ABSTRACT
Mammalian lipopolysaccharide (LPS) binding proteins (LBPs) occur mainly in extracellular fluids and promote LPS delivery to specific host cell receptors. The function of LBPs has been studied principally in the context of host defense; the possible role of LBPs in nonpathogenic host-microbe interactions has not been well characterized. Using the Euprymna scolopes-Vibrio fischeri model, we analyzed the structure and function of an LBP family protein, E. scolopes LBP1 (EsLBP1), and provide evidence for its role in triggering a symbiont-induced host developmental program. Previous studies showed that, during initial host colonization, the LPS of V. fischeri synergizes with peptidoglycan (PGN) monomer to induce morphogenesis of epithelial tissues of the host animal. Computationally modeled EsLBP1 shares some but not all structural features of mammalian LBPs that are thought important for LPS binding. Similar to human LBP, recombinant EsLBP1 expressed in insect cells bound V. fischeri LPS and Neisseria meningitidis lipooligosaccharide (LOS) with nanomolar or greater affinity but bound Francisella tularensis LPS only weakly and did not bind PGN monomer. Unlike human LBP, EsLBP1 did not bind N. meningitidis LOS:CD14 complexes. The eslbp1 transcript was upregulated ~22-fold by V. fischeri at 24 h postinoculation. Surprisingly, this upregulation was not induced by exposure to LPS but, rather, to the PGN monomer alone. Hybridization chain reaction-fluorescent in situ hybridization (HCR-FISH) and immunocytochemistry (ICC) localized eslbp1 transcript and protein in crypt epithelia, where V. fischeri induces morphogenesis. The data presented here provide a window into the evolution of LBPs and the scope of their roles in animal symbioses.
IMPORTANCE
Mammalian lipopolysaccharide (LPS)-binding protein (LBP) is implicated in conveying LPS to host cells and potentiating its signaling activity. In certain disease states, such as obesity, the overproduction of this protein has been a reliable biomarker of chronic inflammation. Here, we describe a symbiosis-induced invertebrate LBP whose tertiary structure and LPS-binding characteristics are similar to those of mammalian LBPs; however, the primary structure of this distantly related squid protein (EsLBP1) differs in key residues previously believed to be essential for LPS binding, suggesting that an alternative strategy exists. Surprisingly, symbiotic expression of eslbp1 is induced by peptidoglycan derivatives, not LPS, a pattern converse to that of RegIIIγ, an important mammalian immunity protein that binds peptidoglycan but whose gene expression is induced by LPS. Finally, EsLBP1 occurs along the apical surfaces of all the host’s epithelia, suggesting that it was recruited from a general defensive role to one that mediates specific interactions with its symbiont.
INTRODUCTION
Over the last decade, several studies have demonstrated that mutualistic and pathogenic associations share a molecular language (for reviews, see references 1 and 2). Key elements of this dialogue are microbe-associated molecular patterns (MAMPs) of microbial cell surfaces, including lipopolysaccharide (LPS) and peptidoglycan (PGN) derivatives, and their cognate host sensors and receptors. The model mutualistic association between the Hawaiian bobtail squid Euprymna scolopes and the bioluminescent marine bacterium Vibrio fischeri has provided an experimental system for the study of these interactions. In the squid-vibrio association, symbiont lipid A, the lipid component of LPS, and the PGN monomer tracheal cytotoxin (TCT) direct various symbiont-induced developmental programs of the squid’s light organ, the set of tissues that harbors the symbionts and modifies their luminescence for use in host behaviors. MAMP-induced morphogenetic programs include the apoptotic loss of the ciliated epithelium that potentiates initial colonization, recruitment of macrophagelike blood cells into the organ, and the transformation of the biochemical environment where the symbionts take up residence in host tissues (for reviews, see references 3 and 4). The luminescence from the V. fischeri monoculture in the light organ allows E. scolopes to match ambient light and thereby disguise its shadow (5). Subsequent studies in mammals have demonstrated that MAMPs also drive development, such as immune system maturation, in complex host-microbial associations, including in the mammalian gut (2). This study uses the squid-vibrio model to provide experimental evidence for the role of lipopolysaccharide-binding protein (LBP), a member of the LBP/BPI (bactericidal/permeability-increasing protein) protein family, as a key player in host responses to symbiont LPS.
As MAMP recognition proteins, mammalian LBP and BPI are closely related, structurally similar proteins that serve complementary roles in innate immunity. LBP relays LPS to host cells, whereas BPI acts as a bactericidal/LPS-neutralizing effector molecule to help clear infection and resolve infection-induced inflammation (6, 7). LBP is present in plasma and tissue fluids, and recent reports have determined that LBP and BPI are expressed in the gut and other epithelia, where they likely mediate responses to the interfacing microbiota (8, 9). During an inflammatory response, the constitutively present levels of LBP are sufficient to catalyze extraction and delivery of individual LPS molecules to CD14 and MD-2/TLR4 to induce potent proinflammatory responses (10–13). LBP levels can increase up to 100-fold during acute-phase reactions following the initial inflammatory responses, promoting, along with BPI, noninflammatory clearance of LPS and eventual resolution of LPS-triggered inflammation (11, 14, 15). Unlike BPI, LBP binding to Gram-negative bacteria (e.g., Escherichia coli) does not produce lethal or sublethal alterations of the bacteria (16).
Despite differences in activity between mammalian LBP and BPI, the structural organization conferring function is generally similar. Thus, the N-terminal domain of each protein binds LPS, whether presented in the form of intact Gram-negative bacteria (16, 17), shed outer membrane blebs, or aggregates of purified LPS (18–20). The C-terminal domain of both LBP and BPI is responsible for delivery of the bound LPS-containing material to host cells (18, 20, 21). Conserved, positively charged residues near the N-terminal tip of both mammalian LBP and BPI are likely important in initial electrostatic interactions with typically polyanionic LPS (22–25). The net cationicity is greater in BPI than in LBP in both the charged N-terminal tip and across the molecule; the isoelectric point (pI) of LBP is near neutral, whereas the BPI pI is ~10. This difference in charge likely accounts for the higher LPS- and bacterial-binding affinity of BPI and the distinct effects that LBP and BPI produce on LPS-rich interfaces, including bacterial outer membranes (21, 26). BPI causes bacterial sublethal and lethal injury, whereas LBP promotes extraction of individual LPS molecules by CD14 at substoichiometric concentrations (12, 19).
Although the gene duplication giving rise to mammalian LBP and BPI is believed to have occurred after the radiation of the mammals (27), related proteins have been reported in several invertebrate groups (28, 29). While a given invertebrate species may have multiple LBP/BPI gene isoforms, an LBP/BPI dichotomy has not been well characterized outside the mammals. Sequence analysis across the animal kingdom shows that these proteins are relatively quickly evolving, with ~21% identity between E. scolopes LBP1 (EsLBP1) and mammalian LBP/BPI proteins, indicating ~1% change in the amino acid sequence every 7 million years. A comparative analysis of structure and function across these deep divergences that have occurred over more than 500 million years offers the opportunity to define biochemical features that are essential for function of these molecules, as well as to explore how evolutionary tinkering can give rise to diversity of function.
Full-length transcripts of four members of the LBP/BPI protein family have been identified in E. scolopes (28, 30). One of these proteins, EsLBP1 (previously called EsLBP [31]), is of particular interest as a candidate for responding to symbiont LPS during development. The pI of EsLBP1 is near neutral, suggesting that it functions similarly to mammalian LBPs, i.e., presenting LPS as a signal molecule. In addition, the gene encoding EsLBP1 increases in expression at 18 h following the onset of host-symbiont interaction, a time when morphogenesis is being signaled by MAMPs (31). In the present study, to provide insight into the possible role of an LBP in inducing an animal developmental program, we sought to characterize the biochemical properties of EsLBP1, comparing them to those of mammalian LBP, and to examine the timing and location of EsLBP1 gene expression and protein production through the early trajectory of host development.
RESULTS
Structural comparison of LPS-binding domain of mammalian LBP and BPI to corresponding region of EsLBP1.
