ABSTRACT
A better understanding of how bacteria resist stresses encountered during the progression of plant-microbe symbioses will advance our ability to stimulate plant growth. Here, we show that the symbiotic system comprising the nitrogen-fixing bacterium Bradyrhizobium diazoefficiens and the legume Aeschynomene afraspera requires hopanoid production for optimal fitness. While methylated (2Me) hopanoids contribute to growth under plant-cell-like microaerobic and acidic conditions in the free-living state, they are dispensable during symbiosis. In contrast, synthesis of extended (C35) hopanoids is required for growth microaerobically and under various stress conditions (high temperature, low pH, high osmolarity, bile salts, oxidative stress, and antimicrobial peptides) in the free-living state and also during symbiosis. These defects might be due to a less rigid membrane resulting from the absence of free or lipidA-bound C35 hopanoids or the accumulation of the C30 hopanoid diploptene. Our results also show that C35 hopanoids are necessary for symbiosis only with the host Aeschynomene afraspera but not with soybean. This difference is likely related to the presence of cysteine-rich antimicrobial peptides in Aeschynomene nodules that induce drastic modification in bacterial morphology and physiology. The study of hopanoid mutants in plant symbionts thus provides an opportunity to gain insight into host-microbe interactions during later stages of symbiotic progression, as well as the microenvironmental conditions for which hopanoids provide a fitness advantage.
IMPORTANCE
Because bradyrhizobia provide fixed nitrogen to plants, this work has potential agronomical implications. An understanding of how hopanoids facilitate bacterial survival in soils and plant hosts may aid the engineering of more robust agronomic strains, especially relevant in regions that are becoming warmer and saline due to climate change. Moreover, this work has geobiological relevance: hopanes, molecular fossils of hopanoids, are enriched in ancient sedimentary rocks at discrete intervals in Earth history. This is the first study to uncover roles for 2Me- and C35 hopanoids in the context of an ecological niche that captures many of the stressful environmental conditions thought to be important during (2Me)-hopane deposition. Though much remains to be done to determine whether the conditions present within the plant host are shared with niches of relevance to the rock record, our findings represent an important step toward identifying conserved mechanisms whereby hopanoids contribute to fitness.
INTRODUCTION
A variety of plants, including leguminous (1), actinorhizal (2), and land-dwelling (3) plants, rely on bacterial symbiotic partners for assimilation of nitrogen, an essential macronutrient. These symbioses require bacterial invasion of plant tissues and adaptation of the bacterial symbiont to the plant host environment, processes in which several microbial membrane lipids play key roles. For instance, phosphatidylcholine is critical for efficient nitrogen fixation in several legume-rhizobial partnerships such as soybean-Bradyrhizobium diazoefficiens (formerly named Bradyrhizobium japonicum [4]) and alfalfa-Sinorhizobium meliloti (5). Similarly, an intact outer membrane (OM) lipopolysaccharide (LPS) is essential for all stages of rhizobial symbiosis, including root hair infection, symbiotic tissue (nodule) establishment, and survival within the plant cell (6). Whether and to what extent other microbial membrane lipids regulate the establishment and maintenance of plant-microbe symbioses are unclear. Here, we consider whether hopanoids (7), steroid-like pentacyclic triterpenoid lipids, support plant-microbe interactions. Hopanoids are the progenitors of hopanes, molecular fossils that exhibit intriguing yet poorly understood abundance patterns in the rock record (8). In part, our interest in hopanoids derives from a desire to interpret ancient biomarkers and the conviction that this requires a nuanced understanding of the biological functions of diverse hopanoids in modern niches.
The capacity for hopanoid biosynthesis is statistically enriched in the (meta-)genomes of bacteria associated with plants (9), and hopanoids have been found in high abundance in membranes of well-studied plant symbionts of the Bradyrhizobium (40% of total lipid extract [TLE]) and Frankia (87%) genera (10, 11). Hopanoids promote membrane rigidity (12) and confer protection against numerous stresses, including acidic or alkaline pH, high temperature, high osmolarity, oxidative stress, detergents, and antibiotics (13–16). They have varied structures, formed via methylation (2Me or 3Me), unsaturation, and/or attachment to a ribose-derived side chain (C35) (Fig. 1A; also see Fig. S1 in the supplemental material) (7). Whether there are functional distinctions under specific environmental conditions for diverse hopanoid types is unclear, yet some evidence suggests that they have nonoverlapping roles. For example, in Rhodopseudomonas palustris (17) and Burkholderia cenocepacia (14), C35 hopanoids are critical for OM stability and for resistance to low pH, detergent (sodium dodecyl sulfate [SDS]), and polymyxin B, respectively. Though the absence of 2Me-hopanoids did not manifest a stress phenotype in previous tests of R. palustris, their biosynthesis is transcriptionally induced under stress (13). This suggests that 2Me-hopanoids might contribute to stress resistance under conditions yet to be identified in this and other organisms; consistent with this notion, 3Me-hopanoids contribute to late-stationary-phase survival in Methylococcus capsulatus (18). In vitro, 2Me-hopanoids rigidify membranes of varied compositions (12). However, until now, no study has explored whether different hopanoids impact fitness in a natural ecological context.
FIG 1 .
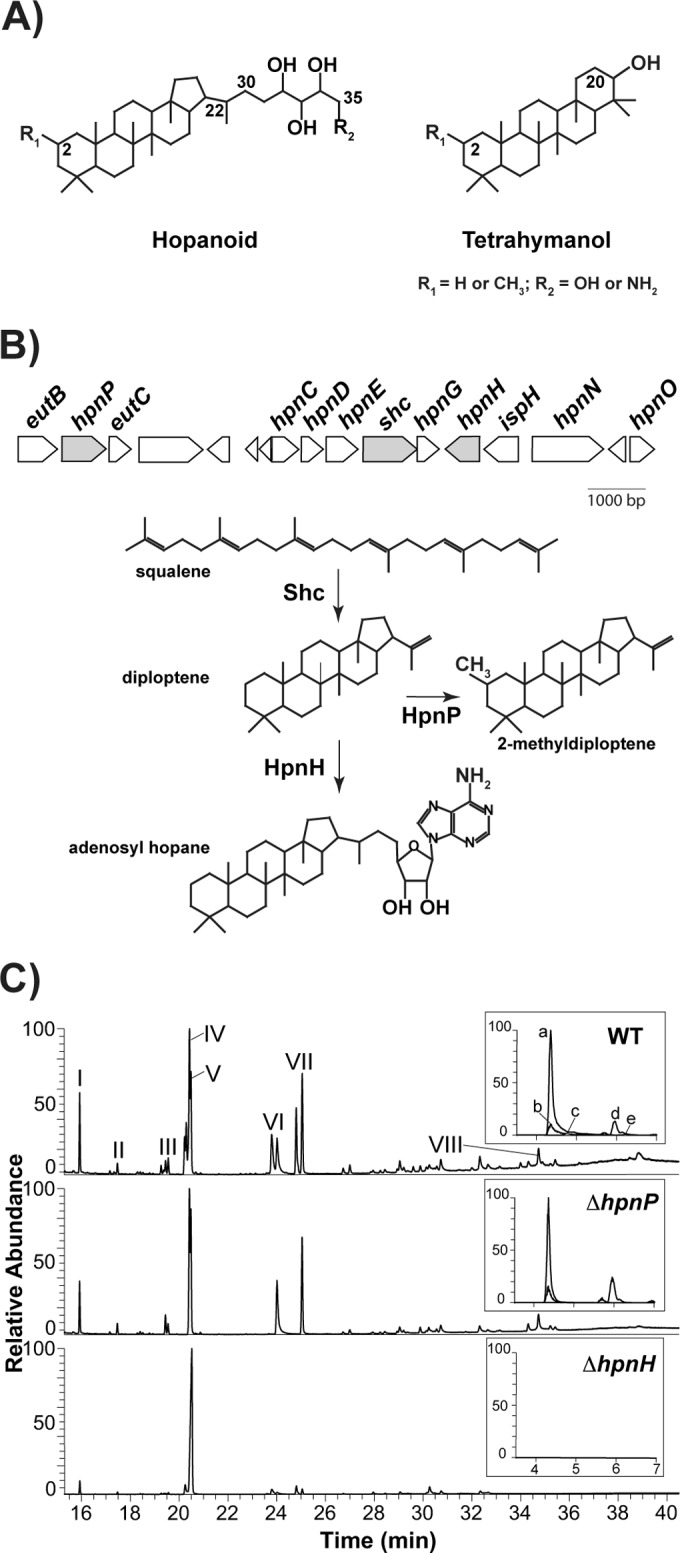
(A) Structures of hopanoid and tetrahymanol. B. diazoefficiens makes C30 hopanoids, such as diploptene (C-22=C-30) and diplopterol (OH at C-22); C35 hopanoids, such as bacteriohopanetetrol (BHT; R2=OH) and aminobacteriohopanetriol (aminotriol; R2=NH2); and tetrahymanol. All these compounds can be methylated at C-2 (2Me, R1=CH3). (B) Hopanoid biosynthetic gene cluster of B. diazoefficiens. In this study, we focused on the genes colored in gray: the shc (squalene hopene cyclase) product catalyzes squalene cyclization to hopene, the first reaction in the hopanoid biosynthetic pathway; the hpnH product catalyzes addition of adenosine to hopene, the first reaction in the synthesis of C35 hopanoids; and the hpnP product catalyzes C-2 methylation. (C) GC-MS and LC-MS (inset) total ion chromatograms of total lipid extracts from aerobically grown B. diazoefficiens strains. For GC-MS, main hopanoid peaks are numbered and the methylated counterparts elute 0.2 to 0.5 min earlier. I, pregnane acetate (standard); II, (2Me) hop-17(21)-ene; III, (2Me) hop-x-ene; IV, (2Me) hop-22(29)-ene (diploptene); V, (2Me) hop-21-ene; VI, (2Me) hopan-22-ol (diplopterol); VII, (2Me and 20Me) tetrahymanol; and VIII, BHP-508. LC-MS: a, aminotriol; b, BHT; c, 2Me-aminotriol; d, adenosylhopane; e, 2Me-BHT. Lipid analysis for each strain was performed in triplicate. For chemical structures of hopanoids, refer to Fig. S1A in the supplemental material.
