Abstract
The evolution of specific appendages is made possible by the ontogenetic deployment of general cell signaling pathways. Many fishes, amphibians and reptiles have unique skin appendages known as barbels, which are poorly understood at the cellular and molecular level. In this study, we examine the cell arrangements, cell division patterns, and gene expression profiles associated with the zebrafish maxillary barbel, or ZMB. The earliest cellular organization of the ZMB is an internal whorl of mesenchymal cells in the dermis of the maxilla; there is no epithelial placode, nor any axially-elongated epithelial cells as expected of an apical ectodermal ridge (AER). As the ZMB develops, cells in S-phase are at first distributed randomly throughout the appendage, gradually transitioning to a proliferative population concentrated at the distal end.
By observing ZMB ontogenetic stages in a Wnt-responsive transgenic reporter line, TCFsiam, we identified a strongly fluorescent mesenchymal cell layer within these developing appendages. Using an in vitro explant culture technique on developing barbel tissues, we co-localized the fluorescent label in these cells with the mitotic marker EdU. Surprisingly, TCF+ cells showed little proliferation, indicating a slow-cycling subpopulation. Transmission electron microscopy of the ZMB located the TCF+ cells in a single, circumferential layer within the barbel’s matrix core. Morphologically, these cells resemble fibroblasts or osteoblasts; in addition to their matrix-bound location, they are identified by their pancake-shaped nuclei, abundant rough endoplasmic reticulum, and cytoplasmic extensions into the surrounding extracellular matrix. Taken together, these features define a novel mesenchymal cell population in zebrafish, the ‘TCF+ core cells.’ A working model of barbel development is proposed, in which these minimally mitotic mesodermal cells produce collagenous matrix in response to ectodermally-derived Wnt signals deployed in a proximal-distal gradient along the appendage. This documents a novel mechanism of vertebrate appendage outgrowth. Similar genetic signals and cell behaviors may be responsible for the independent and repeated evolution of barbel structures in other fish species.
Keywords: appendage, barbel, development, skin, zebrafish
Introduction
Skin is the boundary between an animal and its environment. At this interface, vertebrates have evolved many specialized appendages including scales, fins, teeth, feathers, and hair (Alibardi 2004; Wu et al. 2004; Fraser et al. 2008; Dhouailly 2009). Although anatomically and functionally diverse, all of these structures arise from local interactions between two layers of cells: the epidermis and the dermis (Chuong et al. 2000). Understanding epidermal-dermal interactions at the cellular and molecular levels is of interest for three reasons. Firstly, knowing how skin appendages develop may allow us to infer which are homologous, and how these appendages evolved from those of common ancestors (Musser et al., 2015). Secondly, the developmental mechanisms of appendage outgrowth are the ultimate source of appendage variation, including ontogenetic stages, sexual dimorphisms, or individual phenotypes. Finally, many of the cellular behaviors observed during appendage ontogeny, including cell proliferation, cell migration, and changes in gene expression, are re-used when appendages undergo wound healing or regeneration (Gurtner et al. 2008; Stappenbeck and Miyoshi 2009; Ansell et al. 2012; Murawala et al. 2012; Bielefeld et al. 2013). Therefore the study of epidermal-dermal interactions in the context of appendage development advances at least three inter-related fields simultaneously– those of phylogeny, ontogeny, and physiology.
Of the living vertebrates, fishes have some of the most diverse skin appendages. Within bony fishes, several species are intensively studied, including the zebrafish, medaka, and stickleback (Chen et al. 2004). In zebrafish, skin appendages include pharyngeal teeth (Crucke and Huysseune 2013), elasmoid scales (Mongera et al. 2013), the lateral line (Ghysen and Dambly-Chaudière 2004; Pichon and Ghysen 2004; Piotrowski and Baker 2014), several unpaired fins (Carney et al. 2010), and the nasal and maxillary barbels (LeClair and Topczewski 2010; Duszynski et al. 2013). Among these, the mechanisms of barbel development are of special interest due to the morphological diversity of these structures, temporal variation in their ontogenetic appearance, and multiple gains and losses in fish evolutionary history (Fox 1999). Figure 1 compares the barbels of two fishes, the zebrafish (Danio rerio) and the channel catfish (Ictalurus punctatus). Each fish has a different anatomical arrangement of facial barbels (two pairs vs. four), which appear at different stages of development (juvenile vs. embryo). Histologically, these appendages contain overlapping, but not identical cell types; catfish barbels are supported by cartilaginous rods, whereas zebrafish barbels are not (LeClair and Topczewski 2010; Hawkins 2011). This unique pattern leads to questions of both ontogenetic and phylogenetic processes. Specifically, how do the ectodermal and mesodermal layers of fish skin accomplish the localized extension of these elongated appendages? How often have certain developmental mechanisms been used, within bony fishes, to accomplish so many massively parallel adaptive events?
Figure 1. Evolutionary and developmental context of barbel development in several species of ray-finned fishes (Actinopterygii).
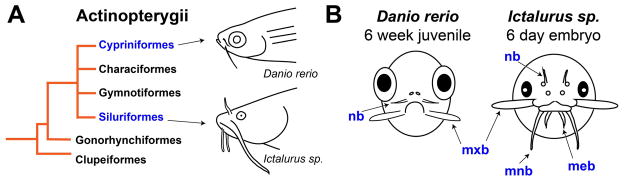
A) Simplified diagram of actinopterygian phylogeny. Two barbelled species within this clade are the zebrafish (Danio rerio) and the channel catfish (Ictalurus punctatus).
B) Comparative schematic of barbel development in D. rerio and I. punctatus. The zebrafish, left, develops two pairs of barbels as a juvenile, approximately 4–6 weeks after fertilization. The catfish, right, develops four pairs of barbels while still an embryo (figure modified from Hawkins, 2011). meb = mental barbels; mnb = mandibular barbels; mxb = maxillary barbels; nb = nasal barbels.
Wnt proteins are highly conserved extracellular ligands that drive many ontogenetic processes, including skin appendage development (Logan and Nusse, 2004; Widelitz, 2008; Chien et al., 2009). Canonical Wnt signaling causes cytoplasmic stabilization and nuclear localization of β-catenin, activating gene transcription through T-cell factor/lymphoid enhancer factor (TCF/LEF) proteins (MacDonald et al., 2009.) Through a different network of cytoplasmic events, Wnts can also signal non-canonically (Sugimura and Li 2010). Experiments in avian and mammalian models have shown that both canonical and non-canonical Wnt signaling have highly conserved roles in dermal differentiation, epidermal-dermal induction, and skin appendage patterning (Huelsken et al., 2001; Andl et al., 2002; Rinn et al., 2008; Widelitz, 2008; Mikkola, 2007; Tran et al., 2010). For example, if extracellular inhibitors of Wnt binding are overexpressed in the epidermis or dermis, abnormal skin and arrested appendages can result (Chodankar et al. 2003; Liu et al. 2007). Likewise, disrupting the intracellular components of canonical Wnt signaling, including adenomatous polyposis coli (APC, Kuraguchi et al., 2006), β-catenin (Huelsken et al., 2001), or TCF/LEFs (Niemann et al., 2002; van Genderen et al., 1994) can also cause skin anomalies. We therefore predicted that Wnt signaling might have a primary role in establishing and maintaining the zebrafish maxillary barbel, affecting dermal cell proliferation and/or matrix production during barbel ontogeny.
Although there has been one recent molecular survey on the ontogeny of catfish barbels (Hawkins 2011), there are no comparable studies for similar structures in zebrafish. To better understand how the maxillary barbel develops, our goal was to study cell shape, cell proliferation and gene expression during outgrowth of this appendage. Our results suggest that, in contrast to other vertebrate appendages, the maxillary barbel forms without any corresponding ectodermal placode or apical ectodermal ridge (AER). Cell division in the developing barbel is also haphazard, showing no localized growth zones. In contrast, we detected a strongly labeled, single-layered subpopulation of TCF-responsive cells within the barbel core. The fluorescent labeling of these cells had a very strong proximal-distal gradient, with maximal signal distally, that was maintained from the juvenile to the adult. Further studies confirmed that these cells, which have not previously been described, were minimally mitotic, but could be uniquely identified by their matrix-bound location, nuclear morphology and cytoplasmic structure. We conclude that we have identified a new cell population in zebrafish, here called the ‘TCF+ core cells’, which may be responsive to canonical Wnt signals. Importantly, this dermal population is uniquely placed to contribute to maxillary barbel outgrowth through enhanced extracellular matrix production.
