Abstract
Endosymbiosis is widespread among cnidarians and is of high ecological relevance. The tropical sea anemone Aiptasia sp. is a laboratory model system for endosymbiosis between reef-building corals and photosynthetic dinoflagellate algae of the genus Symbiodinium. Here we identify the key environmental cues to induce reproducible spawning in Aiptasia under controlled laboratory conditions. We find that simulating a lunar cycle with blue-wavelength light is necessary to promote abundant gamete production and synchronous release in well-fed animals. Sexual reproduction rates are genetically determined and differ among clonal lines under similar conditions. We also find the inverse difference in rates of asexual reproduction. This study provides the requisite basis for further development of the Aiptasia model system, allowing analysis of basic cellular and molecular mechanisms in the laboratory as well as investigations of broad questions of ecological and evolutionary relevance.
Cnidarians exhibit enormous plasticity in their morphologies and life cycles, and have emerged as key models in a broad range of research fields from evolution to ecology. As the sister group to the Bilateria (Fig. 1a), cnidarians are important model systems for studying development1,2, pattern formation3, regeneration4 and stem cell biology5,6 (Fig. 1a). The cnidarians’ evolutionary success over 500 million years has been proposed to be due in part to their ability to form symbioses with prokaryotic and/or eukaryotic microorganisms7. Major symbiotic partners include complex communities of viruses, archaea, bacteria (including cyanobacteria), and eukaryotic algae8,9. More broadly, such symbioses with photosynthetic microbes are also found in mollusks, sponges, acoel flatworms, and vertebrates (salamander) and in all, the translocation of photosynthetically fixed carbon from the symbiont to the host represents a significant energy source10,11.
Figure 1. Symbiosis throughout the cnidarians and the anthozoan life cycle.
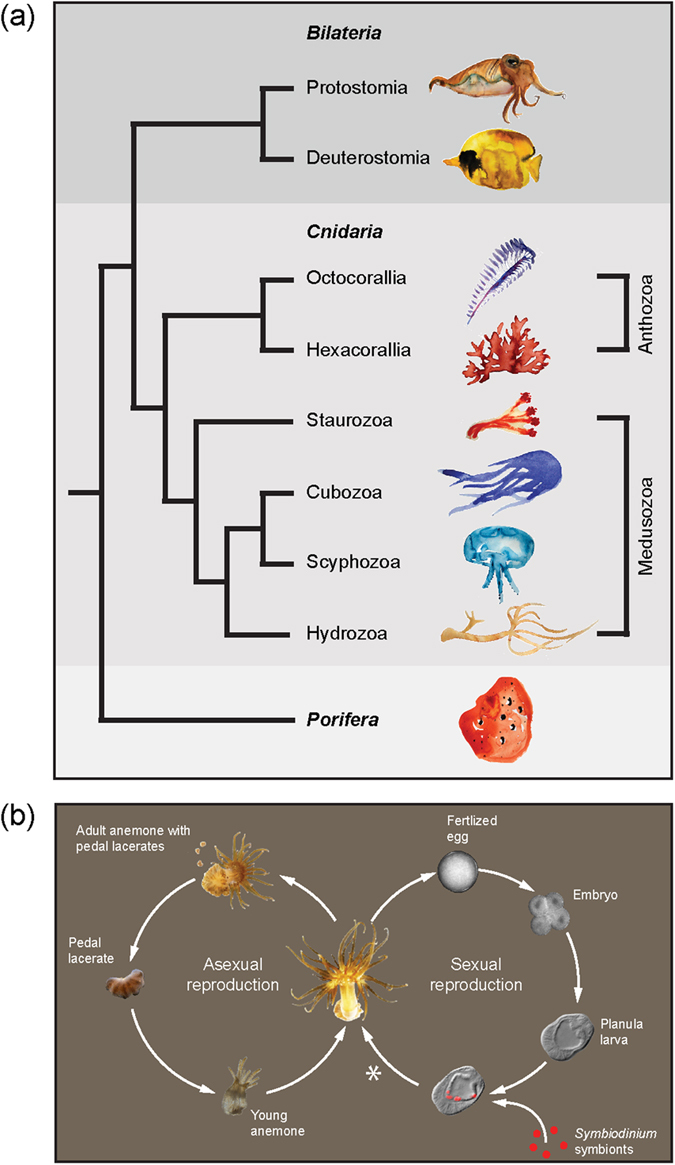
(a) Phylogenetic tree of the major metazoan clades, with phyla shown in bold. Cnidarians represent a sister group to the bilaterians and are at the base of metazoan evolution. Symbiosis with dinoflagellates and green algae occur in species throughout the classes Medusozoa and Anthozoa, the latter of which contains Aiptasia sp. Illustrations were drawn by Stephanie Guse and are used with permission. (b) Overview of Aiptasia life cycle showing dual reproductive modes. *metamorphosis and settlement in the laboratory have not yet been reported, and as such remains an active experimental area.
The most common eukaryotic endosymbiont among the cnidarians is the dinoflagellate Symbiodinium spp.; associations with Symbiodinium are found in many species including the majority of hexacorallia (e.g. reef-building corals, sea anemones), octocorallia (e.g. gorgonians, soft corals, sea pens), hydrozoa (e.g. fire corals) and scyphozoa (e.g. jellyfish)12 (Fig. 1a). Genus Symbiodinium is diverse, containing hundreds of strains worldwide that have been categorized into clades based on ribosomal DNA markers13; species assignments and functional characterizations of strains are an active area of research14. The unicellular Symbiodinium reside intracellularly in cnidarian endodermal tissues and transfer photosynthates to the host. Thus, this widespread phenomenon is likely a key factor in the extensive adaptive radiation of cnidarians in diverse aquatic niches.
The most economically and ecologically critical cnidarian-Symbiodinium symbiosis is that of reef-building corals, which rely so heavily on symbiont-produced nutrition that the relationship is obligatory for the hosts to persist15,16. As such, the breakdown of this symbiosis, termed “coral bleaching”, has become a major threat to coral reefs worldwide17. Despite this importance, much remains unknown about the cellular and molecular basis of the coral-Symbiodinium symbiosis, including its establishment, maintenance, and breakdown in response to stress18. The majority of corals produce symbiont-free planula larvae that must take up symbionts from the environment each generation19. However, most reef-building corals spawn only once per year20, greatly restricting the experimental availability of larvae and thereby severely limiting the systematic study of endosymbiosis establishment.