Recently, it was shown that elimination of hopanoid biosynthesis in photosynthetic Bradyrhizobium strain BTAi1 impairs its symbiosis with the legume Aeschynomene evenia (15). Because the absence of all hopanoids likely had a broad and drastic impact on cellular physiology and hence symbiosis, in this study we tested specific effects of 2Me- and C35 hopanoids (7) using B. diazoefficiens USDA110, the best-studied Bradyrhizobium strain. In addition to C30 and C35 hopanoids (10), B. diazoefficiens makes tetrahymanol, a triterpenoid with a gammacerane skeleton (19) (Fig. 1A; also see Fig. S1 in the supplemental material). Intriguingly, while most hopanoids are thought to occur free within membranes, the C35 hopanoid, (2-Me) 34-carboxyl-bacteriohopane-32,33-diol, was found to be covalently attached to LPS lipidA, a well-established player in a broad range of host-microbe interactions, to form a compound called hopanoid-lipid A (HoLA) (15, 20) (see Fig. S1B).
B. diazoefficiens exhibits two different lifestyles, free living in soil or symbiotic within legume root nodule cells (1, 21). In addition to its native soybean host, B. diazoefficiens can engage in nitrogen-fixing symbioses with the stems and roots of the tropical legume Aeschynomene afraspera (22). In both of the these hosts, development of the symbiosis progresses through a series of defined stages: (i) colonization and invasion of host root tissue; (ii) internalization of bacteria by plant cells to form an organelle-like structure called the symbiosome, comprising endosymbiotic bacterial cells termed “bacteroids” that are surrounded by a plant-derived “peribacteroid” membrane (see Fig. S2 in the supplemental material); and (iii) initiation of nitrogen fixation by bacteroids, during which there is a high rate of nutrient exchange across the symbiosome membranes between plant-supplied carbon sources and fixed atmospheric nitrogen produced by bacterial nitrogenase (21, 23). Supporting such high levels of bacterial nitrogenase activity requires both extensive host control of bacteroid physiology and establishment of a specific host microenvironment, defined by low oxygen, low pH, hyperosmosis, and oxidative stress (24). There is some evidence suggesting that the extent of these environmental stresses varies between plant hosts. For example, A. afraspera was recently shown to produce nodule-specific, cysteine-rich antimicrobial peptides (NCR peptides) that induce differentiation of the bacteroid into an enlarged, elongated, and polyploid state, whereas in the soybean host, NCR peptides are absent and bacteroid morphology and ploidy are similar to those of the free-living state (25). Thus, it is likely that the microbial adaptations required for survival within root nodules are host specific, and we hypothesized that specific hopanoid mutants may exhibit variable phenotypes in diverse plant hosts.
Here, we focus on the phenotypic consequences of the inability of B. diazoefficiens to produce two specific hopanoid classes, 2Me- and C35 hopanoids. We compare its hopanoid-dependent stress phenotypes in the free-living state to those of other hopanoid producers. We further explore the fitness effects of 2Me- or C35 hopanoid production within a natural ecological context: the symbiotic microenvironment of soybean and A. afraspera cells. These studies begin to define the role of 2Me- and C35 hopanoids during the progression of plant-microbe symbioses and provide insight into microbial membrane factors that facilitate adaptations to particular microenvironments.
RESULTS
Shc appears to be essential for the survival of B. diazoefficiens.
To eliminate hopanoid production in B. diazoefficiens and to test whether a requirement for hopanoids in efficient symbiosis is conserved between B. diazoefficiens and Bradyrhizobium strain BTAi1, we first attempted deletion of the gene encoding the enzyme catalyzing the first step in hopanoid biosynthesis, squalene hopene cyclase (Shc) (Fig. 1B). We were unable to isolate a Δshc mutant using either the pK18mobsacB-based markerless gene deletion method (~400 colonies screened) or the gene replacement strategy with pSUP202pol4 (~1,200 colonies screened) (26). This suggests that Shc might be essential either because hopanoids are required for growth and survival of B. diazoefficiens or because squalene, the substrate of Shc (16), accumulates to toxic levels within the Δshc mutant. To rule out the latter possibility, we tried to delete the entire operon encoding squalene-synthesizing enzymes (hpnCDE), shc (hpnF), and hpnG (which catalyzes the second step in the synthesis of C35 hopanoids) (17), but again, we were unable to obtain the ΔhpnCDEFG mutant (~150 colonies screened) (Fig. 1B). These results suggest that hopanoid synthesis is essential for the survival of B. diazoefficiens under the conditions used to select the mutants.
B. diazoefficiens ΔhpnP and ΔhpnH mutants are unable to make 2Me- and C35 hopanoids, respectively.
To eliminate synthesis of 2Me- or C35 hopanoids specifically, we deleted genes predicted to encode the C-2 methylase, hpnP (27), or the first enzyme catalyzing the extension of C30 hopanoids, hpnH (17) (Fig. 1B). As illustrated in Fig. 1C, no methylated hopanoids were detected in ΔhpnP mutant TLE using gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS) (see Tables S1 and S2 in the supplemental material) (28, 29). The ΔhpnH mutant does not make any detectable C35 hopanoids, including aminotriol (a and c), BHP-508 (VIII; degradation product of aminotriol), bacteriohopanetetrol (b and e), and adenosylhopane (d). In addition, the ΔhpnH mutant accumulates a 6-fold excess of the HpnH substrate (17), diploptene (IV; wild type [WT], 18 ± 2 µg/mg TLE; ΔhpnP mutant, 29 ± 6 µg/mg TLE; ΔhpnH mutant, 111 ± 3 µg/mg TLE). We also analyzed the presence of HoLA in the mutants using matrix-assisted laser desorption ionization–mass spectrometry (MALDI-MS) (Fig. 2). WT and ΔhpnP mutant lipidA are composed of a mixture of penta- to hepta-acylated species, whereas ΔhpnH mutant lipidA is mainly hexa-acylated (see Fig. S1B). In WT and ΔhpnP mutant hepta-acylated species, a C35 hopanediolic acid is ester linked to hexa-acylated lipidA, and traces of a second hopanoid substitution are also detected; conversely, the ΔhpnH mutant is missing any lipidA-bound hopanoids. Not only do our results confirm the proposed roles of HpnP and HpnH, but they also show that synthesis of C35 hopanoids is required for HoLA production.
FIG 2 .
