMPER analysis revealed that the abundance of the Massilia genus increased with distance from the influent pipe (average abundances: at 0.2 m, 1.36%; at 0.42 m, 14.44%; at 0.68 m, 23.10%). Massilia species have been isolated from various environmental samples from many sources, including air, dust, soil, roots, and drinking water (21); however, the reason for their dominance away from the influent pipe is unclear.
FIG 3 .
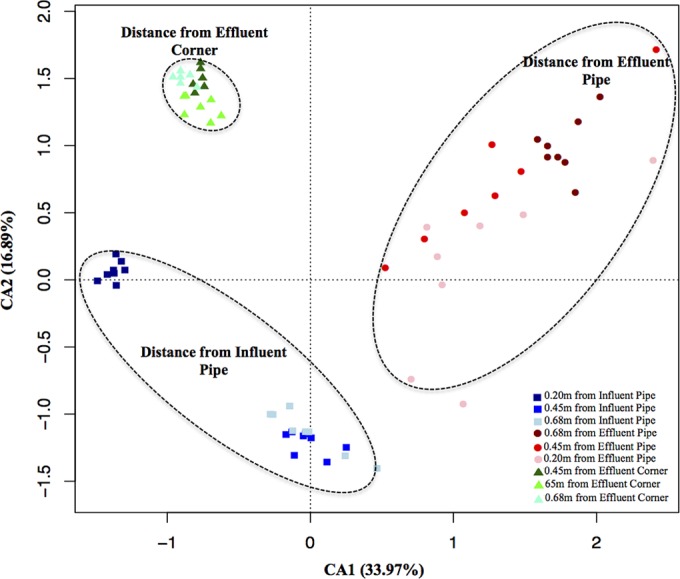
CCA of mesoscale spatial variability in the microbial community in sand samples taken from several locations from the influent and effluent pipes supplying two SSFs.
Correlation between community members and water quality.
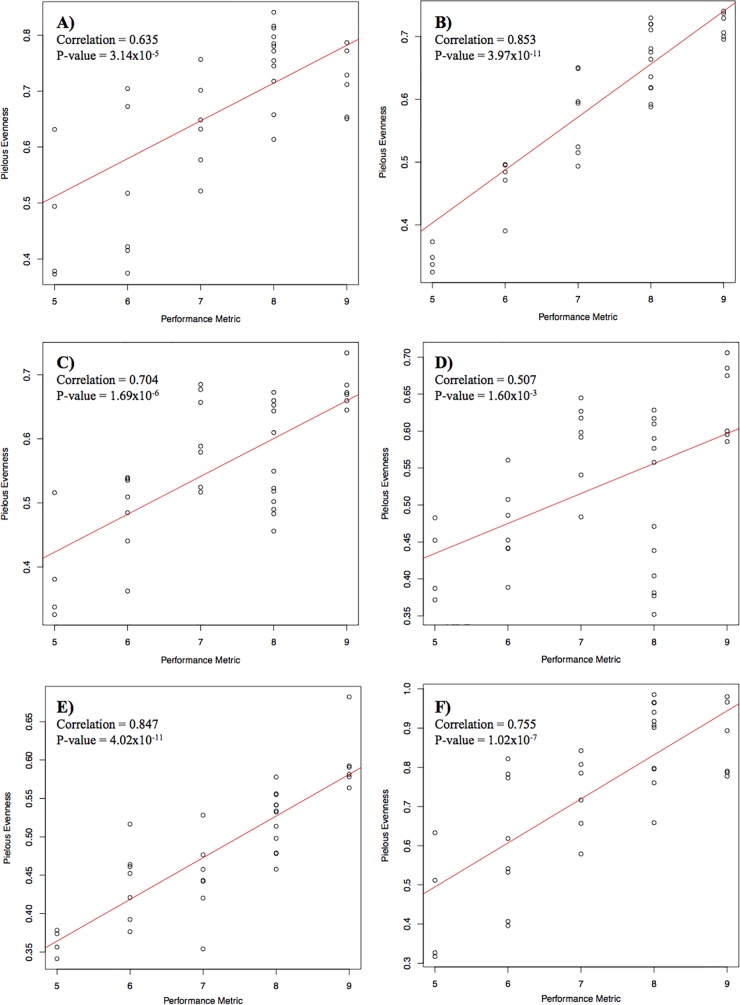
The stepwise multivariate regressions in Table 3 show that the relative proportions of several bacterial families correlate strongly with the removal of particular water quality parameters. These correlations are consistent with the findings of other studies (5, 9). Remarkably, though, we found that the strongest correlation with water quality performance is given not by a subset of particular families but with the overall evenness of the community in the filter; filter age is also important but less so than evenness in a combined regression (Fig. 4; filter age P value, 0.022; evenness P value, 1.620 × 10−4). Previous studies (17, 18) have demonstrated that microbial communities with greater species evenness perform specific functions better than less even communities. This is likely because greater species evenness implies greater robustness and functional stability and therefore a greater ability to adapt to new and fluctuating parameters.
TABLE 3 .