To address this limitation, a practicable symbiotic laboratory model has been developed with the small sea anemone Aiptasia sp. Although Aiptasia is not suited to every application relevant to coral biology (e.g. calcification), it holds many advantages including its symbiotic relationship with the same types of Symbiodinium strains as corals21,22,23. Most importantly, the process of endosymbiosis establishment is similar to that of many reef-building corals: Aiptasia planula larvae are initially non-symbiotic and establish endosymbiosis anew each generation23 (Fig. 1b). Both reef-building corals and Aiptasia live primarily as a sessile polyp stage that switches between asexual reproduction and sexual production of motile planula larvae (Fig. 1b). Based on molecular analysis, Aiptasia sp. is regarded as a single panglobal species with two distinct genetic networks: one on the United States South Atlantic coast and the other consisting of all other Aiptasia sp. sampled worldwide24. Clonal anemone lines can be generated through asexual reproduction via pedal laceration25 (Fig. 1b), and the majority of Aiptasia resources have been developed from clonal line CC7, including transcriptomes26,27 and the genome28. Transcriptomic and genomic resources for many Symbiodinium strains are likewise available29,30,31.
Despite these advantages, a critical aspect remains underdeveloped in Aiptasia: consistent induction of sexual reproduction in the laboratory, a prerequisite to investigations of symbiosis establishment and development of state-of-the-art functional tools such as morpholinos and transgenesis32,33,34. Aiptasia is demonstrably capable of year-round gametogenesis35 and spawning in laboratory conditions23,36, but laboratory-induced spawning has been inefficient and unpredictable and currently no publically available protocols exist to overcome this limitation. Laboratory-induced spawning in the cnidarian Nematostella has been achieved through manipulation of food, temperature, and light alone or in combination37,38. In the wild, coral spawning is correlated with the lunar cycle and blue light appears to play a major role in synchronized spawning39,40,41,42. Similarly, oogenesis in Aiptasia is increased by feeding43 and peaks in relation to the lunar cycle35. Thus, we hypothesized that ample feeding and blue-light cues simulating moonlight may efficiently stimulate laboratory spawning in Aiptasia.
To test this hypothesis, we used three Aiptasia clonal lines used in laboratories worldwide. We first further characterized these clonal lines—CC7 (male) and F003 and H2 (both female)—and used molecular phylogeny to place them into a biogeographical context. Next, we systematically tested different conditions to determine major cues for coordinated and copious gametogenesis and spawning. We found that blue-light cues (presumably mimicking a lunar cycle) together with slight temperature increases produce synchronous and efficient spawning in well-fed CC7 and F003 couples and, to a lesser extent, in CC7 and H2 couples. Quantitative differences of spawning efficiencies between female lines F003 and H2 were inversely correlated to their asexual reproduction rates.
Results
Characterization of Aiptasia clonal lines
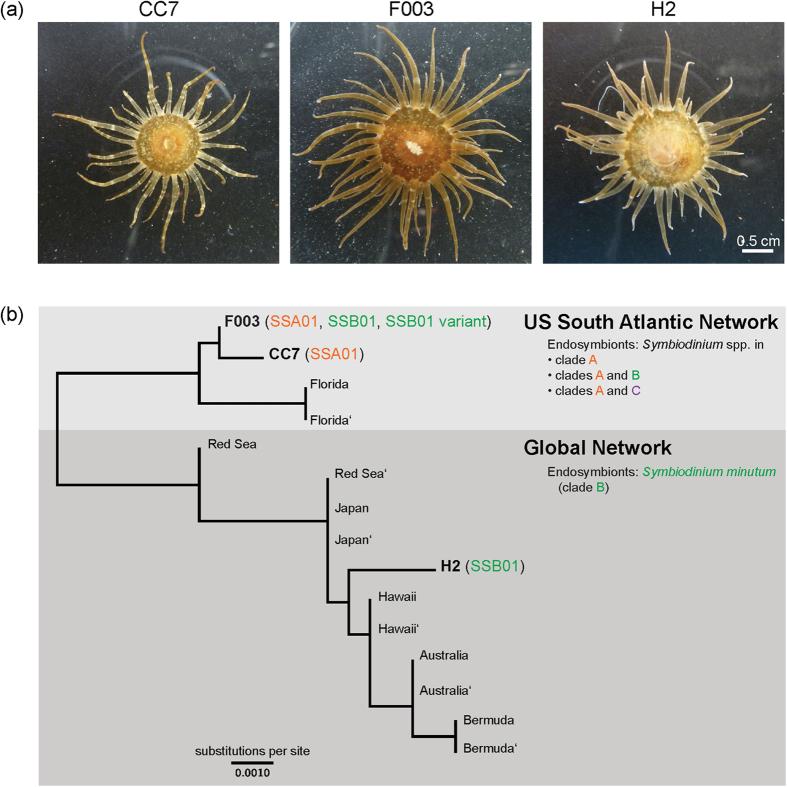
The three clonal lines of Aiptasia CC7, F003, and H2 we routinely maintain in the laboratory (see Aiptasia culture conditions in Methods) appear morphologically similar (Fig. 2a). However, their genetic relationship to each other and to other Aiptasia populations was unclear. We therefore determined the relationship of our lines to wild Aiptasia populations worldwide by sequencing the four independent “SCAR” (“sequence characterized amplified region”) genotyping markers24 of two individuals from each of the three clonal Aiptasia lines. Within each of the lines, both individuals sampled yielded identical sequences for each SCAR marker, demonstrating the continued clonality of the lines. Each clonal line produced a haplotype distinct from the others. In CC7 and F003, two alleles were found in the following three loci and both animals carried the alleles: SCAR3, SCAR4, and SCAR5 (CC7 only). In those cases, the alleles were collapsed together to produce a consensus sequence containing the appropriate base ambiguity. The Maximum Likelihood tree in Fig. 2b shows the relationship of these three clonal lines to twelve globally field-sampled Aiptasia representing eight distinct haplotypes24. As expected, lines CC7 and F003 (founders collected from US South Atlantic coast) and line H2 (founder collected from Hawaii) all cluster closest with individuals collected from the same respective geographic region by Thornhill and colleagues24 (Fig. 2b). Thus, these laboratory clonal lines represent members of both identified Aiptasia genetic networks found worldwide: US South Atlantic network (CC7, F003) and global network (H2).
Figure 2. Characterization of Aiptasia clonal lines CC7 (male), F003 (female), and H2 (female).
(a) Clonal lines appear morphologically similar, with minor variation in coloration and body size. (b) Maximum likelihood tree of ~2 kb concatenated SCAR genotyping markers24 showing the relationship of the three clonal lines to twelve field-sampled Aiptasia sp. individuals24 grouped into two genetic networks. Clonal lines are indicated by name; field-sampled individuals are named by their sampling location, with the second animal from the same location designed by’. Shown also are the endogenous Symbiodinium spp. (clades A-C and strain names) hosted by Aiptasia in the given genetic networks24 and in the three laboratory clonal lines22,26; F003 symbiont characterization from this study.