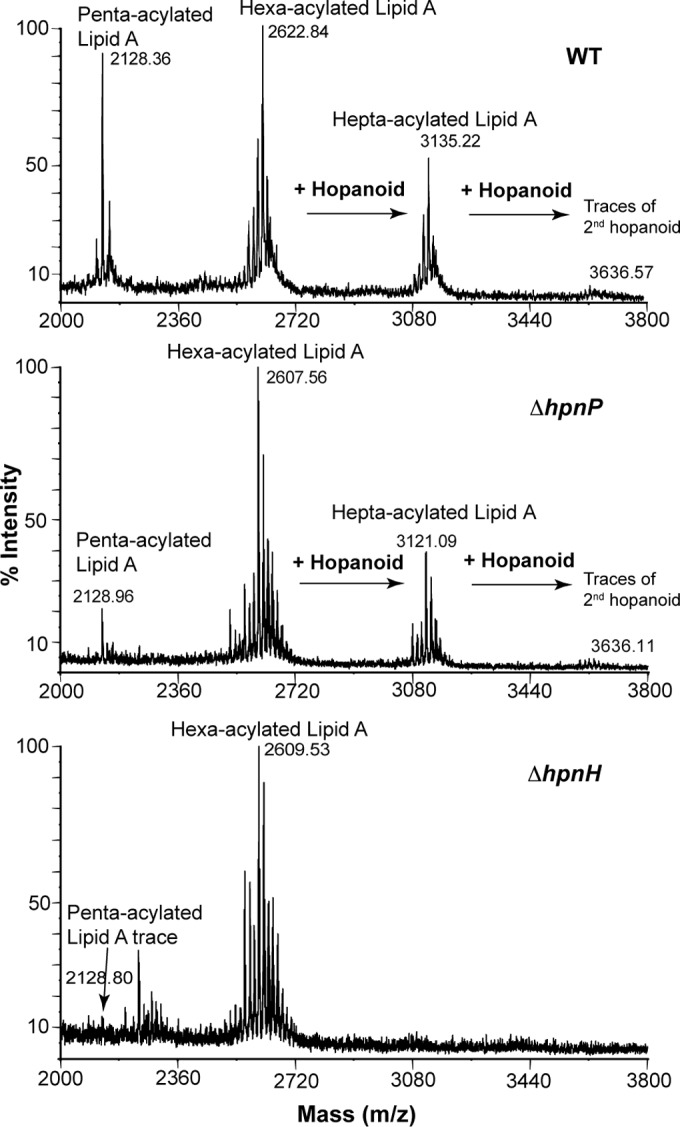
MALDI-MS analysis of lipidA from B. diazoefficiens strains. LipidA from the WT and the ΔhpnP mutant is composed of a mixture of penta-acylated and hexa-acylated species, whereas ΔhpnH mutant lipidA is mainly hexa-acylated. A C35 hopanediolic acid is ester linked to hexa-acylated and hepta-acylated lipid A in the WT and the ΔhpnP mutant. The ΔhpnH mutant does not contain any lipidA-bound hopanoids.
C35 hopanoids contribute to outer membrane rigidity.
We employed a fluorescence polarization method by incubating the dye diphenyl hexatriene (DPH) with whole cells to determine whether 2Me- and C35 hopanoids affect the rigidity of B. diazoefficiens membranes at 25°C and 40°C (Fig. 3). Because previous studies of whole cells of different R. palustris hopanoid mutants indicated that the majority of DPH gets incorporated into the OM, we interpret whole-cell polarization values to reflect the rigidity of the OM (12, 29). Membranes of all strains were less rigid at the higher temperature. The ΔhpnP mutant membrane was as rigid as the WT membrane at both temperatures, whereas the ΔhpnH mutant membrane was less rigid. Thus, C35 hopanoids are important for maintaining membrane rigidity in B. diazoefficiens in vivo, in contrast to R. palustris, where the ΔhpnH mutant membrane showed rigidity similar to that of the WT, despite the capacity of C35 hopanoids to enhance rigidity in vitro (12). This indicates that the fraction of C35 hopanoids or HoLA in the OM may be greater in B. diazoefficiens than R. palustris. Despite the lack of C35 hopanoids, the ΔhpnH mutant membrane is morphologically indistinguishable from the WT membrane, as seen in whole-cell cryo-transmission electron microscopy (cryo-TEM) micrographs (see Fig. S3 in the supplemental material).
FIG 3 .

Whole-cell membrane fluidity measurements by fluorescence polarization show that rigidity decreases for all strains as temperature increases and that the ΔhpnH mutant membrane is less rigid than that of the WT or the ΔhpnP mutant (**, P < 0.01 by Student’s two-tailed t test). Error bars represent the standard deviations from three biological replicates (~22 technical replicates).
C35 hopanoids are important for aerobic growth.
Does a less rigid membrane affect the fitness of ΔhpnH at different temperatures? To address this question, we compared aerobic growth rates of the ΔhpnH mutant at 30°C and 37°C with those of the WT and the ΔhpnP mutant (Fig. 4A and B). The ΔhpnP mutant grows like the WT at both temperatures, whereas the ΔhpnH mutant grows slower at 30°C and is unable to grow at 37°C. These results suggest that C35 hopanoids are important for growth at ambient temperature (30°C) and essential for growth at higher temperature (37°C). As shown in our whole-cell membrane fluidity measurements, the higher the temperature, the less rigid the membrane. This might be the reason why C35 hopanoids are absolutely required to maintain membrane rigidity at 37°C but are dispensable at 30°C. It is important to note that the phenotypic defect of the ΔhpnH mutant could be due to either the absence of C35 hopanoids or the lack of downstream products, such as HoLA, and even accumulation of the HpnH substrate diploptene, or a combination of these factors.
FIG 4 .
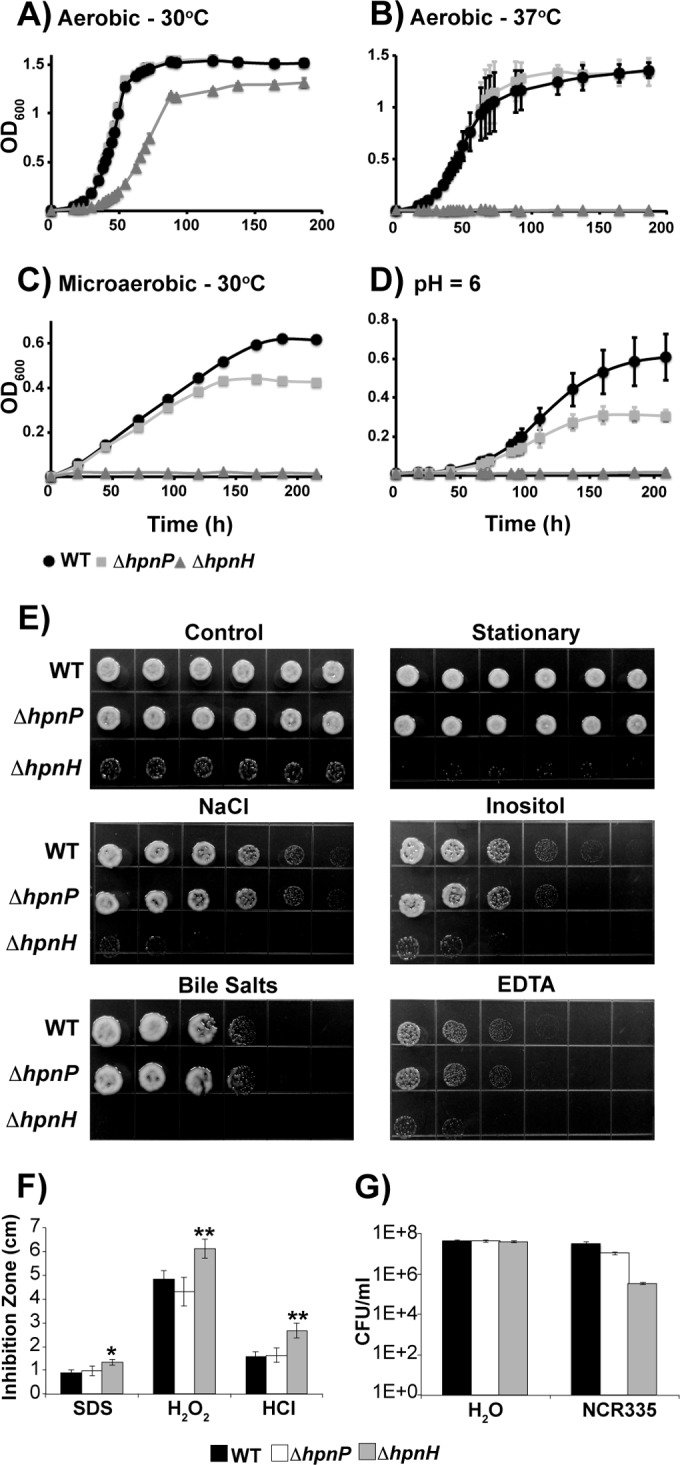
Growth of B. diazoefficiens strains under various stress conditions. (A to D) Growth of the WT (circles), the ΔhpnP mutant (squares), and the ΔhpnH mutant (triangles) was monitored as optical density at 600 nm (OD600) in PSY at 30°C (A), PSY at 37°C (B), microaerobic PSY with 0.5% O2 at 30°C (C), and PSY at pH 6 and 30°C (D). Each curve represents the average of at least three biological replicates, except the microaerobic growth curves, for which a representative data set out of four trials is shown. (E and F) Growth of B. diazoefficiens strains under stress as measured in stressor gradient plates with 50 mM NaCl, 500 mM inositol, 0.4% bile salts, or 1 mM EDTA (E) or by disc diffusion assays with 10% SDS, 5.5 M H2O2, and 2 M HCl (F). Error bars represent standard errors (n = 9). *, P < 0.05, and **, P < 0.01, by Tukey’s honestly significant difference test. (G) NCR335 sensitivity of B. diazoefficiens strains.