Stepwise multivariate regression of water quality parameters and family abundances
| Parameter | Model P value | Adjusted R2 value | Family | P value | Relationship with removal |
|---|---|---|---|---|---|
| Ammonium | 1.562 × 10−5 | 0.4133 | Clade CL500.29 | 0.0399 | + |
| Cellulomonadaceae | 0.0784 | − | |||
| Mycobacteriaceae | 0.0464 | − | |||
| Nocardiaceae | 0.0635 | − | |||
| Carnobacteriaceae | 0.0040 | + | |||
| Rhizobiaceae | 0.0041 | + | |||
| Leuconostocaceae | 0.0094 | − | |||
| Pseudomonadaceae | 0.0286 | − | |||
| Coliforms | 1.837 × 10−6 | 0.5265 | Erysipelotrichaceae | 0.0877 | − |
| Carnobacteriaceae | 0.0653 | − | |||
| Fusobacteriaceae | 0.0418 | − | |||
| “Isosphaeraceae” | 8.23 × 10−5 | + | |||
| Planctomycetaceae | 0.0207 | − | |||
| Desulfobacteraceae | 0.0507 | − | |||
| Sinobacteraceae.1 | 0.0251 | + | |||
| Opitutaceae | 0.0005 | + | |||
| Verrucomicrobia subdivision 3 | 0.0011 | − | |||
| Enterobacteriaceae | 0.0435 | − | |||
| DOC | 2.2 × 10−16 | 0.8583 | Nocardiaceae | 0.0140 | + |
| Promicromonosporaceae | 3.24 × 10−7 | − | |||
| Propionibacteriaceae | 6.59 × 10−9 | − | |||
| Bifidobacteriaceae | 3.00 × 10−7 | + | |||
| Solirubrobacteraceae | 0.0431 | + | |||
| Alicyclobacillaceae | 0.0029 | + | |||
| Pasteuriaceae | 0.0126 | − | |||
| Carnobacteriaceae | 0.0045 | − | |||
| Leuconostocaceae | 7.80 × 10−5 | − | |||
| Sphingomonadaceae | 1.44 × 10−8 | + | |||
| Rhodocyclaceae | 0.0066 | − | |||
| Nitrate | 1.469 × 10−8 | 0.5524 | Brevibacteriaceae | 1.96 × 10−5 | − |
| Dermacoccaceae | 0.0030 | − | |||
| FW | 0.0002 | − | |||
| Rhodobiaceae | 0.1071 | + | |||
| Mycoplasmataceae | 0.0006 | − | |||
| Nitrite | 0.008712 | 0.1952 | Nitrospiraceae | 0.0699 | − |
| Planctomycetaceae | 0.0503 | + | |||
| Hyphomicrobiaceae | 0.0298 | − | |||
| Phyllobacteriaceae | 0.0193 | + | |||
| Rhodobacteraceae | 0.0291 | − | |||
| Xanthobacteraceae | 0.0534 | + | |||
| Performance (▽) | 1.726 × 10−9 | 0.6219 | Holophagaceae | 0.0171 | + |
| Clade CL500.29 | 0.0004 | + | |||
| Kineosporiaceae | 0.0034 | − | |||
| Micrococcaceae | 5.78 × 10−8 | − | |||
| Fusobacteriaceae | 0.0001 | − | |||
| Rhodobiaceae | 0.0197 | − | |||
| Shewanellaceae | 0.0001 | − | |||
| Sphingomonadaceae | 0.0875 | + | |||
| pH | 0.000205 | 0.307 | Dietziaceae | 0.0143 | − |
| Microbacteriaceae | 0.0646 | + | |||
| Micrococcaceae | 0.0002 | + | |||
| Saprospiraceae | 0.0448 | − | |||
| Moraxellaceae | 0.0122 | − | |||
| Phosphate | 9.567 × 10−6 | 0.3917 | Flavobacteriaceae | 1.21 × 10−5 | − |
| Sphingobacteriaceae | 0.0079 | + | |||
| Alicyclobacillaceae | 0.0717 | + | |||
| Carnobacteriaceae | 0.0025 | + | |||
| Leuconostocaceae | 0.0009 | − | |||
| Turbidity | 2.014 × 10−11 | 0.6599 | Actinomycetaceae | 6.55 × 10−6 | + |
| Fusobacteriaceae | 1.55 × 10−11 | − | |||
| “Isosphaeraceae” | 0.0020 | + | |||
| Bradyrhizobiaceae | 0.0007 | − | |||
| Shewanellaceae | 0.0076 | − | |||
| Peptococcaceae | 0.0831 | + | |||
| Total viable bacteria | 2.2 × 10−16 | 0.8407 | Catenulisporaceae | 1.88 × 10−9 | − |
| Rivulariaceae | 2 × 10−16 | − | |||
| Nitrospiraceae | 0.0014 | + | |||
| “Gemmataceae” | 0.0020 | − | |||
| “Pirellulaceae” | 0.0067 | + | |||
| “Procabacteriaceae” | 0.0331 | − |
FIG 4 .
Scatterplots showing the correlation between sand filter performance (▽) and species evenness at different levels of classification. Panels: A, phylum; B, class; C, order; D, family; E, genus; F, OTU. A higher ▽ value corresponds to better water quality performance. Note the different y-axis scales.
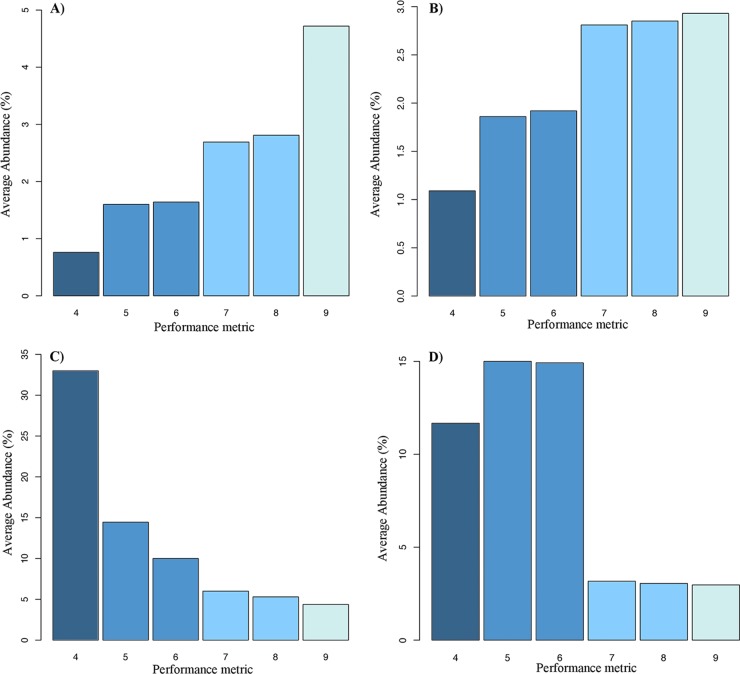
There was a significant relationship between performance classification (poor, average, good, and excellent) and the community composition (MANOVA, P value = 0.01). We determined the major organisms contributing to these performance differences through SIMPER analysis (Table 4). It was found that for excellent performance, an evenly distributed community is required with no overly abundant organisms and that poor performance is due to an uneven community structure, notably, an overabundance of Acidovorax and Sphingobium, and an underabundance of Halomonas and Sphingomonas (Fig. 5), as well as the complete absence of Naxibacter, Streptophyta, and Acinetobacter compared with filters with good or excellent performance. This confirms the relationship between performance and evenness discussed above.
