Abstract
Objectives
Clinical trials in septic shock continue to fail due, in part, to inequitable and sometimes unknown distribution of baseline mortality risk between study arms. Investigators advocate that interventional trials in septic shock require effective outcome risk stratification. We derived and tested a multibiomarker-based approach to estimate mortality risk in adults with septic shock.
Design
Previous genome-wide expression studies identified 12 plasma proteins as candidates for biomarker-based risk stratification. The current analysis used banked plasma samples and clinical data from existing studies. Biomarkers were assayed in plasma samples obtained from 341 subjects with septic shock within 24 hours of admission to the ICU. Classification and regression tree analysis was used to generate a decision tree predicting 28-day mortality based on a combination of both biomarkers and clinical variables. The derived tree was first tested in an independent cohort of 331 subjects, then calibrated using all subjects (n = 672), and subsequently validated in another independent cohort (n = 209).
Setting
Multiple ICUs in Canada, Finland, and the United States.
Subjects
Eight hundred eighty-one adults with septic shock or severe sepsis.
Intervention
None.
Measurements and Main Results
The derived decision tree included five candidate biomarkers, admission lactate concentration, age, and chronic disease burden. In the derivation cohort, sensitivity for mortality was 94% (95% CI, 87–97), specificity was 56% (50–63), positive predictive value was 50% (43–57), and negative predictive value was 95% (89–98). Performance was comparable in the test cohort. The calibrated decision tree had the following test characteristics in the validation cohort: sensitivity 85% (76–92), specificity 60% (51–69), positive predictive value 61% (52–70), and negative predictive value 85% (75–91).
Conclusions
We have derived, tested, calibrated, and validated a risk stratification tool and found that it reliably estimates the probability of mortality in adults with septic shock.
Keywords: biomarkers, decision tree, modeling, outcome, sepsis, stratification
Substantial resources are invested in trying to find new treatments for septic shock, but the vast majority of new treatments fail to demonstrate efficacy during phase 3 trials. There are many reasons for these failures, but one consistent reason is that baseline mortality risk varies widely in patients with septic shock (1). Consequently, current trial designs lead to the inclusion of patients with low mortality risk who are unlikely to benefit from novel therapies beyond standard care as well as patients having an extremely high mortality risk who may be beyond salvage by experimental therapy. This approach can dilute any potential beneficial effect of an experimental therapy for patients with a significant, but modifiable, mortality risk and consequently lead to a negative clinical trial.
Researchers advocate that interventional trials in sepsis must be conducted in the context of effective outcome risk stratification (1–3). A recent trial used a mortality risk scoring system to screen for patients with a high, but potentially modifiable, risk of mortality. The trial failed to demonstrate efficacy, in part, because the actual mortality in the placebo group was far lower than predicted by the scoring system (4). This highlights the importance of developing stratification tools for septic shock to better inform the conduct of clinical trials.
We recently derived and validated a biomarker-based model that reliably predicts 28-day mortality in pediatric septic shock (5). The plasma protein biomarkers were selected objectively based on extensive genome-wide expression studies and predictive modeling (6, 7). Furthermore, the biomarkers were measured from samples obtained during the first 24 hours of admission to the ICU, which is during the time when patients are typically evaluated for inclusion in clinical trials. We hypothesized that the biomarkers used in the pediatric study could also be used to estimate a mortality probability in adults with septic shock. In the current study, we extend our methodology to derive, test, and validate an analogous model in adults with septic shock.
METHODS
Overall Study Design
All analyses of plasma samples and clinical data are based on secondary use of existing data from previous studies, with approval of the respective institutional review boards.
Derivation Cohort
Derivation cohort study subjects (n = 341) were participants in the Vasopressin and Septic Shock Trial (VASST), a randomized, concealed, norepinephrine-controlled trial testing the efficacy of low-dose vasopressin versus norepinephrine in adults with septic shock (Current Controlled Trials number: ISRCTN9485869). The original VASST publication describes all protocol details (8).
Test Cohort
Test cohort study subjects (n = 331) were pooled from two sources. Two hundred and forty-three subjects were participants in a prospective, observational, multicenter cohort study of prevalence and outcome of severe sepsis and septic shock in Finland (FINNSEPSIS) (9). An additional 88 subjects were participants in a single center, observational study at St. Paul’s Hospital in Vancouver, British Columbia (10).
Validation Cohort
Validation cohort study subjects (n = 209) were participants in the Molecular Epidemiology of Severe Sepsis in the Intensive Care Unit study, an ongoing cohort study at the Hospital of the University of Pennsylvania. Eligible patients with septic shock were enrolled in either the emergency department or the medical ICU, and patients or their proxies provided informed consent. Septic shock was defined using published criteria (11).
Candidate Stratification Biomarkers
The 12 candidate biomarkers (gene symbols) included C-C chemokine ligand 3 (CCL3), C-C chemokine ligand 4 (CCL4), neutrophil elastase 2 (ELA2), granzyme B (GZMB), heat shock protein 70 kDa 1B (HSPA1B), interleukin-1α (IL1A), interleukin- 8 (IL8), lipocalin 2 (LCN2), lactotransferrin (LTF), matrix metallopeptidase 8 (MMP8), resistin (RETN), and thrombospondin 1 (THBS1). These biomarkers were selected from 117 gene probes previously shown to have predictive strength for poor outcomes in microarray-based studies involving children with septic shock (6, 7). Final biomarker selection was based on a priori criteria: 1) the gene product (i.e., protein) has biological and mechanistic plausibility regarding the host response to infection, immunity, and/or inflammation, and 2) the gene product is readily measured in the blood compartment.