Endogenous Symbiodinium hosted by Aiptasia clonal lines
Thornhill and colleagues24 found that the two worldwide Aiptasia genetic networks differ in their symbiont composition: the global Aiptasia network harbors only the clade B species Symbiodinium minutum, while members of the US South Atlantic network harbor clade A Symbiodinium strains either solely or, to a lesser extent, in combination with either variants of clade B S. minutum or (rarely) clade C strains (Fig. 2b). Accordingly, clonal line H2 (global network) harbors only the S. minutum strain SSB0122, while clonal line CC7 (US South Atlantic network) hosts a single clade A strain26 designated SSA01 (Fig. 2b). To identify the as-yet-unknown endogenous symbiont(s) of Aiptasia clonal line F003 (US South Atlantic network), we sequenced the chloroplast ribosomal 23S subunit (cp23S) of three independent F003 animals as previously described22. Within each individual, we consistently found three distinct cp23S sequences in relatively constant proportions: ~57% of sequences were a single variant of S. minutum (i.e. identical to strain SSB01 [GenBank #JX221048.1] except for a SNP at SSB01 nt250 and a deletion in SSB01 nt363–nt498); ~35% were identical to SSA01 (GenBank #KT186239); ~8% were identical to S. minutum strain SSB01. The first two sequences are consistent with the typical patterns of the US South Atlantic Aiptasia network24. The third sequence, S. minutum strain SSB01, was not detected during a similar but cursory analysis >1.5 years ago; its current presence may be due to either its previously undetected natural occurrence or to an accidental introduction during extended culture in close proximity to SSB01. Taken together, F003 and CC7 are therefore each representatives of different symbiont patterns typically observed in the US South Atlantic Aiptasia network.
Spawning induction for Aiptasia clonal lines CC7, F003, and H2 under laboratory conditions
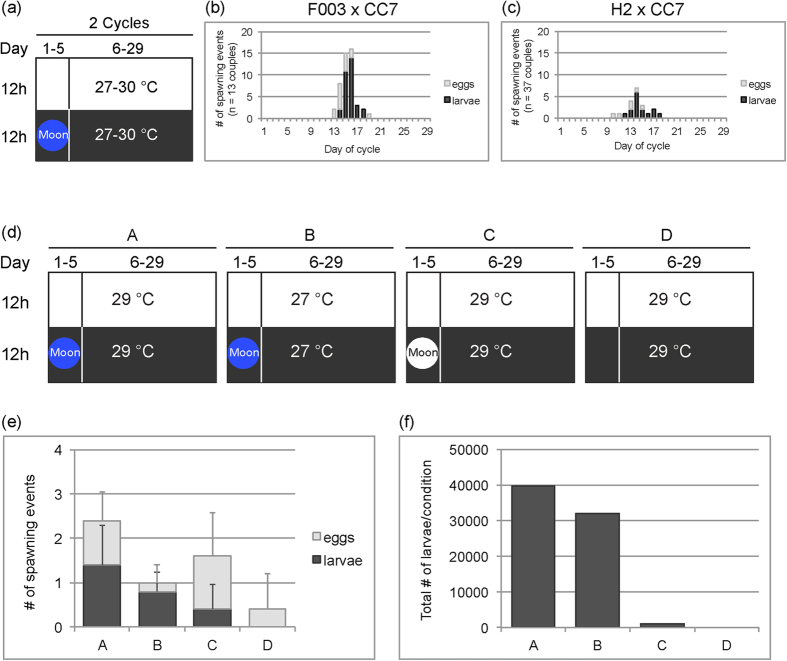
To test whether the simulation of a lunar cycle can trigger spawning in Aiptasia, we subjected spawning couples to a blue-intensive moon cue for five nights per 29-day cycle (Fig. 3a–c and Supplementary Fig. S1 online). Beforehand, animals were fed with Artemia nauplii five times per week for over two months. To avoid competition for food and other resources, animals were cultured in low densities and pedal lacerates were removed on a regular basis (we observed that such treatment markedly increases animal size [Supplementary Fig. S2 online]). We monitored in total 13 couples of [F003xCC7] and 37 couples of [H2xCC7] for two subsequent cycles (Fig. 3a–c). For crosses with F003, spawning was tightly clustered between Day 13 and Day 20 of the 29-day artificial lunar cycle, with peak larvae production on Days 13–17 (Fig. 3b). Spawning in crosses with H2 was likewise synchronous, with a peak of larvae production also around Day 13–18 (Fig. 3c). Overall, couples with F003 were more synchronous and spawned more often than those with H2 (Fig. 3b,c) (see below for more on observed line-specific differences). In both lines, however, the variability of larvae occurrences between couples was relatively high, which may be due to differences in culture conditions prior to spawning induction (Supplementary Fig. S1 online). We monitored couples for only two spawning cycles because spawning frequencies decreased afterwards, somewhat depending on the duration in the pre-induction high-feeding conditions (data not shown).
Figure 3.
Induction of spawning in Aiptasia. (a) Schematic of initial spawning induction conditions during two consecutive 29-day artificial lunar cycles. Temperature and moon cue indicated; color of the circle refers to color of the simulated moon cue (see text). (b,c) Spawning synchronicity and efficiency in [F003 x CC7] couples (b) and [H2 x CC7] couples (c) under the induction conditions in (a).(d) Schematics of various spawning induction conditions A-D for a direct comparison of spawning cues in [F003xCC7] couples. (e) Number of spawning events by five couples in each of the four induction conditions in (d) over one 29-day artificial lunar cycle. Error bars are standard deviations. (f) Quantification of total larvae per condition produced during the larvae spawning events shown in (e).
Because we sought to modify our spawning protocol to maximize spawning efficiency and predictability, during these experiments we intermittently tested other conditions previously known to stimulate spawning in other cnidarians. We compared spawning when animals were fed Artemia nauplii twice weekly versus five times per week; we observed that increased feeding improved spawning, especially for F003-containing couples (Supplementary Fig. S3 online). We tested whether a very low amount of blue light during the pre-spawning-induction period (see Methods) would enhance spawning efficiency during the lunar cycle inductions, yet saw no discernable effect of these treatments (Supplementary Fig. S1 online). Because warming water temperatures appear correlated with gametogenesis and spawning in anthozoans36,38,39,44, we simultaneously tested the effects of temperature shifts during spawning induction. We saw no obvious trend, but interpretation was difficult because the animals in the experiments were subjected to different conditions prior to spawning induction (Supplementary Fig. S1 online). We therefore sought to parse which cues were key to promote efficient and synchronous spawning.
