Abstract
During infection, naïve CD4+ T helper cells differentiate into specialized effector subsets based upon environmental signals propagated by the cytokine milieu. Recently, this paradigm has been complicated by the demonstration that alterations in the cytokine environment can result in varying degrees of plasticity between effector T helper cell populations. Therefore, elucidation of the mechanisms by which cytokines regulate T helper cell differentiation decisions is increasingly important. The gamma common cytokine IL-15 is currently undergoing clinical trials for the treatment of malignancies, due to its well-established role in the regulation of natural killer and CD8+ T cell immune responses. However, the effect of IL-15 signaling on CD4+ T cell activity is incompletely understood. One mechanism by which IL-15 activity is conferred is through trans-presentation via the IL-15 receptor alpha subunit. Here, we demonstrate that differentiated TH1 cells are responsive to trans-presented IL-15. Importantly, while trans-presentation of IL-15 results in STAT5 activation and maintenance of the TH1 gene program, IL-15 treatment alone allows for increased Bcl-6 expression and the upregulation of a TFH-like profile. Collectively, these findings describe a novel role for IL-15 in the modulation of CD4+ T cell responses and provide valuable insight for the use of IL-15 in immunotherapeutic approaches.
An effective immune response requires the orchestrated activity of a cohort of immune cell types. At the center of this coordinated response are CD4+ T helper cells, which provide an instrumental link between the innate and adaptive arms of the immune system. These include T helper 1 (TH1), TH2, TH17, and T follicular helper (TFH) cell populations, all of which arise from naïve CD4+ T cell progenitors1,2,3,4,5. The development of each of these effector T helper cell subpopulations is dictated by the expression of specific transcription factors, termed lineage-defining factors, the expression and activity of which are dependent upon the cytokine environment present during the activation of the naïve CD4+ T cell6,7,8,9,10,11,12. Importantly, each of these populations is responsible for performing unique effector functions that range from the cell-mediated release of inflammatory cytokines during pathogenic infection to aiding in the production of humoral immunity. Despite these distinct roles, numerous studies have recently demonstrated that flexibility, or plasticity, between effector T helper cell populations can occur in response to alterations in the cytokine environment2,13,14,15,16. Based in part upon these insights, novel sets of immunotherapeutic tools are being developed in an effort to manipulate the immune response and capitalize on the flexible nature of T helper cell populations17,18,19,20,21. As such, it is increasingly important to understand how changes to the cytokine environment effect not only the initial differentiation of activated naïve CD4+ T cells, but also the flexibility of established effector T helper cell populations.
The cytokines IL-2 and IL-15 are well-established regulators of immune cell development and function22,23,24. Both IL-2 and IL-15 signal through hetero-trimeric receptors, which are made up of the gamma common (γc) and IL-2Rβ subunits, as well as specificity-conferring IL-2Rα and IL-15Rα subunits, respectively19,22. Like IL-2, IL-15 signals through intracellular domains of the γc and IL-2Rβ subunits, which employ the janus kinase 1 (Jak1) and Jak3 kinases to phosphorylate and activate the transcription factor STAT519,22. However, IL-15 is capable of signaling through a unique “trans-presentation” mechanism, which involves the expression of IL-15Rα in complex with IL-15 on the surface of a presenting cell24,25,26,27,28. This complex interacts with the remaining two thirds of the receptor on a target cell, resulting in the propagation of IL-15 signaling within that cell. Trans-presentation has been shown to increase the affinity of IL-15 for the γc and IL-2Rβ receptor subunits, leading to more robust signaling as compared to IL-15 alone29,30,31.
The role of IL-15 in the stimulation of CD8+ cytotoxic T and natural killer (NK) cell populations is well-characterized and has provided strong pre-clinical support for its use in cancer and vaccine immunotherapeutic approaches24,27,32,33. Currently, IL-15 is undergoing Phase I/II Clinical trials for the treatment of malignant melanoma and renal cell carcinoma32,33,34. However, the effects of IL-15, particularly in the context of trans-presentation, on CD4+ T cell differentiation and effector function are incompletely understood. Therefore, an increased understanding of the effects of IL-15 signaling on the diverse repertoire of CD4+ T cell gene programs is a critical component in not only understanding how this cytokine may affect the natural immune response, but also in helping to unravel its therapeutic potential.
In the present study, we investigated the effect of IL-15 signaling on CD4+ TH1 differentiation, using a well-established model of TH1 cell generation35,36. We demonstrate that exposure of TH1 cells to trans-presented IL-15, but not to IL-15 alone, favors the TH1 gene program. Mechanistically, increased STAT5-activation via trans-presented IL-15 was associated with the expression of TH1-associated genes including Tbx21, Ifng, Prdm1, Il12ra and Il12rb, as well as enhanced IL-12-dependent STAT4 phosphorylation. Strikingly, exposure of TH1 cells to IL-15 in the absence IL-15Rα resulted in decreased STAT5 activation and increased expression of the TFH lineage defining transcription factor Bcl-6. Elevated Bcl-6 expression levels correlated with a decrease in Blimp-1 expression and the upregulation of a TFH-like gene program including Cxcr5, Il6ra, Tnfsf8, and Sh2d1a. Collectively, these results describe previously unappreciated effects of IL-15 signaling on the differential expression of TH1 and TFH gene profiles, and provide added insight into the functional properties of this immunotherapeutic cytokine.