All plasma samples were collected within the first 24 hours of presentation to the ICU. The plasma concentrations of the candidate biomarkers were measured using a multiplex magnetic bead platform (MILLIPLEX MAP, EMD Millipore Corporation, Billerica, MA) and a Luminex 100/200 System (Luminex Corporation, Austin, TX) according to the manufacturers’ specifications. Technical assay performance data were previously published (5).
Additional Stratification Variables
We abstracted available data elements for consideration in the risk modeling that, based on existing literature, we hypothesized could be associated with poor outcomes: serum lactate concentration (mmol/L) at study entry, age, gender, and Acute Physiology and Chronic Health Evaluation (APACHE) II/III score. We also recorded the presence of the following comorbid conditions: New York Heart Association Class IV congestive heart failure, chronic obstructive pulmonary disease, requirement for chronic dialysis, chronic hepatic failure, hematologic or metastatic solid organ malignancy, and requirement for chronic steroids at study entry. We derived a binary “chronic disease” variable to indicate the presence of any one of these comorbidities.
Statistical Analysis
Initially, data are described using medians, interquartile ranges (IQRs), frequencies, and percentages. Comparisons between survivors and nonsurvivors used the Mann-Whitney U test, chi-square test, or Fisher exact test as appropriate. Descriptive statistics and comparisons used SigmaStat Software (Systat Software, San Jose, CA).
All-cause 28-day mortality is the primary outcome variable for the modeling procedures. To derive the decision tree, we employed a classification and regression tree (CART) approach (12, 13). The CART analysis procedure considered all 12 candidate biomarkers as well as other potential clinical predictor variables listed above. The procedure selects cut points and ordering of decision nodes that maximally discriminate between survivors and nonsurvivors. The tree was built using Salford Predictive Modeler v6.6 (Salford Systems, San Diego, CA). Performance of the tree is reported using diagnostic test statistics with 95% CIs computed using the VassarStats Website for Statistical Computation (14). Areas under the receiver operating characteristic (ROC) curves were compared using the method of Hanley and McNeil (15). The net reclassification improvement (NRI) was also used to estimate the incremental predictive ability of the biomarker-based model compared to using APACHE II scores alone (16). The NRI was computed using the R-package Hmisc.
RESULTS
Model Derivation
Table 1 provides the clinical and demographic data for the derivation cohort (n = 341), all of whom had septic shock. The 109 nonsurvivors (32.0%) were older, had a higher median APACHE II score, and a higher proportion had chronic disease at study entry, compared to the 232 survivors. The mean and median times to death in the derivation cohort nonsurvivors were 8.7 ± 7.9 (sd) and 6 days (IQR, 2–13 d), respectively.
Table 1.
Clinical and Demographic Data for the Derivation, Test, and Validation Cohorts
| Derivation Cohort | |||
|---|---|---|---|
| Variable | All | Survivors | Nonsurvivors |
| n | 341 | 232 | 109 |
| Median age (IQR) | 63 (51–73) | 61 (48–72) | 64 (54–76)a |
| Males, n (%) | 201 (58.9) | 142 (61.2) | 59 (54.1) |
| Median APACHE II (IQR)d | 27 (22–32) | 25 (20–30) | 31 (24–35)a |
| Median APACHE III (IQR)d | NA | NA | NA |
| Number of people with chronic disease (%)f | 165 (48.4) | 96 (41.4) | 69 (63.3)c |
| Mean days to death ± sd | NA | NA | 8.7 ± 7.9 |
| Median days to death (IQR) | NA | NA | 6 (2–13) |
| Number of people with septic shock (%) | 341 (100) | 232 (100) | 109 (100) |
| Number of people with a surgical diagnosis (%) | 78 (23) | 56 (24) | 22 (20) |
| Test Cohort | Validation Cohort | ||||
|---|---|---|---|---|---|
| All | Survivors | Nonsurvivors | All | Survivors | Nonsurvivors |
| 331 | 232 | 99 | 209 | 121 | 88 |
| 61 (50–72) | 58 (47–69) | 69 (56–76)a | 62 (51–71) | 63 (50–72) | 62 (53–71) |
| 229 (69.2)b | 164 (70.7) | 65 (59.6)c | 116 (55.5) | 67 (55.3) | 49 (55.7) |
| 23 (18–29)e | 22 (17–27) | 27 (20–32)a | NA | NA | NA |
| NA | NA | NA | 50 (37–79) | 51 (37–76) | 50 (38–82) |
| 95 (28.7)b | 56 (24.1) | 39 (35.8)c | 132 (63.2)b | 68 (56.2) | 64 (74.4)c |
| NA | NA | 11.1 ± 8.0e | NA | NA | 7.5 ± 6.9 |
| NA | NA | 10 (4–17)e | NA | NA | 5 (2–11) |
| 271 (81.9)b | 182 (78.4) | 89 (89.9)c | 209 (100) | 121 (100) | 88 (100) |
| 90 (27) | 68 (29) | 22 (22) | 8 (4)b | 8 (7) | 0 (0)c |
IQR = interquartile range, APACHE = Acute Physiology and Chronic Health Evaluation, NA = not applicable.
p < 0.05 versus respective survivors; Mann-Whitney U test.
p < 0.05 versus derivation cohort; chi-square test.
p < 0.05 versus respective survivors; chi-square test.
The derivation and test cohorts had APACHE II scores recorded, whereas the validation cohort had APACHE III scores recorded.
p < 0.05 versus derivation cohort; Mann-Whitney U test.
The presence of at least one of the following at study entry: New York Heart Association Class 4 congestive heart failure, chronic obstructive pulmonary disease, requirement for chronic dialysis, chronic hepatic failure, hematologic or metastatic solid organ malignancy, or requirement for chronic steroids.