Materials and Methods
Fish lines and husbandry
Three zebrafish strains were used in this study: a wildtype AB strain (ZDB-GENO-960809-7), a transgenic strain expressing membrane-bound green fluorescent protein (Tg(Ola.Actb:Hsa.HRAS-EGFP); ZDB-GENO-061107-1 (Cooper et al. 2005), and a transgenic strain expressing a TCF-responsive nuclear-localized mCherry, an indicator of canonical Wnt signaling (Tg(7xTCF-Xla.Sia:NLS-mCherry); ZDB-GENO-110113-3 (Moro et al. 2012). These strains are hereafter called wild type, mGFP, and TCFsiam, respectively. DePaul University’s Institutional Animal Care and Use Committee (IACUC) approved all experimental protocols.
Juvenile and adult zebrafish were housed in a recirculating system filled with dechlorinated tap water (pH 7.4) at 28–29°C. Light was provided on a 14 hr:10 hr day:night cycle. Sexually mature adults were pair-bred in crossing tanks and fertile eggs collected using standard methods (Westerfield 2000). 6–7 days after hatching, embryos were transferred to ~200 mL static tanks in a 28°C incubator and fed live rotifers and/or larval powder twice daily (Larval Diet LD100; Aquatic Ecosystems). When of sufficient size, larvae were moved to tanks in the recirculating rack and fed live brine shrimp and/or commercial fish flakes (TetraMin Tropical Flakes) for the remainder of the study. For all surgical procedures, fish were anaesthetized in 0.015% Tricaine (MS-222, Fisher Scientific: AC11800-0100) buffered to pH 7.0 in system water. For euthanasia, fully anesthetized fish were submerged in ice water until opercular movements were no longer observed.
Developmental staging and tissue collection
Although there is a single nomenclature for zebrafish embryonic development (Kimmel et al. 1995), there are multiple proposed systems for juveniles and adults (Parichy et al. 2009; Singleman and Holtzman 2014). Postembryonic staging can be complex because there are more structures to observe, as well as increased variability among structures. In this study, we used a simple, straight-line distance measure, standard length (Howe 2002). This variable is rapidly measured, and the relationship between standard length and barbel length has already been mapped in a large (n >100) series of wildtype zebrafish juveniles (LeClair and Topczewski 2010). Because these two morphological variables are strongly correlated, either measure can be used to indicate relative developmental stage.
To obtain a complete developmental series of maxillary barbels including pre-barbels, barbel buds and early-elongating appendages, wildtype and mGFP juveniles were collected at intervals from 4–8 weeks post-fertilization. After euthanasia, they were fixed whole in either Dent’s fixative (80% methanol : 20% DMSO) or 4% paraformaldehyde in phosphate-buffered saline (PF-PBS, pH 7.4) for 2–4 hours at room temperature or overnight at 4°C. After fixation, the specimens were rinsed in 1x phosphate buffered saline (PBS) and each fish was measured under a dissecting microscope from the anteriormost tip of the lower jaw to the posteriormost margin of the trunk musculature along the central axis of the caudal fin. Each fish was then sorted into one of four arbitrary body size classes: 1) pre-barbels (SL < 10 mm), 2) barbel buds (SL = 10–12.5 mm), 3) elongating barbels (SL = 12.5–15 mm) and 4) mature barbels (SL >15 mm). Both maxillary barbels were then dissected off for immediate processing, or dehydrated to 100% methanol for storage at −20°C.
in vitro EdU labeling of zebrafish tissue explants
To identify cells in S-phase during maxillary barbel development, we developed a method for whole-mount EdU labeling of zebrafish tissue explants in vitro. This protocol was adapted from prior studies using EdU to label cell proliferation in whole chick embryos (Warren et al. 2009) and isolated Drosophila brains (Gouge and Christensen 2010). As a positive control for successful mitotic labeling, we also cultured explants of the regenerating adult caudal fin, as its patterns of in vivo cell proliferation have been well described (Iovine 2007; Kizil et al. 2009). A detailed description of our control and experimental treatments is given below.
To prepare the control tissue, we excised the distal caudal fins from groups of 6 adult wildtype fish on each of 4 consecutive days, designated days 3, 2, 1 and 0. Fish prepared on days 3, 2 and 1 were returned to the recirculating system and allowed to regenerate until day 0 tissue collection. Fish prepared on day 0 were collected while still anaesthetized, immediately after the tail excision surgery. Overall, 24 adult tails were examined. Also on day 0, we collected 12 wildtype siblings. These fish had standard lengths of 10–15 mm, and each provided two maxillary barbels in various stages of development, for a total of 24 barbel appendages. Using sterile technique in a flow-through biocontainment hood, the regenerating blastemas of the adult tails and the maxillary barbels of the juveniles were dissected off and placed, 2–3 tissue pieces per well, in a 48-well plastic tissue culture plate. Each experimental well held 200 μL of pre-warmed (28°C) sterile Leibovitz-15 culture media (pH 7.4; VWR: 89222-116), supplemented with 3% fetal bovine serum (FBS) and antibiotics (100 U/mL penicillin and 100 ug/mL streptomycin). This mixture was called ‘complete L-15’. Next, a 2x working solution of EdU was freshly prepared by thoroughly mixing 1 part EdU stock solution (10 mM) with 98 parts complete L-15 and 1 part DMSO (= 1000 μM EdU, 1% DMSO). Finally, 200 μL of this 2x EdU solution was added to each experimental well holding the tissue. The final volume in each well was therefore 400 μL, and the final concentration was 500 μM EdU : 0.5% DMSO. As a negative control, the EdU working solution was not added to several experimental wells containing either barbel or tail tissue.. To allow EdU uptake, the covered, sterile plate was placed in a dark, humidified air incubator (28°C) for 4 hours. After this interval, most of the solution in each well was carefully removed from around the tissue fragments with a P-1000 pipettor and a narrow gel-loading tip. Each well was then washed two times with 0.5 mL of 1x PBS. Finally, 0.5 mL of freshly thawed 4% PF-PBS fixative was added to each well and the plate was placed at 4°C overnight.
Whole-mount detection of the EdU-labeled explants followed the manufacturer’s instructions (Click-iT EdU Imaging Kit, Invitrogen: C10339). The fixed tissues in each well were washed 2–3 times in 0.5 mL blocking buffer (3% BSA in PBS), soaked 1 hour in 0.5 mL permeabilization buffer (1x PBS + 0.5% Triton-X), and rinsed 2–3 times in 0.5 mL blocking buffer. After excess wash was removed, 300 μL of freshly prepared ‘Click-iT’ reaction mix was added to each well. In this step, copper-catalyzed ‘click’ reaction occurs in which a fluorophore-conjugated azide binds irreversibly to an alkyne group of EdU (Chehrehasa et al. 2009). To insure adequate mixing during this light-sensitive step, the plate was wrapped in foil and gently agitated on a titer-plate shaker for 30 minutes at room temperature. The reaction cocktail was then removed. After two washes in 0.5 mL blocking buffer and one wash in 0.5 mL PBS, nuclei were then counterstained by adding 250 μL of Hoechst 33342 solution to the PBS in each well (final concentration = ~5 μg/mL). After another 30 minutes of agitation in the dark, excess Hoechst was removed by 2–3 additional PBS rinses. Finally, the tissue fragments were pipetted out of the wells, cleared in 70% glycerol, mounted on microscope slides and imaged using confocal microscopy. Emission/excitation wavelengths were 590/615 nm for the EdU fluorophore (Alexa Fluor 594) and 350/461 nm for the Hoechst nuclear label, respectively.