Results
Exposure to IL-15 alone is insufficient to maintain expression of the TH1 gene program
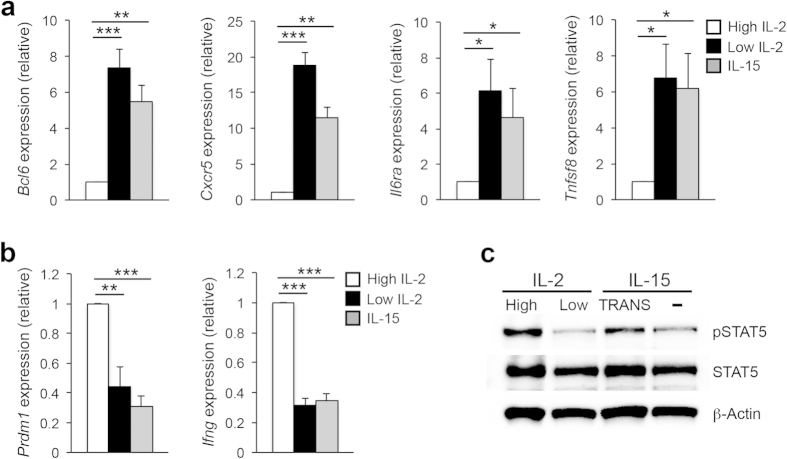
It has become increasingly evident that mature effector CD4+ T cells are responsive to alterations in environmental stimuli in the form of cytokines13,14,15,37,38,39. The adaptability of effector T helper cell populations allows these cells to respond to the environment in real-time, and is particularly important in clinical applications where the use of cytokines or cytokine neutralizing antibodies is employed to promote or abrogate specific immune responses. For example, previous work has demonstrated that signaling through the gamma common (γc) cytokine IL-2 is indispensable for the promotion and maintenance of the TH1 cell fate while, conversely, the withdrawal of IL-2 results in a loss of TH1 cell identity and the up-regulation of a TFH-like gene expression profile36,40. Interestingly, a second γc cytokine, IL-15, is currently undergoing Phase I/II clinical trials for the treatment of many types of cancer33,34,41. As the downstream signaling events of IL-15 are similar to those of IL-2, we hypothesized that IL-15 may maintain the TH1 profile at the expense of TFH gene expression. To begin to test this hypothesis, we examined whether IL-15 treatment alone would be sufficient to both promote TH1 gene expression patterns and repress the induction of the TFH gene program following IL-2 withdrawal from differentiated TH1 cells. As with our previous study, cells exposed to high IL-2 environments repressed the TFH gene program and maintained TH1 expression patterns while, conversely, IL-2 withdrawal from TH1 cells resulted in a significant increase in Bcl6 expression and the induction of a TFH-like profile (Fig. 1a,b)36. Interestingly, treatment with IL-15 similarly resulted in a significant increase in Bcl6 expression compared to that observed in high IL-2 cells (Fig. 1a). Importantly, the increase in Bcl6 expression was associated with an increase in the TFH-associated genes Cxcr5, Il6ra, and Tnfsf8. Furthermore, while the TFH-associated gene program was upregulated, the expression of the TH1 genes Ifng and Prdm1 was decreased (Fig. 1b). One possibility that we considered was that at day 3 (the time-point of IL-15 administration), our ex vivo generated TH1 cells were not fully committed to the TH1 phenotype and therefore maintained a higher degree of flexibility with alternative T helper cell identities. To examine this possibility, we performed a similar experiment, but added IL-15 on day 5 of the TH1 culture (rather than day 3). Importantly, a similar induction of the TFH-like profile was observed with TH1 cells exposed to standard (3 days) or extended (5 days) TH1 culturing conditions (Supplementary Fig. S1). Collectively, these results indicate that IL-15 treatment alone is insufficient to support TH1 gene expression patterns and instead results in the upregulation of Bcl-6 expression and a TFH-like program.
Figure 1. IL-15 treatment alone is insufficient to activate STAT5 and the TH1 program.
(a,b) Primary CD4+ T cells were isolated from C57BL/6 mice and stimulated on plate-bound αCD3 and αCD28 for 3 days under TH1-polarizing conditions. On day 3, cells were split and cultured with either high IL-2 (250 U/ml), low IL-2 (10 U/ml), or IL-15 (15 ng/ml). Two days following cytokine treatment, RNA was isolated to determine transcript levels for the indicated TFH and TH1-associated genes. Samples were normalized to Rps18 as a control. Data are presented relative to the high IL-2-treated sample. Three (a,b) independent experiments were performed with the error bars representing SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t-test). (c) Cells were treated and harvested as in “a” with the exception that an additional sample was treated with 15 ng/ml of trans-presented IL-15 (IL-15, TRANS). Following cell isolation, an immunoblot assay was performed to assess STAT5 activation levels. Total STAT5 and β-Actin are shown as controls. The image shown is representative of three independent experiments performed.