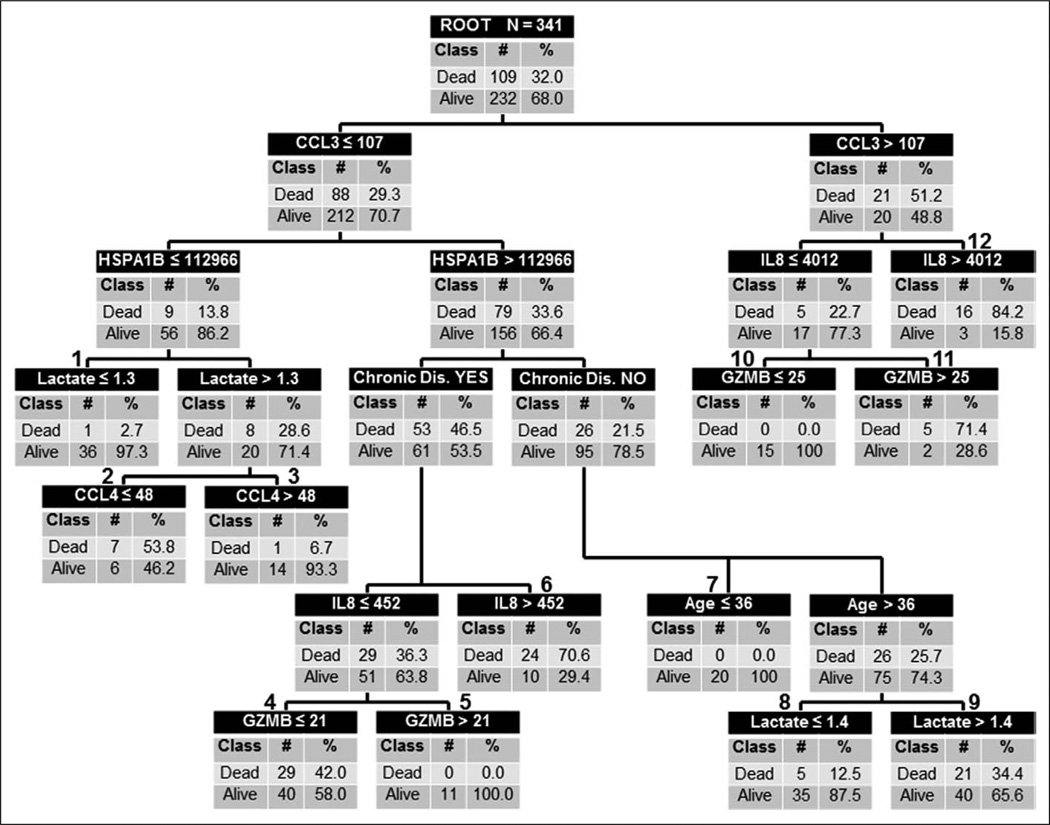
Supplemental Table 1 (Supplemental Digital Content 1, http://links.lww.com/CCM/A829) shows the performance of the individual biomarkers. Figure 1 shows the derived decision tree. Maximum accuracy was achieved with five of the 12 candidate stratification biomarkers: CCL3, HSPA1B, IL8, GZMB, and CCL4. Serum lactate concentration at study entry, age, and presence of chronic disease further improved predictive accuracy. The individual comorbid conditions did not improve predictive capacity. There were six low-risk terminal nodes (0.0–12.5% risk of death; terminal nodes 1, 3, 5, 7, 8, and 10) and six high-risk terminal nodes (34.4–84.2% risk of death; terminal nodes 2, 4, 6, 9, 11, and 12). Of the 138 subjects classified as low risk, 131 survived (94.9%) and 7 (5.1%) had died by 28 days. Of the 203 subjects classified as high risk, 102 (50.2%) had died by 28 days. Table 2 shows the diagnostic test characteristics of the decision tree in the derivation cohort.
Figure 1.
Classification tree from the derivation cohort (n = 341). The classification tree consists of 11 decision points and 22 daughter nodes. The classification tree includes five of the 12 candidate stratification biomarkers: C-C chemokine ligand 3 (CCL3), heat shock protein 70 kDa 1B (HSPA1B), interleukin-8 (IL8), granzyme B (GZMB), and C-C chemokine ligand 4 (CCL4). For consistency, the serum concentrations of all candidate stratification biomarkers are provided in pg/mL. The classification tree also includes serum lactate concentrations (mmol/L), age (yr), and the presence/absence of chronic disease as defined in the Methods section. The root node provides the total number of patients in the derivation cohort and the number of survivors and nonsurvivors with the respective rates. Each daughter node provides the respective decision rule criterion and the number of survivors and nonsurvivors with the respective rates. The numbers above daughter nodes designate terminal nodes. Terminal nodes 1, 3, 5, 7, 8, and 10 are considered low-risk terminal nodes, whereas terminal nodes 2, 4, 6, 9, 11, and 12 are considered high-risk terminal nodes.
Table 2.
Diagnostic Test Characteristics of the Decision Trees
| Variable | Derivation Cohort | Test Cohort | Calibration Cohort (Combined Derivation and Test Cohorts) |
Validation Cohort |
|---|---|---|---|---|
| No. of subjects | 341 | 331 | 672 | 209 |
| No. of true positives | 102 | 87 | 182 | 75 |
| No. of true negatives | 131 | 114 | 291 | 73 |
| No. of false positives | 101 | 118 | 173 | 48 |
| No. of false negatives | 7 | 12 | 26 | 13 |
| Sensitivity | 94% (87–97)a | 88% (79–93) | 88% (82–92) | 85% (76–92) |
| Specificity | 56% (50–63) | 49% (43–56) | 63% (58–67) | 60% (51–69) |
| Positive predictive value | 50% (43–57) | 42% (36–50) | 51% (46–57) | 61% (52–70) |
| Negative predictive value | 95% (89–98) | 90% (84–95) | 92% (88–94) | 85% (75–91) |
| Positive likelihood ratio | 2.1 (1.8–2.5) | 1.7 (1.5–2.0) | 2.3 (2.1–2.7) | 2.1 (1.7–2.7) |
| Negative likelihood ratio | 0.1 (0.06–0.2) | 0.2 (0.1–0.4) | 0.2 (0.1–0.3) | 0.2 (0.1–0.4) |
| Area under the curve | 0.834 (0.792–0.875) | 0.720 (0.661–0.780) | 0.793 (0.758–0.823) | 0.726 (0.660–0.792) |
Numbers in parentheses represent 95% CIs.