TABLE 4 .
SIMPER analysis of the top 15 taxa accounting for the majority of the dissimilarities between SSFs producing different levels of water quality
| Taxon | Genus | Contribution (%) | Avg % with performance rating of: |
||
|---|---|---|---|---|---|
| Poor | Avg | Excellent | |||
| Alphaproteobacteria | Sphingobium | 12.61 | 14.96 | 2.97 | |
| Gammaproteobacteria | Pseudomonas | 4.55 | 5.72 | 3.52 | |
| Betaproteobacteria | Acidovorax | 10.27 | 12.23 | 4.37 | |
| Bacteroidetes | Flavobacterium | 6.64 | 6.51 | 6.07 | |
| Alphaproteobacteria | Methylobacterium | 2.37 | 1.81 | 1.14 | |
| Gammaproteobacteria | Halomonas | 5.72 | 1.62 | 6.34 | |
| Alphaproteobacteria | Sphingomonas | 3.11 | 1.89 | 2.93 | |
| Betaproteobacteria | Naxibacter | 3.63 | 3.18 | 2.55 | |
| Betaproteobacteria | Massilia | 1.44 | 0.7 | 1.4 | |
| Betaproteobacteria | Polynucleobacter | 0.005 | 1.88 | 0.84 | |
| Alphaproteobacteria | Novosphingobium | 2.17 | 1.98 | 1.53 | |
| Bacteroidetes | Arcicella | 1.4 | 0.7 | 1.02 | |
| Gammaproteobacteria | Nevskia | 0.002 | 0.13 | 0.36 | |
| Streptophyta | Streptophyta | 0.19 | 1.11 | 0.11 | |
| Gammaproteobacteria | Acinetobacter | 0.86 | 0.89 | 0.24 | |
| Alphaproteobacteria | Sphingobium | 17.16 | 24.33 | 38.73 | |
| Gammaproteobacteria | Pseudomonas | 12.61 | 14.96 | 2.97 | |
| Betaproteobacteria | Acidovorax | 4.55 | 5.72 | 3.52 | |
| Bacteroidetes | Flavobacterium | 10.27 | 12.23 | 4.37 | |
| Alphaproteobacteria | Methylobacterium | 6.64 | 6.51 | 6.07 | |
| Gammaproteobacteria | Halomonas | 2.37 | 1.81 | 1.14 | |
| Alphaproteobacteria | Sphingomonas | 5.72 | 1.62 | 6.34 | |
| Betaproteobacteria | Naxibacter | 3.11 | 1.89 | 2.93 | |
| Betaproteobacteria | Massilia | 3.63 | 3.18 | 2.55 | |
| Betaproteobacteria | Polynucleobacter | 1.44 | 0.7 | 1.4 | |
| Alphaproteobacteria | Novosphingobium | 0.005 | 81.8 | 0.84 | |
| Bacteroidetes | Arcicella | 2.17 | 1.98 | 1.53 | |
| Gammaproteobacteria | Nevskia | 1.4 | 0.7 | 1.02 | |
| Streptophyta | Streptophyta | 0.002 | 0.13 | 0.36 | |
| Gammaproteobacteria | Acinetobacter | 0.19 | 1.11 | 0.11 | |
| Alphaproteobacteria | Sphingobium | 4.14 | 3.11 | 2.97 | |
| Gammaproteobacteria | Pseudomonas | 6.41 | 8.68 | 3.52 | |
| Betaproteobacteria | Acidovorax | 5.5 | 5.65 | 4.37 | |
| Bacteroidetes | Flavobacterium | 6.13 | 5.51 | 6.07 | |
| Alphaproteobacteria | Methylobacterium | 3.82 | 4.31 | 1.14 | |
| Gammaproteobacteria | Halomonas | 5.64 | 2.75 | 6.34 | |
| Alphaproteobacteria | Sphingomonas | 4.57 | 2.83 | 2.93 | |
| Betaproteobacteria | Naxibacter | 2.82 | 2.21 | 2.55 | |
| Betaproteobacteria | Massilia | 2.92 | 2.85 | 1.4 | |
| Betaproteobacteria | Polynucleobacter | 1.33 | 1.26 | 0.84 | |
| Alphaproteobacteria | Novosphingobium | 1.78 | 1.12 | 1.53 | |
| Bacteroidetes | Arcicella | 2.04 | 1.44 | 1.02 | |
| Gammaproteobacteria | Nevskia | 1.35 | 1.71 | 0.36 | |
| Streptophyta | Streptophyta | 0.85 | 0.98 | 0.11 | |
| Gammaproteobacteria | Acinetobacter | 1.32 | 1.19 | 0.24 | |
FIG 5 .
Bar graphs of the average percent abundance of four key genera at different levels of water quality. Panels: A, Halomonas; B, Sphingomonas; C, Acidovorax; D, Sphingobium. Note the different y-axis scales.
DISCUSSION
SSFs host diverse bacterial communities.
The first studies to attempt to characterize the microbial ecology of SSFs were performed several decades ago (22, 23). They concluded that the diversity of the bacterial communities in these filters was low. However, that work was carried out with conventional plating and isolation techniques, which are known to underestimate the true diversity. Since that initial work, several studies have been published (5, 7, 9, 11–13, 24, 25) that have begun to use more modern molecular methods in order to answer the same questions as Brink and Lloyd. Those studies have all found that SSF communities are extremely diverse both metabolically and phylogenetically (24) However, with the exception of references 12 and 13, all of those studies were of SSFs used to purify wastewater or storm water (rather than drinking water, as in this study) or only of samples from the Schmutzdecke and not from various depths, as in this study.