A key finding from our previous work was that the IL-2-dependent phosphorylation status of STAT5 inversely correlated with the expression of Bcl-636. As IL-15 also signals via the activation of STAT5, we hypothesized that IL-15 treatment alone may not be sufficient to phosphorylate STAT5, thus resulting in increased Bcl-6 expression. To test this possibility, we performed an immunoblot analysis to assess the degree of STAT5 phosphorylation in the high IL-2-, low IL-2-, and IL-15-treated TH1 cells. While high IL-2 cells displayed elevated levels of STAT5 phosphorylation (pSTAT5), there was very little STAT5 activation in either the low IL-2- or IL-15-treated populations (Fig. 1c). It has been demonstrated that IL-15 is capable of signaling through a unique “trans-presentation” mechanism, which involves the expression of IL-15Rα in complex with IL-15 on the surface of a second presenting cell. Thus, although CD4+ T cells have variable expression of IL-15Rα, this suggests that IL-15Rα expression is dispensable for IL-15-responsiveness as trans-presentation provides IL-15Rα in the form of a presenting complex27. Therefore, we sought to determine whether TH1 cells would display enhanced STAT5 activation in response to trans-presented IL-15. Indeed, we observed increased STAT5 phosphorylation upon exposure of TH1 cells to IL-15 in complex with a recombinant IL-15Rα (IL-15Trans) (Fig. 1c). Collectively, these data suggest that IL-15 signaling is a potential differential regulator of the opposing TH1 and TFH gene programs.
IL-15 signaling via trans-presentation represses Bcl-6 expression in TH1 cells
The demonstration that trans-presentation of IL-15 results in increased STAT5 activation prompted us to more thoroughly explore the effect of IL-15 signaling on the expression of the TFH-lineage defining transcription factor Bcl-6. We exposed TH1 cells to two different concentrations of IL-15 (75 ng/ml and 15 ng/ml) either alone or in the trans-presented form. Interestingly, upon exposure of cells to a high environmental concentration of IL-15, Bcl6 expression remained relatively low (Fig. 2a). Conversely, a significant increase in Bcl6 expression was observed when TH1 cells were exposed to a low concentration of IL-15 alone. Importantly, there was a ~6 fold decrease in Bcl6 expression when the cells were exposed to the same low concentration of IL-15 in the trans-presented form (Fig. 2a). To determine whether Bcl-6 protein expression was consistent with the observed changes in transcript, we performed an immunoblot analysis. Indeed, while there was an increase in Bcl-6 expression at the low IL-15 concentration, we did not observe an induction of Bcl-6 expression in the trans-presented IL-15 sample (Fig. 2b). Importantly, the increase in Bcl-6 expression inversely correlated with the level of IL-15-dependent STAT5 phosphorylation. Collectively, these data indicate that differentiated CD4+ TH1 cells are capable of responding to IL-15 signaling, particularly during trans-presentation, and that increased IL-15 signaling results in the repression of Bcl-6 expression.
Figure 2. IL-15 signaling represses the expression of Bcl-6.
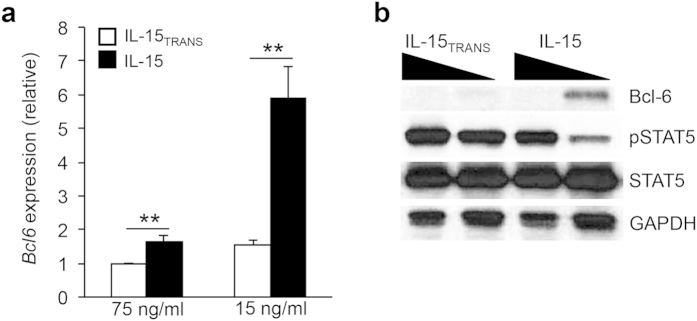
(a) Primary CD4+ T cells were isolated from C57BL/6 mice and stimulated on plate-bound αCD3 and αCD28 for 3 days under TH1-polarizing conditions. On day 3, cells were split and cultured with either IL-15TRANS or IL-15 (75 ng/ml or 15 ng/ml). Two days following cytokine treatment, RNA was isolated to determine the transcript level for Bcl6. Samples were normalized to an Rps18 control and data are represented relative to the IL-15TRANS (75 ng/ml) sample. Five independent experiments were performed with the error bars representing SEM. **P < 0.01 (unpaired Student’s t-test). (b) Samples were treated and harvested as in “a”. Two days following cytokine treatment, cells were harvested for each condition and an immunoblot analysis was carried out to determine the effect of IL-15 signaling on Bcl-6 expression and STAT5 activation. Total STAT5 and β-Actin are shown as controls. The displayed image is representative of three independent experiments performed.