We focused on the LPS-binding N-terminal domain of human LBP/BPI (hLBP/hBPI) proteins for comparison with EsLBP1 by primary sequence alignment (Fig. 1A). Overall, the degree of conservation with human LBP/BPI proteins is limited; EsLBP1 is 23.6% identical to hBPI and 25.7% identical to hLBP within this region. In contrast to the extensive differences in overall primary structure, the polycationic region of mammalian BPI and LBP most strongly implicated in LPS binding (residues 86 to 102) (22–24) also represents the most cationic local region of EsLBP1 (Fig. 1A). The sequence alignment of human and squid proteins predicts that the disulfide bond of the mammalian LBP/BPI family within the N-terminal domain (32) is conserved in EsLBP1 (Fig. 1A). The tertiary structure of EsLBP1, as derived by SWISS-MODEL, is most similar to the 2.40-Å structure of human BPI (SMTL accession number 1BP1.1.A) (22). The QMEAN4 score, a protein model quality measure based on scoring functions like torsion and solvation that are related to model geometry (33), is −5.63 (Fig. 1B). Thus, the overall model quality is low, i.e., confidence that the three-dimensional (3-D) structure of BPI can predict that of EsLBP1 is limited. However, it should be noted that the model quality varies greatly by region (Fig. 1B), conforming well in the extended protein regions that define the unique boomeranglike configuration of mammalian LBP and BPI (22, 34) but deviating greatly in regions in both the N- and C-terminal domains that impart interactive properties specific to LBP or BPI. For example, the highest net concentration of positive charge present in mammalian BPI and LBP, at the tip region of the N-terminal domain (Fig. 1D and E), is also manifest in EsLBP1 (Fig. 1C) but in an area of high divergence from the template. The residues predicted by ClustalW2 alignment to be involved in the conserved disulfide bond (Fig. 1A) are in an area of relatively good fit to the BPI template and in close proximity to each other, as in human BPI (Fig. 1F and G). The choice of template structure, human BPI or mouse LBP, for the EsLBP1 model did not affect these general characteristics.
FIG 1 .
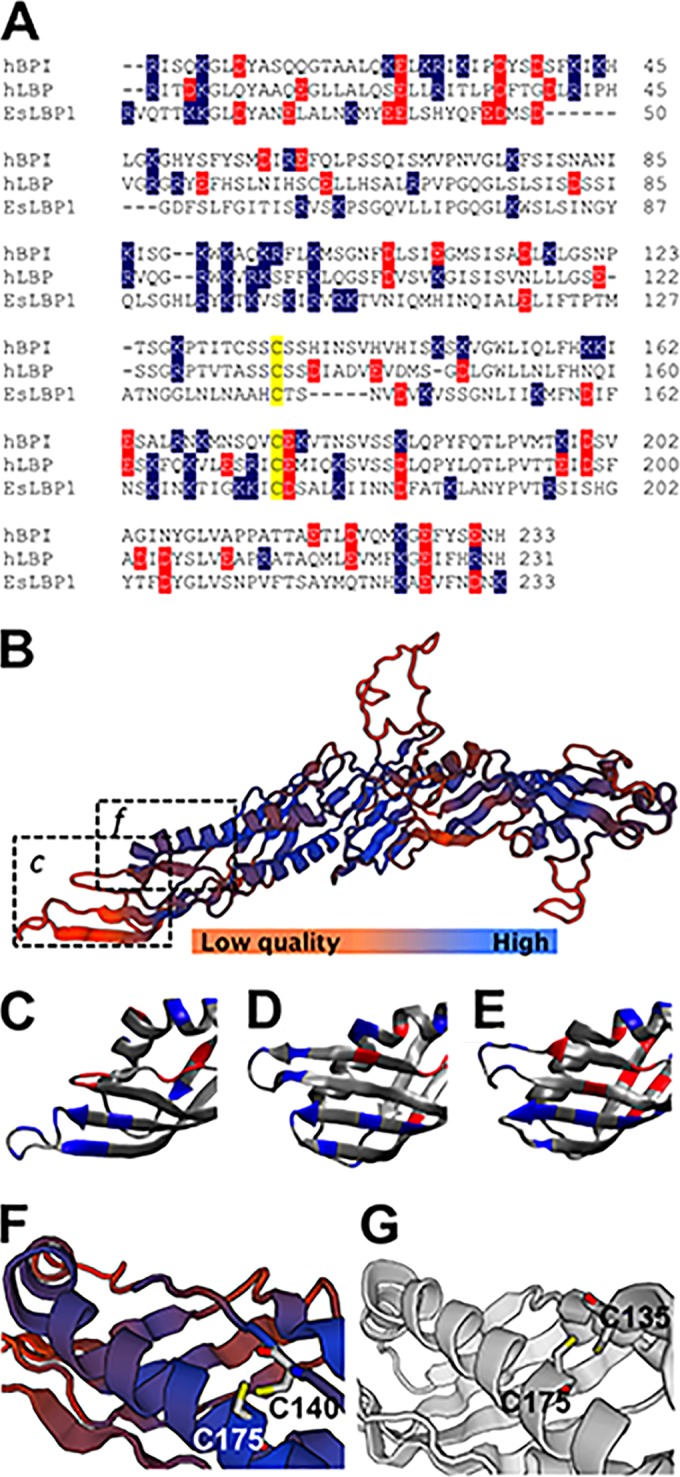
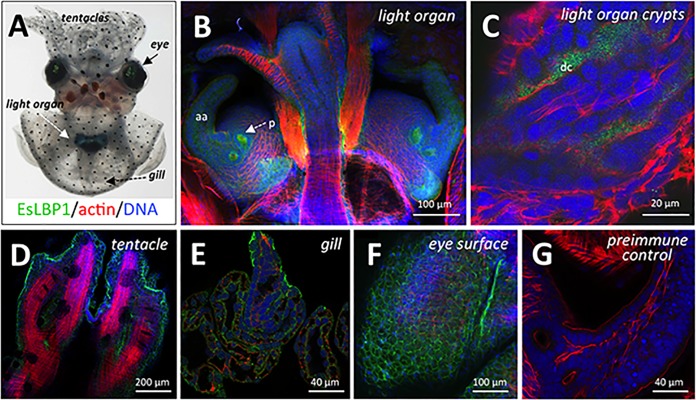
Characteristics of the EsLBP1 protein. (A) Amino acid sequence alignment of N-terminal LBP/BPI domains of human BPI (hBPI), human LBP (hLBP), and EsLBP1. Cysteine residues of the conserved disulfide bond (32) are color coded in yellow, negatively charged residues (Glu, asp) in red, and positively charged residues (Arg, Lys) in blue. Numbering for human LBP and BPI is based on the system in reference 32. (B) SWISS-MODEL-predicted 3-D structure of EsLBP1 based on human BPI template (SMTL accession number 1BP1.1.A). Color gradient shown in key indicates quality of structure as determined by local QMEAN4 score. Dashed boxes show regions depicted in panels C and F. (C) N-terminal tip of EsLBP1 model from panel B. Charged amino acids are color coded as blue for positive residues (Arg, Lys) and red for negative residues (Glu, Asp). (D) Corresponding N-terminal region in human BPI structure, marked for charge as described for panel C. (E) Corresponding N-terminal region in mouse LBP (SMTL accession number 4m4d.1), marked for charge as described for panel C. (F) Region of predicted disulfide bond in EsLBP1 structure, with the positions of side chains of residues C140 and C175 indicated. Colors of backbone illustration depict QMEAN4 score as shown by the key in panel B. (G) Region of disulfide bond in human BPI structure, with the positions of side chains of residues C135 and C175 indicated.
LOS-/LPS-binding properties of EsLBP1.
The structural comparisons described above are consistent with the LPS-binding properties of EsLBP1. To test this hypothesis more directly, we assayed the binding of metabolically labeled (3H) meningococcal lipooligosaccharide (LOS) to recombinant His-tagged EsLBP1. Binding was measured by quantifying cocapture of the radiolabeled LOS by nickel beads to which His-tagged proteins bind. Cocapture of [3H]LOS was dependent on the dose of conditioned medium containing EsLBP1. Control conditioned medium lacking EsLBP1 produced significantly lower cocapture of [3H]LOS, without dose dependency. High levels of cocapture occurred following incubations of 1 nM LOS with ~0.1 to 1 nM EsLBP1, indicating very-high-affinity binding of meningococcal LOS to the squid protein (Fig. 2A). Preincubation of EsLBP1 with comparable amounts of unlabeled Neisseria meningitidis or V. fischeri LOS/LPS (up to 100-fold excess) prior to its addition to and incubation with [3H]LOS reduced cocapture of [3H]LOS radiolabeled substrate. Francisella tularensis LPS, which binds human LBP poorly (35), only caused this decrease in capture when added in 100-fold excess of the [3H]LOS (Fig. 2B).