2Me- and C35 hopanoids are important for microaerobic growth, and C35 hopanoids are involved in stress tolerance.
Hopanoids have been shown to contribute to stress tolerance in diverse organisms (13–16). We speculated that such protection would also be seen in B. diazoefficiens. To test this hypothesis, we challenged ΔhpnP and ΔhpnH mutants with a variety of stressors that are relevant during the initiation and progression of symbiosis, such as hypoxia, acidic pH, high osmolarity, reactive oxygen species, and peptide antibiotics (24, 25).
Under hypoxic conditions with 0.5% oxygen, the ΔhpnP mutant is unable to attain growth yields as high as those of the WT and the ΔhpnH mutant fails to grow (Fig. 4C). This indicates that in the free-living state 2Me-hopanoids contribute to microaerobic growth and C35 hopanoids are essential. Using GC-MS, we determined the abundance of these hopanoid types in the WT (see Table S3 in the supplemental material). The amount of 2Me-hopanoids dramatically increased from 33% ± 2% TLE under oxic conditions to 77% ± 2% TLE under hypoxic conditions. This is consistent with hopanoid methylation being important to sustain WT levels of microaerobic growth. The only C35 hopanoid detectable by GC-MS, BHP-508, increased in abundance from 3% ± 1% TLE for cells grown aerobically to 21% ± 1% TLE when grown microaerobically, in agreement with a microaerobic growth defect for the ΔhpnH mutant.
Under acidic conditions (pH 6), the ΔhpnH mutant is unable to grow (Fig. 4D). The ΔhpnH mutant is also more prone to stationary-phase stress, osmotic stressors (NaCl and inositol), and membrane destabilizers (bile salts and EDTA [17]) than the WT, as evidenced by a reduction in ΔhpnH mutant growth on stressor gradient plates (Fig. 4E). Additionally, disc diffusion assays showed that the ΔhpnH mutant is more sensitive to oxidative (hydrogen peroxide [H2O2]), detergent (SDS), and acidic (hydrochloric acid [HCl]) stresses than the WT (Fig. 4F). Because B. diazoefficiens is exposed to NCR peptides in A. afraspera, we also tested the sensitivity of the ΔhpnH mutant to two antimicrobial peptides, polymyxin B (30) and NCR335 from the legume Medicago truncatula (31). The ΔhpnH mutant displayed a 10-fold-lower MIC (48 µg/ml) for polymyxin B than did the WT (512 µg/ml). In addition, the ΔhpnH mutant was found to be 100-fold more susceptible than the WT to NCR335 (Fig. 4G). The ΔhpnP mutant withstood all of the abovementioned stressors as well as did the WT, with the exception of acidic stress, where it grew slower than the WT.
C35 hopanoids are required to establish an efficient symbiosis with A. afraspera but not with soybean.
Because hopanoids are required for microaerobic growth and stress tolerance in the free-living state, we hypothesized that they would aid survival within the plant microenvironment. To test this, we analyzed the symbiotic phenotypes of ΔhpnP and ΔhpnH mutants on two host plants, soybean and A. afraspera. On soybean, at 21 days post inoculation (dpi), both mutants induced fewer nodules and displayed slightly reduced nitrogenase activity as estimated by the acetylene reduction assay (ARA) relative to the WT (see Fig. S4A to C in the supplemental material). However, these differences were not statistically significant, and cytological analyses done by confocal microscopy and TEM revealed no differences between WT and mutant nodules (see Fig. S4M to X). Thus, under the conditions of these assays, neither 2Me- nor C35 hopanoids appear to be important for symbiosis between B. diazoefficiens and soybean.
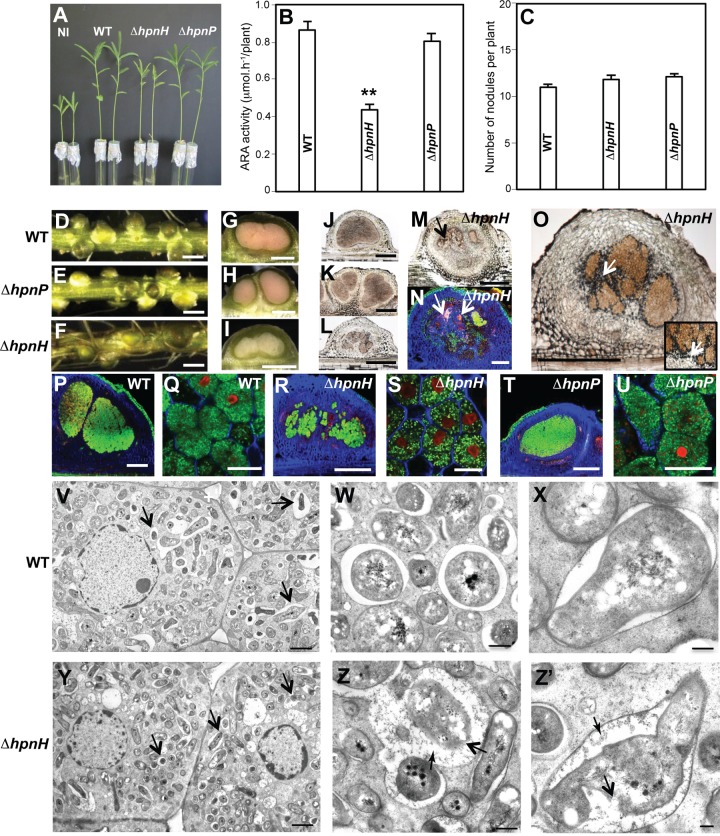
In contrast, a clear effect of the hpnH mutation could be observed when B. diazoefficiens infected A. afraspera. Plants inoculated with the ΔhpnH mutant displayed typical nitrogen starvation symptoms, including foliage chlorosis, reduced plant growth, and half the acetylene reduction activity of WT- and ΔhpnP mutant-infected plants (Fig. 5A and B). Reduced nitrogen fixation was not due to a decrease in the numbers of nodules, which were comparable in WT- and ΔhpnH mutant-infected plants at 9, 14, and 21 dpi, suggesting that the hpnH mutation does not affect nodule organogenesis (Fig. 5C; see also Fig. S5 in the supplemental material). However, cytological analyses revealed that ΔhpnH mutant nodules displayed several disorders in comparison to WT and ΔhpnP mutant nodules (Fig. 5D to Z′).
FIG 5 .
B. diazoefficiens ΔhpnH mutant is impaired in symbiosis with A. afraspera at 21 dpi. (A) Comparison of growth of plants, noninoculated (NI) or inoculated with WT or ΔhpnH or ΔhpnP mutant. (B) Quantification of acetylene reduction activity (ARA) in plants inoculated with WT or ΔhpnH or ΔhpnP mutant. Error bars represent standard errors (n = 10). **, P < 0.01 by Tukey’s honestly significant difference test. (C) Number of nodules per plant elicited by WT and ΔhpnH and ΔhpnP mutants. (D to M) Aspect of the nodules elicited by WT (D, G, and J), ΔhpnP mutant (E, H, and K), and ΔhpnH mutant (F, I, L, and M). (D to F) Whole roots; bars, 1 mm. (G to I) Cross section of live nodules; bars, 500 µm. (J to L) Nodule thin sections viewed by bright-field microscopy; bars, 500 µm. (M) The black arrow shows plant defense reactions (necrotic plant cells); bar, 500 µm. (N) Aspect of the nodules elicited by ΔhpnH mutant as observed by confocal microscopy using the live-dead kit; bar, 200 µm. White arrows show plant defense reactions. (O) Aspect of the nodules elicited by ΔhpnH mutant stained with Lugol; bar, 500 µm. White arrows point to starch granules in black. (P to U) Confocal microscopy observations of nodules elicited by WT (P and Q), ΔhpnH (R and S), and ΔhpnP (T and U) strains and stained using Syto9 (green; healthy bacteroids), calcofluor (blue; plant cell wall), and propidium iodide (red; infected plant nuclei and bacteroids with compromised membranes); bars, 200 µm (P R, and T) and 20 µm (Q, S, and U). (V to Z′) TEM of nodules elicited by WT (V, W, and X) and ΔhpnH mutant (Y, Z, and Z′). (V and Y) Black arrows show symbiososmes. (Z) Cell envelope of some ΔhpnH mutant bacteroids is not well delineated (boldface black arrow), and some deposits of cellular material can be observed in the peribacteroid space (lightface black arrow). (Z′) The boldface black arrow shows bacteroid wall breakdown. The lightface black arrow shows cellular material of unknown origin. Bars, 2 µm (V and Y), 0.5 µm (W and Z), and 0.2 µm (X and Z′).