IL-15 signaling regulates the balance of the transcriptional repressors Bcl-6 and Blimp-1
There is a well-established antagonistic relationship between the transcriptional repressors Bcl-6 and Blimp-1 (encoded by the gene Prdm1) that guides a number of B and T lymphocyte developmental stages42,43. For example, previous work from our lab and that of others has demonstrated that Blimp-1 antagonizes TFH cell differentiation9,44. In the context of TH1 cell development, Blimp-1 inhibits the TFH program by directly repressing TFH-associated genes including Cxcr5, Il6ra, and Tnfsf8. Therefore, we hypothesized that IL-15 signaling, similar to that of IL-2, may differentially regulate the expression of these opposing transcriptional regulators and ultimately dictate the establishment of a TH1 or TFH gene program. Indeed, TH1 cells exposed to trans-presented IL-15 maintained elevated expression of Prdm1, and importantly, increased Blimp-1 protein expression was also observed with trans-presented IL-15 regardless of high or low environmental concentrations of the cytokine (Fig. 3a,b). Conversely, IL-15 cytokine treatment alone resulted in a decrease in Blimp-1 expression. Importantly, the reduction in Blimp-1 expression was coincident with a decrease in STAT5 phosphorylation, and a corresponding increase in Bcl-6 expression (Fig. 3b).
Figure 3. IL-15 trans-presentation maintains Blimp-1 expression.
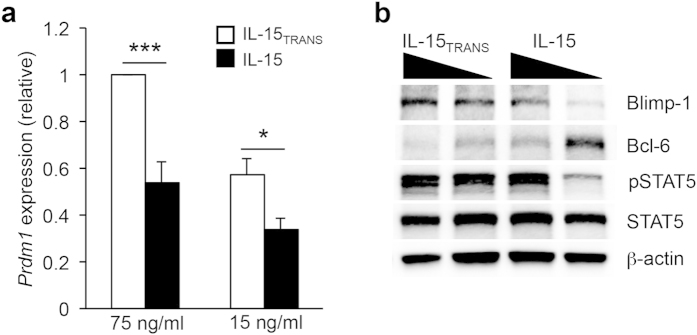
(a) Primary CD4+ T cells were cultured as described in Fig. 2. Two days following the indicated IL-15 cytokine treatment, cells were harvested for each condition and RNA was isolated to determine transcript levels for Prdm1 under IL-15 or IL-15TRANS (75 ng/mL or 15 ng/mL) conditions as indicated. Samples were normalized to Rps18 as a control. Graphical representations are normalized to the IL-15TRANS (75 ng/ml) sample. (b) An immunoblot analysis was performed to examine the effect of IL-15 signaling on Blimp-1, Bcl-6, p-STAT5, and STAT5 expression. β-Actin was monitored to ensure equal protein loading. For (a), figures are representative of five independent experiments. Error bars represent SEM. *P < 0.05, ***P < 0.001 (unpaired Student’s t-test). For (b), the image is representative of three independent experiments.
Strength of IL-15 signaling differentially regulates TH1 and TFH gene expression patterns
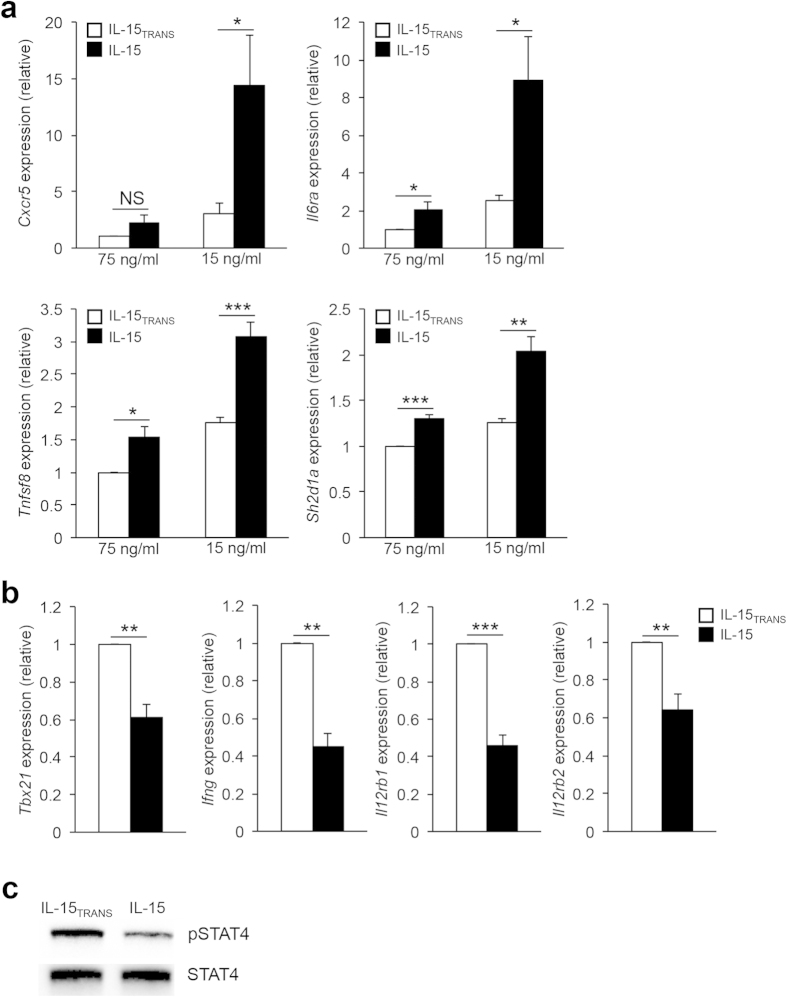
In our previous study, we found that the TFH-associated genes Cxcr5, Il6ra, and Tnfsf8 are all direct targets of Blimp-1-mediated repression36. Therefore, Bcl-6 activates the TFH gene profile, at least in part, through the repression of a second transcriptional repressor (Blimp-1). Given the upregulation of Blimp-1 in response to IL-15 signaling, we hypothesized that the trans-presentation of IL-15 may also lead to the repression of the TFH gene program. To investigate this possibility, we examined the expression of the TFH-associated genes Cxcr5, Il6ra, Tnfsf8, and Sh2d1a under different IL-15 signaling conditions. Indeed, IL-15 signaling was associated with decreased expression of all four genes, in both dose- and trans-presentation-dependent manners (Fig. 4a). However, under low IL-15 signaling conditions, the expression of each gene was significantly induced. Once again, treating the cells with the trans-presented form of IL-15, even at low concentrations, was sufficient to maintain inhibition of Cxcr5, Il6ra, Tnfsf8, and Sh2d1a expression, most likely due to the increase in Blimp-1 expression (Fig. 