Microscopy, photography, image processing and video formatting
To detect and colocalize fluorescent signals, tissue samples were examined using an epifluorescent microscope (Nikon Eclipse 50i) and/or a laser scanning confocal (Leica Microsystems SPE; Model # DMI4000). Images were exported and archived as individual TIF files. Single TIFs were processed in Adobe Photoshop CS5 using standard tools to adjust image sharpness, brightness, and contrast. Z-stacks of TIFs were imported into FluoRender, a free confocal imaging program (www.sci.utah.edu/software/fluorender.html), and three-dimensionally rendered using the software tools available. Step-by step Fluorender workflow protocols are available on request. To produce animations, rendered images series were exported from Fluorender as JPEG files and assembled into annotated movies using Movavi Video Suite.
Results
Barbel outgrowth: changes in cell shape and distribution
Skin appendage morphogenesis involves regulated cell rearrangements of both epithelium and mesenchyme (Osterfield et al. 2013) Certain embryonic structures are found repeatedly in vertebrate ontogeny, and represent current archetypes of how appendages are organized. Examples include epithelial placodes, which are important for the location of superficial structures such as hair, teeth, feather tracts and sensory ganglia (Widelitz and Chuong 1999; Chodankar et al. 2003; Mikkola and Millar 2006; Nechiporuk et al. 2007) and apical ectoderm ridges (AERs), which are found on developing tetrapod limb buds and fish fins (Saunders 1948; Kosher et al. 1979; Nomura et al. 2006; Yano et al. 2012). In this study, we used transgenic zebrafish expressing membrane-bound green fluorescent protein (mGFP) to observe changes in cell shape and distribution during ZMB morphogenesis. GFP expression in this line is randomly mosaic; for these experiments, fish lines were screened for strong expression in the epidermis, as opposed to the dermis. This allowed us to distinguish the two layers, and, in combination with nuclear staining, observe the general shape and distribution of epidermal cells. Specifically, we sought to identify any epithelial reorganization, such as a placode or ridge, which would precede barbel outgrowth.
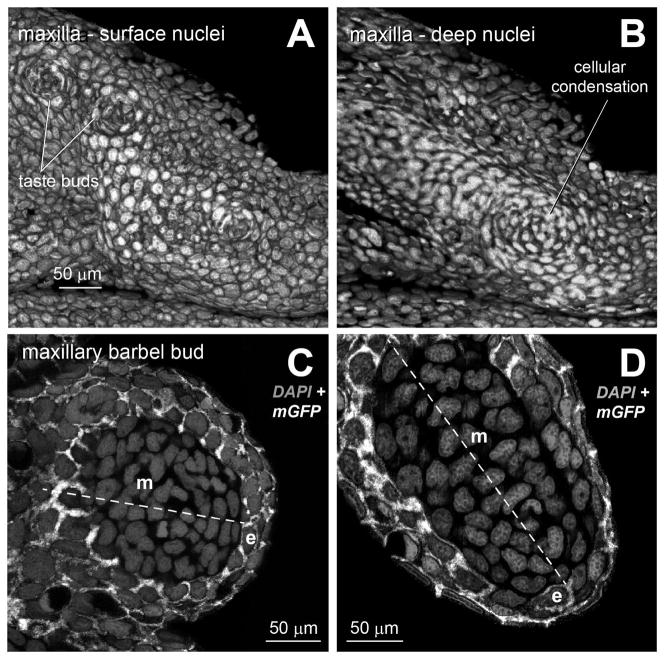
Forty-eight mGFP zebrafish were sorted into four categories of standard length (SL): <10 mm (n = 16), 10–12.5 mm (n = 14), 12.5–15 mm (n = 12), and >15 mm SL (n = 6). Budding of the ZMB is first observed at approximately 10 mm SL; therefore, the majority of specimens were at this stage or earlier. In juvenile zebrafish without external protrusions of the barbels (SL < 10 mm), the lateral surface of DAPI-stained maxillae appeared relatively featureless (Fig. 2A). Apart from superficial taste bud rosettes and epidermal cells concentrically arranged around mucous glands, there were no obvious epithelial specializations. Using confocal microscopy, however, we identified in the dermis of the maxilla a distinct whorl of nuclei (Fig. 2B). This cellular condensation may represent the first sign of the future appendage. As the maxillary barbel bud extends, the epidermal cells form a stratified layer 1–2 cells thick. These cells are round or slightly flattened, as indicated by the membrane-bound GFP signal (Fig. 2C). In larger barbel buds (~300 microns long), the most distal epidermal cells are nearly flat, with nuclei arranged tangential to the convex dermal core (Fig. 2D). In mature barbels (>1 mm long), the epidermal cells form a stratified cuboidal epithelium with 3–4 layers of cells throughout, as previously described (LeClair and Topczewski 2010). At no stage did we observe any local thickening of the epidermis, typical of an epithelial placode, nor any collection of distally elongated epithelial cells, characteristic of an AER.
Figure 2. Ectodermal and mesodermal arrangements in the maxillary barbel bud.
All images are 3D renderings of confocal Z-stacks from fixed tissues. All cells are stained with DAPI (gray) to show nuclear morphology and cell arrangement. In all panels, anterior/proximal is to the left.
A) Lateral view of the surface of a developing maxilla from a juvenile zebrafish (standard length < 10 mm) at the prospective site of barbel outgrowth. Taste buds appear as cellular rosettes.
B) Deep confocal slice of the same specimen. The presumptive barbel is marked by a cellular condensation of dermal mesenchyme.
C) Optical section through an early maxillary barbel bud (~100 μm). The proximal-distal axis is dotted, with distal to the lower right. In this transgenic specimen, epithelial cells (e) express membrane-bound EGFP (white). The epithelial layer of the barbel bud (e) contains 2–3 layers of cuboidal or slightly flattened cells. The mesenchymal cells (m) form a whorl around the bud center.
D) Optical section through an elongating barbel bud (~300 μm). At this stage, the distalmost epithelial cells (e) are more squamous, with tangentially flattened nuclei. e = epithelial cells; m = mesenchymal cells
Barbel outgrowth: patterns of cell proliferation
Appendage outgrowth is the net result of cell distribution and cell proliferation (Zeller et al. 2009). Cell proliferation is required for organized growth; however, there are different spatial patterns in which proliferation occurs. An apical model predicts that the most distal cells will divide most rapidly, (Boehm et al. 2010), whereas a basal model predicts the opposite (Milán et al. 1996). A distinct sub-apical region of concentrated cell division can be called a “progress zone” (Gibson-Brown et al. 1998); alternatively, or cell divisions can be spatially random (Baena-López et al. 2005). Each patterns of proliferation suggests a corresponding distribution of developmental signals locally controlling the cell cycle.
To analyze cell proliferation in the developing ZMB, we collected barbel tissues from three body size classes (10–12.5, 12.5–15, and >15 mm SL) and explanted them for 4 hours in a sterile culture medium containing EdU (see Methods). After detection, we were able to map the labeled cells in this outgrowing structure, a phenomenon not previously described. Similarly-treated positive control tissues (the distal ends of regenerating adult caudal fins) were also successfully labeled, showing intense signal in nuclei of the blastema (not shown). In early-developing maxillary barbels (10–12.5 mm SL) we saw scattered EdU labeling in both epithelial and mesenchymal layers throughout the appendage (Fig. 3A). These labeled cells had a haphazard or random pattern. Similarly, barbels from slightly more developed fish (12.5–15 mm SL) had both proximal and distal regions labeled with dispersed, EdU-positive cells (Fig. 3B). We noted slightly increased mitotic labeling on the ventral surface of the barbel, where there are numerous epithelial taste-bud clusters. In the largest barbels examined (>15 mm SL), this dispersed pattern of cell labeling was essentially unchanged (Fig. 3C). Overall, the gross morphology of the cultured explants appeared excellent, and optical sectioning of the stained specimens showed bright, crisp nuclear labeling in both superficial and deep cell populations. This indicates that, during the 4-hour culture period, a large number of cells remained viable and were able to complete S-phase. Further, it shows that the EdU label effectively penetrated the skin and basement membrane, even in the absence of physiological circulation or mechanical fluid mixing.
Figure 3. Patterns of cell division in the developing zebrafish maxillary barbel.