FIG 2 .
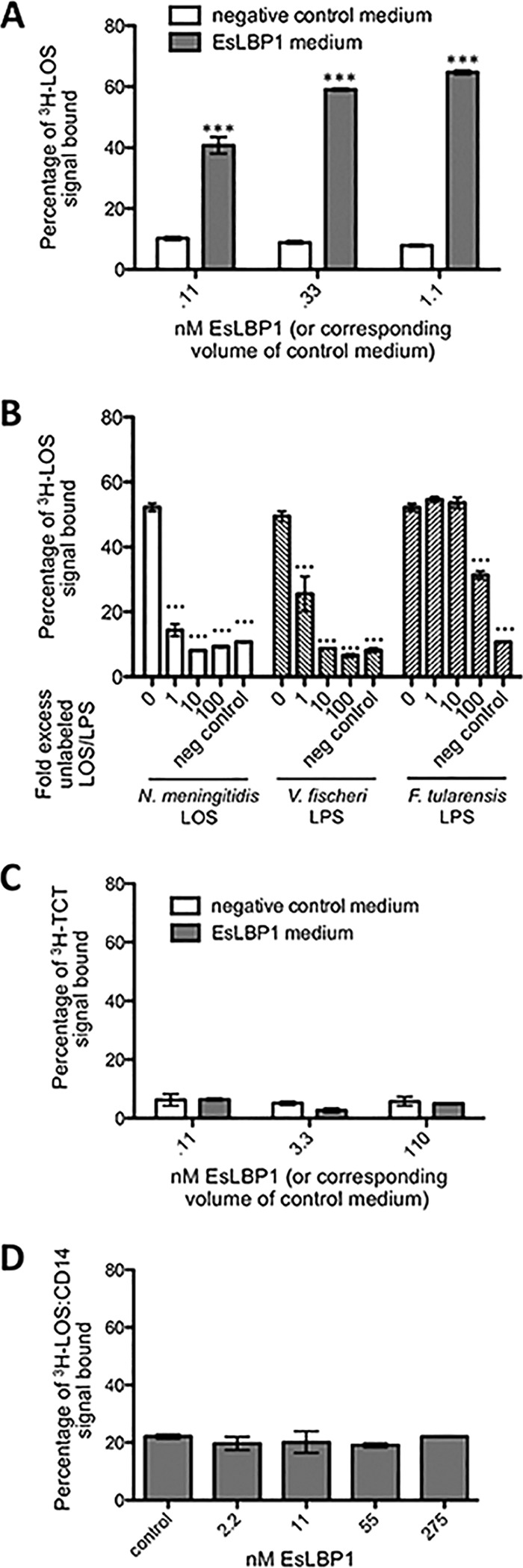
Binding of EsLBP1-His6 to MAMP substrates. (A) Results of capture assay with EsLBP1-His6 medium or control medium incubated with 5,000 cpm 3H-labeled Neisseria LOS and pelleted to nickel beads. (B) Results of competition experiment for 0.3 nM EsLBP1-His6 or control medium, 5,000 cpm 3H-labeled Neisseria LOS, and 0 to 100-fold excess by weight of unlabeled N. meningitidis LOS, V. fischeri LPS, or F. tularensis LPS. (C) As described for panel A but measuring capture of 1,000 cpm 3H-labeled TCT. (D) As described for panel A but measuring capture of 5,000 cpm [3H]LOS:CD14 aggregates; EsLBP1-His6 was diluted with control medium such that all treatments had a constant total volume of conditioned medium. Error bars indicate standard errors of the means (SEM) of three technical replicates. Statistical comparisons by ANOVA and pairwise tests compare capture by EsLBP1-His6 medium to corresponding volume of control medium (A or C) or the sole control reaction (D); in panel B, comparisons are to the capture assay with LBP1-His6 medium and no competing unlabeled endotoxin (bars labeled 0). ***, P < 0.001.
As mammalian LBP has a role in binding pneumococcal cell wall fragments (36), we sought to determine whether EsLBP1 binds the peptidoglycan monomer, TCT. Additionally, human LBP reacts with monomeric endotoxin:CD14 complexes to form supramolecular complexes containing LBP, endotoxin, and CD14 (J. P. Weiss unpublished data). We adapted the LOS/LPS capture assay described above to measure binding to [3H]TCT and to [3H]LOS:human CD14 complexes, but in neither case was EsLBP1-dependent cocapture observed (Fig. 2C and D). Thus, we found no evidence for EsLBP1 binding of TCT or LOS:CD14 complexes.
EsLBP1 does not kill Escherichia coli.
The pI of EsLBP1 suggests that it more likely functions like LBP and not BPI. To test this hypothesis more directly, we assayed the effects of EsLBP1 and, for comparison, recombinant human LBP and BPI-21 (the LPS-binding and bactericidal N-terminal Mr 21,000 fragment of human BPI [37]) on the viability of E. coli. As shown previously (16, 17), BPI but not LBP produced killing of E. coli, as manifested by reduced CFU (Fig. 3). At the same protein concentration tested, EsLBP1 had no effect on bacterial viability, resembling mammalian LBP.
FIG 3 .

Effect of EsLBP1 on viability of E. coli. Shown are CFU of E. coli strain PL2 after treatment of log-phase culture for 1 h with 30 nM hBPI-21 (N-terminal fragment of human BPI), 30 nM hLBP (human LBP), or 30 nM EsLBP1 or no protein. Error bars indicate SEM of 5 matched biological replicates. Significant differences found by ANOVA and pairwise tests are indicated as follows: **, P < 0.01; *, P <0.05.
eslbp1 transcript expression is induced by symbiosis and TCT but not by the binding partner, LPS.
Previous microarray results (31) showed that EsLBP1 is upregulated at 18 h following colonization of E. scolopes by V. fischeri, i.e., following the first full colonization of host crypts. Here, we used quantitative reverse transcriptase PCR (qRT-PCR) with gene-specific primer sets (see Table S1 in the supplemental material) to establish the time course of eslbp1 expression over the trajectory of early development. We also investigated the role of symbiont MAMPs in the induction of eslbp1. A reproducible, significant, ~3-fold difference in eslbp1 expression between aposymbiotic and symbiotic light organs was first observed at 12 h postinoculation; the degree of upregulation in symbiotic over aposymbiotic light organs at 24 h varied but was typically greater than 20-fold (Fig. 4A). Lipid A, the LPS derivative involved in symbiont-induced development, from either V. fischeri or E. coli was used at 10 ng/ml, a level that optimally induces host cell phenotypes, but it did not induce changes in eslbp1 expression. However, TCT added at 1 µM was shown to increase eslbp1 ~11-fold over the background level; the TCT effect was not further amplified by the addition of lipid A (Fig. 4B and C).
FIG 4 .

eslbp1 transcript induction by V. fischeri colonization or MAMPs. (A) Normalized eslbp1 transcript levels in E. scolopes light organs at 4, 8, 12, and 24 h postinoculation with V. fischeri are represented as the ratio of transcript levels in symbiotic (Sym) light organs and control, aposymbiotic (Apo) light organs. ***, P < 0.001. (B) Normalized eslbp1 transcript levels in light organs of aposymbiotic and symbiotic animals at 24 h postinoculation as in the experiment whose results are shown in panel A, along with the levels in uninoculated animals treated for 24 h with lipid A (10 ng/ml) from V. fischeri (Vf) or E. coli (Ec). (C) eslbp1 transcript levels in light organs of aposymbiotic and symbiotic animals at 24 h postinoculation as described for panel A, along with the levels in uninoculated animals treated for 24 h with lipid A (10 ng/ml) from V. fischeri and/or TCT (1 µM). (B and C) Small letters indicate statistically indistinguishable groups of treatments as determined by ANOVA and pairwise comparison (P < 0.01 in all cases). Error bars indicate SEM of three replicates (A) or four replicates (B and C).
eslbp1 transcripts localize to light organ tissues interfacing with V. fischeri.