3). Interestingly, not all TFH-associated genes responded similarly to IL-15 signaling. For example, the expression of Pdcd1, Icos, and Il21 did not appear to be augmented by changes to IL-15 concentration or trans-presentation (Supplementary Fig. S2). These data strongly suggest that IL-15 signaling is not a comprehensive regulator of the entire TFH gene program. This is, perhaps, not surprising, as it is well-established that there are required roles for B cell interactions, as well as pro-TFH cytokines such as IL-6 and IL-21, in the full commitment to the TFH cell fate17. Thus, it is likely that the ex vivo setting lacks at least some of the necessary environmental factors required to promote the induction of a complete TFH gene profile.
Figure 4. IL-15 signaling regulates the expression of TFH-associated genes.
(a) Primary CD4+ T cells were cultured as described in Fig. 2. RNA was isolated to determine transcript levels for Cxcr5, Il6ra, Tnfsf8, and Sh2d1a. Samples were normalized to Rps18 as a control and the data are represented relative to the IL-15TRANS (75 ng/ml) sample. (b) Primary CD4+ T cells were cultured as described as in “a”. RNA was isolated to determine transcript levels for Tbx21, Ifng, Il12rb1, and Il12rb2. Samples were normalized to Rps18 as a control. Data are presented relative to the IL-15TRANS (75 ng/ml) sample. Four (a,b) independent experiments were performed with the error bars representing SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t-test). (c) Cells were treated and harvested as in “a”. Following cell isolation, an immunoblot assay was performed to assess STAT4 activation levels. Total STAT4 is shown as a control. The image shown is representative of three independent experiments performed.
As our data suggest that enhanced IL-15 signaling is capable of maintaining TFH gene repression even in the absence of IL-2, we hypothesized that trans-presented IL-15 may also be sufficient to promote the expression of TH1-associated genes. Indeed, treatment with trans-presented IL-15 resulted in increased expression of both the TH1 lineage-defining transcription factor Tbx21, and canonical cytokine Ifng, when compared to IL-15 treatment alone (Fig. 4b). Furthermore, cells that were exposed to trans-presented IL-15 expressed higher levels of both Il12rb1 and Il12rb2 compared to cells that received IL-15 alone (Fig. 4b). Consistent with the observed increase in Il12rb1 and Il12rb2 expression, we found that exposure of cells to trans-presented IL-15 also resulted in an increase in IL-12-dependent STAT4 activation (Fig. 4c). Taken together, these findings suggest that intracellular signaling induced by trans-presented IL-15 can promote additional cytokine-mediated responses critical for the promotion or repression of the TH1 and TFH gene programs, respectively.
Discussion
CD4+ T cell differentiation from the naïve cell state into any of several effector cell types is dependent upon the cytokine environment in which the cell is located during its activation. Until recently, it was believed that CD4+ T cell differentiation into an effector subset was a terminal event, but a significant body of work has established that differentiated CD4+ T cells are capable of modulating their effector functions in response to changes in the cytokine environment15,38,39,45,46. This revelation may be particularly impactful given the use of cytokines and cytokine neutralizing antibodies as approaches to regulate the immune response in a variety of immunotherapeutic strategies.