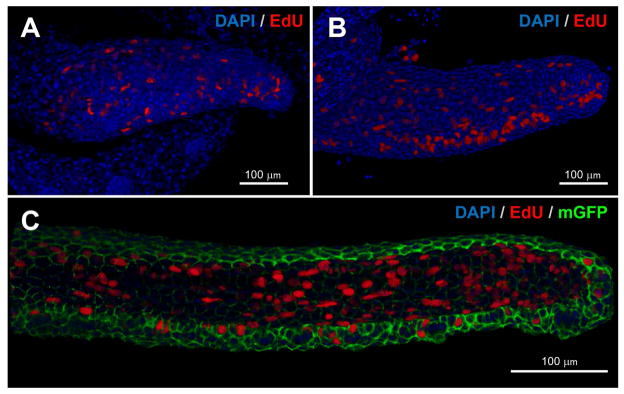
Whole-mount in vitro mitotic labeling of explanted maxillary barbel buds. Nuclei are stained blue; cells in S-phase (EdU+) are also red. Panels A and B are depth-sensitive full-thickness image renderings, in which superficial nuclei appear brighter than deeper ones. Panel C is a similar rendering; for clarity, the membrane-GFP signal has been removed from the superficial layers.
A) Mitotic labeling of a wildtype juvenile barbel (~300 μm). Dividing cells are scattered throughout the appendage.
B) Mitotic labeling of a slightly longer barbel appendage (~500 μm). Clusters of dividing cells on the ventral side of the barbel correspond to the developing taste buds.
C) Mitotic labeling of an elongated juvenile barbel (~1 mm) from a membrane-EGFP transgenic zebrafish. Labeled nuclei are found in both the epithelial and mesenchymal layers throughout the appendage.
A Wnt/TCF reporter line labels a subset of ZMB core cells
Wnt signaling is crucial for many developmental processes, but particularly dermal differentiation (Widelitz 2008; Tran et al. 2010). Several zebrafish lines have been engineered to express fluorescent reporters downstream of Wnt pathway activation (Dorsky et al. 2002; Moro et al. 2013). Of these, we chose the recently-developed TCFsiam:mCherry line, which provides a readout of canonical Wnt signaling, through TCF activity in affected nuclei (Moro et al. 2012). This line faithfully mimics all known embryonic Wnt signaling domains; however, there are no studies using this line in juveniles or adults. By observing this line at various stages of barbel outgrowth, we sought to detect any local activation of this key genetic pathway.
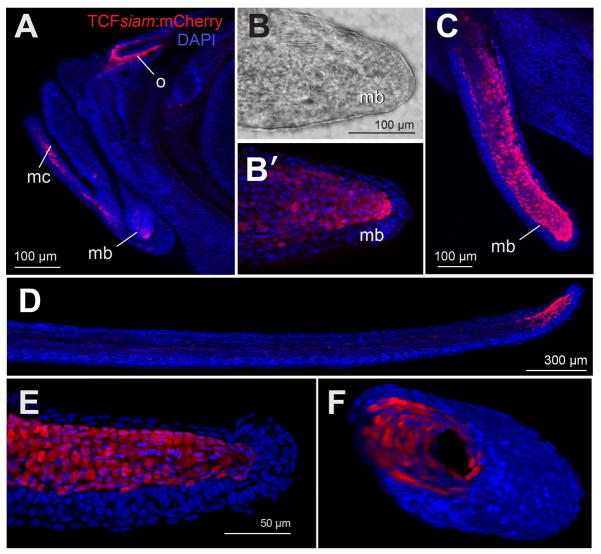
We raised hemizygous TCFsiam zebrafish to adulthood and outcrossed them to wild types. At 24 hours post-fertilization, we screened the progeny for strong mCherry expression in the tailbud nuclei, a known domain of Wnt activity. All confirmed mCherry+ transgenics were then allowed to grow for at least 4 weeks prior to collection, screening, and counterstaining of the maxillary barbel tissue. Juvenile TCFsiam zebrafish had strong nuclear mCherry in cells of the olfactory pit, mandibular cartilage, and maxillary barbel bud (Fig. 4A). Magnification of the bud domain showed several dozen mCherry+ nuclei arranged in a cone-shaped sleeve over the barbel core (Fig. 4B, B′). At later stages of barbel outgrowth, a similar distribution of nuclei was observed within the distal two-thirds of the appendage (Fig. 4C). In adult maxillary barbels, there was little or no fluorescent signal along the barbel shaft; however, a strongly-labeled subpopulation remained at the distal end (Fig. 4D). Volume rendering showed that, at all stages examined, the mCherry+ nuclei were radially flattened, circumferentially distributed, and occupied a single, sub-epidermal layer (Fig. 4E, 4F, and Movie S1). These signals identify a novel subpopulation of maxillary barbel cells that is continuously responsive to TCF, a component of canonical Wnt signaling. These cells are present from the early bud stage to the adult appendage.
Figure 4. TCFsiam:mCherry reporter expression in zebrafish maxillary barbel tissues.
A) Overview of a juvenile zebrafish head. All nuclei are blue; TCF+ nuclei are also red. Strong nuclear mCherry signal was detected in cells of the olfactory pit (o) and Meckel’s cartilage (mc). A third small domain is in the area of the early maxillary barbel bud (mb).
B) Transmitted light micrograph of an early maxillary barbel bud (mb).
B′) The same specimen as B, showing a subepithielial cone of red fluorescent nuclei.
C) An elongated juvenile maxillary barbel (mb). A “sleeve” of red nuclei occupies the distal two-thirds of the appendage. Signal is weaker in the proximal regions.
D) An adult maxillary barbel. mCherry+ cells are highly concentrated at the distal tip just under the epidermis. Only scattered proximal cells express the label.
E) Magnification of the distal tip of a juvenile barbel. The mCherry+ nuclei are found in a single layer underneath the epidermis.
F) Volume rendering of the mCherry+ cells in a juvenile maxillary barbel. In this panel, the volume is rotated and cleaved to show an oblique cross-section. Red nuclei are arranged in a single, circumferential layer just underneath the barbel epithelium (blue), and directly surrounding the acellular barbel core (black).
TCF+ cells of the ZMB are not highly mitotic
Wnt ligands can have multiple cellular effects, inducing coordinated changes in gene expression, cell morphology and behavior. Having identified the TCF+ core cells by their reporter gene activity, we next wanted to understand their mitotic behavior. Specifically, we wished to explore if these were rapidly-cycling cells that might provide a pool of undifferentiated precursors to the developing appendage. To test this, we repeated our in vitro EdU-labeling technique on freshly explanted barbel tissues from this transgenic line. In these experiments, the dissection and labeling procedures were identical to those previously performed, except that EdU detection was performed with an AlexaFluor 488-conjugated azide, staining S-phase nuclei green. All double-labeled cells (mCherry+/488+) would indicate TCF+ cells that were also in S-phase during the 4-hour labeling interval.
Confocal imaging of the stained, transgenic barbels confirmed a single, deep layer of mCherry+ nuclei circumferentially arranged around the central core (Fig. 5A). We also detected intense EdU labeling in many cells, but most of these were green-only and were more superficially located. These abundant, superficial EdU+ cells likely represent basal epithelial cells, a rapidly-dividing subpopulation of the epidermis. Some of the mCherry+ cells were double-labeled (mCherry+/EdU+, Fig. 5B, C and D); however, these were a small minority of the total population. We conclude that the TCF+ barbel cells have some mitotic activity during the stages examined. However, relative to surrounding cells, they infrequently divide.
Figure 5. Co-localization of a canonical Wnt reporter (TCFsiam:mCherry) with EdU-488 mitotic labeling using in vitro explant cultured maxillary barbels/.