Blue light is the key cue to induce efficient and synchronous spawning to produce larvae
To determine the distinct effects of temperature and simulated moon cue to effectively induce spawning, we conducted a side-by-side comparison of different spawning cues using only couples of [F003xCC7] originating from identical pre-induction culture conditions. Couples were grown at 26 °C and fed well for seven months prior to exposure to the following conditions: (A) blue-light simulated moon at 29 °C; (B) blue-light simulated moon at 27 °C; (C) white-light simulated moon at 29 °C; (D) no simulated moon (Fig. 3d). Blue light appeared to be the most important cue, as these conditions (A and B) produced efficient and synchronized spawning of larvae (Fig. 3e) as well as copious production of tens of thousands of larvae (Fig. 3f). The temperature increase from growth conditions to 29 °C (Conditions A, C, and D) appeared to be beneficial to egg release (Fig. 3e), although this temperature shift alone or with a white-light cue results in inefficient larvae production, with either no or very low numbers of larvae generated (Fig. 3f). White light, which in this case is composed of ~20% blue wavelengths (see Methods), does stimulate some spawning, but mostly eggs (Condition C, Fig. 3e) and over a wider range of days in the cycle (Days 16–21, Supplementary Fig. S4 online). This poor synchronization and low output resulted in very little larvae, especially when compared to condition A in which the only difference is the blue spectrum of the light (Fig. 3f). All data for these experiments are found in Supplementary Fig. S4 online.
Quantification of line-specific differences in larvae production
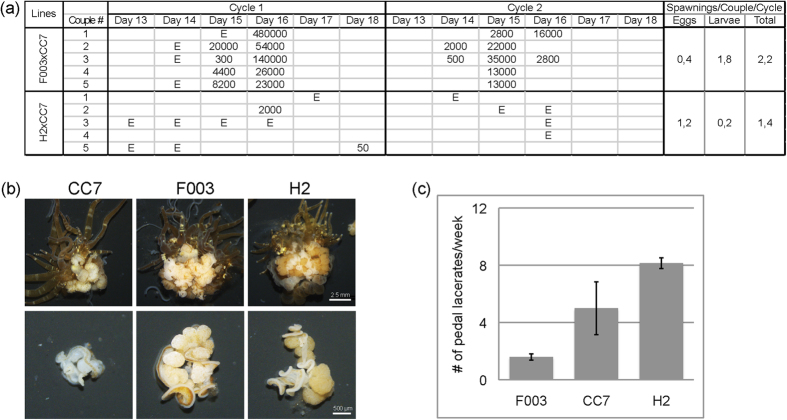
To confirm and quantify the differences we observed in spawning efficiencies between females of clonal lines F003 and H2 (Fig. 3b,c), we performed additional experiments under the best spawning induction parameters from above. We chose F003, H2, and CC7 animals of similar body size and cultured them in low-density cultures that were fed five times a week for five months, after which five couples of [F003xCC7] and five couples of [H2xCC7] were subjected to the blue-light lunar cycle cue (Condition A, Fig. 3d) and larval output was quantified. Consistent with our observations in the earlier experiments (Fig. 3b,c), we again found that couples with F003 spawned much more efficiently than those with H2: crosses with F003 produced larvae on average 1.8 times per couple per cycle, whereas H2 produced larvae only an average of 0.2 times per couple per cycle (Fig. 4a). Reproductive output for F003 was also far higher, typically with over 10,000 larvae per event, than for H2-containing couples that only spawned small numbers of larvae or, in most cases, eggs (Fig. 4a). Nevertheless, spawning synchrony was high in crosses with both female lines, falling between Days 13 and 18 (Fig. 4a).
Figure 4. Line-specific differences in reproduction between Aiptasia female clonal lines.
(a) Quantification of larvae and egg production by [F003 x CC7] and [H2 x CC7] couples in two cycles of spawning induction in condition A from Fig. 3d. E denotes occurrence of eggs; numbers indicate approximate quantification of larvae. (b) Cross-sections of longitudinally dissected anemones from the three clonal lines shows many bulbous gonads within mesentery tissues. (c) Weekly quantification of asexual reproduction via pedal laceration in the three clonal lines over five weeks. n = five individuals per clonal line; error bars are standard deviations.
Line-specific differences in gonad development
In Aiptasia sp. gonad development occurs within the mesentery tissue in the interior body column and is directly dependent upon food uptake43. We dissected well-fed CC7, F003, and H2 animals and indeed found many gonads within mesentery tissue (Fig. 4b, upper row). CC7 male gonads were typically smaller and lighte167 r in comparison to the bigger, darker gonads from females in lines F003 and H2 (Fig. 4b, lower row). However, qualitative visual assessments consistently showed that F003 animals contained higher ratios of distinct gonads to non-gonad mesentery than H2 animals (Fig. 4b). In both F003 and H2 gonads, we observed immature oocytes with prominent germinal vesicles, in contrast to released eggs during spawning (Supplementary Fig. S5 online). Sperm isolated from CC7 gonads were morphologically similar to those naturally released during spawning (Supplementary Fig. S5 online).
Line-specific differences in asexual reproduction
A previous study of a wild clonal Aiptasia sp. population found no apparent tradeoff between reproductive modes, yet it was postulated that genotypes may allocate resources between reproduction modes differently, perhaps as an adaptation to the local environment43. To assess whether such tradeoffs or differences occur in various genotypes of Aiptasia, particularly in light of the observed differences in sexual reproduction (see above), we monitored the asexual reproduction rates of clonal lines CC7, F003, and H2. Asexual reproduction is achieved via pedal laceration25, wherein symbiont-containing lacerates bud off the pedal disc and migrate away from the parental animal over the course of several days, after which they develop tentacles (Fig. 1b). We quantified the number of pedal lacerates (pl) produced per animal per week and found that under standard growth conditions, H2 produces significantly more pedal lacerates (8 pl/week) than F003 (1.7 pl/week) (Fig. 4c). Male clonal line CC7 produces on average 5 pl/week (Fig. 4c). Thus, the rates of asexual reproduction differ greatly between genotypes with no clear correlation to the sex of the clonal line; within females, H2 asexually reproduces faster than F003 in these conditions.