Model Testing
The test cohort consisted of 331 subjects with septic shock (81.9%) or severe sepsis (18.1%), of whom 99 (29.9%) did not survive to 28 days. Table 1 provides the clinical and demographic data. Compared to the derivation cohort, the test cohort had a higher proportion of male subjects, a lower median APACHE II score, a lower proportion of subjects with chronic disease, and a lower proportion of subjects with septic shock. The mortality rate of the test cohort (29.9%) was not significantly different compared to the derivation cohort (32.0%). Within the test cohort, nonsurvivors had a higher median age, a lower proportion of male subjects, a higher median APACHE II score, and a higher proportion of subjects with either chronic disease or septic shock, compared to the survivors. The mean and median times to death in the test cohort nonsurvivors were 11.1 ± 8.0 and 10 days (IQR, 4–17 d), respectively, both of which were significantly greater compared to the derivation cohort.
Supplemental Figure 1 (Supplemental Digital Content 2, http://links.lww.com/CCM/A830) shows the classification of the test cohort subjects according to the decision tree. The algorithm from the derivation cohort (Fig. 1) was applied to the test cohort with no modifications. One hundred and twenty-six test cohort subjects were classified as low risk (terminal nodes 1, 3, 5, 7, 8, and 10), whereas 205 were classified as high risk (terminal nodes 2, 4, 6, 9, 11, and 12). Among the low-risk subjects, the mortality rate was 9.5%, whereas among the high- risk subjects, the mortality rate was 42.4%. Table 2 shows the diagnostic test characteristics of the decision tree in the test cohort. The model did not perform differently when applied against only the test cohort subjects with septic shock (n = 271, data not shown).
Comparison With APACHE II
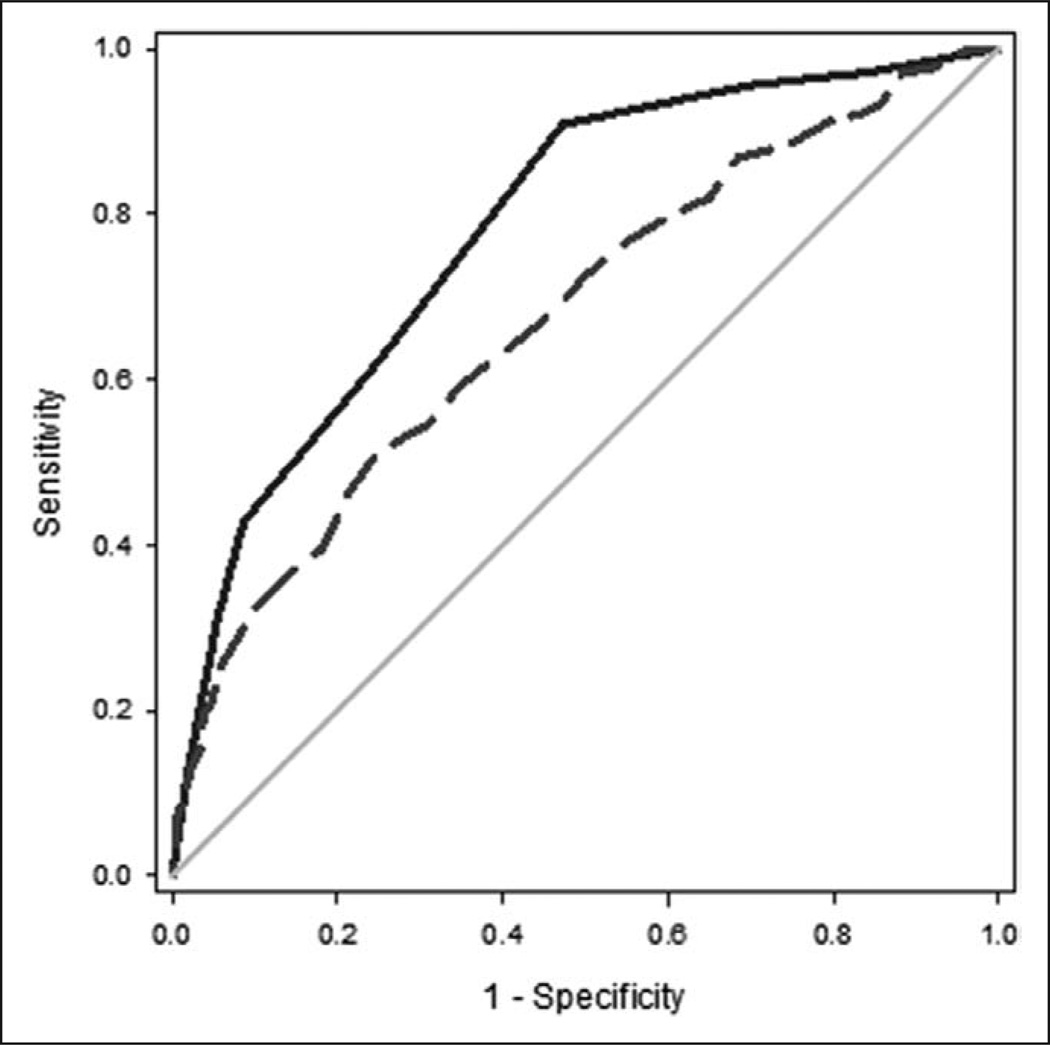
We compared the performance of the biomarker-based model with that of APACHE II for all subjects in the derivation and test cohorts (n = 672). Figure 2 shows the ROC curves for the biomarker-based model and APACHE II. The area under the curve (AUC) for the biomarker-based model (0.784; 95% CI, 0.747–0.820) was superior to that of APACHE II (0.676; 95% CI, 0.632–0.721; p = 0.0001).