We found the microbial diversity in our two SSFs to be far larger than previously reported, with 36 phyla and 239 families observed, compared to the 21 phyla and 149 families found by Wakelin et al. (11) in an Australian SSF. Such differences in diversity might be explained by the contrasting methodological approaches and primers used, as well as the different water sources and, perhaps more significantly, the different depths sampled within the SSF. Wakelin et al. (11) used storm water and sampled only the Schmutzdecke. However, as in references 11 and 26, Proteobacteria was found to be the dominant phylum. We also observed a significant difference between the sand community and the influent and effluent water. This is not surprising, given the differences between these environments and those observed in other sediment systems (27). More interestingly, the small OTU overlap between our sand and water samples may suggest that other sources could be important to the community, with the caveats of sampling and sequencing noise discussed above.
Reproducibility of filter performance and microbial community composition.
The microbial community compositions of the SSFs were significantly different, depending upon the status (operational or drained), filter age, sample location, month of sample collection, and the distances from the influent and effluent pipes and the depths at which samples were taken (Table 3). This is a novel observation. The age of the filter was the most significant parameter in explaining both changes in the microbial community and a water quality variable, which is not surprising, as it is widely documented by operators that SSF performance improves with maturity (2). Additionally, the increase in the abundance of Ruminococcaceae and the presence of the nine other families in older filters (see Table S1 in the supplemental material) can be explained by the fact that they are all either facultative or strict anaerobes commonly found in wastewater and sewage (28). Their increased abundance is likely due to prolonged exposure to feces from wildlife (i.e., birds) surrounding the filters and similar exposure at the reservoir feeding the filters.
Surprisingly, filter identity did not impact microbial community structure. This suggests that the communities at this site are highly reproducible and that a characteristic microbiota is present; the extent to which this is true at other sites is an interesting open question. We also observed similar water quality production by the filters, which may be linked to the similarity of the microbiota. Another surprising finding was that depth was of only marginal significance in explaining differences in community composition. This contrasts with many other freshwater studies (29), where depth has been shown to be extremely important, as it is linked to chemical gradients driving changes in community composition. Although it was surprising to find depth a marginally significant variable, this is not the first study to do so. Recently Röske et al. (30) showed that depth was not significant in explaining community composition in sediment from the Saidenbach drinking water reservoir in Germany. Regardless of this, future work should focus on determining whether such water chemistry gradients exist and if they affect or shape the microbial communities of SSFs.
Although vertical (depth) spatial differences in the SSF microbial community were marginal, lateral differences (side and distances from the influent and effluent pipes) were highly significant. Both can be a consequence of habitat heterogeneities imposed by differences in physicochemical characteristics (31) such as partially filled or unfilled voids between sand grains that would disperse nutrients and microbes or the dilution of components away from the influent pipe, creating nutritional gradients. Perhaps such a dispersal of nutrients occurs faster and more easily along the surface of SSFs rather than vertically, and thus, this may account for the higher significance of lateral than vertical spatial differences.
Species evenness is critical to performance.
Stepwise multivariate regression showed that the water quality performance of SSFs significantly correlates with both the age and species evenness of the filters (Table 3), with better-performing filters having greater evenness (Fig. 4). This is the first study, to our knowledge, to correlate bacterial species evenness with the differing levels of performance of water filters (Fig. 4). Greater evenness has been linked to greater robustness and functional stability and therefore the ability to adapt to new and fluctuating parameters such as those brought by weather events (e.g., storms) that would impact the composition of the influent water feeding the filters (17). Therefore, the increased species evenness and richness found in excellently performing filters is additional confirmation of the “insurance hypothesis” conceived by Yachi and Loreau (32), which proposes that both functional redundancy and evenness are necessary for functionally robust ecosystems.
The importance of species evenness is further emphasized by the dramatic effect seen during draining events compared to operational times, in particular, the overabundance of Firmicutes and Planctomycetes. This dominance may be directly related to the fact that Firmicutes bacteria are known to produce endospores during periods of starvation or stress (i.e., during SSF draining periods, when organisms in the sand are exposed to temperature, pH, oxygen, nutrition, and UV fluctuations). The dominance of Planctomycetes can be explained by the increased exposure to sunlight (due to reduced water depth), resulting in heightened algal growth, which has been shown to promote increased growth of Planctomycetes bacteria (33). Likewise, a similar effect occurs during chlorination, with the decline in evenness being attributed to an increase in Deltaproteobacteria. Deltaproteobacteria are widely documented as being capable of reductive dechlorination or halorespiration, the process of using halogenated compounds, such as sodium hypochlorite, as terminal electron acceptors in anaerobic respiration (34). This may explain their dominance after chlorination.
Nonparametric MANOVA revealed that, at different levels of performance (excellent, average, and poor), the composition of the microbial community is different. During periods of excellent performance, there is an increased abundance of Sphingomonas and Acinetobacter (Table 4), two genera known to be capable of the biodegradation and metabolism of a wide range of chemicals, e.g., polycyclic aromatic hydrocarbons (PAHs), cyanotoxins, endocrine disruptors, and herbicides (35–39), all of which would likely be present in the reservoir water feeding the SSFs. Moreover, Innerebner et al. (40) recently showed that Sphingomonas exerts a striking plant-protective effect by suppressing disease symptoms and diminishing pathogen growth. Therefore, within SSFs, this recent finding may help to explain why their abundance is greater in filters with excellent performance, which typically have no or low pathogen counts. Likewise, the increased abundance of Halomonas in filters with excellent performance may be explained by the recent discovery that several members of this genus can produce bioflocculants capable of a >80% turbidity reduction (41).
Conversely, in filters showing poor performance, the overabundance of Acidovorax and Sphingobium may be explained by niche competition; both of these genera are known to be capable of processes similar to those of Sphingomonas and Acinetobacter, which are found in greater abundance in filters displaying excellent performance. The antagonistic effects of such competition between members of the Sphingomonadaceae family (Sphingomonas, Sphingobium, Novosphingobium, and Sphingopyxis) has been examined (42) and has been shown to have effects on PAH removal from contaminated soils. However, it is important to note that it is impossible to determine if differences in filter performance are due to the microbial community or if it is the performance of the filters that shapes the community. Speculatively, the former seems most plausible, as SSFs function predominantly via biological mechanisms. Additionally, little change in the characteristics of the influent water feeding the filters was found (see Fig. S1 in the supplemental material), and therefore, performance differences must be attributed to the microbial community. Overall, these findings show that greater species evenness is integral to excellent SSF performance and for the first time associate specific genera with differing levels of water quality performance.