Using hybridization chain reaction-fluorescent in situ hybridization (HCR-FISH), we localized the expression of eslbp1 within the light organ to determine whether the gene is expressed in proximity to V. fischeri and to ascertain the steps of the establishment of symbiosis in which EsLBP1 may play a role. We evaluated the locations of transcripts in light organs at 24 h postinoculation using gene-specific probes for eslbp1, for hsp90 (as a counterstain for E. scolopes tissue), and for V. fischeri 16S ribosomal subunit (to label the symbionts) (see Table S2 in the supplemental material). eslbp1 transcript signals in symbiotic light organs were elevated relative to the levels in nonsymbiotic light organs throughout the epithelial surfaces closely associated with V. fischeri, including the pores, ducts, and crypts of the organ (Fig. 5).
FIG 5 .
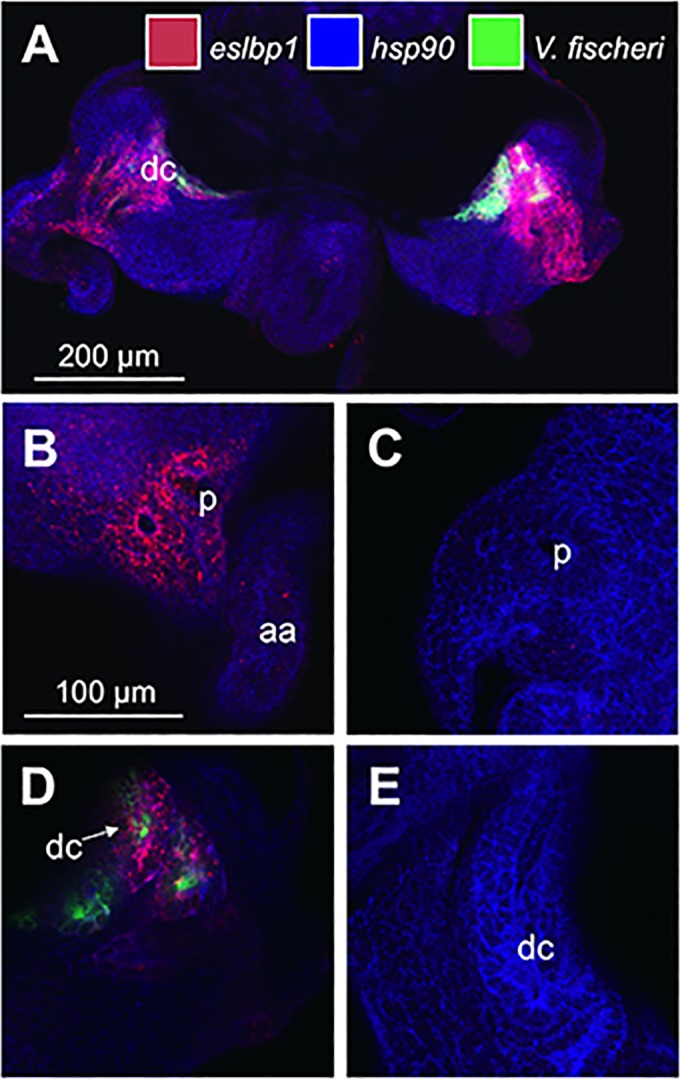
Localization of eslbp1 transcripts in light organs after 24 h of colonization with V. fischeri. (A) Overview of a symbiotic light organ at 24 h. (B) Micrograph of pore/anterior appendage region of symbiotic light organ at 24 h. (C) Pore/anterior appendage region of aposymbiotic light organ at 24 h. (D) Deep crypt region of symbiotic light organ at 24 h. (E) Deep crypt region of aposymbiotic light organ at 24 h. aa, anterior appendage; dc, deep crypt; p, pore; red, eslbp1 HCR probe; blue, hsp90; green, V. fischeri 16S ribosomal subunit.
EsLBP1 protein is present at epithelial surfaces that directly associate with bacteria and with the environment.
We compared the data on the localization of eslbp1 transcripts and of protein. Using chicken anti-EsLBP1 antibody, we confirmed and expanded upon previously reported EsLBP1 localization at 18 h postinoculation in symbiotic animals (31). EsLBP1 was highly abundant throughout the light organ (Fig. 6B). The antibody cross-reactivity was high in anterior-appendage epithelial cells, as well as the apical surfaces of pore and duct cells. EsLBP1 protein was also abundant in the extracellular crypt spaces of symbiotic light organs, the site of long-term colonization by V. fischeri (Fig. 6C).
FIG 6 .
Localization of EsLBP1 protein in light organs at 18 h. Juvenile E. scolopes squid were colonized with V. fischeri for 18 h and then probed with chicken anti-EsLBP1 antibody. (A) E. scolopes hatchling under white light, ventral view Light organ is transparent structure immediately ventral to ink sac. Bottom, key to color coding for labeling of tissues in panels B to G: green, anti-LBP1 antibody or preimmune control (detected by goat anti-chicken FITC secondary antibody); red, actin (rhodamine-phalloidin); blue, DNA (TOTO-3). (B) Immunocytochemistry micrograph overview of a symbiotic light organ and closely associated viscera at 18 h, highlighting anti-EsLBP1 antibody staining. p, pore; aa, anterior appendage. (C) Higher-magnification micrograph of deep crypt spaces of symbiotic light organ. dc, deep crypt lumen. (D to F) High-magnification micrographs of non-light organ tissues: tentacles, gills, and outer surface of eye, respectively. (G) Preimmune IgY control for EsLBP1 antibody at anterior appendage.
To determine whether EsLBP1 protein is specific to the light organ, we also analyzed other epithelial tissues of the squid that interact with environmental bacteria, including tentacles, gills, and eyes (Fig. 6D to F). We found that the protein is not exclusive to the light organ but is abundant along the apical surfaces of many or perhaps all epithelia but absent in deeper tissues, such as muscle. The signal was not seen in light organs or other tissues treated with preimmune serum (Fig. 6G).
DISCUSSION
In this study, we provide evidence based on both functional assays and structural modeling that is consistent with EsLBP1 functioning as an LBP-like protein. Most notably, EsLBP1 binds Gram-negative bacterial LOS and LPS with nanomolar or higher avidity under in vitro conditions, i.e., when LPS/LOS is presented as part of supramolecular assemblies containing LPS-rich lipid-water interfaces, as in aggregates of purified LPS/LOS. eslbp1 gene expression is regulated by exposure to the peptidoglycan monomer TCT, which synergizes with LPS in the triggering of V. fischeri-induced morphogenesis of the host symbiotic tissues. The gene is expressed and the protein produced across the organ’s epithelia—from the point where V. fischeri initially gathers, along the path of its migration, to where it takes up permanent residence in the crypts. The protein is also abundant along the apical surfaces of other epithelial tissues, where colonization by bacteria does not occur.
Although EsLBP1 has only ~25% primary structure identity with mammalian LBPs in the N-terminal domain (Fig. 1A) and ~21% overall, several key structural features likely important for LPS-related functions are predicted to be conserved. These features include (i) the localization of the most cationic region of these proteins within the predicted outer tip of the N-terminal domain of these proteins (Fig. 1C to E), albeit with substitutions in some specific cationic residues that are conserved in mammalian LBP and BPI, (ii) the extended configuration of the holoproteins, possibly important for the coordinated binding of LPS-rich supramolecular assemblies by the N-terminal domain and delivery of LPS to specific host targets mediated by the C-terminal domain, and (iii) the presence of conserved cysteines (140 and 175) within the N-terminal domain that form a disulfide linkage necessary for protein stability and function (38). Replacements of specific cationic residues conserved in mammalian LBP and BPI are common in proteins of this family in many nonmammalian vertebrates and invertebrates, including the Atlantic cod Gadus morhua and mollusks Crassostrea gigas (oyster) and Biomphalaria glabrata (snail) (28, 39, 40), reinforcing the notion that the overall charge of the region is critical. Remarkably, the much lower avidity of mammalian LBP and BPI for F. tularensis LPS compared to their avidity for meningococcal LOS (35) is also manifest with EsLBP1 (Fig. 2B). These data suggest similar determinants of LPS interaction with the mammalian and squid proteins. In addition, they are consistent with a key role of electrostatic interactions between cationic regions of the proteins and anionic moieties at or closely neighboring the lipid A region of most LPS/LOS species that are less prominent in F. tularensis LPS (41). It is noteworthy that the quality of the 3-D model of EsLBP1 is strongest within the core of the boomerang-shaped structure that is unique to the LBP/BPI family (22, 34). Regions of EsLBP1 that diverge from the human BPI 3-D structure template are generally at the periphery of the molecule. Divergence at the N terminus could reflect greater intrinsic mobility of the LPS-binding region to optimize interactions with diverse LPS-rich bacterial interfaces, and divergence at the C terminus may correspond to functional regions that have evolved to recognize different (e.g., host) targets. It is also noteworthy that the predicted structure of EsLBP1 within the C-terminal domain resembles that of mammalian LBP (34) more than that of BPI (22, 42), lacking the A′ helix present in BPI and instead forming a loop in this region that is displaced relative to the corresponding region of BPI (Fig. 1B).