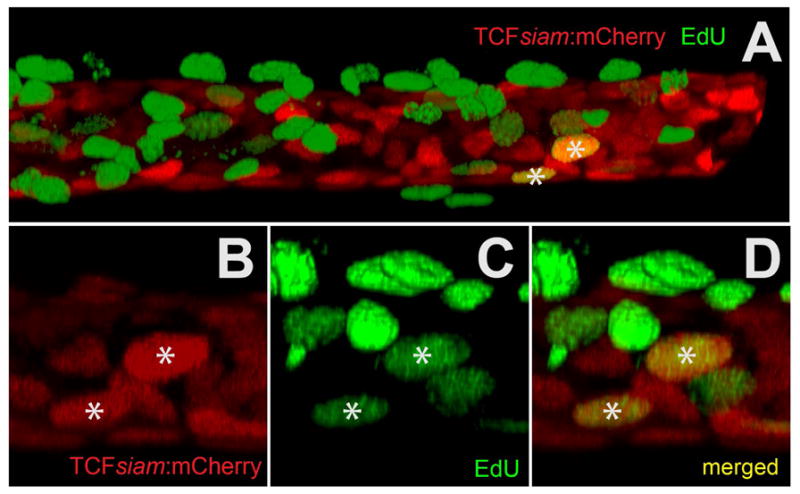
A) Two-channel rendering of a developing zebrafish maxillary barbel; the distal end, which is digitally truncated, points right. mCherry+ cells (red) occupy a cylindrical, mesenchymal sheath surrounding the barbel core. Numerous EdU+ cells (green) are superficial to this layer, indicating maximal cell division in the overlying epithelium. Only two ventral cells are double-labeled (mCherry+/EdU+).
B) mCherry+ cells from a second double-labeled specimen.
C) EdU+ cells (green) from the same region as B.
D) Merged image of B and C. The two yellow nuclei (*) are double-labeled.
Ultrastructure of the TCF+ core cells
Using nuclear-localized reporter genes and mitotic staining methods, we identified a TCF+, slow-dividing cell population within the ZMB. This nuclear staining alone, however, did not provide a complete picture of this new cell type or its developmental role. As previously described, ZMB extension involves the accumulation of dense, non-mineralized matrix fibers within the dermal core of the appendage (LeClair and Topczewski 2010). These condense to form the central rod, which is the anatomical supporting structure and mechanical lever arm for the appendage Histologically, the central rod appears largely acellular, and the cell population that forms it is not known. We therefore wanted to locate the TCF+ cells with respect to other barbel structures, particularly the central rod and the epidermal-dermal boundary.
To obtain maximum resolution for these studies, we performed transmission electron microscopy on cross-sections of adult barbel tissue. Within these sections, we attempted to identify the TCF+ population based on our prior observation of the sub-epithelial, circumferential arrangement and strongly flattened nuclear morphology of these cells. In each highly magnified maxillary barbel cross-section, we observed a ring of cells with these features. Assuming that these are the same cells identified by the TCFsiam:mCherry nuclear label, we will refer to them as the ‘TCF+ cells.’ In the following description we review the structural features of the maxillary barbel and the histological context of this novel cell type.
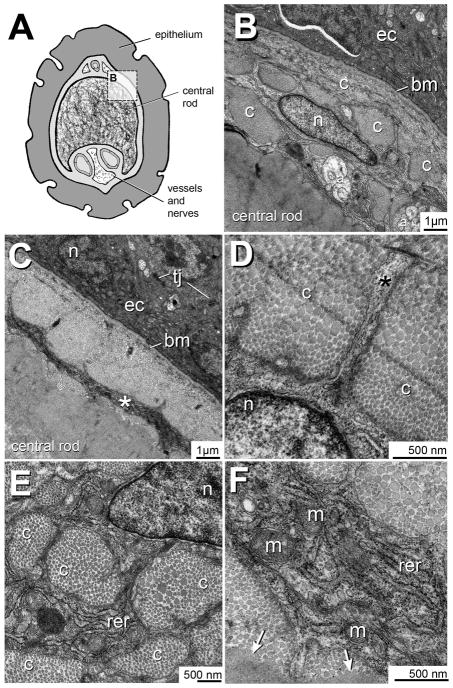
The ZMB in cross-section presents a dermal core of cells and matrix surrounded by a multilayered epithelium (Fig. 6A). The epithelial layer features an electron-dense cytoplasm and abundant tight junctions between adjacent cells (Fig. 6B). These cells rest on a prominent basement membrane with a thick collagen underlayer. This membrane defines the epidermal-dermal boundary. The TCF+ cells occupy the subadjacent dermal compartment and are easily identified by their large, flat nuclei. Surrounding each nucleus are ribbons of convoluted cytoplasm in close contact with surrounding collagen bundles. Notably, each of the TCF+ cells is entirely embedded within the matrix of the central rod, approximately 2–3 microns below the rod surface.
Figure 6. Transmission electron microcopy of the TCF+ ZMB core cells.
A) Cross-sectional overview of the adult zebrafish maxillary barbel. The rectangular boundary indicates the region magnified in panel B.
B) The maxillary barbel epidermal-dermal boundary. Ectodermal cells (ec) are electron-dense and rest on a prominent basement membrane (bm). Approximately 2 microns below this membrane lies the radially-flattened, pancake-shaped nucleus (n) of a TCF+ core cell. Adjacent to the cell are several collagen bundles (c). Deep to the cell are several axons (a) and the acellular matrix of the barbel’s central rod.
C) Micrograph of a similar region. The electron-dense epithelial cells (ec) are connected by tight junctions (tj). Approximately 2–3 microns below the basement membrane is a layer of cell cytoplasm (*) that appears as a continuous, circumferential ring.
D) A cytoplasmic projection (asterisk) extends from a matrix-bound cell through a dense field of collagen fibers (c).
E) Several cross-cut collagen bundles (c) near a TCF+ cell. The heterochromatic cell nucleus (n) is at the upper right. Between the bundles are ribbons of cytoplasm enriched in rough endoplasmic reticulum (rer). The small, dark nodules are individual ribosomes.
F) Magnification of a cytoplasmic ribbon shows extensive rough endoplasmic reticulum (rer) and intracellular vesicles. The double-wrapped ovoid organelles are mitochondria (m). Collagen fibers close to the cell appear well separated, showing individual ovoid cross-sections. In contrast, the collagenous matrix farther away from the cell appears hyperpolymerized (arrows), with no individual fibrils present.
a = axon; bm = basement membrane; c = collagen bundle; ec = ectodermal cell; m = mitochondria; n = nucleus; rer = rough endoplasmic reticulum; tj = tight junction.
In sections not intersecting TCF+ cell nuclei, we observed a circumferential ring of matrix-bound cytoplasm at a similar depth (Fig. 7B). Within this ring, membrane-localized tight junctions indicate overlapping cytoplasmic projections from multiple cells. Although most of these projections were circumferential, there are occasional radial or oblique extensions. These blunt or tapered arms extend up to one micron into the surrounding matrix (Fig. 6D). Within each TCF+ cell we observed a dense granular cytoplasm enriched with rough endoplasmic reticulum (Fig. 6E). Finally, deep to the TCF+ cells lies a broad region of acellular collagenous matrix, representing the remainder of the barbel core. Interestingly, the matrix immediately adjacent to the TCF+ cells appeared more electrolucent, in contrast to this central region, which was more electron-dense. Collagen bundles near the TCF+ cells appeared loosely packed, with well-separated circular cross-sections through individual triple helices. In contrast, the matrix distant from the cells appeared more homogeneous with few or no inter-fibrillar gaps (Fig. 6F). This may indicate hyperpolymerization of collagen towards the more central regions of the appendage.
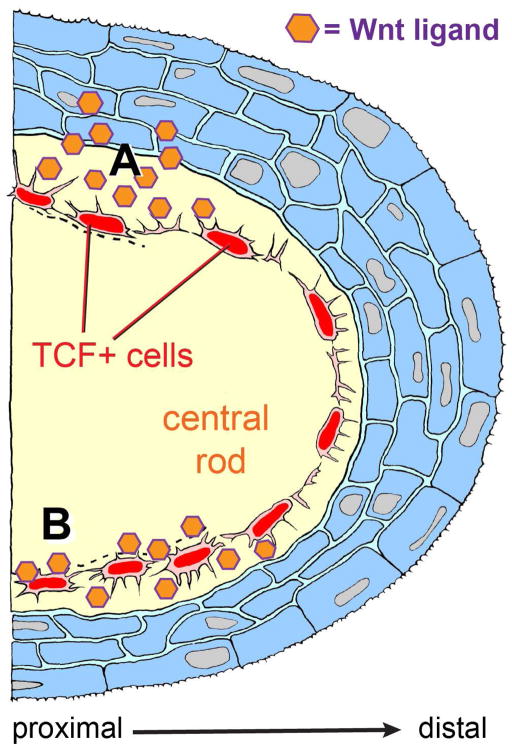
Figure 7. Making maxillary barbels with Wnt signals: location of a new dermal cell population, and alternative models of appendage outgrowth.