Conclusion.
In summary, this study is the first to provide a functionally and operationally relevant spatial and temporal characterization of the microbial community structures of two full-scale SSFs. We conclude that the microbial diversity of SSFs is far greater than previously documented and that in terms of community composition and performance, the two SSFs sampled were indistinguishable and highly reproducible. Both filters produced high-quality drinking water, with quality improving as the filters matured. The month, filter age, and side and the distances from the influent and effluent pipes and depths at which the samples were taken significantly impacted the microbial community in SSFs, with filter age being the most significant variable. As the filters aged, both the number and density of OTUs increased, as did species evenness. Further, Illumina sequencing indicated that the abundance of various members of the microbial community, specifically, Acidovorax, Halomonas, Sphingobium, and Sphingomonas, was important for performance. More significantly, it was found that increased species evenness was critical for excellent filter performance. Decreased species evenness was found in drained and early-stage SSFs, coinciding with an increased abundance of Planctomycetes bacteria, likely induced by additional exposure to sunlight. Future work should investigate the impact of reducing the drainage period or the effects of covering filters during draining and scraping events on species evenness and the abundance of Planctomycetes bacteria. Such work could significantly reduce the period of time SSFs are nonoperational because of poor performance and hence have economic benefits.
Together, the results of this study provide the most detailed characterization of the functional microbial community found in SSFs to date and provide a framework for future ecological and physiological microbial research on these systems. This study is the first to provide insight into the importance of specific taxa and community evenness to performance. However, quantification of the extent of their importance versus other abiotic and biotic factors, such as the role played by protozoa and fungi, will require additional field-based studies, as well as ecophysiological studies under carefully controlled laboratory conditions.
MATERIALS AND METHODS
Operation and setup of SSFs.
Two-dimensionally identical full-scale SSFs (filters A and B) at Scottish Water’s Fairmilehead Water Treatment Works in Edinburgh were sampled approximately monthly from April until November 2011 (see Fig. S1 in the supplemental material), with the filters being decommissioned by the addition of chlorine in November 2011. The filters differed only in age (days since scraped). The Fairmilehead site has seven SSFs that receive raw water from several reservoirs in southern Scotland. The filters have a bed depth of 1 m and a surface area of approximately 1,800 m2. Additional to the monthly sampling, an 8-week intensive sampling strategy was used from May to June. The purpose of the intensive sampling program was to allow the SSF community to be monitored more closely during a time hypothesized to be more microbially active. In total, 16 sampling sessions were conducted, providing data from representative points in the life cycle of the filters (scraped, juvenile, mature, clogged, and drained; Fig 1). It should be noted that drained filters were sampled 20 h after draining had occurred. Further, the first sampling points taken during decommissioning were collected 20 h after chlorine delivery; both filters remained operational, with the water they produced entering the distribution system until November 2011.
Sampling of SSFs.
Sampling entailed the collection of one 50-cm sand core from each side of both filter beds with a multistage sediment sampler (AMS, American Falls, ID). Cores were taken at various locations on the three separate accessible sides of the filters (see Fig. S1 in the supplemental material). These undisturbed cores were sectioned at eight depths (0, 4, 10, 15, 20, 30, 40, and 50 cm), and 0.5 g of each subsample was used for DNA extraction with FastDNA spin kits for soil (MP Bio-Medical, Cambridge, United Kingdom) in accordance with the manufacturer’s instructions. Furthermore, in order to gain a better understanding of the spatial variation within the community on a more microbially realistic scale, six cores were taken at each side of both full-scale SSFs on 21 June 2011 (see Fig. S1C in the supplemental material).
On each sampling occasion, 2-liter samples of influent and effluent water were collected from the two filters. Water temperature, dissolved oxygen, and pH were measured on site with portable meters. Water samples were processed in triplicate for turbidity, DOC, specific UV absorbency, chemical oxygen demand, nitrate, nitrite, ammonia, phosphate, and total coliforms by the methodology outlined in water industries standard methods (43); additionally, total viable bacterial counts at 13°C and 30°C were performed as described in reference 9. To evaluate overall filter performance, the newly created aggregate performance metric ▽ (discussed in reference 12) was used. This parameter assigned the effluent of each filter a number from 0 to 10 based on the number of water quality parameters outlined in reference 1 it fulfilled. The ranking is as follows: 0 to 4 is designated poor performance, 5 or 6 is average, 7 or 8 is good, and 9 or 10 is excellent.
Illumina 16S rRNA amplicon sequencing.
The 16S rRNA gene amplicons of 674 full-scale SSF samples (56 water and 618 sand samples), representing different depths, filters, filter ages, and levels of filter performance were processed by the Earth Microbiome Project with primers 515F (GTGCCAGCMGCCGCGGTAA) and 806R (GGACTACHVGGGTWTCTAAT) in accordance with the protocol outlined in reference 44. Amplified samples were pooled (equimolar concentrations) and sequenced on an Illumina HiSeq 2000. Quality filtering of reads was applied as described previously (44). Reads were assigned to OTUs (cutoff of 97% sequence identity) by using a closed reference OTU-picking protocol with QIIME version 5 and the Greengenes database (version 13.5) (49). Additionally, two mock communities processed in triplicate were included in order to act as positive controls. Both the raw and processed reads can be found at http://qiita.microbio.me/study/description/755. Furthermore, to verify that the pipeline and database used did not bias the results, the data were additionally processed with Mothur (version 1.36.1) and the SILVA database (release 119).
Statistics.