Discussion
Here we have developed a robust protocol to induce spawning in three clonal lines of the model symbiotic cnidarian Aiptasia, thereby opening new possibilities in analysis of symbiosis establishment, early development, and other key processes. Similar to the model cnidarian Nematostella, we find that extensive feeding of Aiptasia over multiple months in low-density culture is an important prerequisite for subsequent efficient spawning, especially for females. However, in resemblance to reef-building corals rather than to Nematostella, we have identified the key trigger to induce the synchronous release of gametes in well-fed Aiptasia animals to be the simulation of full moon by blue light during five nights within an artificial lunar monthly cycle. Interestingly, the wavelength of the applied light is consequential: LEDs exclusively emitting light between 400–460 nm effectively stimulate the release of gametes, while LEDs with white light, even when enriched in the blue wavelengths (such as actinic spectra) have minimal effect. This indicates that Aiptasia, similar to reef-building corals, may have specific blue-light-sensing photoreceptors (e.g. cryptochromes) that are thought to mediate synchronous mass spawning events of corals worldwide40,41. Indeed, we identified in the Aiptasia genome28 a homolog of the Acropora millepora blue-light cryptochrome receptor cry2 (data not shown) that is expressed in correlation with coral spawning41. Many reef-building corals release gametes synchronously after the increase in water temperature (occurring during spring) in correlation to the last full moon and sunset39. Accordingly, we observed that a modest temperature shift (+3 °C) is not sufficient to induce efficient spawning in Aiptasia alone, but in combination with the simulation of full moon appears to be beneficial. However, Aiptasia collected in the wild has been proposed to spawn year-round in correlation with the lunar cycle35, suggesting no strict inherent temperature limitations, although influences of seasonal changes in spawning efficiency cannot be ruled out36. Taken together, our data support the idea that the blue light is the key spawning cue with minor influences of water temperature.
In studies of gonad development of Aiptasia populations in the wild, Chen and colleagues found that Aiptasia eggs appeared to be released about 8–14 days after the natural full moon35. In the studies herein, we observe the synchronous release of gametes on Day 13–17 within a 29-day cycle; because our simulated full-moon cues occurs on Days 1–5, this spawning corresponds to 8–12 days after the moon cue (i.e. peak of moonlight followed by increasingly darkening conditions). Thus, our studies match up remarkably well, with the timing of gamete release nearly identical between natural Aiptasia populations and our populations kept in the laboratory for over five years and stimulated by an artificial light cue. The persistence of this innate ability to respond to the stimuli indicates that synchronized spawning is strongly genetically encoded and shared worldwide throughout Aiptasia sp. including across the major global genetic networks. Moreover, we observe that Aiptasia gametes are released a few hours after diurnal onset of darkness (data not shown), which is likewise identical to reef-building corals that spawn a few hours after sunset, varying by species45,46. Taken together, the monthly timing (i.e. new moon) and diurnal timing (i.e. after sunset) ensure spawning during a period of the darkest possible conditions. This is hypothesized to not only aid in predation avoidance but also to ensure embryos pass through fragile cleavage stages prior to exposure to potentially damaging UV radiation during daylight47.
Interestingly, we find that the two female lines F003 and H2 differ in their reproductive strategies: under standard growths conditions and extensive feeding, the H2 colony expands more efficiently by asexual pedal laceration than F003, which in turn appears to invest a major part of incoming nutrients into oocyte production. The consistency of these responses indicates that they stem from genetically encoded differences24,48. It has been hypothesized that distinct genotypes with different allocation ratios between asexual and sexual reproduction may be favored in different habitats49. Consistent with the high asexual reproduction rates in H2, Thornhill and colleagues24 propose that efficient clonal propagation and associated vertical symbiont transmission of members of the global Aiptasia network primarily led to its worldwide distribution and status as an invasive species in aquaria and in the wild35,50,51, and the low-level genetic population structure of its associated endosymbiont S. minutum. Such extensive distribution therefore likely occurred quickly, via anthropogenic vectoring such as ballast waters, fouling of ships, aquaculture, or the aquarium trade24. In contrast, its lower asexual reproduction rates apparently restrict F003 to a more local distribution, yet its enhanced sexual reproduction rates would increase the genotypic diversity of its community. Indeed, this is recapitulated in the diversity of the sequenced SCAR markers: we found that F003 and CC7 showed greater genotypic diversity, with two alleles at several loci, whereas H2 was monoallelic at all four loci investigated. Such genotypic diversity might lead to the observed greater flexibility with associated symbionts24, and indeed we find that F003 hosts multiple endogenous Symbiodinium strains from different clades.
This study opens the door to a steady supply of Aiptasia larvae for a number of crucial purposes, not least of which is addressing what may be considered one of the final steps in the establishment of the Aiptasia model system: closing the life cycle in the laboratory via metamorphosis and settlement of planula larvae. To date this has not been reported, although metamorphosis and settlement under laboratory conditions has been established for corals52,53,54, providing vital guidance on how to approach this in Aiptasia. The protocol provided here allows sufficient Aiptasia larvae to systematically test and thereby identify metamorphosis and settlement cues in the laboratory. Although larvae production allows new techniques to be developed and important biological questions to be addressed in areas like development, endosymbiosis establishment and photobiology, microscopic imaging, gene knockdown, and transformation, settlement will further this by opening other paths like classical genetics and generation of transgenic lines.
These developments will bring Aiptasia fully into the stable of cnidarian laboratory models, which have proven to be vital in gaining basic biological insights. Yet Aiptasia also represents a new type of model system because of its additional feature of an ecologically relevant endosymbiosis. That photosynthetic symbioses appear in organisms across the tree of life makes comparative analyses of these critical relationships interesting in evolutionary and ecological contexts. The symbioses inherent in nearly all higher metazoans (including humans) have been increasingly recognized in the molecular age, and particularly in the global context of accelerating environmental change, Aiptasia will contribute to opening new paths in emerging fields that seek to describe the ever-more-recognized complexity of organisms in a changing world.
Methods
Aiptasia clonal lines
The founder animals for male line CC726 and female line F003 (courtesy of the John Pringle lab) were originally collected by Carolina Biological Supply Company (#162865; Burlington, USA) from an area approximately centered on Wilmington, North Carolina, USA. The founder animal for line H2 was collected off Coconut Island, Hawaii22.
Genotypic characterization of Aiptasia clonal lines
To genotypically characterize the three clonal lines CC7, F003, and H2, we used the SCAR markers developed by Thornhill and colleagues24. Genomic DNA was extracted from two representative animals of each line with the DNeasy Blood and Tissue Kit (#69504; Qiagen, Venlo, Netherlands) according to the manufacturer’s protocol for animal tissue. The gDNA of each animal was then amplified with each of the four SCAR marker primer pairs24: ApSCAR3F and ApSCAR3R; ApSCAR4F and ApSCAR4R; ApSCAR5F and ApSCAR5R; ApSCAR7F and ApSCAR7R2. Amplification reactions of 50 μL contained 2 U Taq polymerase, 0.2 μM of each primer, 200 μM dNTPs, 20 mM Tris-HCl (pH 8.8), 10 mM (NH4)SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton®X-100, and 150 ng gDNA template. Amplification conditions were as follows: initial denaturation at 95 °C for two min; 30 cycles of denaturation at 95 °C, annealing at 58 °C for 45 s, extension at 60 °C for 1.5 min; final extension at 68 °C for 5 min. Products were purified with the Wizard SV Gel and PCR Clean-Up System (#A9281; Promega, Madison, USA), cloned, and four or more positive bacterial clones were sequenced with the M13F (−21) or M13R (−26) universal primers.