Diagrammatic section through a developing maxillary barbel bud, distal end to the right. An outer later of stratified epithelium (blue) covers a convex dermal core. Within the core is a mass of acellular matrix (yellow), in which is embedded arranged a single layer of Wnt-responsive cells (the TCF+ core cells, red). These cells contact each other circumferentially via overlapping cytoplasmic bridges; smaller cytoplasmic projections extend radially into the surrounding collagenous material. Activated TCF+ cells divide rarely, but increase secretory activity, depositing collagenous matrix to widen and lengthen the appendage. A) Paracrine activation: Wnt ligands secreted by the overlying epidermis activate the TCF+ core cells. B) Autocrine activation: TCF+ cells maintain their own activation by secreting Wnt ligand(s) that are bound to the surrounding matrix. For simplicity, other cell types within the dermal compartment (e.g., blood vessels, peripheral nerves, and melanophores) are not shown.
Discussion
Evolution and morphogenesis of skin appendages
In this study we provide several new observations on the development of barbels, a widely distributed structure in amphibians, reptiles, and fishes (Fox 1999; Volff 2005; Kasumyan 2011). Urodele tadpoles, for example, have a pair of rod-like structures, known as balancers, which develop in the branchial region and are used to attach to underwater substrates (Brunelli et al. 2007). Erpeton tentaculus, an aquatic snake, has a pair of highly innervated cranial tentacles that sense prey (Quint et al. 2002; Catania et al. 2010). Barbel-like appendages are also common in teleost fishes, with almost half of the existing orders having one or more barbelled species (Hawkins, 2011). Such barbels can be paired or unpaired, can extend from different regions of the head or body, and can be sexually dimorphic (Fox,1999). Within the Cyprinidae, the clade to which zebrafish belong, some species have paired barbels and others lack them. These appendages are thought to have evolved repeatedly, representing either independent adaptations, or convergent regressions to an ancestral condition (Briolay et al. 1998). Although variability in barbel number, position, and function is well documented, there are few studies on the ontogenetic mechanisms by which these structures emerge. An understanding of how barbels originate at the cellular and molecular levels, using zebrafish as a model, may help to explain the diversity of barbel distribution and composition within cyprinids as a whole. Comparative developmental studies of other barbelled fishes, including both laboratory- and commercially-reared fish species, may illuminate more generally the mechanisms by which barbels have evolved repeatedly in many lineages of fish phylogeny.
Changes in cell shape and distribution
Local and regional cell-shape changes are often critical first steps in the organization of appendages. The classic discovery of epithelial-mesenchymal organizing centers, such as the apical ectoderm ridge, or AER, strongly inform research and reviews of appendage outgrowth (Widelitz et al. 1997). A similar mechanism has been observed during zebrafish fin development, in which an apical ectodermal ridge becomes an apical fold (AF), an organizing structure in the fin bud (Yano et al. 2012). Removal of the chick AER causes limb truncations (SAUNDERS 1948), whereas removal of the zebrafish AF causes mesenchymal elongation. These and other experiments have shown that epithelial ridges and folds are key molecular centers for appendage development, and that ectopic expression, genetic mutation, or chemical disruption can strongly affect the morphological result. Most broadly, the reciprocal interactions at these influential sites are thought to represent ancestral genetic “cassettes” or regulatory “modules” that have been used repeatedly in vertebrate phylogeny. It is important to note, however, that alternative methods of appendage development may occur, particularly in structures not homologous to limbs.
In this study, we observed that the first morphological sign of maxillary barbel outgrowth is a mesenchymal condensation within the dermis of the maxilla. However, there is no corresponding epidermal structure, such as a placode or AER. Rather, the distal epithelial cells of the barbel remain rounded or become progressively flattened as the appendage grows. Outgrowth of the ZMB thus differs from vertebrate limb development, but appears morphologically similar to the barbels of the channel catfish (Hawkins 2011) or the balancers of urodele tadpoles (Brunelli et al. 2007). These latter structures have a rounded epithelial cover at the slightly bulbous or cone-shaped distal end, but no particular ridge or other apical structure. How, then, do these structures receive the signal to grow? Lacking an AER, these appendages may receive local genetic signals that are functionally equivalent. Gene expression studies in catfish barbels, for example, demonstrate that transcripts for FGFs, Shh, Dlx and other key ligands can be spatially restricted even within the continuous epithelium of the developing appendage (Hawkins 2011). Therefore one or more signaling molecules may yet be discovered that instruct the areas from which zebrafish barbels eventually grow.
Patterns of cell proliferation
Appendages reach their final shape and size through coordinated cell division, cell migration, and cell death. Of these, we examined the cell division patterns in the maxillary barbel to determine whether they were concentrated proximally or distally, restricted to a “progress zone”, or randomly arranged. Similar techniques have been used on other developing zebrafish structures, leading to more precise models of their growth mechanics. Using 5′-bromo-2′-deoxyuridine (BrdU) to label dividing cells during juvenile caudal fin development, (Goldsmith et al. 2006) showed that cells in the dorsal and ventral fin rays proliferate faster than cells in the central fin rays. This different in rate causes the blunt-ended larval structure to grow into the characteristic bi-lobed shape of the adult. More recently, in vivo EdU labeling was used to locate mitotic cells in the embryonic zebrafish notochord, which are concentrated at the distal end (Sugiyama et al. 2009). Mapping local patterns of cell division can thus identify highly mitotic precursors that contribute maximally to appendage outgrowth.
In this report, we used an in vitro explant culture system to label patterns of cell proliferation in the developing ZMB. In contrast to the zebrafish caudal fin and notochord, in which there is a strong positional gradient of cells in S-phase, there is no similar pattern in the maxillary barbel. Over a tenfold increase in barbel length– from 100 microns to 1 mm– labeled nuclei appeared evenly distributed in proximal and distal areas of this appendage. Most of these cell divisions were in the basal layer of the epithelium, as expected for a healthy layer of skin. Occasional divisions were detected in deeper cells, including the novel “core cells” described here. However, these were not localized along any appendage axis. This suggests that the developing maxillary barbel has a “pan-appendage” growth pattern.
An alternative method of EdU labeling in zebrafish is peritoneal injection of the compound. However, this requires more expertise in animal handling, two handling procedures per fish (once at administration and once at collection) and more EdU per fish to label all body tissues, not all of which are collected or used. In our short-term plate-based assay, we achieved good EdU labeling of small, whole tissue fragments using minimal reagent, while exposing each fragment to the same concentration of label. Tissue integrity was maintained well, and all fragments yielded good quality nuclear staining. Although the long-term physiology of explanted tissues may not be identical to the corresponding tissues in vivo, the short-term capacity to see and manipulate cellular processes is enhanced. Historically, L-15 explant culture has been used to maintain fragments of fish and amphibian tissues– including gonad (Bonnin 1975), thyroid (Bonnin 1971), pancreas (Pouyet and Beaumont 1975), and gill (Avella et al. 1994)– for periods of days to weeks. Similar techniques may be of interest to researchers wishing to simultaneously label and/or visualize cellular events in a convenient plate-based format.