The regulation of lbp gene expression by TCT and not LPS was unexpected, given that in mammalian systems, LPS increases lbp expression (43, 44); we are not aware of experiments showing the effect of peptidoglycan treatment on mammalian lbp transcript or protein levels. Recent studies have indicated that LPS preparations are often contaminated with trace amounts of peptidoglycan, which can actually be the element that is active (45). In light of the results in the squid system, a revisit of the MAMP induction of genes encoding mammalian LBP/BPI may be fruitful. Regulation of an LBP by TCT is of added interest, as it is the converse of the regulation of expression of a well-studied C-type lectin, RegIIIγ. This protein is expressed in the intestinal epithelium of mice, binds peptidoglycan, and is preferentially bactericidal against Gram-positive bacteria (46, 47). However, the expression of the gene encoding RegIIIγ is induced by the presence of Gram-negative bacteria or LPS (47, 48).
Cross talk and synergy between MAMPs and their receptors have been observed elsewhere in mammalian systems. TCT and lipid A act synergistically in the case of nitric oxide synthase induction during Bordetella pertussis infection of hamster tracheal cells (49, 50); in this case, TCT and lipid A are thought to deliver their signal through parallel pathways. In contrast, a sequential priming effect of peptidoglycan on the LPS response has been noted in human blood; the administration of staphylococcal peptidoglycan increases the response to subsequent LPS treatment, apparently through upregulation of such factors as CD14 and Toll-like receptor 4 (TLR4) on monocytes (51). Our experimental results suggest a similar mechanistic relationship in the squid, wherein increased eslbp1 gene expression induced by TCT promotes developmental responses induced by LPS and thus results in the LPS-peptidoglycan synergy in squid-vibrio symbiosis that has been described (4). This synergy may be particularly important given the relatively low potency of the V. fischeri LPS (52), presumably related to its unusual acyl chain and O-antigen structure (53, 54), which may serve to protect the host from endotoxicity. TCT’s induction of eslbp1 is another example of peptidoglycan products from beneficial bacteria influencing host immune development. In mammals, the diverse effects attributed to the peptidoglycan of the microbiota include maturation of the gut-associated lymphoid tissues (reviewed in reference 55).
Although the irreversible signal for light organ morphogenesis is delivered by MAMP-host cell interactions in the crypt spaces, cell death does not occur in these epithelia. Instead, at ~12 h, the MAMPS remotely trigger an irreversible program of cell death that results in a 4- to 5-day regression of the superficial ciliated epithelia, which are several cell layers away from the crypts (56). eslbp1 is significantly upregulated at this 12-h time point (Fig. 4A), and thus, its expression is plausibly timed to play a role in the transmission of this signal. Furthermore, the crypt epithelia have an abundance of this protein (Fig. 6C), a finding reminiscent of the observation of LBP in the mucus of mouse intestines (9).
Our studies of EsLBP1 show that it is not exclusively a light organ protein but, rather, is abundant in most if not all epithelia (Fig. 6D to F). These data suggest that the light organ has recruited LBP as a protein to signal the presence of a mutualistic partner or control its population rather than to respond to a pathogen. The hypothesized signaling role for EsLBP1 is unproven but is supported by EsLBP1’s structural and functional properties, which resemble mammalian LBP more closely than BPI. These features include its predicted near-neutral isoelectric point (28) and the absence of bactericidal activity (Fig. 3), which are distinguishing features of mammalian LBP compared to BPI (12).
In mammals, LBP can either promote LPS-triggered inflammation or blunt it by promoting noninflammatory clearance mechanisms (6, 11–14, 57). The former seems entirely CD14 dependent, whereas the latter is largely CD14 independent. No CD14 has been detected among E. scolopes transcripts, although transcripts with an MD-2-related lipid-recognition (ML) domain have been noted (30). The inability of EsLBP1, unlike mammalian LBP, to interact with monomeric LOS:soluble CD14 complexes (Fig. 2D) leaves open the possibility of an alternative target of EsLBP1-LPS complexes. In mammals, up to 100-fold increases in extracellular LBP levels can promote “silent uptake” of LPS rather than activation of inflammation (10, 11, 14). Thus, future studies in the squid will also need to investigate whether induced increases in EsLBP1 levels also have more complex effects on the evolution of the symbiotic response and relationship.
The presence of multiple proteins in the LBP/BPI family in E. scolopes that are predicted to have various biochemical properties (28) raises the possibility that the squid, like mammals, expresses both LBP-like and BPI-like proteins whose expression and functions are differentiated in a way to best coordinate host responses to Gram-negative bacterial interaction, in this case leading to E. scolopes-Vibrio fischeri symbiosis. If so, this system would provide the first example outside mammals of the coexistence of both LBP- and BPI-like family members in a single species. Future studies of this family of lipid (LPS)-binding proteins during E. scolopes-V. fischeri symbiosis should advance the understanding of both the evolution and structure/function of the LBP/BPI family and its role in mutualism. LBP may also play important roles, as yet mostly unknown, in others of the innumerable beneficial animal-bacterial associations.
MATERIALS AND METHODS
Alignment of EsLBP1 with mammalian LBP and BPI.
Human BPI (NCBI accession no. CAD99178.1), human LBP (AAB31143.1), and EsLBP1 (JF514880.1) sequences were aligned using ClustalW2 (58, 59). LBP/BPI family N-terminal domains were defined by SMART (60). A 3-D model of EsLBP1 was formed with SWISS-MODEL (61–64); the model with the best QMEAN4 score (33) was chosen. Images were generated with PV, a Java Script Protein Viewer (http://dx.doi.org/10.5281/zenodo.12620), and VMD 1.9.2 (65).
Expression of EsLBP1 in insect cells.
The complete open reading frame of EsLBP1 was amplified from E. scolopes cDNA using primers LBP1pBAC3F (5′ATACACCATGGTAATGTCTTGCCCCACTCAA3′) and LBP1pBAC3R (5′TATCACTCGAGAATAGATGTAATTGCCAAGTC3′). Two single-nucleotide polymorphism (SNPs) resulting in amino acid substitutions relative to the published EsLBP1 sequence, T253S (a change of T to S at position 253) and A218T, were consistently noted in this cDNA preparation and were included in the expressed recombinant EsLBP1. The PCR product was digested with EcoRI and XhoI and ligated into the plasmid pBAC-3 (EMD Millipore, Billerica, MA), adding a leader peptide and His6 tag, and sequenced. The construct was transfected into and expressed from Sf9 insect cells by Kinnakeet Biotechnology (Midlothian, VA). Conditioned medium from these cells was blotted with anti-tetra-His antibodies (Qiagen), by which means the concentration of EsLBP1 in the undiluted medium was estimated at 2 µM. Control medium including His-tagged E. coli β-glucuronidase was prepared from cells transfected with the BacMagic3 transfection control plasmid (EMD Millipore).
Preparation of MAMPs.
[3H]LOS (5,000 cpm/ng LOS) and [14C]LOS (6 cpm/ng LOS) were extracted and purified from metabolically labeled Neisseria meningitidis and used as aggregates of purified LOS as described previously (66). The [14C]LOS was used as unlabeled LOS in competition experiments with [3H]LOS (Fig. 2B). Metabolic labeling with radiolabeled acetate yielded equivalent radiolabeling of all LOS molecules (67). Unlabeled lipopolysaccharide and lipid A from wild-type V. fischeri, F. tularensis, and N. meningitidis were prepared by the water-phenol method (68); diphosporyl E. coli lipid A was purchased from Sigma Aldrich (St. Louis, MO). LPS and LPS derivative stock solutions were sonicated before use as previously described (52); preparations used in animal experiments were initially solubilized at 1 mg/ml in 10-mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer, pH 6.3, before dilution. Unlabeled TCT was purified from Bordetella pertussis (69); endotoxin contamination was undetectable as assayed with Pyrochrome chromogenic reagent (Associates of Cape Cod, East Falmouth, MA). To prepare [3H]TCT, peptidoglycan of N. gonorrhoeae strain kh619 (MS11 ldcA) was metabolically labeled using [6-3H]-glucosamine (70) and sacculi were purified as described previously (71). [3H]TCT was produced by digestion of sacculi with gonococcal LtgD, and TCT was purified by reversed-phase high-performance liquid chromatography (HPLC) (71).