Sequences were processed in Geneious version 6.1.855 [ http://www.geneious.com] by initially aligning to the reference sequences from Thornhill et al. and trimming and concatenating accordingly24. In cases where a given marker in an anemone line produced two alleles (see Results), the alleles were collapsed to create a consensus sequence containing the appropriate base ambiguity. The three clonal line sequences and the twelve reference sequences from Thornhill et al.24 were aligned with ClustalW56 and a maximum likelihood phylogenetic tree was generated using PhyML57 with default parameters and the general time-reversible (GTR) nucleotide substitution model. Tree topology searches chose the best topologies of both Nearest Neighbor Interchanges (NNI) and Subtree Pruning and Regrafting (SPR) topological moves. Topology, branch length, and substitution rate were optimized, and branch support was estimated by bootstrap analysis of 100 replicates.
Identification of endogenous Symbiodinium in F003 clonal line
To determine which symbiont strain(s) are endogenous to the Aiptasia F003 line, we used the Symbiodinium Domain V chloroplast large subunit ribosomal DNA (cp23S) marker for phylogenetic analysis as previously described22,58. Three adult individuals were each homogenized with a MICCRA D1 homogenizer (Miccra, Müllheim, Germany) and algal genomic DNA was extracted using the DNeasy Plant Mini Kit (#69104; Qiagen, Venlo, Netherlands) following the manufacturer’s instructions. The cp23S region was amplified from each gDNA sample with the 23S4F and 23S7R primers58. Amplification reactions of 50 μl contained Phusion polymerase, 0.1 μM of each primer, 200 μM dNTPs, 1X Phusion HF buffer (#B0518S; NEB, Ipswich, USA), and ∼50–100 ng gDNA. Amplification conditions were as follows: initial denaturation at 94 °C for 5 min; 35 cycles of 94 °C for 30 s, 50 ˚C for 1 min, 72 °C for 1.5 min; and final extension at 72 °C for 5 min. For each amplification reaction, products (∼650 bp) were purified with the Wizard SV Gel and PCR Clean-Up System, cloned, and 18 or more positive bacterial clones were sequenced with the M13F (−21) universal primer. In total, 60 high quality cp23s sequences (18, 21, and 21 sequences per animal) were aligned in Geneious version 7.1.755 [ http://www.geneious.com] and compared to cp23S sequences from SSB0122 (GenBank Accession #JX221048.1) and the CC7 endogenous clade A strain26 SSA01 (GenBank Accession #KT186239), which was sequenced as described above.
Aiptasia culture conditions
General anemone stocks were maintained at a density of 30–40 animals (unless stated otherwise) per tank in medium-sized food-grade translucent polycarbonate tanks (GN 1/4–100 cm height, #44 CW; Cambro, Huntington Beach, USA) filled with artificial seawater (ASW) (Coral Pro Salt; Red Sea Aquatics Ltd, Houston, USA) at 31–34 ppt salinity. Stock tanks were kept in Intellus Ultra Controller Incubators (Model I-36LL4LX; Percival, Perry, USA) at 26 °C on a diurnal 12L:12D cycle (12 h light:12 h dark) under white fluorescent bulbs with an intensity of ~20–25 μmol m−2 s−1 of photosynthetically active radiation (PAR), as measured with an Apogee PAR quantum meter (MQ-200; Apogee, Logan, USA). Animals were fed two to five times per week with freshly hatched Artemia nauplii. Seawater in the tanks was exchanged two to three times per week and tanks were cleaned with cotton-tip swabs as required.
Spawning induction of Aiptasia
To induce Aiptasia gamete release following a simulated full moon cue, individuals with oral disk diameter ~0.7 cm or greater were combined into couples ([H2xCC7] or [F003xCC7]), transferred to ASW in small food-grade translucent polycarbonate tanks (GN 1/9–65 cm height, #92 CW; Cambro, Huntington Beach, CA, USA) and kept in standard cell culture incubators (Heracell 150i; Thermo Scientific, Waltham, USA) at 27 °C–30 °C (see temperatures indicated in Results). Tanks were fed Artemia nauplii five times per week and kept on a diurnal 12L:12D cycle at intensities of 20–30 μmol m−2 s−1 provided by white LED lights, which were comprised of two types of LEDs—those with wavelengths of 425 nm–705 nm (color temperature 15,000 Kelvin) and those with wavelength 460 nm – at a ratio of 5:1 respectively (SolarStinger SunStrip “Marine”; Econlux, Cologne, Germany). A 29-day lunar cycle was simulated by subjecting the tanks to constant blue light during the 12 h “dark” period in the first five nights of the cycle (Day 1–5). Blue LED lights were comprised of LEDs with wavelengths 400 nm, 420 nm, 440 nm, and 460 nm at a ratio of 1:1:1:1 (SolarStinger SunStrip “Deep Blue”; Econlux, Cologne, Germany) and were 10–16 μmol m−2 s−1 in intensity. The presence of gametes or planula larvae was checked by examining the tanks with a Leica S8APO stereoscope.
Isolation of larvae for quantification
Larvae were isolated from the spawning tanks by stepwise filtration: tank contents were first passed through a 70 or 100 μm cell strainer to withhold coarse particles and permit larval passage, after which larvae were concentrated by passage through a 40 μm cell strainer and then rinsed into glass beakers with filter-sterilized ASW. For approximate quantification, beakers were mixed well to evenly distribute larvae and six 10 μl drops were pipetted onto a glass slide. The average number of larvae per drop was used to extrapolate the approximate total.
Dissection of gonads and gametes from Aiptasia
To promote gonad growth in anemones, 20–25 small anemones per clonal line (~0.5 cm height and no apparent gonads) were taken from stock tanks and combined into a medium-sized food-grade translucent polycarbonate tank, with two tanks per line. Animals were kept at 27 °C, fed five times per week with Artemia nauplii, and sampled after seven months. Anemones were transferred to a standard petri dish containing ASW and dissected in half longitudinally with a scalpel; mucus, acontia, and remaining food particles were removed with forceps. Animals were transferred to new petri dishes with ASW and images were captured using a Leica S8APO binocular (top illumination) equipped with a Leica MC170 HD color camera. Individual gonads were then detached from the anemones with forceps, transferred to new petri dishes, and imaged under identical parameters as above but with a higher magnification.