Figure 2.
Comparison of receiver operating characteristic (ROC) curves for the biomarker-based model and Acute Physiology and Chronic Health Evaluation (APACHE) II. The ROC curves are calculated based on the respective mortality probabilities and 28-day all-cause mortality and are based on all subjects in the combined derivation and test cohorts (n = 672). The ROC curve for the biomarker-based model (solid line) yielded an area under the curve (AUC) of 0.784 (0.747–0.820), whereas the ROC curve for APACHE II (dashed line) yielded an AUC of 0.676 (0.632–0.721).
When adding the information from the biomarker-based model to the information in APACHE II, the NRI was 0.576 (95% CI, 0.341–0.812). The NRI is a measure of how much the accuracy of predicted outcomes is improved when adding information (16). The NRI ranges between −2 and +2. A score of –2 indicates that all true positives are reclassified as false negatives and all true negatives are reclassified as false positives, and no false classifications are reclassified as true classifications. Conversely, when the score is 2, adding the information correctly reclassifies every case. Our results demonstrate that the biomarker-based model provides additional classification value beyond the information included in APACHE II.
Model Calibration
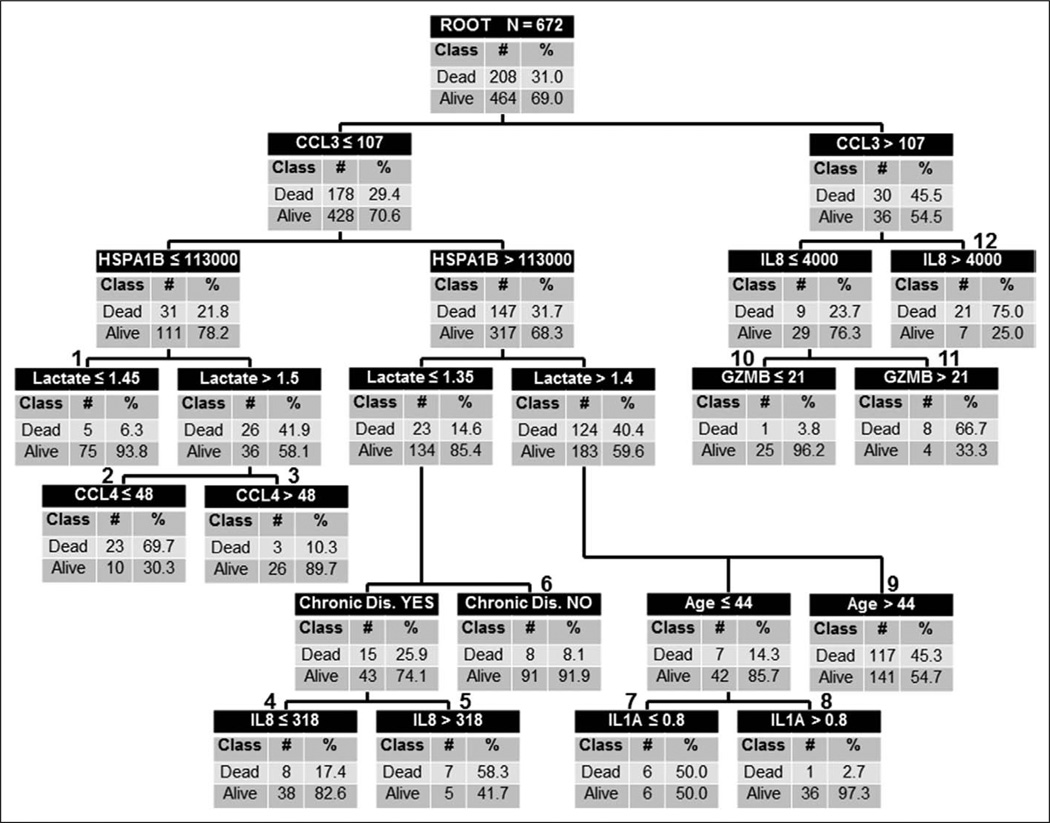
Assuming that a larger sample size could generate a more generalizable model, the decision tree was calibrated by combining all subjects in the derivation and test cohorts (n = 672) and repeating the CART analysis. The calibrated decision tree is shown in Figure 3. Notable changes included the addition of IL1A as a lower level decision leading to terminal nodes 7 and 8, and the replacement of a GZMB-based decision leading to terminal nodes 4 and 5 with an IL8-based decision. In addition, the decisions in the center of the tree based on lactate and chronic disease status changed their relative level positions. The calibrated tree contained six low-risk terminal nodes (2.7–17.4% risk of death; terminal nodes 1, 3, 4, 6, 8, and 10) and six high-risk terminal nodes (45.3–75.0% risk of death; terminal nodes 2, 5, 7, 9, 11, and 12). Of the 317 subjects classified as low risk, 291 survived (91.8%) and 26 (8.9%) had died by 28 days. Of the 355 subjects classified as high risk, 182 (51.3%) had died by 28 days. Table 2 shows the diagnostic test characteristics of the calibrated decision tree.
Figure 3.