One such example is the therapeutic use of the cytokine IL-15, which is currently under evaluation in clinical trials for the treatment of a number of types of cancer32,33,34. IL-15 may be an attractive alternative to IL-2-based therapies, as it is able to promote many of the same therapeutic effects due to similar downstream signaling pathways47. However, unlike IL-2, IL-15 does not support regulatory T cell populations or promote activation induced cell death (AICD), both of which may inhibit anti-cancer immune responses48. Although the effects of IL-15 signaling have been well characterized in CD8+ T and NK cell populations, the effect of IL-15 on the CD4+ T cell effector population has remained relatively unknown49. Thus, as the application of this promising cytokine increases, it will be crucial to understand its effects on these vital cell populations in a clinical setting.
Here, our data demonstrate that CD4+ T cells are capable of responding to IL-15 treatment, particularly when IL-15 is trans-presented in complex with the IL-15Rα subunit. Subsequently, IL-15 signaling and the concurrent activation of the transcription factor STAT5 are associated with the positive regulation of TH1 genes including Tbx21, Ifng, and Prdm1. Additionally, we observed increased expression of the IL-12 receptor subunits (Il12rb1 and Il12rb2), as well as STAT4 activation, in response to IL-15 trans-presentation. This data is in agreement with previous work demonstrating a similar role for IL-2 in the positive regulation of the components of the IL-12R, and suggests that IL-2- and IL-15-signaling, in the context of trans-presentation, share at least some downstream effects40.
We also found that there is an IL-15 trans-presentation-dependent inhibitory effect upon the expression of genes associated with the TFH cell fate, including the expression of the transcriptional repressor Bcl-6. Our findings indicate that there is an inverse relationship between STAT5 activation and Bcl-6 expression, hinting at a potential regulatory mechanism whereby STAT5 may directly repress Bcl-6. As these data are similar to previous studies examining the repressive effect of IL-2 signaling on the TFH gene program, they suggest that the combined action of STAT5 and Blimp-1 may be a conserved regulatory mechanism utilized by IL-2 and IL-15 signaling to promote the TH1 cell fate36,50,51. Further investigation will be necessary to fully elucidate whether there are specific transcriptional mechanisms unique to IL-15 signaling that both inhibit and promote the expression of the TFH and TH1 gene programs, respectively.
Despite the apparent antagonistic nature of Bcl-6 expression and IL-15 signaling, it is interesting to note that both factors have been shown to positively influence the formation of T cell memory. Bcl-6 expression has been linked to the promotion of the effector-to-memory cell transition in both CD4+ and CD8+ T cells, while IL-15 is critical for the homeostatic maintenance of memory cell populations42,52,53. Future work will be necessary to dissect how the findings presented in this study may translate into an increased understanding of the interplay between these intrinsic and extrinsic factors that govern memory cell fate.
Collectively, this study highlights a novel role for IL-15 signaling in modulating the gene expression profiles of CD4+ T helper cell populations. Consequently, these changes in gene expression dictate both the fate and function of the responding cells. As such, an increased understanding of the implications of cytokine signaling, including that of IL-15, on the plasticity of CD4+ T cell populations will be critical as these environmental modulators continue to be utilized in novel immunotherapeutic strategies.
Methods
Primary cells and cell culture
Primary CD4+ T cells were isolated from the spleens and lymph nodes of 5–8 week old C57BL/6 mice using the R&D Systems MagCellect CD4+ T cell Isolation Kit according to the manufacturer’s instructions. Cells were cultured in complete Iscove’s Modified Dulbecco’s medium [cIMDM; IMDM (12440053, Life Technologies), 10% FBS (26140079, Life Technologies), 1% Penicillin-Streptomycin (15140122, Life Technologies), 0.05% BME (BP176, Fisher Scientific)] and plated on αCD3 (BDB553057, Fisher Scientific) and αCD28 (BDB553294, Fisher Scientific) under TH1 polarizing conditions (IL-12, αIL-4, and IL-2) as described previously36,54. After three days (or five, as in Supplementary Fig. 1), cells were split and maintained for an additional two days in IL-12 and αIL-4 and either high (75 ng/ml) or low (15 ng/ml) concentrations of IL-15, or a stable complex of IL-15 and its receptor (referred to in this manuscript as trans-presented IL-15, or IL-15TRANS), as indicated. The Institutional Animal Care and Use Committee of Virginia Tech approved all experimentation involving the use of mice. All methods were performed in accordance with the approved guidelines.
Generation of the IL-15/IL15Rα complex
A stable complex of IL-15/IL-15Rα was generated by combining soluble IL-15 (447-ML, R&D Systems) with a recombinant mouse IL-15Rα Fc chimera (551-MR, R&D Systems) at a ratio of 2:1. The combination was incubated at 37 °C for 45 minutes immediately prior to use, as described previously29,41.