To isolate and image gametes, small fractions of male and female gonads in ASW were opened with scalpel and forceps. Gonads were placed by forceps (female) or pipette (male) onto glass microscope slides and covered with glass coverslip. Representative images were taken with a Nikon Eclipse 80i microscope equipped with a Nikon DS-1QM monochrome camera (sperm) or a Nikon DS-1U color camera (female gonad).
Quantification of asexual reproduction rates
To determine the rate of asexual reproduction, five animals approximately three to four months old from each clonal line were individually placed into ASW in small food-grade translucent polycarbonate tanks and fed five times per week with Artemia nauplii. Pedal laceration was analyzed for each individual at the end of every week for five consecutive weeks. The number of pedal lacerates were counted by eye or with a stereoscope (Leica S8APO), and after counting all pedal lacerates were removed.
Additional Information
How to cite this article: Grawunder, D. et al. Induction of Gametogenesis in the Cnidarian Endosymbiosis Model Aiptasia sp. Sci. Rep. 5, 15677; doi: 10.1038/srep15677 (2015).
Supplementary Material
Acknowledgments
Funding was provided to A.G. by the Emmy-Noether-Programme of the German Research Foundation (DFG) (grant no. GU 1128/3–1) and by a Marie Curie Career Integration Grant (CIG) under the FP7-PEOPLE-2013-CIG program, European Commission; and to I.W. by a PhD fellowship from the Graduate Program in Areas of Basic and Applied Biology (GABBA) of University of Porto. We thank John Pringle, Santiago Perez, and Pringle lab members for providing Aiptasia lines and advice; Tingting Xiang and Arthur Grossman for Symbiodinium strains; Dan Thornhill for Aiptasia SCAR sequences; Steffen Lemke, Thomas Holstein, and Suat Özbek for advice and comments.
Footnotes
Author Contributions D.G., E.A.H. and A.G. designed the experiments; D.G., E.A.H., M.B., I.W. and N.B. performed the experiments. E.A.H. and A.G. wrote the manuscript. All authors reviewed the manuscript.
References
- Darling J. A. et al. Rising starlet: the starlet sea anemone, Nematostella vectensis. Bioessays 27, 211–221 (2005). [DOI] [PubMed] [Google Scholar]
- Genikhovich G. & Technau U. The starlet sea anemone Nematostella vectensis: an anthozoan model organism for studies in comparative genomics and functional evolutionary developmental biology. Cold Spring Harb. Protoc. 2009 9, (2009). [DOI] [PubMed] [Google Scholar]
- Bode H. R. Axial patterning in Hydra. Cold Spring Harbor Perspect. Biol. 2009 1, a00463 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstein T. W., Hobmayer E. & Technau U. Cnidarians: an evolutionarily conserved model system for regeneration? Dev. Dynam. 226, 257–267 (2003). [DOI] [PubMed] [Google Scholar]
- David C. N. & Murphy S. Characterization of interstitial stem cells in Hydra by cloning. Dev. Biol. 58, 372–383 (1977). [DOI] [PubMed] [Google Scholar]
- Bosch T. C. G. Hydra and the evolution of stem cells. Bioessays 31, 478–486 (2009). [DOI] [PubMed] [Google Scholar]
- Bosch T. C. G. et al. How do environmental factors influence life cycles and development? An experimental framework for early-diverging metazoans. Bioessays 36, 1185–1194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E., Koren O., Reshef L., Efrony R. & Zilber-Rosenberg E. The role of microorganisms in coral health, disease, and evolution. Nat. Rev. Microbiol. 5, 355–362 (2007). [DOI] [PubMed] [Google Scholar]
- Augustin R., Fraune S., Franzenburg S. & Bosch T. C. Where simplicity meets complexity: Hydra, a model for host-microbe interactions. Adv. Exp. Med. Biol. 710, 71–81 (2011). [DOI] [PubMed] [Google Scholar]
- Rumpho M. E., Pelletreau K. N., Moustafa A. & Bhattacharya D. The making of a photosynthetic animal. J. Exp. Biol. 214, 303–311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham E. R., Fay S. A., Davey A. & Sanders R. W. Intracapsular algae provide fixed carbon to developing embryos of the salamander Ambystoma maculatum. J. Exp. Biol. 216, 452–459 (2013). [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lanetty M. Evolving lineages of Symbiodinium-like dinoflagellates based on ITS1 rDNA. Mol. Phylogenet. Evol. 28, 152–168 (2003). [DOI] [PubMed] [Google Scholar]
- Pochon X. & Gates R. D. A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai’i. Mol. Phylogenet. Evol. 56, 492–497 (2010). [DOI] [PubMed] [Google Scholar]
- Lajeunesse T. C., Parkinson J. E. & Reimer J. D. A genetics-based description of Symbiodinium minutum sp. nov. and S. psygmophilum sp. nov. (Dinophyceae), two dinoflagellates symbiotic with Cnidaria. J. Phycol. 48, 1380–1391 (2012). [DOI] [PubMed] [Google Scholar]
- Muscatine L. The role of symbiotic algae in carbon and energy flux in coral reefs in Coral Reefs (ed. Zubinsky Z. ) 75–87 (Elsevier, 1990). [Google Scholar]
- Yellowlees D., Rees T. A. V. & Leggat W. Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ. 31, 679–694 (2008). [DOI] [PubMed] [Google Scholar]
- Hoegh-Guldberg O. et al. Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742 (2007). [DOI] [PubMed] [Google Scholar]
- Davy S. K., Allemand D. & Weis V. M. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76, 229–261 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harii S., Yasuda N., Rodriguez-Lanetty M., Irie T. & Hidaka M. Onset of symbiosis and distribution patterns of symbiotic dinoflagellates in the larvae of scleractinian corals. Mar. Biol. 156, 1203–1212 (2009). [Google Scholar]
- Harrison P. L. Sexual reproduction of scleractinian corals in Coral Reefs: an Ecosystem in Transition (eds. Zubinsky Z. & Stambler N. ) 59–85 (Springer, 2011). [Google Scholar]
- Weis V. M., Davy S. K., Hoegh-Guldberg O., Rodriguez-Lanetty M. & Pringle J. R. Cell biology in model systems as the key to understanding corals. Trends Ecol. Evol. 23, 369–376 (2008). [DOI] [PubMed] [Google Scholar]
- Xiang T., Hambleton E. A., DeNofrio J. C., Pringle J. R. & Grossman A. R. Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity. J. Phycol. 49, 447–458 (2013). [DOI] [PubMed] [Google Scholar]