At the cellular level, ΔhpnH mutant nodules were smaller (Fig. 5D to F) and had pink or, in ~30% of cases, even white central tissue in contrast to WT and ΔhpnP mutant nodules, which were dark pink due to the accumulation of the O2 carrier leghemoglobin (Fig. 5G to I). The central symbiotic tissue of ΔhpnH mutant nodules was often disorganized and partially infected (Fig. 5L, M, and R), as opposed to the fully occupied tissue of WT and ΔhpnP mutant nodules (Fig. 5J, K, P, and T). In some ΔhpnH mutant nodules, the presence of necrotic regions—characterized by the accumulation of autofluorescent brown compounds—could be seen (Fig. 5M and N). These are likely polyphenol compounds whose production is associated with plant defense responses (32). Within ΔhpnH mutant nodules, iodine staining also revealed accumulation of starch granules in the noninfected cells surrounding the symbiotic tissue, whereas such granules were rarely observed in WT and ΔhpnP mutant nodules (Fig. 5O). Starch accumulation is indicative of an imbalance between the photosynthates furnished by the plant and the ability of the bacterium to metabolize them, a typical feature of nonfixing or underperforming strains (33, 34).
To determine whether ΔhpnH mutant symbiotic defects stem from a problem in the bacterial differentiation process or are due to a damaged membrane, we examined nodule sections by confocal microscopy using live-dead staining (35) and TEM to analyze the ultrastructure of bacteroids. Confocal microscopy revealed that all strains, including the ΔhpnH mutant, differentiated properly into elongated bacteroids, which were, for the majority, viable, as indicated by the green Syto9 staining (Fig. 5P to U). However, TEM analysis showed that the cell envelope of some ΔhpnH mutant bacteroids was not well delineated and, in a few cases, even broken (Fig. 5Y and Z′). Similar damage was seen in the peribacteroid membrane that surrounds bacteroids. Deposits of cellular material, possibly resulting from the release of plant or bacterial cytoplasm, were also observed in the peribacteroid space, suggesting a beginning of senescence or perhaps necrosis of symbiotic bacterial cells (Fig. 5Z and Z′). Such defects were not observed in the WT (Fig. 5V to X) or ΔhpnP nodules. Taken together, our data indicate that under these conditions, C35 hopanoids, but not 2Me-hopanoids, play an important role in facilitating the fitness of B. diazoefficiens in symbiosis with A. afraspera.
DISCUSSION
Bacteria that provide plants with fixed nitrogen represent an attractive agronomical alternative to nitrogen fertilizers (36). Recently, we found a statistically significant correlation between the presence of hopanoid biosynthetic genes and organisms, metabolisms, and environments known to support plant-microbe interactions (9). This raised the hypothesis that hopanoids could support bacterial fitness in the context of symbiosis for certain organisms. Building on our recent observation that hopanoids promote symbiosis between the legume A. evenia and the Bradyrhizobium BTAi1 strain (15), we explored the generality of this finding by studying the phylogenetically different and better-known Bradyrhizobium species B. diazoefficiens, focusing on the roles of two dominant hopanoid classes. In the free-living state of B. diazoefficiens, 2Me-hopanoids contribute to growth under microaerobic and acidic conditions and C35 hopanoids are required for microaerobic growth and tolerance to diverse stresses found in the symbiotic microenvironment. Consistent with these phenotypes, C35 hopanoids are critical for symbiosis between B. diazoefficiens and A. afraspera, and yet they are dispensable for symbiosis with soybean. This intriguing finding suggests that the microenvironment encountered by plant symbionts varies between hosts.
Bradyrhizobium strain BTAi1 and B. diazoefficiens differ physiologically in important ways. Shc proteinss from these two strains fall in distinct phylogenetic clades (15). Unlike Bradyrhizobium strain BTAi1, B. diazoefficiens is unable to photosynthesize (37, 38). Moreover, B. diazoefficiens infects plants via a Nod factor-dependent pathway, whereas Bradyrhizobium strain BTAi1 uses alternate symbiotic strategies (39). Our inability to delete shc in B. diazoefficiens suggests that hopanoids are essential in this species, in contrast to Bradyrhizobium strain BTAi1, where shc mutants are viable. This fundamental difference between the species likely reflects differences in the niches that they inhabit as a consequence of their metabolic differences and what is required for survival therein.
What insights can we gain about 2Me- and C35 hopanoid functions in natural contexts based on ex planta experiments? This is the first study to identify conditions under which synthesis of 2Me-hopanoids is important for any cell type. These include hypoxic and acidic conditions that B. diazoefficiens possibly perceives as stress, thus upregulating 2Me-hopanoid production as has been seen in R. palustris (13) to enhance membrane rigidity and stability (12). However, it is puzzling that 2Me-hopanoids are dispensable under similar conditions within the plant cell, and a priority for future work will be to explain this paradox. Prior work in B. cenocepacia has shown that C35 hopanoids promote stress tolerance and antibiotic resistance (14). Similarly, we found that synthesis of C35 hopanoids is important for growth under oxic and hypoxic conditions and for tolerance to diverse stressors. Consistent with our whole-cell membrane rigidity measurements, these phenotypes could be due to higher membrane fluidity resulting from the absence of C35 hopanoids, the inability to make HoLA, and/or the accumulation of the C30 hopanoid diploptene. We hope to tease apart these possibilities going forward. Intriguingly, the abundance of 2Me-hopanes in ancient sediments peaks during oceanic anoxic events (8), and today, C35 hopanoids appear to be enriched in hypoxic regions of the oceans (40).
Why do we observe context-dependent hopanoid phenotypes in planta? The phenotypic difference of the requirement for C35 hopanoids in A. afraspera but not in soybean likely stems from differences between the plants’ intracellular environments. In both plants, the bacterium is exposed to a variety of stresses, including oxidative, osmotic, and acidic stresses, within the microaerobic niche of the infected plant cell (24). Although such an environment is less than ideal for the ΔhpnH mutant, it is able to colonize as evidenced by its successful symbiosis with the soybean plant. However, unlike soybean, in A. afraspera, B. diazoefficiens undergoes terminal differentiation due to the action of NCR peptides (25). Because the ΔhpnH mutant is highly sensitive to NCR peptides, exposure within the host might reduce the viable mutant population by causing cell death or increasing susceptibility to this and other plant defense mechanisms. Consistent with this hypothesis, the ΔhpnH mutant only partially infects the A. afraspera nodule tissue.
In A. afraspera, synthesis of C35 hopanoids is critical for several aspects of the symbiosis, including evasion of plant defense reactions, efficient utilization of plant photosynthates, and nitrogen fixation. Two reasons why the plant host mounts an immune response against the ΔhpnH mutant may be that the altered mutant surface layer, as seen in TEM images, is unable to suppress this response (41) and/or the host induces nodule senescence prematurely on detecting an underproductive symbiont (42). Consistent with this, nitrogenase activity is reduced in the ΔhpnH mutant relative to the WT, a likely consequence of poor cell viability. Similarly, the buildup of plant carbon as starch in ΔhpnH mutant nodules might indicate slow metabolism and/or perturbation of membrane transport processes that facilitate bacteroid carbon acquisition.