The calibrated classification tree based on the combination of all subjects in the derivation and test cohorts (calibration cohort, n = 672). The classification tree consists of 11 decision points and 22 daughter nodes. The classification tree includes six of the 12 candidate stratification biomarkers: C-C chemokine ligand 3 (CCL3), heat shock protein 70 kDa 1B (HSPA1B), interleukin-8 (IL8), granzyme B (GZMB), C-C chemokine ligand 4 (CCL4), and interleukin-1α (IL1A). The classification tree also includes serum lactate concentrations (mmol/L), age (yr), and the presence/absence of chronic disease as defined in the Methods section. The conventions of the calibrated classification tree are the same as that described for Figure 1. Terminal nodes 1, 3, 4, 6, 8, and 10 are considered low-risk terminal nodes, whereas terminal nodes 2, 5, 7, 9, 11, and 12 are considered high-risk terminal nodes.
Validating the Calibrated Decision Tree
The calibrated decision tree was validated in a cohort of 209 subjects with septic shock, of whom 88 (42.1%) did not survive to 28 days. Table 1 provides the clinical and demographic data. Compared with the derivation cohort, the validation cohort had a higher mortality rate, a higher proportion of subjects with chronic disease, and a lower proportion of subjects with a surgical diagnosis. Within the validation cohort, nonsurvivors had a higher proportion of subjects with chronic disease, and a lower proportion of subjects with a surgical diagnosis, compared to the survivors. The mean and median times to death in the validation cohort nonsurvivors were 7.5 ± 6.9 and 5 days (IQR, 2–11 d), respectively.
Supplemental Figure 2 (Supplemental Digital Content 3, http://links.lww.com/CCM/A831) shows the classification of the validation cohort subjects according to the calibrated decision tree. The calibrated algorithm (Fig. 2) was applied to the validation cohort with no modifications. Eighty-six subjects were classified as low risk (terminal nodes 1, 3, 4, 6, 8, and 10), whereas 123 were classified as high risk (terminal nodes 2, 5, 7, 9, 11, and 12). Among the low-risk subjects, the mortality rate was 15.1%, whereas among the high-risk subjects, the mortality rate was 60.9%. Table 2 shows the performance of the calibrated decision tree in the validation cohort.
Since the validation cohort had APACHE III data available, we compared the performance of the calibrated model with that of APACHE III. In the validation cohort, the AUC for the calibrated model was 0.726 (95% CI, 0.660–0.792), whereas the AUC for APACHE III was 0.514 (95% CI, 0.434–0.595; p < 0.0001).
DISCUSSION
We have derived, tested, calibrated, and validated a risk stratification tool for adult septic shock that estimates the risk of 28-day mortality. A panel of biomarkers measured during the initial presentation to the ICU with septic shock, as well as admission lactate concentrations, age, and chronic disease status, form the basis of the model. On testing and validation, we observe good performance of the model. For all subjects in the study (n = 881), the high-risk terminal nodes of the calibrated model identified a cohort with a mortality rate of 56%, whereas the low-risk terminal nodes identified a cohort with a mortality rate of 11%. This dichotomous interpretation demonstrates that the model can partition a heterogeneous cohort of patients into two broad groups having an approximately five-fold difference in mortality risk. A more comprehensive interpretation allows assigning risk based on the respective terminal nodes, and this partitions patients across a clinically relevant range of mortality probabilities. The negative predictive value and the negative likelihood ratio of the model indicate that it may be most reliable as a “rule-out tool.”
A strength of our modeling is the initial approach to deriving the candidate stratification biomarkers. Using our extensive genome-wide expression databank, we identified 117 gene probes possibly associated with outcome in children with septic shock (6, 7, 17–24). From these, we selected the 12 biomarkers using a priori criteria.
The modeling process considered the candidate stratification biomarkers and clinical variables potentially associated with outcome. Interestingly, the biomarkers dominated the upper level decision rules, whereas the clinical variables contributed to either the lower level decision rules or not at all. This suggests that the biomarkers contribute significant and consistent stratification information that adds to stratification based on clinical findings; the NRI supports this assertion. The pediatric modeling procedures yielded similar results (5). In fact, the upper level decision rules of the pediatric and adult models consisted of the same three biomarkers (i.e., CCL3, HSPA1B, and IL8), albeit with different cutoff values. This suggests consistent utility for these three particular stratification biomarkers. We do note a limitation of our current study in that we did not consider other biomarkers having potential prognostic utility in adult populations (25), nor could we consider all possible clinical variables potentially associated with outcome due to reliance on secondary data sources.
The 2008 Surviving Sepsis Campaign International Guidelines recommend a serum lactate concentration greater than 4 mmol/L as a threshold indicator of tissue hypoperfusion warranting initiation of protocolized, quantitative resuscitation (26). The 2012 guidelines provide the same recommendation based on a reported mortality of 46% in septic patients with both hypotension and serum lactate concentration greater than 4 mmol/L (27, 28). Our calibrated decision tree indicates that serum lactate concentrations below these levels are associated with increased risk of mortality, which is consistent with two recent studies (29, 30).
The available data allowed a comparison of the biomarker- based model performance with both APACHE II and APACHE III, and the biomarker-based model outperformed APACHE II and III in these cohorts. We note that the Mortality Probability Model II at 24 hours (MPM II24), customized for patients with sepsis, yielded an AUC of 0.826 in a previous study (31). MPM II24 may perform better than our biomarker-based model, but MPM II24 data were not recorded in the studies from which our data were pooled.
There are important differences between the cohorts included in this study. First, the initial derivation cohort was drawn from participants in an interventional clinical trial. They may represent a more selected population of patients than the subjects in the test and validation cohorts who were drawn from observational databases. Second, the cohorts were drawn from three different healthcare systems, each with their own healthcare practices, cultures, and demographics. Third, while all subjects in the derivation and validation cohorts met criteria for septic shock, the subjects in the test cohort met criteria for either septic shock (81.9%) or severe sepsis (19.1%). Fourth, the cohorts differed in age, gender, illness severity, time to death, surgical status, and chronic disease burden. All of these differences represent potential confounders, yet the models performed consistently well across cohorts.