RNA isolation and qRT-PCR
Two days following IL-15 cytokine treatment, cells were harvested for each condition and RNA was isolated using the Nucleospin RNA II kit from Macherey-Nagel (740955.5) per the manufacturer’s protocol. cDNA was generated using the Superscript First Strand Synthesis Kit (11904018, Life Technologies). Triplicate qRT-PCR reactions using 10–20 ng of template were performed in 20μL reactions with gene-specific primers (Supplementary Table S1) and iTaq Universal SYBR Green Supermix (172-5124, Bio-Rad). All samples were normalized to the ribosomal protein S18 (Rps18) control. For IL-2-treated samples, expression levels were normalized to the high IL-2-treated sample. For IL-15-treated samples, expression levels were normalized to the high IL-15TRANS-treated sample.
Immunoblot analysis
To examine the effect of IL-15 and IL-15TRANS at the protein level, primary T cells for each condition were harvested 2 days following IL-2, IL-15, or IL-15TRANS treatment. Antibodies used were as follows: pSTAT5 (611964, BD Biosciences), STAT5 (sc-482X, Santa Cruz), Bcl-6 (561520, BD Biosciences), pSTAT4 (5267S, Cell Signaling), STAT4 (sc-486X, Santa Cruz), Blimp-1 (A01647-40, Genscript), GAPDH (sc-25778, Santa Cruz), β-Actin (A00730-40, Genscript). Further details regarding the clone and/or dilution of antibodies used can be found in Supplementary Table S2).
Statistical analysis
All data represent at least three independent experiments. Error bars represent the standard error of the mean (SEM). For statistical analysis, unpaired t tests were performed using GraphPad Prism online software. P values < 0.05 were considered statistically significant.
Additional Information
How to cite this article: Cooley, I. D. et al. Trans-presentation of IL-15 modulates STAT5 activation and Bcl-6 expression in TH1 cells. Sci. Rep. 5, 15722; doi: 10.1038/srep15722 (2015).
Supplementary Material
Acknowledgments
The authors would like to thank members of the Oestreich Lab for critical reading of the manuscript. The authors would also like to acknowledge the faculty of VTCRI for thoughtful discussions. This work was supported by start-up funds from VTCRI.
Footnotes
Author Contributions I.D.C. and K.A.R. assisted with the design of the study, performed experiments, analyzed data, and wrote the manuscript. K.J.O. supervised the research, designed the study, analyzed data, and wrote the manuscript.
References
- Crotty S. Follicular Helper CD4 T Cells (T(FH)). Annu Rev Immunol 29, 621–663 (2011). [DOI] [PubMed] [Google Scholar]
- O’Shea J. J. & Paul W. E. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327, 1098–1102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt R. L., Kang S. J., Liang H. E. & Locksley R. M. T helper cell effector fates—who, how and where? Curr Opin Immunol 18, 271–277 (2006). [DOI] [PubMed] [Google Scholar]
- Zhu J. & Paul W. E. CD4 T cells: fates, functions, and faults. Blood 112, 1557–1569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudensky A. Y. Regulatory T cells and Foxp3. Immunol Rev 241, 260–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J. D., Gavin M. A. & Rudensky A. Y. Foxp3 programs the development and function of CD4+ CD25+ regulatory T cells. Nat Immunol 4, 330–336 (2003). [DOI] [PubMed] [Google Scholar]
- Hori S., Nomura T. & Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003). [DOI] [PubMed] [Google Scholar]
- Ivanov II et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006). [DOI] [PubMed] [Google Scholar]
- Johnston R. J. et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325, 1006–1010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R. I. et al. Bcl6 mediates the development of T follicular helper cells. Science 325, 1001–1005 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo S. J. et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669 (2000). [DOI] [PubMed] [Google Scholar]
- Yu D. et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31, 457–468 (2009). [DOI] [PubMed] [Google Scholar]
- Bonelli M. et al. Helper T cell plasticity: impact of extrinsic and intrinsic signals on transcriptomes and epigenomes. Curr Top Microbiol Immunol 381, 279–326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geginat J. et al. Plasticity of human CD4 T cell subsets. Front Immunol 5, 630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Chong M. M. & Littman D. R. Plasticity of CD4+ T cell lineage differentiation. Immunity 30, 646–655 (2009). [DOI] [PubMed] [Google Scholar]
- Oestreich K. J. & Weinmann A. S. Master regulators or lineage-specifying? Changing views on CD4(+) T cell transcription factors. Nat Rev Immunol 12, 799–804 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V. & Glimcher L. H. T-bet in disease. Nat Immunol 12, 597–606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W., Lin J. X. & Leonard W. J. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 38, 13–25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifka M. K. & Amanna I. How advances in immunology provide insight into improving vaccine efficacy. Vaccine 32, 2948–2957 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye S. G., Ma C. S., Brink R. & Deenick E. K. The good, the bad and the ugly—TFH cells in human health and disease. Nat Rev Immunol 13, 412–426 (2013). [DOI] [PubMed] [Google Scholar]