The global agricultural economy is largely based on nitrogen fertilizers, with the United States alone consuming 13,000 tons per annum (43). However, the usage of nitrogen fertilizers comes with a price, as their production requires burning of fossil fuels and their runoff from soils leads to surface and groundwater contamination (44). Rhizobia, which naturally fix nitrogen in association with common crops, are an environmentally and economically feasible alternative to nitrogen fertilizers. Our results reveal that hopanoids affect the ability of B. diazoefficiens to cope with environmental stresses as well as its nitrogen fixation efficiency in a plant host-dependent manner. This observation is relevant to interpreting ancient patterns of hopane deposition, which correlate with paleoenvironmental conditions where nitrogen fixation may have provided a selective advantage (8). In addition, understanding the roles of hopanoids in bacterial stress resistance and how they facilitate nitrogen fixation may enable the engineering of agronomically useful strains with enhanced tolerance to rising temperature and salinity.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table S4 in the supplemental material. Escherichia coli strains were grown in lysogeny broth (LB) (45) at 37°C. B. diazoefficiens strains were grown at 30°C in either rich medium (peptone-salts-yeast extract medium with 0.1% arabinose [PSY] [46] or yeast extract-mannitol medium [YM] [47]) or minimal medium (buffered nodulation medium [BNM] [48]).
DNA methods, plasmid construction, and transformation.
All plasmid constructions and primers used in this study are described in Table S4 in the supplemental material. Standard methods were used for plasmid DNA isolation and manipulation in E. coli (49, 50). DNA sequences of all cloning intermediates were confirmed by sequencing at Retrogen (San Diego, CA). Plasmids were mobilized by conjugation from E. coli S17-1 into B. diazoefficiens strains as previously described (51).
Mutant construction.
The markerless genetic exchange method described in reference 26 was employed to delete hpnP (blr2995) and hpnH (bll3006) (26) (Fig. 1B). Attempts to delete shc (hpnF, blr3004) and the hpnCDEFG (blr3001 to blr3005) operon have been described in Text S1 in the supplemental material.
Hopanoid analysis.
Lipid extraction was performed by the Bligh-Dyer method followed by hopanoid analysis using high-temperature GC-MS (16) and ultraperformance LC-MS (29) as previously described. Growth of bacterial cultures and analysis methods have been described in detail in Text S1 in the supplemental material.
LipidA analysis.
Isolation, purification, and characterization of lipidA from B. diazoefficiens strains were performed by gas-liquid chromatography–MS (GLC-MS) and MALDI-time of flight (TOF) MS as described in reference 15 and in Text S1 in the supplemental material.
Membrane rigidity.
For whole-cell membrane rigidity measurements, we used the fluorescence polarization method using the dye DPH described in reference 12.
Growth curves, stress assays, and MIC determinations.
The growth curve assays, stress assays, and MIC determinations were performed as outlined in references 13 and 15 and have been described in detail in Text S1 in the supplemental material.
Plant cultivation, symbiotic analysis, and microscopy.
Soybean and A. afraspera plants were grown hydroponically as described in reference 22. The plant infection assays and cytological analyses methods are outlined in Text S1 in the supplemental material.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download
(A) Chemical structures of isolated hopanoids from B. diazoefficiens. (B) Cell envelope of a Gram-negative bacterium comprises an inner membrane (IM) and an outer membrane (OM). Hopanoids are found within both membranes. They are either “free” or covalently “bound” to lipidA (HoLA), which is present in the outer leaflet of the OM. As seen in the expanded view of the OM, B. diazoefficiens makes short (C30) hopanoids such as diploptene and extended (C35) hopanoids such as bacteriohopanetetrol (BHT) and aminotriol. Penta- and hexa-acylated lipidA contain 5 and 6 fatty acyl chains, respectively. Hepta-acylated lipidA contains the C35 hopanoid 34-carboxyl-bacteriohopane-32,33-diol, covalently attached to hexa-acylated lipidA. B. diazoefficiens makes another triterpenoid with a gammacerane skeleton called tetrahymanol. With the exception of HoLA, hopanoid positioning in the inner versus outer leaflet has not been established, and example structures are placed randomly. Dark gray and light gray shading represent hydrophilic and hydrophobic regions, respectively. Download
Endosymbiotic context of B. diazoefficiens within root nodules of A. afraspera. B. diazoefficiens exists as a bacteroid, a terminally differentiated enlarged, elongated, and polyploid state, within infected plant cortical cells. In addition to its own membrane, each bacteroid is surrounded by a peribacteroid plant-derived membrane. The double-layered bacteroid is called a symbiosome. The infected plant cell niche is characterized by low O2, low pH, hyperosmosis, and oxidative stress (21, 24). Download
Cryo-transmission electron microscopy (cryo-TEM) micrographs show intact outer and inner membranes in all B. diazoefficiens strains. Bar, 200 nm. Download
B. diazoefficiens ΔhpnH and ΔhpnP mutants are unimpaired in symbiosis with soybean at 21 dpi. (A) Comparison of growth of plants, noninoculated (NI) or inoculated with WT or ΔhpnH or ΔhpnP mutant. (B) Quantification of acetylene reduction activity (ARA) in plants inoculated with WT and ΔhpnH and ΔhpnP mutants. (C) Nodulation efficiency of WT and ΔhpnH and ΔhpnP mutants on plants. Error bars in panels B and C represent standard errors (n = 10). Based on Tukey’s honestly significant difference (HSD) test, differences between strains were found to be insignificant (P > 0.05). (D to L) Aspects of nodules elicited by WT (D, G, and J), ΔhpnH mutant (E, H, and K), and ΔhpnP mutant (F, I, and L). (D to F) Whole roots; bar, 4 mm, (G to I) Cross section of live nodules; bar, 1 mm. (J to L) Nodule thin sections viewed by bright-field microscopy; bar, 1 mm. (M to R) Observation of nodules elicited by WT (M and P), ΔhpnH (N and Q), and ΔhpnP (O and R) strains by confocal microscopy using Syto9 (green; healthy bacteroids), calcofluor (blue; plant cell wall), and propidium iodide (red,; infected plant nuclei and bacteroids with compromised membranes). Bars, 300 µm (M to O) and 50 µm (P to R). (S to W) Transmission electron micrographs of nodules elicited by WT (S and V), ΔhpnH mutant (T and W), and ΔhpnP mutant (U and X). Bars, 1 µm (S to U) and 0.2 µm (V to X). Download
Kinetics of nodulation and nitrogen fixation of A. afraspera plants inoculated with B. diazoefficiens. (A) Number of nodules elicited by WT and ΔhpnH and ΔhpnP mutants on plants at 9, 14, and 21 days postinoculation (dpi). (B) Acetylene-reducing activity (ARA) in plants inoculated with WT and ΔhpnH and ΔhpnP mutants at 9, 14, and 21 dpi. Error bars represent standard errors (n = 10). Asterisks above the error bars indicate significant differences at P < 0.05 (*) and P < 0.01 (**) (Tukey’s honestly significant difference [HSD] test). Download
Compounds identified by high-temperature GC-MS.
Compounds identified by ultraperformance LC-MS.
Hopanoid and tetrahymanol quantification by GC-MS using pregnane-acetate as standard.
Strains, plasmids, and primers used in this study.
ACKNOWLEDGMENTS
This work was supported by awards from NASA (NNX12AD93G), the National Science Foundation (1224158), and the Howard Hughes Medical Institute (HHMI) to D.K.N.; the Agence Nationale de la Recherche, grant “BugsInACell” no. ANR-13-BSV7-0013, to E.G.; and the Italian Ministry of Education, Universities and Research (PRIN) and Mizutani Foundation for Glycoscience 2014 to A.M. and A.S. D.K.N. is an HHMI Investigator. The EM facility where the cryo-TEM micrographs were collected is supported by the Agouron and Beckman foundations.
We thank Raphael Ledermann and Hans-Martin Fischer for providing strains and protocols and for performing confirmatory soybean symbiosis assays; Alasdair McDowell in Grant J. Jensen’s lab for collecting cryo-TEM micrographs at the EM facility; and Luisa Sturiale and Domenico Garozzo for recording the MS spectra. We are grateful to Newman lab members for constructive comments on the manuscript.
Footnotes
Citation Kulkarni G, Busset N, Molinaro A, Gargani D, Chaintreuil C, Silipo A, Giraud E, Newman DK. 2015. Specific hopanoid classes differentially affect free-living and symbiotic states of Bradyrhizobium diazoefficiens. mBio 6(5):e01251-15. doi:10.1128/mBio.01251-15.