EsLBP1—endotoxin, TCT, and LOS:CD14 complex binding assays.
Conditioned medium containing EsLBP1 was used in parallel with equal volumes of control (β-glucuronidase-containing) conditioned medium (Fig. 2A and B) or diluted with control conditioned medium (Fig. 2D) to permit the testing of a range of EsLBP1 concentrations. Medium containing EsLBP1 or control medium was mixed with a 3H-radiolabeled N. meningitidis MAMP, either 5,000 cpm of LOS (Fig. 2A) or LOS complexed with human CD14 (13) (Fig. 2C), or 1,000 cpm TCT (Fig. 2D), along with 0.1% human serum albumin in Dulbecco’s phosphate-buffered saline (PBSA) in 0.2-ml reaction mixture volumes and incubated for 30 min at 27°C. Ten microliters of Ni2+-agarose resin was added, the reaction mixtures were brought to 0.5 ml total with PBSA, and the reaction mixtures were incubated on a rotating wheel for an additional 30 min. The beads were pelleted by gentle centrifugation for 1 min and washed twice with PBSA for 5 min each. Recovery of radiolabeled material in each of the recovered fractions was measured by liquid scintillation spectroscopy. Cocapture of radiolabeled material was calculated as the percentage of the total recovered radioactive material present in beads. For competition experiments (Fig. 2B), unlabeled LOS/LPS was preincubated with 0.33 nM EsLBP1 for 30 min at 27°C before the addition of 3H-LOS and incubation and cocapture as described above.
Assay for bactericidal activity of proteins.
Cultures of E. coli strain PL2 were grown in tryptic soy broth (TSB) at 37°C to mid-log phase and then diluted 100-fold in TSB with no recombinant protein or 30 nM of one of the following: hBPI-21 (N-terminal fragment of human BPI), hLBP (human LBP) (both from Xoma, Berkeley, CA), or EsLBP1. Cultures were incubated for a further 60 min at 37°C. Aliquots of each culture were diluted 1:6,250, and 10-µl amounts were plated in triplicate on tryptic soy agar and incubated overnight at 37°C, and then CFU were counted.
General procedures for animal experiments.
Adult E. scolopes animals were collected from the sand flats of O’ahu, HI, and transported and maintained as described in previous publications (72); experiments used newly hatched juveniles in artificial seawater collected from the table on which they hatched. Symbiotic animals were exposed to ~5,000 CFU/ml of V. fischeri strain ES114 (73); aposymbiotic animals were not. Symbiosis was verified with luminescence using a TD-20/20 luminometer (Turner Designs, Sunnydale, CA). In MAMP treatment experiments, lipid A was used at 10 ng/ml and TCT at 1 µM.
qRT-PCR.
We stabilized light organ tissues, extracted RNA, prepared cDNA, and conducted quantitative reverse transcriptase PCR (qRT-PCR) experiments in accordance with MIQE guidelines (74), as described previously (75), except that qRT-PCR used the gene-specific primers given in Table S1 in the supplemental material and the protocol used was 3 min at 94°C and 40 cycles of 15 s at 94°C, 20 s at 59°C, 20 s at 68°C. We used the comparative quantification cycle (ΔΔCq) method to determine expression levels (76). eslbp1 levels were normalized to the mean levels of control transcripts for the 40S ribosomal subunit and serine hydroxymethyltransferase (HMT).
HCR-FISH.
Hybridization chain reaction-fluorescent in situ hybridization (HCR-FISH) was used to visualize transcripts of E. scolopes and V. fischeri genes and was performed according to established protocols (77). All animals examined were collected at 24 h after exposure to V. fischeri strain ES114. The E. scolopes transcripts probed were those of eslbp1 and hsp90, used as a counterstain for E. scolopes tissue. 16S ribosomal subunit transcripts were probed to counterstain for V. fischeri. Probe sequences are given in Table S2 in the supplemental material.
Immunocytochemistry.
Immunocytochemistry for EsLBP1 in 18-h postinoculation hatchlings was performed with chicken anti-EsLBP1 antibody and viewed with an LSM510 laser-scanning confocal microscope (Zeiss, Thornwood, NY) as described previously (31), except that the primary antibody incubation was reduced to 7 days. The fluorophores included goat anti-chicken fluorescein isothiocyanate (FITC) antibody (for EsLBP1), rhodamine-phalloidin (actin cytoskeleton), and TOTO-3 (DNA).
Statistics.
For experiments with quantitative comparisons, with the sole exception of comparison of CFU levels (Fig. 3), data were log transformed to provide for normality prior to statistical analysis. In the time course experiment, the highest eslbp1 level from each treatment was removed as an outlier (Fig. 4A). Comparisons between treatments were made with analysis of variance (ANOVA) (repeated measures ANOVA for CFU), followed by post hoc pairwise comparisons with Tukey multiple comparisons of means.
SUPPLEMENTAL MATERIAL
Primers used for qRT-PCR experiments.
Probe sequences for HR-FISH experiments.
ACKNOWLEDGMENTS
We thank T. Gioannini, A. Teghanemt, K. Nikolakakis, and E. Ruby for technical assistance and advice on experimental design and on the manuscript.
This work was supported by the National Institutes of Health through grants R01 AI050661 and OD11024 to M.J.M.-N, R01 AI097157 to J.P.D., and R01 AI059372 to J.P.W., by a VA merit award to J.P.W., and by National Research Service Award Institutional Research Training Grant awards T32 AI007414 to the University of Wisconsin Cellular and Molecular Parasitology Training Program, T32 AI55397 to the University of Wisconsin Microbes in Health and Disease training program, and T32 GM008692 to the University of Wisconsin Medical Scientist training program.
Footnotes
Citation Krasity BC, Troll JV, Lehnert EM, Hackett KT, Dillard JP, Apicella MA, Goldman WE, Weiss JP, McFall-Ngai MJ. 2015. Structural and functional features of a developmentally regulated lipopolysaccharide-binding protein. mBio 6(5):e01193-15. doi:10.1128/mBio.01193-15.