Correlations between water quality parameters were explored by using the nonparametric Kendall τ procedure. Taxonomic and OTU tables generated for the samples were used to calculate pairwise dissimilarities between samples based on the Bray-Curtis dissimilarity index. The resulting matrices were examined for temporal and spatial patterns in the bacterial community structure by NMDS as implemented in the Vegan package (45). Significant differences in the microbial community compositions of filters, filter ages, depths, locations of the cores, seasons, and addition of chlorine were determined by nonparametric MANOVA (46). To determine the contributions of individual taxa to differences in filter performance, SIMPER analysis (47) was used. SIMPER analysis is a useful way to measure the magnitudes of differences; however, in order to decide whether a taxon differed significantly, pairwise t tests (Kendall nonparametric) adjusted for multiple comparisons by the Benjamini-Hochberg false-discovery method (48) were performed. Only taxa with a false-discovery rate of <5% were reported. Shannon diversity indices, Chao’s richness, Pielou’s evenness, and rarefaction curves were calculated on rarefied samples at a 3% genetic distance. The relationships between environmental variables and patterns in bacterial community structure were examined by CCA with significance tested by ANOVA after reducing the overall suite of environmental variables with a stepwise Akaike information criterion model. Additionally, the functional relationships between water quality parameters and bacterial groups were analyzed by stepwise multivariate forward/reverse regression analysis. All statistical analysis was performed in R (R Development Core Team, 2011).
SUPPLEMENTAL MATERIAL
Schematic of Fairmilehead filter plant (A). Shown are sampling locations within the filter bed, where numbers represent the sampling order (B), and mesoscale sampling locations (C). Download
Summary of physical and chemical characteristics of influent water. Filter age is the number of days since the filter was scraped. Shown are the temperature (TEMP) in degrees Celsius, pH, turbidity (NTU), DO in milligrams per liter, DOC in milligrams per liter, UV absorbance at 254 nm, specific UV absorbance (SUVA) in liters per milligram-meter, chemical oxygen demand (COD) in milligrams per liter, phosphate (PO4) in milligrams per liter, nitrate (NO3) in milligrams per liter, nitrite (NO2) in milligrams per liter, ammonium (NH4) in milligrams per liter, total viable bacteria (TVB) grown at 30°C, TVB grown at 13°C (TVB13°C), and coliforms in CFU per milliliter. Download
Summary of physical and chemical characteristics of effluent water. Filter age is the number of days since the filter was scraped. Parameter names and units are the same as in Figure S2. Download
Summary of the percent removal of the physical and chemical characteristics. Filter age is the number of days since the filter was scraped. Parameter names and units are the same as in Figure S2. Download
Kendall correlation tests showing the relationships between the percent removal values of the various water quality parameters. Red indicates positive correlations, and blue indicates negative correlations. P values: period, 0.5; *, 0.1; **, 0.001; ***, 0. Temperature, pH, and UV were left out of the analysis. Download
Bar graphs showing the actual numbers of OTUs (light blue) and the numbers of sequenced OTUs (dark blue) in two mock communities. (a) A SSF mock community created from the clone library mentioned in reference 12. (b) The mock community created by Shakya et al. (M. Shakya, C. Quince, J. H. Campbell, Z. K. Yang, C. W. Schadt, and M. Podar, Environ Microbiol 15:1882–1892, 2013). Download
Average relative abundances of the top 18 families in SSFs in different filter age bins (early, 0 to 4 weeks after scraping; mid, 5 to 8 weeks after scraping; late, >10 weeks after scraping). Download
Heat maps showing the temporal and spatial changes in species evenness in SSFs A and B. Panels: a, filter A length 1; b, filter A length 2; c, filter A length 3; e, filter B length 1; f, filter B length 3. Panel d is an aerial picture showing lengths and filter locations. Download
Stacked bar graphs depicting the percent abundances of each phylum at different depths and locations in filters A (panels A and B) and B (panels C and D) over time. Download
Average percent relative abundances of the nine families found only in late filter age-binned SSFs.
ACKNOWLEDGMENTS
S.H. is supported by a Lord Kelvin/Adam Smith Research scholarship from the University Of Glasgow. C.Q. is funded through an MRC fellowship (MR/M50161X/1) as part of the Cloud Infrastructure for Microbial Bioinformatics (CLIMB) consortium (MR/L015080/1).
Special thanks to the Earth Microbiome Project, Scottish Water, operators at the Fairmilehead water treatment works, Ian Scouller, Stuart McLean, and Robert Boyd, without whom this work would not have been possible.
Footnotes
Citation Haig S-J, Quince C, Davies RL, Dorea CC, Collins G. 2015. The relationship between microbial community evenness and function in slow sand filters. mBio 6(5):e00729-15. doi:10.1128/mBio.00729-15.