REFERENCES
- 1.Masson-Boivin C, Giraud E, Perret X, Batut J. 2009. Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol 17:458–466. doi: 10.1016/j.tim.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Benson DR, Silvester WB. 1993. Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol Res 57:293–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meeks JC, Elhai J. 2002. Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol Mol Biol Rev 66:94–121. doi: 10.1128/MMBR.66.1.94-121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delamuta JRM, Ribeiro RA, Ormeno-Orrillo E, Melo IS, Martinez-Romero E, Hungria M. 2013. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int J Syst Evol Microbiol 63:3342–3351. doi: 10.1099/ijs.0.049130-0. [DOI] [PubMed] [Google Scholar]
- 5.Aktas M, Wessel M, Hacker S, Klüsener S, Gleichenhagen J, Narberhaus F. 2010. Phosphatidylcholine biosynthesis and its significance in bacteria interacting with eukaryotic cells. Eur J Cell Biol 89:888–894. doi: 10.1016/j.ejcb.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Carlson RW, Forsberg LS, Kannenberg EL. 2010. Lipopolysaccharides in Rhizobium-legume symbioses. Subcell Biochem 53:339–386. doi: 10.1007/978-90-481-9078-2_16. [DOI] [PubMed] [Google Scholar]
- 7.Rohmer M. 1993. The biosynthesis of triterpenoids of the hopane series in the eubacteria—a mine of new enzyme reactions. Pure Appl Chem 65:1293–1298. [Google Scholar]
- 8.Knoll AH, Summons RE, Waldbauer JR, Zumberge JE. 2007. The geological succession of primary producers in the oceans, p 133–163. In Falkowski PG, Knoll AH (ed), Evolution of primary producers in the sea. Elsevier, San Diego, CA. [Google Scholar]
- 9.Ricci JN, Coleman ML, Welander PV, Sessions AL, Summons RE, Spear JR, Newman DK. 2014. Diverse capacity for 2-methylhopanoid production correlates with a specific ecological niche. ISME J 8:675–684. doi: 10.1038/ismej.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kannenberg EL, Perzl M, Hartner T. 1995. The occurrence of hopanoid lipids in Bradyrhizobium bacteria. FEMS Microbiol Lett 127:255–261. doi: 10.1111/j.1574-6968.1995.tb07482.x. [DOI] [PubMed] [Google Scholar]
- 11.Nalin R, Putra SR, Domenach AM, Rohmer M, Gourbiere F, Berry AM. 2000. High hopanoid/total lipids ratio in Frankia mycelia is not related to the nitrogen status. Microbiology 146:3013–3019. doi: 10.1099/00221287-146-11-3013. [DOI] [PubMed] [Google Scholar]
- 12.Wu CH, Bialecka-Fornal M, Newman DK. 2015. Methylation at the C-2 position of hopanoids increases rigidity in native bacterial membranes. eLife doi: 10.7554/eLife.05663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulkarni G, Wu CH, Newman DK. 2013. The general stress response factor EcfG regulates expression of the C-2 hopanoid methylase HpnP in Rhodopseudomonas palustris TIE-1. J Bacteriol 195:2490–2498. doi: 10.1128/JB.00186-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmerk CL, Welander PV, Hamad MA, Bain KL, Bernards MA, Summons RE, Valvano MA. 2015. Elucidation of the Burkholderia cenocepacia hopanoid biosynthesis pathway uncovers functions for conserved proteins in hopanoid-producing bacteria. Environ Microbiol 17:735–750. doi: 10.1111/1462-2920.12509. [DOI] [PubMed] [Google Scholar]
- 15.Silipo A, Vitiello G, Gully D, Sturiale L, Chaintreuil C, Fardoux J, Gargani D, Lee HI, Kulkarni G, Busset N, Marchetti R, Palmigiano A, Moll H, Engel R, Lanzetta R, Paduano L, Parrilli M, Chang WS, Holst O, Newman DK, Garozzo D, D’Errico G, Giraud E, Molinaro A. 2014. Covalently linked hopanoid-lipid A improves outer-membrane resistance of a Bradyrhizobium symbiont of legumes. Nat Commun 5:5106. doi: 10.1038/ncomms6106. [DOI] [PubMed] [Google Scholar]
- 16.Welander PV, Hunter RC, Zhang L, Sessions AL, Summons RE, Newman DK. 2009. Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1. J Bacteriol 191:6145–6156. doi: 10.1128/JB.00460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welander PV, Doughty DM, Wu CH, Mehay S, Summons RE, Newman DK. 2012. Identification and characterization of Rhodopseudomonas palustris TIE-1 hopanoid biosynthesis mutants. Geobiology 10:163–177. doi: 10.1111/j.1472-4669.2011.00314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welander PV, Summons RE. 2012. Discovery, taxonomic distribution, and phenotypic characterization of a gene required for 3-methylhopanoid production. Proc Natl Acad Sci U S A 109:12905–12910. doi: 10.1073/pnas.1208255109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bravo J, Perzl M, Härtner T, Kannenberg EL, Rohmer M. 2001. Novel methylated triterpenoids of the gammacerane series from the nitrogen-fixing bacterium Bradyrhizobium japonicum USDA 110. Eur J Biochem 268:1323–1331. doi: 10.1046/j.1432-1327.2001.01998.x. [DOI] [PubMed] [Google Scholar]
- 20.Komaniecka I, Choma A, Mazur A, Duda KA, Lindner B, Schwudke D, Holst O. 2014. Occurrence of an unusual hopanoid-containing lipid A among lipopolysaccharides from Bradyrhizobium species. J Biol Chem 289:35644–35655. doi: 10.1074/jbc.M114.614529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oldroyd GE, Murray JD, Poole PS, Downie JA. 2011. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45:119–144. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- 22.Renier A, Maillet F, Fardoux J, Poinsot V, Giraud E, Nouwen N. 2011. Photosynthetic Bradyrhizobium sp. strain ORS285 synthesizes 2-O-methylfucosylated lipochitooligosaccharides for nod gene-dependent interaction with Aeschynomene plants. Mol Plant Microbe Interact 24:1440–1447. doi: 10.1094/MPMI-05-11-0104. [DOI] [PubMed] [Google Scholar]
- 23.Prell J, Poole P. 2006. Metabolic changes of rhizobia in legume nodules. Trends Microbiol 14:161–168. doi: 10.1016/j.tim.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Gibson KE, Kobayashi H, Walker GC. 2008. Molecular determinants of a symbiotic chronic infection. Annu Rev Genet 42:413–441. doi: 10.1146/annurev.genet.42.110807.091427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czernic P, Gully D, Cartieaux F, Moulin L, Guefrachi I, Patrel D, Pierre O, Fardoux J, Chaintreuil C, Nguyen P, Gressent F, Dasilva C, Poulain J, Wincker P, Rofidal V, Hem S, Barrière Q, Arrighi J, Mergaert P, Giraud E. 2015. Convergent evolution of endosymbiont differentiation in Dalbergioid and IRLC legumes mediated by nodule-specific cysteine-rich peptides. Plant Physiol doi: 10.1104/pp.15.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masloboeva N, Reutimann L, Stiefel P, Follador R, Leimer N, Hennecke H, Mesa S, Fischer H. 2012. Reactive oxygen species-inducible ECF sigma factors of Bradyrhizobium japonicum. PLoS One 7:e43421. doi: 10.1371/journal.pone.0043421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welander PV, Coleman ML, Sessions AL, Summons RE, Newman DK. 2010. Identification of a methylase required for 2-methylhopanoid production and implications for the interpretation of sedimentary hopanes. Proc Natl Acad Sci U S A 107:8537–8542. doi: 10.1073/pnas.0912949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sessions AL, Zhang L, Welander PV, Doughty D, Summons RE, Newman DK. 2013. Identification and quantification of polyfunctionalized hopanoids by high temperature gas chromatography-mass spectrometry. Org Geochem 56:120–130. doi: 10.1016/j.orggeochem.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neubauer C, Dalleska NF, Cowley ES, Shikuma NJ, Wu CH, Sessions AL, Newman DK. 2015. Lipid remodeling in Rhodopseudomonas palustris TIE-1 upon loss of hopanoids and hopanoid methylation. Geobiology 13:443–453. doi: 10.1111/gbi.12143. [DOI] [PubMed] [Google Scholar]
- 30.Newton BA. 1956. Properties and mode of action of the polymyxins. Bacteriol Rev 20:14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiricz H, Szucs A, Farkas A, Pap B, Lima RM, Maroti G, Kondorosi E, Kereszt A. 2013. Antimicrobial nodule-specific cysteine-rich peptides induce membrane depolarization-associated changes in the transcriptome of Sinorhizobium meliloti. Appl Environ Microbiol 79:6737–6746. doi: 10.1128/AEM.01791-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasse J, Debilly F, Truchet G. 1993. Abortion of infection during the Rhizobium meliloti-alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. Plant J 4:555–566. doi: 10.1046/j.1365-313X.1993.04030555.x. [DOI] [Google Scholar]