We propose that the primary potential application of the biomarker-based model is to enhance patient selection for interventional clinical trials. Excluding the lowest risk patients who are likely to survive without experimental intervention reduces their exposure to risk of adverse effects from the new intervention. Furthermore, the approach may increase the likelihood of positive trial findings; excluding the highest risk patients unlikely to survive with any therapy removes patients who may be too sick to respond to treatment, which could enhance the potential for measurable risk reduction for new therapy among moderate or high-risk patients with modifiable outcomes. Application of the model in this manner would require the development of a rapid, multiassay platform and computer support to reliably apply the decision rule. Although neither of these currently exist, the technologies to develop them are routinely available.
In conclusion, we have derived, tested, and validated a multibiomarker-based risk model that estimates mortality probability in adults with septic shock. Favorable comparisons to existing scoring systems and good performance in the context of potentially profound confounding factors support the generalizability and utility of the model. We propose that the model may enhance clinical trial design. Another potential application of the model may include serving as a benchmarking metric for quality improvement efforts.
Acknowledgments
Dr. Wong conceived and developed the study, obtained funding for the study, directly took part in the analyses, and wrote the article. Dr. Lindsell collaborated with Dr. Wong in the initial design of the study and in obtaining funding, oversaw the statistical analyses, and edited the article. Ms. Thair, Dr. Russell, Dr. Fjell, Dr. Boyd, and Dr. Walley are Vasopressin and Septic Shock Trial investigators and edited the article. Drs. Pettilä, Karlsson, and Ruokonen are FINNSEPSIS investigators and edited the article. Drs. Meyer, Shashaty, and Christie are Molecular Epidemiology of Severe Sepsis in the Intensive Care Unit investigators and edited the article. Ms. Hart assisted in data management and statistical analysis and edited the article. Mr. Lahni conducted all biomarker assays and edited the article.
Supported, in part, by the National Institutes of Health (NIH) (RC1HL100474, RO1GM064619, and RO1GM099773), an Innovation Award from the Center for Technology Commercialization at the Cincinnati Children’s Hospital Research Foundation, and an Institutional Clinical and Translational Science Award (NIH/National Center for Research Resources) (8UL1 TR000077).
Dr. Wong and the Cincinnati Children’s Hospital Research Foundation have submitted a provisional patent application for the stratification model.
Dr. Wong’s institution received grant support from the National Institutes of Health (NIH). Dr. Wong has a patent pending for a biomarker model with U.S. Patent Office and received support for article research from the NIH. Dr. Lindsell’s institution received grant support from the NIH (his contributions to this work were supported, in part, by an institutional Clinical and Translational Science Award [CTSA] from the NIH). Dr. Lindsell and his institution have a patent (he was named as coinventor on a patent for a multibiomarker- based risk stratification model for pediatric sepsis). Dr. Lindsell received support for article research from the NIH. Dr. Meyer’s institution received grant support from the NIH (HL102254 and Submitted R01) and GlaxoSmithKline (research grant to support plasma collection). Dr. Meyer received support for article research from the NIH. Ms. Thair consulted for LKL Consulting and received grant support from University of British Columbia (grant funding for PhD graduate studies). Dr. Russell served as board member for Sirius Genomics; consulted for Ferring, Astra Zeneca, Medimmune, Grifols, and Sirius Genomics; has a patent with the University of British Columbia; and has stock options with Sirius Genomics. Dr. Russel’s institution received grant support from Ferring, Astra Zeneca, and Sirius Genomics. Dr. Shashaty’s institution received grant support from the NIH (Career Development Award to study the association of adiposity with acute kidney injury in critically ill trauma patients). Dr. Christie provided expert testimony for various law firms (expert testimony on asbestos litigation in brake workers) and received support for article research from the NIH (salary is supported by NIH HL115354, HL081619, HL087115). Dr. Christie’s institution received grant support from the NIH and Glaxo- SmithKline. Ms. Hart received support for article research from the NIH. Mr. Lahni received support for article research from the NIH. Dr. Walley is employed by the University of British Columbia and received grant support from Canadian Institutes of Health Research.
Footnotes
Supplemental digital content is available for this article.