REFERENCES
- 1.Patten DA, Collett A. 2013. Exploring the immunomodulatory potential of microbial-associated molecular patterns derived from the enteric bacterial microbiota. Microbiology 159:1535–1544. doi: 10.1099/mic.0.064717-0. [DOI] [PubMed] [Google Scholar]
- 2.Eberl G, Boneca IG. 2010. Bacteria and MAMP-induced morphogenesis of the immune system. Curr Opin Immunol 22:448–454. doi: 10.1016/j.coi.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 3.McFall-Ngai M, Nyholm SV, Castillo MG. 2010. The role of the immune system in the initiation and persistence of the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol 22:48–53. doi: 10.1016/j.smim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McFall-Ngai MJ. 2014. The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol 68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones BW, Nishiguchi MK. 2004. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar Biol 144:1151–1155. doi: 10.1007/s00227-003-1285-3. [DOI] [Google Scholar]
- 6.Zweigner J, Schumann RR, Weber JR. 2006. The role of lipopolysaccharide-binding protein in modulating the innate immune response. Microbes Infect 8:946–952. doi: 10.1016/j.micinf.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Schultz H, Weiss JP. 2007. The bactericidal/permeability-increasing protein (BPI) in infection and inflammatory disease. Clin Chim Acta 384:12–23. doi: 10.1016/j.cca.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy O, Canny G, Serhan CN, Colgan SP. 2003. Expression of BPI (bactericidal/permeability-increasing protein) in human mucosal epithelia. Biochem Soc Trans 31(Pt 4):795–800. doi: 10.1042/bst0310795. [DOI] [PubMed] [Google Scholar]
- 9.Vreugdenhil AC, Snoek AM, Greve JW, Buurman WA. 2000. Lipopolysaccharide-binding protein is vectorially secreted and transported by cultured intestinal epithelial cells and is present in the intestinal mucus of mice. J Immunol 165:4561–4566. doi: 10.4049/jimmunol.165.8.4561. [DOI] [PubMed] [Google Scholar]
- 10.Hailman E, Vasselon T, Kelley M, Busse LA, Hu MC, Lichenstein HS, Detmers PA, Wright SD. 1996. Stimulation of macrophages and neutrophils by complexes of lipopolysaccharide and soluble CD14. J Immunol 156:4384–4390. [PubMed] [Google Scholar]
- 11.Kitchens RL, Thompson PA. 2005. Modulatory effects of sCD14 and LBP on LPS-host cell interactions. J Endotoxin Res 11:225–229. doi: 10.1179/096805105X46565. [DOI] [PubMed] [Google Scholar]
- 12.Weiss J. 2003. Bactericidal/permeability-increasing protein (BPI) and lipopolysaccharide-binding protein (LBP): structure, function and regulation in host defence against gram-negative bacteria. Biochem Soc Trans 31(Pt 4):785–790. doi: 10.1042/bst0310785. [DOI] [PubMed] [Google Scholar]
- 13.Gioannini TL, Teghanemt A, Zhang D, Prohinar P, Levis EN, Munford RS, Weiss JP. 2007. Endotoxin-binding proteins modulate the susceptibility of bacterial endotoxin to deacylation by acyloxyacyl hydrolase. J Biol Chem 282:7877–7884. doi: 10.1074/jbc.M605031200. [DOI] [PubMed] [Google Scholar]
- 14.Hamann L, Alexander C, Stamme C, Zähringer U, Schumann RR. 2005. Acute-phase concentrations of lipopolysaccharide (LPS)-binding protein inhibit innate immune cell activation by different LPS chemotypes via different mechanisms. Infect Immun 73:193–200. doi: 10.1128/IAI.73.1.193-200.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunzendorfer S, Lee HK, Soldau K, Tobias PS. 2004. TLR4 is the signaling but not the lipopolysaccharide uptake receptor. J Immunol 173:1166–1170. doi: 10.4049/jimmunol.173.2.1166. [DOI] [PubMed] [Google Scholar]
- 16.Prohinar P. 2003. Responses of Escherichia coli to the neutrophil bactericidal/permeability-increasing protein. Ph.D. thesis, University of Iowa, Iowa City, IA. [Google Scholar]
- 17.Iovine NM, Elsbach P, Weiss J. 1997. An opsonic function of the neutrophil bactericidal/permeability-increasing protein depends on both its N- and C-terminal domains. Proc Natl Acad Sci U S A 94:10973–10978. doi: 10.1073/pnas.94.20.10973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han J, Mathison JC, Ulevitch RJ, Tobias PS. 1994. Lipopolysaccharide (LPS) binding protein, truncated at Ile-197, binds LPS but does not transfer LPS to CD14. J Biol Chem 269:8172–8175. [PubMed] [Google Scholar]
- 19.Gazzano-Santoro H, Parent JB, Conlon PJ, Kasler HG, Tsai CM, Lill-Elghanian DA, Hollingsworth RI. 1995. Characterization of the structural elements in lipid A required for binding of a recombinant fragment of bactericidal/permeability-increasing protein rBPI23. Infect Immun 63:2201–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theofan G, Horwitz AH, Williams RE, Liu PS, Chan I, Birr C, Carroll SF, Mészáros K, Parent JB, Kasler H.. 1994. An amino-terminal fragment of human lipopolysaccharide-binding protein retains lipid A binding but not CD14-stimulatory activity. J Immunol 152:3623–3629. [PubMed] [Google Scholar]
- 21.Iovine N, Eastvold J, Elsbach P, Weiss JP, Gioannini TL. 2002. The carboxyl-terminal domain of closely related endotoxin-binding proteins determines the target of protein-lipopolysaccharide complexes. J Biol Chem 277:7970–7978. doi: 10.1074/jbc.M109622200. [DOI] [PubMed] [Google Scholar]
- 22.Beamer LJ, Carroll SF, Eisenberg D. 1997. Crystal structure of human BPI and two bound phospholipids at 2.4-angstrom resolution. Science 276:1861–1864. doi: 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- 23.Lamping N, Hoess A, Yu B, Park TC, Kirschning CJ, Pfeil D, Reuter D, Wright SD, Herrmann F, Schumann RR. 1996. Effects of site-directed mutagenesis of basic residues (Arg 94, Lys 95, Lys 99) of lipopolysaccharide (LPS)-binding protein on binding and transfer of LPS and subsequent immune cell activation. J Immunol 157:4648–4656. [PubMed] [Google Scholar]
- 24.Reyes O, Vallespi MG, Garay HE, Cruz LJ, González LJ, Chinea G, Buurman W, Araña MJ. 2002. Identification of single amino acid residues essential for the binding of lipopolysaccharide (LPS) to LPS binding protein (LBP) residues 86–99 by using an Ala-scanning library. J Pept Sci 8:144–150. doi: 10.1002/psc.375. [DOI] [PubMed] [Google Scholar]
- 25.Weiss J, Victor M, Elsbach P. 1983. Role of charge and hydrophobic interactions in the action of the bactericidal/permeability-increasing protein of neutrophils on gram-negative bacteria. J Clin Invest 71:540–549. doi: 10.1172/JCI110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tobias PS, Soldau K, Iovine NM, Elsbach P, Weiss J. 1997. Lipopolysaccharide (LPS)-binding proteins BPI and LBP form different types of complexes with LPS. J Biol Chem 272:18682–18685. doi: 10.1074/jbc.272.30.18682. [DOI] [PubMed] [Google Scholar]
- 27.Inagawa H, Honda T, Kohchi C, Nishizawa T, Yoshiura Y, Nakanishi T, Yokomizo Y, Soma G. 2002. Cloning and characterization of the homolog of mammalian lipopolysaccharide-binding protein and bactericidal permeability-increasing protein in rainbow trout Oncorhynchus mykiss. J Immunol 168:5638–5644. doi: 10.4049/jimmunol.168.11.5638. [DOI] [PubMed] [Google Scholar]
- 28.Krasity BC, Troll JV, Weiss JP, McFall-Ngai MJ. 2011. LBP/BPI proteins and their relatives: conservation over evolution and roles in mutualism. Biochem Soc Trans 39:1039–1044. doi: 10.1042/BST0391039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beamer LJ, Fischer D, Eisenberg D. 1998. Detecting distant relatives of mammalian LPS-binding and lipid transport proteins. Protein Sci 7:1643–1646. doi: 10.1002/pro.5560070721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kremer N, Philipp EE, Carpentier MC, Brennan CA, Kraemer L, Altura MA, Augustin R, Häsler R, Heath-Heckman EA, Peyer SM, Schwartzman J, Rader BA, Ruby EG, Rosenstiel P, McFall-Ngai MJ. 2013. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 14:183–194. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chun CK, Troll JV, Koroleva I, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, Bonaldo MeF, Casavant TL, Soares MB, Ruby EG, McFall-Ngai MJ. 2008. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc Natl Acad Sci U S A 105:11323–11328. doi: 10.1073/pnas.0802369105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beamer LJ, Carroll SF, Eisenberg D. 1998. The BPI/LBP family of proteins: a structural analysis of conserved regions. Protein Sci 7:906–914. doi: 10.1002/pro.5560070408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benkert P, Biasini M, Schwede T. 2011. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckert JK, Kim YJ, Kim JI, Gürtler K, Oh DY, Sur S, Lundvall L, Hamann L, van der Ploeg A, Pickkers P, Giamarellos-Bourboulis E, Kubarenko AV, Weber AN, Kabesch M, Kumpf O, An HJ, Lee JO, Schumann RR. 2013. The crystal structure of lipopolysaccharide binding protein reveals the location of a frequent mutation that impairs innate immunity. Immunity 39:647–660. doi: 10.1016/j.immuni.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Barker JH, Weiss J, Apicella MA, Nauseef WM. 2006. Basis for the failure of Francisella tularensis lipopolysaccharide to prime human polymorphonuclear leukocytes. Infect Immun 74:3277–3284. doi: 10.1128/IAI.02011-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber JR, Freyer D, Alexander C, Schröder NW, Reiss A, Küster C, Pfeil D, Tuomanen EI, Schumann RR. 2003. Recognition of pneumococcal peptidoglycan: an expanded, pivotal role for LPS binding protein. Immunity 19:269–279. doi: 10.1016/S1074-7613(03)00205-X. [DOI] [PubMed] [Google Scholar]