Fishing for Wnts; identification of a novel, TCF+ mesenchymal cell population in maxillary barbel development
All appendages arise from interactions between two adjacent layers: an outer epithelium and an inner mesenchyme. A key evolutionary question, then, is how the same two layers interact to produce diverse skin appendages in different animal species. Embryologically, the trunk and tail dermis comes from the dermatomal layer of the somite (Widelitz 2008), whereas the head dermis, which forms anterior appendages such as barbels, comes from cranial neural crest cells (Tran et al. 2010). In general, the dermis directs the size, shape, and distribution of skin appendages by exchanging signals with the epidermis (Olivera-Martinez et al. 2004). Highly conserved molecular pathways such as wingless (Wnt), fibroblast growth factor (FGF), bone morphogenetic protein (BMP), and Hedgehog (Hh) mediate these dermal-epidermal interactions and are essential for the proper formation of mammalian hairs (Alibardi 2005; Sennett and Rendl 2012), avian feathers (Ny et al. 2005), and reptilian scales (Chang et al. 2009). Therefore, it is thought that variations in the timing and expression of these signals produce an “appendage spectrum” of novelty and diversity across animal taxa.
The transparent ZMB is an ideal context in which to observe the genetic signaling of these adjacent cell layers, using the optical tools of the zebrafish model. In this study, we used the TCFsiam transgenic zebrafish line, which was designed to provide a visible readout of the canonical Wnt pathway. Specifically, this line displays intense nuclear mCherry expression in cells expressing TCF, a DNA-binding component of canonical Wnt signaling. Our analysis of this line indicates a strong TCF+ cell population within early-budding, elongating and adult barbel tissue. Co-localization of the reporter-labeled nuclei with EdU labeling shows that the TCF+ cells are occasionally mitotic, but not highly proliferative relative to surrounding cells. Using TEM, we further located the TCF+ cells to a single, circumferential layer underneath the basement membrane and within the dermal core. Similar to osteoblasts, these barbel “core cells” are entirely encased in surrounding matrix, but connect to each other via small cytoplasmic projections. In addition to their unique location, cells within this layer are identified by their pancake-shaped nuclei and abundant RER.
How to build a barbel: ultimate and proximate mechanisms
The ultimate mechanism of barbel development may be the global metamorphic transformation of the larval zebrafish to a juvenile, a process occurring 3–4 weeks post-fertilization (Parichy et al., 2009), This period includes multiple morphological and physiological changes, including alterations in fin morphology, external pigmentation, and circulating hormone levels (McMenamin and Parichy, 2013).. Considering the proximate mechanisms for barbel outgrowth, the TCF+ cells within this appendage likely play a key role. Specifically, we speculate that their function is not to divide, but to produce collagenous matrix in response to Wnt signals, which are present in a proximal-distal gradient along the appendage. There are two possibilities for the source of the putative Wnt ligand. In a paracrine mechanism, the source is the adjacent epithelium (Fig. 7, top). In this model, sufficient ligand must pass through the basement membrane and subadjacent collagenous layers for the matrix-bound mesenchymal cells to detect it, possibly via their short cytoplasmic projections. In an autocrine mechanism (Fig. 7, bottom), the TCF+ cells are themselves the ligand source, cooperatively maintaining a Wnt-enriched environment.
In conclusion, we propose a model of barbel outgrowth consistent with our collective observations. In response to unspecified proximal signals from the epidermis, a pre-barbel condensation forms within the dermis lateral to the maxillary bone. Cells within this whorl divide, extending a dermal bud. Specific cells within this bud become entrained in a Wnt-responsive signal loop, and begin to secrete the collagenous matrix that will form the barbel core. The secretory subpopulation forms a radially flattened, circumferentially arranged “growth ring”, which can gradually add to barbel length and girth throughout the lifespan. Cells within the ring contact each other through cytoplasmic bridges, and divide only when needed to maintain a single-cell layer. As the barbel matures, the matrix-producing cells at the proximal end decrease or halt decreasing matrix production. Cells at the tip, however, maintain Wnt-responsiveness, constantly contributing to the most recently assembled part of the appendage. Although much of this model remains unexplored, similar gene, expression, cell behavior and matrix mechanics may be responsible for the independent and repeated evolution of barbel structures in other fish species.
Supplementary Material
S1. 3D rotation of DAPI-stained barbel bud epithelium
S2. 3D rotation of TCFsiam:mCherry+ maxillary barbel core cells.
Acknowledgments
Research funding was provided by NIH R15-HD064169 (to E.E.L.). A portion of this work was performed in partial fulfillment of the academic requirements for the Master of Science at DePaul University (S.S.). Confocal reconstruction was made possible in part by FluoRender software from the University of Utah, funded by NIH R01-GM098151-01. Transmission electron microscopy was performed at the Northwestern University Center for Advanced Microscopy, which is generously supported by NCI CCSG P30 CA060553, awarded to the Robert H. Lurie Comprehensive Cancer Center. Lennell Reynolds provided expert TEM specimen preparation. We thank Jacek Topczewski and two anonymous reviewers for critical comments on the manuscript.
References
- Alibardi L. Dermo-epidermal interactions in reptilian scales: Speculations on the evolution of scales, feathers, and hairs. J Exp Zool B Mol Dev Evol. 2004;302 (4):365–83. doi: 10.1002/jez.b.20028. [DOI] [PubMed] [Google Scholar]
- Alibardi L. Proliferation in the epidermis of chelonians and growth of the horny scutes. J Morphol. 2005;265 (1):52–69. doi: 10.1002/jmor.10337. [DOI] [PubMed] [Google Scholar]
- Ansell DM, Holden KA, Hardman MJ. Animal models of wound repair: Are they cutting it? Exp Dermatol. 2012;21 (8):581–5. doi: 10.1111/j.1600-0625.2012.01540.x. [DOI] [PubMed] [Google Scholar]
- Avella M, Berhaut J, Payan P. Primary culture of gill epithelial cells from the sea bass Dicentrarchus labrax. In Vitro Cell Dev Biol Anim. 1994;30A (1):41–9. doi: 10.1007/BF02631417. [DOI] [PubMed] [Google Scholar]
- Baena-López LA, Baonza A, García-Bellido A. The orientation of cell divisions determines the shape of Drosophila organs. Curr Biol. 2005;15 (18):1640–4. doi: 10.1016/j.cub.2005.07.062. [DOI] [PubMed] [Google Scholar]
- Bielefeld KA, Amini-Nik S, Alman BA. Cutaneous wound healing: recruiting developmental pathways for regeneration. Cell Mol Life Sci. 2013;70 (12):2059–81. doi: 10.1007/s00018-012-1152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm B, Westerberg H, Lesnicar-Pucko G, Raja S, Rautschka M, Cotterell J, Swoger J, Sharpe J. The role of spatially controlled cell proliferation in limb bud morphogenesis. PLoS Biol. 2010;8 (7):e1000420. doi: 10.1371/journal.pbio.1000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin J. Cultures organotypiques de thyroïdes d’un Poisson Téléostéen marin: Gobius niger L. Effets de la TSH et de la prolactine. Comptes Rendues Soc Biol Filiales. 1971;1971 (2 Juin):1284–1291. [PubMed] [Google Scholar]
- Bonnin J. Association en culture organotypique d’hypophyses et de tissu glandularie du testicule de Gobius niger L. Comptes Rendues Soc Biol Filiales. 1975:919–923. [PubMed] [Google Scholar]
- Briolay J, Galtier N, Brito R, Bouvet Y. Molecular phylogeny of Cyprinidae inferred from cytochrome b DNA sequences. Mol Phylogenet Evol. 1998;9 (1):100–8. doi: 10.1006/mpev.1997.0441. [DOI] [PubMed] [Google Scholar]
- Brunelli E, Perrotta I, Bonacci A, Tripepi S. The balancers of Triturus italicus: an ultrastructural approach. Italian Journal of Zoology. 2007;74 (2):135–142. [Google Scholar]