REFERENCES
- 1.European Union Council 1998. Council directive 98/83/EC. Relative to the quality of drinking water. European Union; Council, Brussels, Belgium: http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:31998L0083. [Google Scholar]
- 2.Huisman L, Wood W. 1974. Slow sand filtration. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Haig SJ, Collins G, Davies RL, Dorea CC, Quince C. 2011. Biological aspects of slow sand filtration: past, present and future. Water Sci Technol 11:468–472. doi: 10.2166/ws.2011.076. [DOI] [Google Scholar]
- 4.Arndt R, Wagner E. 2004. Rapid and slow sand filtration techniques and their efficacy at filtering triactinomyxons of Myxobolus cerebralis from contaminated water. N Am J Aquacult 66:261–270. doi: 10.1577/A04-004.1. [DOI] [Google Scholar]
- 5.Calvo-Bado LA, Pettitt TR, Parsons N, Petch GM, Morgan JAW, Whipps JM. 2003. Spatial and temporal analysis of the microbial community in slow sand filters used for treating horticultural irrigation water. Appl Environ Microbiol 69:2116–2125. doi: 10.1128/AEM.69.4.2116-2125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urbonas BR. 1999. Design of a sand filter for stormwater quality enhancement. Water Environ Res 71:102–113. doi: 10.2175/106143099X121625. [DOI] [Google Scholar]
- 7.Ramond J, Welz PJ, Tuffin MI, Burton SG, Cowan DA. 2013. Assessment of temporal and spatial evolution of bacterial communities in a biological sand filter mesocosm treating winery wastewater. J Appl Microbiol 115:91–101. doi: 10.1111/jam.12203. [DOI] [PubMed] [Google Scholar]
- 8.Weber-Shirk M, Dick R. 1997. Biological mechanisms in slow sand filters. J Am Water Works Assoc 89:72–83. [Google Scholar]
- 9.Bahgat M, Dewedar A, Zayed A. 1999. Sand-filters used for wastewater treatment: buildup and distribution of microorganisms. Water Res 33:1949–1955. doi: 10.1016/S0043-1354(98)00290-5. [DOI] [Google Scholar]
- 10.Aslan S. 2008. Biological nitrate removal in a laboratory-scale slow sand filter. Water S Afr 34:99–105. http://www.wrc.org.za/Knowledge%20Hub%20Documents/Water%20SA%20Journals/Manuscripts/2008/01/WaterSA_2008_01_2041.pdf. [Google Scholar]
- 11.Wakelin S, Page D, Dillon P, Pavelic P, Abell GCJ, Gregg AL, Brodie E, DeSantis TZ, Goldfarb KC, Anderson G. 2011. Microbial community structure of a slow sand filter Schmutzdecke: a phylogenetic snapshot based on rRNA sequence analysis. Water Sci Technol 11:426–436. doi: 10.2166/ws.2011.063. [DOI] [Google Scholar]
- 12.Haig S, Quince C, Davies RL, Dorea CC, Collins G. 2014. Replicating the microbial community and water quality performance of full-scale slow sand filters in laboratory-scale filters. Water Res 61:141–151. doi: 10.1016/j.watres.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Haig S, Schirmer M, D’Amore R, Gibbs J, Davies RL, Collins G, Quince C. 2015. Stable-isotope probing and metagenomics reveal predation by protozoa drives E. coli removal in slow sand filters. ISME J 9:797–808. doi: 10.1038/ismej.2014.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashsham SA, Fernandez AS, Dollhopf SL, Dazzo FB, Hickey RF, Tiedje JM, Criddle CS. 2000. Parallel processing of substrate correlates with greater functional stability in methanogenic bioreactor communities perturbed by glucose. Appl Environ Microbiol 66:4050–4057. doi: 10.1128/AEM.66.9.4050-4057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK. 2005. The contribution of species richness and composition to bacterial services. Nature 436:1157–1160. doi: 10.1038/nature03891. [DOI] [PubMed] [Google Scholar]
- 16.Allison SD, Martiny JBH. 2008. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A 105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, De Vos P, Verstraete W, Boon N. 2009. Initial community evenness favours functionality under selective stress. Nature 458:623–626. doi: 10.1038/nature07840. [DOI] [PubMed] [Google Scholar]
- 18.Werner JJ, Knights D, Garcia ML, Scalfone NB, Smith S, Yarasheski K, Cummings TA, Beers AR, Knight R, Angenent LT. 2011. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc Natl Acad Sci U S A 108:4158–4163. doi: 10.1073/pnas.1015676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schirmer M, Ijaz UZ, D’Amore R, Hall N, Sloan WT, Quince C. 2015. Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res 43:e37. doi: 10.1093/nar/gku1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Pryor MA, Edwards MA, Falkinham JO III, Pruden A. 2013. Effect of GAC pre-treatment and disinfectant on microbial community structure and opportunistic pathogen occurrence. Water Res 47:5760–5772. doi: 10.1016/j.watres.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 21.Gallego V, Sánchez-Porro C, García MT, Ventosa A. 2006. Massilia aurea sp. nov., isolated from drinking water. Int J Syst Evol Microbiol 56(Pt 10):2449–2453. [DOI] [PubMed] [Google Scholar]
- 22.Brink N. 1967. Ecological studies in biological filters. Int Rev Gesamte Hydrobiol Hydographie 52:51–122. doi: 10.1002/iroh.19670520104. [DOI] [Google Scholar]
- 23.Lloyd B. 1974. The functional microbial ecology of slow, sand filters. Ph.D. thesis. University of Surrey, Surrey, United Kingdom. [Google Scholar]
- 24.Eighmy T, Collins R, Spanos M, Fenstermacher J. 1992. Microbial populations, activities and carbon metabolism in slow sand filters. Water Res 26:1319–1328. doi: 10.1016/0043-1354(92)90126-O. [DOI] [Google Scholar]
- 25.Bai Y, Liu R, Liang J, Qu J. 2013. Integrated metagenomic and physiochemical analyses to evaluate the potential role of microbes in the sand filter of a drinking water treatment system. PLoS One 8:e61011. doi: 10.1371/journal.pone.0061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petry-Hansen H, Steele H, Grooters M, Wingender J, Flemming H. 2006. Feacal contamination indicator organisms in slow sand filters, p. 143–151. In Gimbel R, Graham NJD, Collins MR (ed), Slow sand filtration: recent developments in water treatment technology. IWA Publishing, London, United Kingdom. [Google Scholar]
- 27.Gobet A, Böer SI, Huse SM, van Beusekom JEE, Quince C, Sogin ML, Boetius A, Ramette A. 2012. Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J 6:542–553. doi: 10.1038/ismej.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitman WB, Goodfellow MK, Kämpfer P, Busse HJ, Trujillo ME, Ludwig W. 2012. Bergey’s manual of systematic bacteriology of systematic bacteriology, vol 5: the Actinobacteria, part A and B. Springer, New York, NY. [Google Scholar]
- 29.Lin X, McKinley J, Resch CT, Kaluzny R, Lauber CL, Fredrickson J, Knight R, Konopka A. 2012. Spatial and temporal dynamics of the microbial community in the Hanford unconfined aquifer. ISME J 6:1665–1676. doi: 10.1038/ismej.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Röske K, Sachse R, Scheerer C, Röske I. 2012. Microbial diversity and composition of the sediment in the drinking water reservoir Saidenbach (Saxonia, Germany). Syst. Appl. Microbiol 35: 35–44. [DOI] [PubMed] [Google Scholar]