- 37.Giroir BP, Quint PA, Barton P, Kirsch EA, Kitchen L, Goldstein B, Nelson BJ, Wedel NJ, Carroll SF, Scannon PJ. 1997. Preliminary evaluation of recombinant amino-terminal fragment of human bactericidal/permeability-increasing protein in children with severe meningococcal sepsis. Lancet 350:1439–1443. doi: 10.1016/S0140-6736(97)06468-4. [DOI] [PubMed] [Google Scholar]
- 38.Capodici C, Weiss J. 1996. Both N- and C-terminal regions of the bioactive N-terminal fragment of the neutrophil granule bactericidal/permeability-increasing protein are required for stability and function. J Immunol 156:4789–4796. [PubMed] [Google Scholar]
- 39.Stenvik J, Solstad T, Strand C, Leiros I, Jørgensen TT. 2004. Cloning and analyses of a BPI/LBP cDNA of the Atlantic cod (Gadus morhua L.). Dev Comp Immunol 28:307–323. doi: 10.1016/j.dci.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez M, Gueguen Y, Destoumieux-Garzón D, Romestand B, Fievet J, Pugnière M, Roquet F, Escoubas JM, Vandenbulcke F, Levy O, Sauné L, Bulet P, Bachère E. 2007. Evidence of a bactericidal permeability increasing protein in an invertebrate, the Crassostrea gigas Cg-BPI. Proc Natl Acad Sci U S A 104:17759–17764. doi: 10.1073/pnas.0702281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barker JH, Kaufman JW, Zhang DS, Weiss JP. 2014. Metabolic labeling to characterize the overall composition of Francisella lipid A and LPS grown in broth and in human phagocytes. Innate Immun 20:88–103. doi: 10.1177/1753425913485308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleiger G, Beamer LJ, Grothe R, Mallick P, Eisenberg D. 2000. The 1.7-A crystal structure of BPI: a study of how two dissimilar amino acid sequences can adopt the same fold. J Mol Biol 299:1019–1034. doi: 10.1006/jmbi.2000.3805. [DOI] [PubMed] [Google Scholar]
- 43.Geller DA, Kispert PH, Su GL, Wang SC, Di Silvio M, Tweardy DJ, Billiar TR, Simmons RL. 1993. Induction of hepatocyte lipopolysaccharide binding protein in models of sepsis and the acute-phase response. Arch Surg 128:22–28. doi: 10.1001/archsurg.1993.01420130026005. [DOI] [PubMed] [Google Scholar]
- 44.Wan Y, Freeswick PD, Khemlani LS, Kispert PH, Wang SC, Su GL, Billiar TR. 1995. Role of lipopolysaccharide (LPS), interleukin-1, interleukin-6, tumor necrosis factor, and dexamethasone in regulation of LPS-binding protein expression in normal hepatocytes and hepatocytes from LPS-treated rats. Infect Immun 63:2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, Harley W, Fox A, Golenbock D, Silverman N. 2004. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20:637–649. doi: 10.1016/S1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- 46.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. 2011. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cash HL, Whitham CV, Behrendt CL, Hooper LV. 2006. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. 2008. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flak TA, Heiss LN, Engle JT, Goldman WE. 2000. Synergistic epithelial responses to endotoxin and a naturally occurring muramyl peptide. Infect Immun 68:1235–1242. doi: 10.1128/IAI.68.3.1235-1242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flak TA, Goldman WE. 1999. Signalling and cellular specificity of airway nitric oxide production in pertussis. Cell Microbiol 1:51–60. doi: 10.1046/j.1462-5822.1999.00004.x. [DOI] [PubMed] [Google Scholar]
- 51.Hadley JS, Wang JE, Foster SJ, Thiemermann C, Hinds CJ. 2005. Peptidoglycan of Staphylococcus aureus upregulates monocyte expression of CD14, Toll-like receptor 2 (TLR2), and TLR4 in human blood: possible implications for priming of lipopolysaccharide signaling. Infect Immun 73:7613–7619. doi: 10.1128/IAI.73.11.7613-7619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foster JS, Apicella MA, McFall-Ngai MJ. 2000. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev Biol 226:242–254. doi: 10.1006/dbio.2000.9868. [DOI] [PubMed] [Google Scholar]
- 53.Phillips NJ, Adin DM, Stabb EV, McFall-Ngai MJ, Apicella MA, Gibson BW. 2011. The lipid A from Vibrio fischeri lipopolysaccharide: a unique structure bearing a phosphoglycerol moiety. J Biol Chem 286:21203–21219. doi: 10.1074/jbc.M111.239475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Post DM, Yu L, Krasity BC, Choudhury B, Mandel MJ, Brennan CA, Ruby EG, McFall-Ngai MJ, Gibson BW, Apicella MA. 2012. O-antigen and core carbohydrate of Vibrio fischeri lipopolysaccharide: composition and analysis of their role in Euprymna scolopes light organ colonization. J Biol Chem 287:8515–8530. doi: 10.1074/jbc.M111.324012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wheeler R, Chevalier G, Eberl G, Gomperts Boneca I. 2014. The biology of bacterial peptidoglycans and their impact on host immunity and physiology. Cell Microbiol 16:1014–1023. doi: 10.1111/cmi.12304. [DOI] [PubMed] [Google Scholar]
- 56.Doino Lemus J, McFall-Ngai MJ. 2000. Alterations in the proteome of the Euprymna scolopes light organ in response to symbiotic Vibrio fischeri. Appl Environ Microbiol 66:4091–4097. doi: 10.1128/AEM.66.9.4091-4097.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horwitz AH, Williams RE, Nowakowski G. 1995. Human lipopolysaccharide-binding protein potentiates bactericidal activity of human bactericidal/permeability-increasing protein. Infect Immun 63:522–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 59.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Letunic I, Doerks T, Bork P. 2012. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guex N, Peitsch MC, Schwede T. 2009. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30(Suppl 1):S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 63.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. 2009. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 65.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 66.Giardina PC, Gioannini T, Buscher BA, Zaleski A, Zheng DS, Stoll L, Teghanemt A, Apicella MA, Weiss J. 2001. Construction of acetate auxotrophs of Neisseria meningitidis to study host-meningococcal endotoxin interactions. J Biol Chem 276:5883–5891. doi: 10.1074/jbc.M009273200. [DOI] [PubMed] [Google Scholar]
- 67.Post DM, Zhang D, Weiss JP, Gibson BW. 2006. Stable isotope metabolic labeling of Neisseria meningitidis lipooligosaccharide. J Endotoxin Res 12:93–98. doi: 10.1179/096805106X102156. [DOI] [PubMed] [Google Scholar]
- 68.Apicella MA, Griffiss JM, Schneider H. 1994. Isolation and characterization of lipopolysaccharides, lipooligosaccharides, and lipid A. Methods Enzymol 235:242–252. [DOI] [PubMed] [Google Scholar]
- 69.Cookson BT, Cho HL, Herwaldt LA, Goldman WE. 1989. Biological activities and chemical composition of purified tracheal cytotoxin of Bordetella pertussis. Infect Immun 57:2223–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cloud KA, Dillard JP. 2002. A lytic transglycosylase of Neisseria gonorrhoeae is involved in peptidoglycan-derived cytotoxin production. Infect Immun 70:2752–2757. doi: 10.1128/IAI.70.6.2752-2757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kohler PL, Hamilton HL, Cloud-Hansen K, Dillard JP. 2007. AtlA functions as a peptidoglycan lytic transglycosylase in the Neisseria gonorrhoeae type IV secretion system. J Bacteriol 189:5421–5428. doi: 10.1128/JB.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McFall-Ngai M, Montgomery MK. 1990. The anatomy and morphology of the adult bacterial light organ of Euprymna scolopes Berry (Cephalopoda:Sepiolidae). Biol Bull 179:332–339. doi: 10.2307/1542325. [DOI] [PubMed] [Google Scholar]
- 73.Boettcher KJ, Ruby EG. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 172:3701–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 75.Heath-Heckman EA, Peyer SM, Whistler CA, Apicella MA, Goldman WE, McFall-Ngai MJ. 2013. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-vibrio symbiosis. mBio 4(2):e00167-13. doi: 10.1128/mBio.00167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nikolakakis K, Lehnert E, McFall-Ngai MJ, Ruby EG. 2015. Use of hybridization chain reaction-fluorescent in situ hybridization to track gene expression by both partners during initiation of symbiosis. Appl Environ Microbiol 81:4728–4735. doi: 10.1128/AEM.00890-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for qRT-PCR experiments.
Probe sequences for HR-FISH experiments.