- Liao W., Lin J. X. & Leonard W. J. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr Opin Immunol 23, 598–604 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochman Y., Spolski R. & Leonard W. J. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol 9, 480–490 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo E. F. & Schluns K. S. Regulating the immune system via IL-15 transpresentation. Cytokine 59, 479–490 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois S., Mariner J., Waldmann T. A. & Tagaya Y. IL-15Ralpha recycles and presents IL-15 In trans to neighboring cells. Immunity 17, 537–547 (2002). [DOI] [PubMed] [Google Scholar]
- Sandau M. M., Schluns K. S., Lefrancois L. & Jameson S. C. Cutting edge: transpresentation of IL-15 by bone marrow-derived cells necessitates expression of IL-15 and IL-15R alpha by the same cells. J Immunol 173, 6537–6541 (2004). [DOI] [PubMed] [Google Scholar]
- Stonier S. W. & Schluns K. S. Trans-presentation: a novel mechanism regulating IL-15 delivery and responses. Immunol Lett 127, 85–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluns K. S., Klonowski K. D. & Lefrancois L. Transregulation of memory CD8 T-cell proliferation by IL-15Ralpha+ bone marrow-derived cells. Blood 103, 988–994 (2004). [DOI] [PubMed] [Google Scholar]
- Rubinstein M. P. et al. Converting IL-15 to a superagonist by binding to soluble IL-15R{alpha}. Proc Natl Acad Sci USA 103, 9166–9171 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanick N. A. et al. Elucidation of the interleukin-15 binding site on its alpha receptor by NMR. Biochemistry 46, 9453–9461 (2007). [DOI] [PubMed] [Google Scholar]
- Mortier E., Bernard J., Plet A. & Jacques Y. Natural, proteolytic release of a soluble form of human IL-15 receptor alpha-chain that behaves as a specific, high affinity IL-15 antagonist. J Immunol 173, 1681–1688 (2004). [DOI] [PubMed] [Google Scholar]
- Conlon K. C. et al. Redistribution, hyperproliferation, activation of natural killer cells and CD8 T cells, and cytokine production during first-in-human clinical trial of recombinant human interleukin-15 in patients with cancer. J Clin Oncol 33, 74–82 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. IL-15 Agonists: The Cancer Cure Cytokine. J Mol Genet Med 7, 85 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann T. A. et al. Phase 1 trial of IL-15 trans presentation blockade using humanized Mikbeta1 mAb in patients with T-cell large granular lymphocytic leukemia. Blood 121, 476–484 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich K. J., Huang A. C. & Weinmann A. S. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J Exp Med 208, 1001–1013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich K. J., Mohn S. E. & Weinmann A. S. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol 13, 405–411 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R., Hatton R. D. & Weaver C. T. The Th17 family: flexibility follows function. Immunol Rev 252, 89–103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K. M. & Stockinger B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nat Immunol 11, 674–680 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. & Paul W. E. Heterogeneity and plasticity of T helper cells. Cell Res 20, 4–12 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W., Lin J. X., Wang L., Li P. & Leonard W. J. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol 12, 551–559 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoklasek T. A., Schluns K. S. & Lefrancois L. Combined IL-15/IL-15Ralpha immunotherapy maximizes IL-15 activity in vivo. J Immunol 177, 6072–6080 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S., Johnston R. J. & Schoenberger S. P. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol 11, 114–120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins G. & Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol 26, 133–169 (2008). [DOI] [PubMed] [Google Scholar]
- Oestreich K. J. & Weinmann A. S. T-bet employs diverse regulatory mechanisms to repress transcription. Trends Immunol 33, 78–83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirahara K. et al. Helper T-cell differentiation and plasticity: insights from epigenetics. Immunology 134, 235–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayamada S., Takahashi H., Kanno Y. & O’Shea J. J. Helper T cell diversity and plasticity. Curr Opin Immunol 24, 297–302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim G. C. & Radvanyi L. The IL-2 cytokine family in cancer immunotherapy. Cytokine Growth Factor Rev 25, 377–390 (2014). [DOI] [PubMed] [Google Scholar]
- Steel J. C., Waldmann T. A. & Morris J. C. Interleukin-15 biology and its therapeutic implications in cancer. Trends Pharmacol Sci 33, 35–41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A., Sullivan L. & Caligiuri M. A. Molecular pathways: interleukin-15 signaling in health and in cancer. Clinical Cancer Res 20, 2044–2050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. J., Choi Y. S., Diamond J. A., Yang J. A. & Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med 209, 243–250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R. I. et al. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J Biol Chem 287, 11234–11239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasper D. J., Tejera M. M. & Suresh M. CD4 T-cell memory generation and maintenance. Crit Rev Immunol 34, 121–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S. M. & Cui W. Transcriptional control of effector and memory CD8(+) T cell differentiation. Nat Rev Immunol 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich K. J. et al. Bcl-6 directly represses the gene program of the glycolysis pathway. Nat Immunol 15, 957–964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.