- 31.Deschesne A, Pallud C, Grundmann GL. 2007. Spatial distribution of bacteria at the microscale in soil, p 87–107. In Franklin RB, Mills AL (ed), The spatial distribution of microbes in the environment. Springer, New York, NY. [Google Scholar]
- 32.Yachi S, Loreau M. 1999. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc Natl Acad Sci U S A 96:1463–1468. doi: 10.1073/pnas.96.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pizzetti I, Fuchs BM, Gerdts G, Wichels A, Wiltshire KH, Amann R. 2011. Temporal variability of coastal planctomycetes clades at Kabeltonne Station, North Sea. Appl Environ Microbiol 77:5009–5017. doi: 10.1128/AEM.02931-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson RE. 2013. Genomic insights into organohalide respiration. Curr Opin Biotechnol 24:498–505. doi: 10.1016/j.copbio.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 35.Shi T, Fredrickson JK, Balkwill DL. 2001. Biodegradation of polycyclic aromatic hydrocarbons by Sphingomonas strains isolated from the terrestrial subsurface. J Ind Microbiol Biotechnol 26:283–289. doi: 10.1038/sj.jim.7000130. [DOI] [PubMed] [Google Scholar]
- 36.Bending GD, Lincoln SD, Sørensen SR, Morgan JAW, Aamand J, Walker A. 2003. In-field spatial variability in the degradation of the phenyl-urea herbicide isoproturon is the result of interactions between degradative Sphingomonas spp. and soil pH. Appl Environ Microbiol 69:827–834. doi: 10.1128/AEM.69.2.827-834.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valeria AM, Ricardo EJ, Stephan P, Alberto WD. 2006. Degradation of microcystin-RR by Sphingomonas sp. CBA4 isolated from San Roque reservoir (Cordoba-Argentina). Biodegradation 17:447–455. doi: 10.1007/s10532-005-9015-9. [DOI] [PubMed] [Google Scholar]
- 38.Fang HHP, Liang D, Zhang T. 2007. Aerobic degradation of diethylphthalate by Sphingomonas sp. Bioresour Technol 98:717–720. doi: 10.1016/j.biortech.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 39.Williams PA, Ray C. 2008. Catabolism of aromatic compounds by Acinetobacter, p 99–117. In Gerischer U (ed), Acinetobacter molecular biology. Caister Academic Press, United Kingdom. [Google Scholar]
- 40.Innerebner G, Knief C, Vorholt JA. 2011. Sphingomonas strains protect Arabidopsis thaliana against leaf pathogenic Pseudomonas syringae in a controlled model system. Appl Environ Microbiol 77:3202–3210. doi: 10.1128/AEM.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cosa S, Ugbenyen AM, Mabinya LV, Rumbold K, Okoh AI. 2013. Characterization and flocculation efficiency of a bioflocculant produced by a marine Halobacillus. Environ Tech 34:2671–2679. [DOI] [PubMed] [Google Scholar]
- 42.Cunliffe M, Kertesz MA. 2006. Effect of Sphingobium yanoikuyae B1 inoculation on bacterial community dynamics and polycyclic aromatic hydrocarbon degradation in aged and freshly PAH-contaminated soils. Environ Pollut 144:228–237. doi: 10.1016/j.envpol.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 43.Clesceri L, Greenberg A, Eaton A, Association APH (ed). 2005. Standard methods for the examination of water and wastewater, 21st ed. American Public Health Association, Washington, DC. [Google Scholar]
- 44.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oksanen J, Blanchet, Kindt FG, Legendre R, Minchin P, O’Hara PR, O’Hara RB. 2012. Vegan: community ecology package, R package version 2.0-3. https://cran.r-project.org/web/packages/vegan/index.html.
- 46.Anderson M. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46. [Google Scholar]
- 47.Clarke KR. 1993. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 48.Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann Stat 29: 1165–1188. [Google Scholar]
- 49.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Keller EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic of Fairmilehead filter plant (A). Shown are sampling locations within the filter bed, where numbers represent the sampling order (B), and mesoscale sampling locations (C). Download
Summary of physical and chemical characteristics of influent water. Filter age is the number of days since the filter was scraped. Shown are the temperature (TEMP) in degrees Celsius, pH, turbidity (NTU), DO in milligrams per liter, DOC in milligrams per liter, UV absorbance at 254 nm, specific UV absorbance (SUVA) in liters per milligram-meter, chemical oxygen demand (COD) in milligrams per liter, phosphate (PO4) in milligrams per liter, nitrate (NO3) in milligrams per liter, nitrite (NO2) in milligrams per liter, ammonium (NH4) in milligrams per liter, total viable bacteria (TVB) grown at 30°C, TVB grown at 13°C (TVB13°C), and coliforms in CFU per milliliter. Download
Summary of physical and chemical characteristics of effluent water. Filter age is the number of days since the filter was scraped. Parameter names and units are the same as in Figure S2. Download
Summary of the percent removal of the physical and chemical characteristics. Filter age is the number of days since the filter was scraped. Parameter names and units are the same as in Figure S2. Download
Kendall correlation tests showing the relationships between the percent removal values of the various water quality parameters. Red indicates positive correlations, and blue indicates negative correlations. P values: period, 0.5; *, 0.1; **, 0.001; ***, 0. Temperature, pH, and UV were left out of the analysis. Download
Bar graphs showing the actual numbers of OTUs (light blue) and the numbers of sequenced OTUs (dark blue) in two mock communities. (a) A SSF mock community created from the clone library mentioned in reference 12. (b) The mock community created by Shakya et al. (M. Shakya, C. Quince, J. H. Campbell, Z. K. Yang, C. W. Schadt, and M. Podar, Environ Microbiol 15:1882–1892, 2013). Download
Average relative abundances of the top 18 families in SSFs in different filter age bins (early, 0 to 4 weeks after scraping; mid, 5 to 8 weeks after scraping; late, >10 weeks after scraping). Download
Heat maps showing the temporal and spatial changes in species evenness in SSFs A and B. Panels: a, filter A length 1; b, filter A length 2; c, filter A length 3; e, filter B length 1; f, filter B length 3. Panel d is an aerial picture showing lengths and filter locations. Download
Stacked bar graphs depicting the percent abundances of each phylum at different depths and locations in filters A (panels A and B) and B (panels C and D) over time. Download
Average percent relative abundances of the nine families found only in late filter age-binned SSFs.