- 33.Finan TM, Wood JM, Jordan DC. 1983. Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J Bacteriol 154:1403–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lodwig EM, Hosie AHF, Bourdès A, Findlay K, Allaway D, Karunakaran R, Downie JA, Poole PS. 2003. Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422:722–726. doi: 10.1038/nature01527. [DOI] [PubMed] [Google Scholar]
- 35.Haag AF, Baloban M, Sani M, Kerscher B, Pierre O, Farkas A, Longhi R, Boncompagni E, Hérouart D, Dall’angelo S, Kondorosi E, Zanda M, Mergaert P, Ferguson GP. 2011. Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol 9:e1001169. doi: 10.1371/journal.pbio.1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bohlool BB, Ladha JK, Garrity DP, George T. 1992. Biological nitrogen fixation for sustainable agriculture: a perspective. Plant Soil 141:1–11. [Google Scholar]
- 37.Fleischman D, Kramer D. 1998. Photosynthetic rhizobia. Biochim Biophys Acta 1364:17–36. doi: 10.1016/S0005-2728(98)00011-5. [DOI] [PubMed] [Google Scholar]
- 38.Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K, Kohara M, Matsumoto M, Shimpo S, Tsuruoka H, Wada T, Yamada M, Tabata S. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res 9:189–197. doi: 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- 39.Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, Avarre JC, Jaubert M, Simon D, Cartieaux F, Prin Y, Bena G, Hannibal L, Fardoux J, Kojadinovic M, Vuillet L, Lajus A, Cruveiller S, Rouy Z, Mangenot S, Segurens B, Dossat C, Franck WL, Chang WS, Saunders E, Bruce D, Richardson P, Normand P, Dreyfus B, Pignol D, Stacey G, Emerich D, Vermeglio A, Medigue C, Sadowsky M. 2007. Legumes symbioses: absence of nod genes in photosynthetic bradyrhizobia. Science 316:1307–1312. doi: 10.1126/science.1139548. [DOI] [PubMed] [Google Scholar]
- 40.Kharbush JJ, Ugalde JA, Hogle SL, Allen EE, Aluwihare LI. 2013. Composite bacterial hopanoids and their microbial producers across oxygen gradients in the water column of the California Current. Appl Environ Microbiol 79:7491–7501. doi: 10.1128/AEM.02367-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kannenberg EL, Carlson RW. 2001. Lipid A and O-chain modifications cause Rhizobium lipopolysaccharides to become hydrophobic during bacteroid development. Mol Microbiol 39:379–391. doi: 10.1046/j.1365-2958.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 42.West SA, Kiers ET, Simms EL, Denison RF. 2002. Sanctions and mutualism stability: why do rhizobia fix nitrogen? Proc Biol Sci 269:685–694. doi: 10.1098/rspb.2001.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.US Department of Agriculture 2013. Table 1. U.S. consumption of nitrogen, phosphate, and potash, 1960–2011. http://www.ers.usda.gov/data-products/fertilizer-use-and-price.aspx.
- 44.Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai Z, Freney JR, Martinelli LA, Seitzinger SP, Sutton MA. 2008. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320:889–892. doi: 10.1126/science.1136674. [DOI] [PubMed] [Google Scholar]
- 45.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 46.Mesa S, Hauser F, Friberg M, Malaguti E, Fischer H, Hennecke H. 2008. Comprehensive assessment of the regulons controlled by the FixLJ-FixK2-FixK1 cascade in Bradyrhizobium japonicum. J Bacteriol 190:6568–6579. doi: 10.1128/JB.00748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vincent JM. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific Publications, Oxford, United Kingdom. [Google Scholar]
- 48.Podlesakova K, Fardoux J, Patrel D, Bonaldi K, Novak O, Strnad M, Giraud E, Spichal L, Nouwen N. 2013. Rhizobial synthesized cytokinins contribute to but are not essential for the symbiotic interaction between photosynthetic bradyrhizobia and Aeschynomene legumes. Mol Plant Microbe Interact 26:1232–1238. doi: 10.1094/MPMI-03-13-0076-R. [DOI] [PubMed] [Google Scholar]
- 49.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1992. Current protocols in molecular biology. John Wiley & Sons, New York, NY. [Google Scholar]
- 50.Gibson DG, Young L, Chuang R, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 51.Hahn M, Meyer L, Studer D, Regensburger B, Hennecke H. 1984. Insertion and deletion mutations within the nif region of Rhizobium japonicum. Plant Mol Biol 3:159–168. doi: 10.1007/BF00016063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download
(A) Chemical structures of isolated hopanoids from B. diazoefficiens. (B) Cell envelope of a Gram-negative bacterium comprises an inner membrane (IM) and an outer membrane (OM). Hopanoids are found within both membranes. They are either “free” or covalently “bound” to lipidA (HoLA), which is present in the outer leaflet of the OM. As seen in the expanded view of the OM, B. diazoefficiens makes short (C30) hopanoids such as diploptene and extended (C35) hopanoids such as bacteriohopanetetrol (BHT) and aminotriol. Penta- and hexa-acylated lipidA contain 5 and 6 fatty acyl chains, respectively. Hepta-acylated lipidA contains the C35 hopanoid 34-carboxyl-bacteriohopane-32,33-diol, covalently attached to hexa-acylated lipidA. B. diazoefficiens makes another triterpenoid with a gammacerane skeleton called tetrahymanol. With the exception of HoLA, hopanoid positioning in the inner versus outer leaflet has not been established, and example structures are placed randomly. Dark gray and light gray shading represent hydrophilic and hydrophobic regions, respectively. Download
Endosymbiotic context of B. diazoefficiens within root nodules of A. afraspera. B. diazoefficiens exists as a bacteroid, a terminally differentiated enlarged, elongated, and polyploid state, within infected plant cortical cells. In addition to its own membrane, each bacteroid is surrounded by a peribacteroid plant-derived membrane. The double-layered bacteroid is called a symbiosome. The infected plant cell niche is characterized by low O2, low pH, hyperosmosis, and oxidative stress (21, 24). Download
Cryo-transmission electron microscopy (cryo-TEM) micrographs show intact outer and inner membranes in all B. diazoefficiens strains. Bar, 200 nm. Download
B. diazoefficiens ΔhpnH and ΔhpnP mutants are unimpaired in symbiosis with soybean at 21 dpi. (A) Comparison of growth of plants, noninoculated (NI) or inoculated with WT or ΔhpnH or ΔhpnP mutant. (B) Quantification of acetylene reduction activity (ARA) in plants inoculated with WT and ΔhpnH and ΔhpnP mutants. (C) Nodulation efficiency of WT and ΔhpnH and ΔhpnP mutants on plants. Error bars in panels B and C represent standard errors (n = 10). Based on Tukey’s honestly significant difference (HSD) test, differences between strains were found to be insignificant (P > 0.05). (D to L) Aspects of nodules elicited by WT (D, G, and J), ΔhpnH mutant (E, H, and K), and ΔhpnP mutant (F, I, and L). (D to F) Whole roots; bar, 4 mm, (G to I) Cross section of live nodules; bar, 1 mm. (J to L) Nodule thin sections viewed by bright-field microscopy; bar, 1 mm. (M to R) Observation of nodules elicited by WT (M and P), ΔhpnH (N and Q), and ΔhpnP (O and R) strains by confocal microscopy using Syto9 (green; healthy bacteroids), calcofluor (blue; plant cell wall), and propidium iodide (red,; infected plant nuclei and bacteroids with compromised membranes). Bars, 300 µm (M to O) and 50 µm (P to R). (S to W) Transmission electron micrographs of nodules elicited by WT (S and V), ΔhpnH mutant (T and W), and ΔhpnP mutant (U and X). Bars, 1 µm (S to U) and 0.2 µm (V to X). Download
Kinetics of nodulation and nitrogen fixation of A. afraspera plants inoculated with B. diazoefficiens. (A) Number of nodules elicited by WT and ΔhpnH and ΔhpnP mutants on plants at 9, 14, and 21 days postinoculation (dpi). (B) Acetylene-reducing activity (ARA) in plants inoculated with WT and ΔhpnH and ΔhpnP mutants at 9, 14, and 21 dpi. Error bars represent standard errors (n = 10). Asterisks above the error bars indicate significant differences at P < 0.05 (*) and P < 0.01 (**) (Tukey’s honestly significant difference [HSD] test). Download
Compounds identified by high-temperature GC-MS.
Compounds identified by ultraperformance LC-MS.
Hopanoid and tetrahymanol quantification by GC-MS using pregnane-acetate as standard.
Strains, plasmids, and primers used in this study.