The remaining authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.Marshall JC. Sepsis: Rethinking the approach to clinical research. J Leukoc Biol. 2008;83:471–482. doi: 10.1189/jlb.0607380. [DOI] [PubMed] [Google Scholar]
- 2.Opal SM, LaRosa SP. Recombinant human activated protein C as a therapy for severe sepsis: Lessons learned? Am J Respir Crit Care Med. 2013;187:1041–1043. doi: 10.1164/rccm.201303-0505ED. [DOI] [PubMed] [Google Scholar]
- 3.Osuchowski MF, Connett J, Welch K, et al. Stratification is the key: Inflammatory biomarkers accurately direct immunomodulatory therapy in experimental sepsis. Crit Care Med. 2009;37:1567–1573. doi: 10.1097/CCM.0b013e31819df06b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Opal SM, Laterre PF, Francois B, et al. ACCESS Study Group. Effect of eritoran, an antagonist of MD2-TLR4, on mortality in patients with severe sepsis: The ACCESS randomized trial. JAMA. 2013;309:1154–1162. doi: 10.1001/jama.2013.2194. [DOI] [PubMed] [Google Scholar]
- 5.Wong HR, Salisbury S, Xiao Q, et al. The pediatric sepsis biomarker risk model. Crit Care. 2012;16:R174. doi: 10.1186/cc11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan JM, Wong HR. Biomarker discovery and development in pediatric critical care medicine. Pediatr Crit Care Med. 2011;12:165–173. doi: 10.1097/PCC.0b013e3181e28876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong HR. Genome-wide expression profiling in pediatric septic shock. Pediatr Res. 2013;73:564–569. doi: 10.1038/pr.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell JA, Walley KR, Singer J, et al. VASST Investigators. Vasopressin versus norepinephrine infusion in patients with septic shock. N Engl J Med. 2008;358:877–887. doi: 10.1056/NEJMoa067373. [DOI] [PubMed] [Google Scholar]
- 9.Karlsson S, Varpula M, Ruokonen E, et al. Incidence, treatment, and outcome of severe sepsis in ICU-treated adults in Finland: The Finnsepsis study. Intensive Care Med. 2007;33:435–443. doi: 10.1007/s00134-006-0504-z. [DOI] [PubMed] [Google Scholar]
- 10.Nakada TA, Russell JA, Boyd JH, et al. beta2-Adrenergic receptor gene polymorphism is associated with mortality in septic shock. Am J Respir Crit Care Med. 2010;181:143–149. doi: 10.1164/rccm.200903-0332OC. [DOI] [PubMed] [Google Scholar]
- 11.Levy MM, Fink MP, Marshall JC, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med. 2003;31:1250–1256. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- 12.Che D, Liu Q, Rasheed K, et al. Decision tree and ensemble learning algorithms with their applications in bioinformatics. Adv Exp Med Biol. 2011;696:191–199. doi: 10.1007/978-1-4419-7046-6_19. [DOI] [PubMed] [Google Scholar]
- 13.Muller R, Möckel M. Logistic regression and CART in the analysis of multimarker studies. Clin Chim Acta. 2008;394:1–6. doi: 10.1016/j.cca.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 14.VassarStats: Website for Statistical Computation. [Accessed March 6, 2013]; Available at: http://faculty.vassar.edu/lowry/VassarStats.html. [Google Scholar]
- 15.Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148:839–843. doi: 10.1148/radiology.148.3.6878708. [DOI] [PubMed] [Google Scholar]
- 16.Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: A framework for traditional and novel measures. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cvijanovich N, Shanley TP, Lin R, et al. Genomics of Pediatric SIRS/Septic Shock Investigators. Validating the genomic signature of pediatric septic shock. Physiol Genomics. 2008;34:127–134. doi: 10.1152/physiolgenomics.00025.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanley TP, Cvijanovich N, Lin R, et al. Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol Med. 2007;13:495–508. doi: 10.2119/2007-00065.Shanley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong HR, Cvijanovich N, Allen GL, et al. Genomics of Pediatric SIRS/Septic Shock Investigators. Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum. Crit Care Med. 2009;37:1558–1566. doi: 10.1097/CCM.0b013e31819fcc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong HR, Cvijanovich N, Lin R, et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 2009;7:34. doi: 10.1186/1741-7015-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong HR, Shanley TP, Sakthivel B, et al. Genomics of Pediatric SIRS/Septic Shock Investigators. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30:146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong HR. Genetics and genomics in pediatric septic shock. Crit Care Med. 2012;40:1618–1626. doi: 10.1097/CCM.0b013e318246b546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong HR. Clinical review: Sepsis and septic shock—The potential of gene arrays. Crit Care. 2012;16:204. doi: 10.1186/cc10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wynn JL, Cvijanovich NZ, Allen GL, et al. The influence of developmental age on the early transcriptomic response of children with septic shock. Mol Med. 2011;17:1146–1156. doi: 10.2119/molmed.2011.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall JC, Reinhart K International Sepsis Forum. Biomarkers of sepsis. Crit Care Med. 2009;37:2290–2298. doi: 10.1097/CCM.0b013e3181a02afc. [DOI] [PubMed] [Google Scholar]
- 26.Dellinger RP, Levy MM, Carlet JM, et al. International Surviving Sepsis Campaign Guidelines Committee; American Association of Critical- Care Nurses; American College of Chest Physicians; American College of Emergency Physicians; Canadian Critical Care Society; European Society of Clinical Microbiology and Infectious Diseases; European Society of Intensive Care Medicine; European Respiratory Society; International Sepsis Forum; Japanese Association for Acute Medicine; Japanese Society of Intensive Care Medicine; Society of Critical Care Medicine; Society of Hospital Medicine; Surgical Infection Society; World Federation of Societies of Intensive and Critical Care Medicine. Surviving Sepsis Campaign: International guidelines for management of severe sepsis and septic shock: 2008. Crit Care Med. 2008;36:296–327. [Google Scholar]
- 27.Dellinger RP, Levy MM, Rhodes A, et al. Surviving sepsis campaign: International guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med. 2013;41:580–637. doi: 10.1097/CCM.0b013e31827e83af. [DOI] [PubMed] [Google Scholar]
- 28.Levy MM, Dellinger RP, Townsend SR, et al. Surviving Sepsis Campaign. The Surviving Sepsis Campaign: Results of an international guideline-based performance improvement program targeting severe sepsis. Crit Care Med. 2010;38:367–374. doi: 10.1097/CCM.0b013e3181cb0cdc. [DOI] [PubMed] [Google Scholar]
- 29.Wacharasint P, Nakada TA, Boyd JH, et al. Normal-range blood lactate concentration in septic shock is prognostic and predictive. Shock. 2012;38:4–10. doi: 10.1097/SHK.0b013e318254d41a. [DOI] [PubMed] [Google Scholar]
- 30.Nichol AD, Egi M, Pettila V, et al. Relative hyperlactatemia and hospital mortality in critically ill patients: A retrospective multi-centre study. Crit Care. 2010;14:R25. doi: 10.1186/cc8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arabi Y, Al Shirawi N, Memish Z, et al. Assessment of six mortality prediction models in patients admitted with severe sepsis and septic shock to the intensive care unit: A prospective cohort study. Crit Care. 2003;7:R116–R122. doi: 10.1186/cc2373. [DOI] [PMC free article] [PubMed] [Google Scholar]