- Hambleton E. A., Guse A. & Pringle J. R. Similar specificities of symbiont uptake by adults and larvae in an anemone model system for coral biology. J. Exp. Biol. 217, 1613–1619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill D. J., Xiang Y., Pettay D. T., Zhong M. & Santos S. R. Population genetic data of a model symbiotic cnidarian system reveal remarkable symbiotic specificity and vectored introductions across ocean basins. Mol. Ecol. 22, 4499–4515 (2013). [DOI] [PubMed] [Google Scholar]
- Clayton W. S. Jr. Pedal laceration by the anemone Aiptasia pallida. Mar. Ecol. Prog. Ser. 21, 75–80 (1985). [Google Scholar]
- Sunagawa S. et al. Generation and analysis of transcriptomic resources for a model system on the rise: the sea anemone Aiptasia pallida and its dinoflagellate endosymbiont. BMC Genomics 10, 258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert E. M., Burriesci M. S. & Pringle J. R. Developing the anemone Aiptasia as a tractable model for cnidarian-dinoflagellate symbiosis: the transcriptome of aposymbiotic A. pallida. BMC Genomics 13, 271 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten S. et al. The genome of Aiptasia, a sea anemone model for coral symbiosis. P. Natl. Acad. Sci. USA Advance online publication, doi: 10.1073/pnas.1513318112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoguchi E. et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23, 1399–1408 (2013). [DOI] [PubMed] [Google Scholar]
- Bayer T. et al. Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS ONE 7, e35269 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T., Nelson W., Rodriguez J., Tolleter D. & Grossman A. R. Symbiodinium transcriptome and global responses of cells to immediate changes in light intensity when grown under autotrophic or mixotrophic conditions. Plant J. 82, 67–80 (2015). [DOI] [PubMed] [Google Scholar]
- Wittlieb J., Khalturin K., Lohmann J. U., Anton-Erxleben F. & Bosch T. C. G. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. P. Natl. Acad. Sci. USA 103, 6208–6211 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose T. & Houliston E. Two oppositely localised Frizzled RNAs as axis determinants in a cnidarian embryo. PLoS Biol. 5, e70 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikmi A., McKinney S. A., Delventhal K. M. & Gibson M. C. TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis. Nat. Commun. 5, 5486 (2014). [DOI] [PubMed] [Google Scholar]
- Chen C., Soong K. & Chen C. A. The smallest oocytes among broadcast-spawning actiniarians and a unique lunar reproductive cycle in a unisexual population of the sea anemone, Aiptasia pulchella (Anthozoa: Actiniaria). Zool. Stud. 47, 37–45 (2008). [Google Scholar]
- Schlesinger A., Kramarsky-Winter E., Rosenfeld H., Armoza-Zvoloni R., Loya Y. & Earley R. Sexual plasticity and self-fertilization in the sea anemone. Aiptasia diaphana. PLoS ONE 5, e11874 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand C. & Uhlinger K. R. The culture, sexual and asexual reproduction, and growth of the sea anemone Nematostella vectensis. Biol. Bull. 182, 169–172 (1992). [DOI] [PubMed] [Google Scholar]
- Fritzenwanker J. H. & Technau U. Induction of gametogenesis in the basal cnidarian Nematostella vectensis (Anthozoa). Dev. Genes Evol. 212, 99–103 (2002). [DOI] [PubMed] [Google Scholar]
- Ryland J. S. Reproduction in Zoanthidea (Anthozoa: Hexacorallia). Invertebr. Reprod. Dev. 31, 177–188 (1997). [Google Scholar]
- Gorbunov M. Y. & Falkowski P. G. Photoreceptors in the cnidarian hosts allow symbiotic corals to sense blue moonlight. Limnol. Oceanogr. 47, 309–315 (2002). [Google Scholar]
- Levy O. et al. Light-responsive cryptochromes from a simple multicellular animal, the coral Acropora millepora. Science 3184, 67–470 (2007). [DOI] [PubMed] [Google Scholar]
- Guest J. R., Baird A. H., Goh B. P. & Chou L. M. Seasonal reproduction in equatorial reef corals. Invertebr. Reprod. Dev. 48, 207–218 (2005). [Google Scholar]
- Chen C., Chang H.-Y. & Soong K. No tradeoff between sexual and asexual investments in the sea anemone Aiptasia pulchella (Anthozoa: Actiniaria). Zool. Stud. 51, 996–1005 (2012). [Google Scholar]
- Siebert A. E. Jr. A description of the embryology, larval development, and feeding of the sea anemones Anthopleura elegantissima and A. xanthogrammica. Can. J. Zool. 52, 1383–1388 (1974). [DOI] [PubMed] [Google Scholar]
- Vize P. D., Hilton J. D., Brady A. K. & Davies S. W. Light sensing and the coordination of coral broadcast spawning behavior. Proc. 11th Int. Coral Reef Symp. 378–381 (2008). [Google Scholar]
- Brady A. K., Hilton J. D. & Vize P. D. Coral spawn timing is a direct response to solar light cycles and is not an entrained circadian response. Coral Reefs 28, 677–680 (2009). [Google Scholar]
- Gilbert S. F. & Epel D. Ecological Developmental Biology: Integrating Epigenetics, Medicine, and Evolution. 1–462 (Sinauer, 2008). [Google Scholar]
- Via S. et al. Adaptive phenotypic plasticity: consensus and controversy. Trends Ecol. Evol. 10, 212–217 (1995). [DOI] [PubMed] [Google Scholar]
- Roff D. A. The Evolution of Life Histories: Theory and Analysis. 1–535 (Chapman & Hall, 1992). [Google Scholar]
- Mito T. & Uesugi T. Invasive alien species in Japan: the status quo and the new regulation for prevention of their adverse effects. Global Environmental Research 8, 171–191 (2004). [Google Scholar]
- Rhyne A. L., Lin J. & Deal K. J. Biological control of aquarium pest anemone Aiptasia pallida Verrill by peppermint shrimp Lysmata Risso. J. Shellfish Res. 23, 227–229 (2004). [Google Scholar]
- Iwao K., Fujisawa T. & Hatta M. A cnidarian neuropeptide of the GLWamide family induces metamorphosis of reef-building corals in the genus Acropora. Coral Reefs 21, 127–129 (2002). [Google Scholar]
- Takahashi T. & Hatta M. The importance of GLWamide neuropeptides in cnidarian development and physiology. J. Amino Acids 2011, 1–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebben J. et al. Chemical mediation of coral larval settlement by crustose coralline algae. Sci. Rep. 5, 10803 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. et al. New algorithms and methods to estimate Maximum-Likelihood Phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- Pochon X., Montoya-Burgos J. I., Stadelmann B. & Pawlowski J. Molecular phylogeny, evolutionary rates, and divergence timing of the symbiotic dinoflagellate genus Symbiodinium. Mol. Phylogenet. Evol. 38, 20–30 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.