- Carney TJ, Feitosa NM, Sonntag C, Slanchev K, Kluger J, Kiyozumi D, Gebauer JM, Coffin Talbot J, Kimmel CB, Sekiguchi K, Wagener R, Schwarz H, Ingham PW, Hammerschmidt M. Genetic analysis of fin development in zebrafish identifies furin and hemicentin1 as potential novel fraser syndrome disease genes. PLoS Genet. 2010;6 (4):e1000907. doi: 10.1371/journal.pgen.1000907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania KC, Leitch DB, Gauthier D. Function of the appendages in tentacled snakes (Erpeton tentaculatus) J Exp Biol. 2010;213 (3):359–67. doi: 10.1242/jeb.039685. [DOI] [PubMed] [Google Scholar]
- Chehrehasa F, Meedeniya AC, Dwyer P, Abrahamsen G, Mackay-Sim A. EdU, a new thymidine analogue for labelling proliferating cells in the nervous system. J Neurosci Methods. 2009;177 (1):122–30. doi: 10.1016/j.jneumeth.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Ortí G, Meyer A. Novel evolutionary relationship among four fish model systems. Trends Genet. 2004;20 (9):424–31. doi: 10.1016/j.tig.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Chodankar R, Chang CH, Yue Z, Jiang TX, Suksaweang S, Burrus L, Chuong CM, Widelitz R. Shift of localized growth zones contributes to skin appendage morphogenesis: role of the Wnt/beta-catenin pathway. J Invest Dermatol. 2003;120 (1):20–6. doi: 10.1046/j.1523-1747.2003.12008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong C, Chodankar R, Widelitz R, Jiang T. Evo-devo of feathers and scales: building complex epithelial appendages. Curr Opin Genet Dev. 2000;10 (4):449–56. doi: 10.1016/s0959-437x(00)00111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MS, Szeto DP, Sommers-Herivel G, Topczewski J, Solnica-Krezel L, Kang HC, Johnson I, Kimelman D. Visualizing morphogenesis in transgenic zebrafish embryos using BODIPY TR methyl ester dye as a vital counterstain for GFP. Dev Dyn. 2005;232 (2):359–68. doi: 10.1002/dvdy.20252. [DOI] [PubMed] [Google Scholar]
- Crucke J, Huysseune A. Unravelling the blood supply to the zebrafish pharyngeal jaws and teeth. J Anat. 2013;223 (4):399–409. doi: 10.1111/joa.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhouailly D. A new scenario for the evolutionary origin of hair, feather, and avian scales. J Anat. 2009;214 (4):587–606. doi: 10.1111/j.1469-7580.2008.01041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky R, Sheldahl L, Moon R. A transgenic Lef1/beta-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev Biol. 2002;241 (2):229–37. doi: 10.1006/dbio.2001.0515. [DOI] [PubMed] [Google Scholar]
- Duszynski RJ, Topczewski J, Leclair EE. Divergent requirements for fibroblast growth factor signaling in zebrafish maxillary barbel and caudal fin regeneration. Dev Growth Differ. 2013;55 (2):282–300. doi: 10.1111/dgd.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox H. Barbels and barbel-like tentacular structures in sub-mammalian vertebrates: A review. Hydrobiologia. 1999;403 (0):153–193. [Google Scholar]
- Fraser GJ, Bloomquist RF, Streelman JT. A periodic pattern generator for dental diversity. BMC Biol. 2008;6:32. doi: 10.1186/1741-7007-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghysen A, Dambly-Chaudière C. Development of the zebrafish lateral line. Curr Opin Neurobiol. 2004;14 (1):67–73. doi: 10.1016/j.conb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Silver LM, Niswander L, Papaioannou VE. Involvement of T-box genes Tbx2-Tbx5 in vertebrate limb specification and development. Development. 1998;125 (13):2499–509. doi: 10.1242/dev.125.13.2499. [DOI] [PubMed] [Google Scholar]
- Goldsmith MI, Iovine MK, O’Reilly-Pol T, Johnson SL. A developmental transition in growth control during zebrafish caudal fin development. Dev Biol. 2006;296 (2):450–7. doi: 10.1016/j.ydbio.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Gouge C, Christensen T. Detection of S phase in multiple Drosophila tissues utilizing the EdU labeling technique. Drosophila Information Service. 2010;93:203–212. [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453 (7193):314–21. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- Hawkins M. Master’s Thesis, Ecology and Evolutionary Biology. University of Colorado at Boulder; Ann Arbor, USA: 2011. The development and evolutionary origin of barbels in the channel catfish Ictalurus punctatus (Siluriformes: Ictaluridae) [Google Scholar]
- Howe JC. Standard length: Not quite so standard. Fisheries Research (Amsterdam) 2002;56 (1):1–7. [Google Scholar]
- Iovine MK. Conserved mechanisms regulate outgrowth in zebrafish fins. Nat Chem Biol. 2007;3 (10):613–8. doi: 10.1038/nchembio.2007.36. [DOI] [PubMed] [Google Scholar]
- Kasumyan A. Tactile reception and behavior of fish. Journal of Icthyology. 2011;51 (11):1035–1103. [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203 (3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kizil C, Otto GW, Geisler R, Nusslein-Volhard C, Antos CL. Simplet controls cell proliferation and gene transcription during zebrafish caudal fin regeneration. Dev Biol. 2009;325 (2):329–40. doi: 10.1016/j.ydbio.2008.09.032. [DOI] [PubMed] [Google Scholar]
- Kosher RA, Savage MP, Chan SC. In vitro studies on the morphogenesis and differentiation of the mesoderm subjacent to the apical ectodermal ridge of the embryonic chick limb-bud. J Embryol Exp Morphol. 1979;50:75–97. [PubMed] [Google Scholar]
- LeClair E, Topczewski J. Development and regeneration of the zebrafish maxillary barbel: a novel study system for vertebrate tissue growth and repair. PLoS One. 2010;5 (1):e8737. doi: 10.1371/journal.pone.0008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Thirumangalathu S, Gallant NM, Yang SH, Stoick-Cooper CL, Reddy ST, Andl T, Taketo MM, Dlugosz AA, Moon RT, Barlow LA, Millar SE. Wnt-beta-catenin signaling initiates taste papilla development. Nat Genet. 2007;39 (1):106–12. doi: 10.1038/ng1932. [DOI] [PubMed] [Google Scholar]
- McMenamin SK, Parichy DM. Metamorphosis in teleosts. Current Topics in Developmental Biology. 2013;103:127–165. doi: 10.1016/B978-0-12-385979-2.00005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola ML, Millar SE. The mammary bud as a skin appendage: unique and shared aspects of development. J Mammary Gland Biol Neoplasia. 2006;11 (3–4):187–203. doi: 10.1007/s10911-006-9029-x. [DOI] [PubMed] [Google Scholar]
- Milán M, Campuzano S, García-Bellido A. Cell cycling and patterned cell proliferation in the wing primordium of Drosophila. Proc Natl Acad Sci U S A. 1996;93 (2):640–5. doi: 10.1073/pnas.93.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongera A, Singh AP, Levesque MP, Chen YY, Konstantinidis P, Nüsslein-Volhard C. Genetic lineage labeling in zebrafish uncovers novel neural crest contributions to the head, including gill pillar cells. Development. 2013;140 (4):916–25. doi: 10.1242/dev.091066. [DOI] [PubMed] [Google Scholar]
- Moro E, Ozhan-Kizil G, Mongera A, Beis D, Wierzbicki C, Young RM, Bournele D, Domenichini A, Valdivia LE, Lum L, Chen C, Amatruda JF, Tiso N, Weidinger G, Argenton F. In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev Biol. 2012;366 (2):327–40. doi: 10.1016/j.ydbio.2012.03.023. [DOI] [PubMed] [Google Scholar]
- Moro E, Vettori A, Porazzi P, Schiavone M, Rampazzo E, Casari A, Ek O, Facchinello N, Astone M, Zancan I, Milanetto M, Tiso N, Argenton F. Generation and application of signaling pathway reporter lines in zebrafish. Mol Genet Genomics. 2013;288 (5–6):231–42. doi: 10.1007/s00438-013-0750-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawala P, Tanaka EM, Currie JD. Regeneration: The ultimate example of wound healing. Semin Cell Dev Biol. 2012;23(9):954–962. doi: 10.1016/j.semcdb.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Musser JM, Wagner GP, Prum RO. Nuclear β-catenin localization supports homology of feathers, avian scutate scales, and alligator scales in early development. Evolution and Development. 2015;17(3):185–194. doi: 10.1111/ede.12123. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Linbo T, Poss KD, Raible DW. Specification of epibranchial placodes in zebrafish. Development. 2007;134 (3):611–23. doi: 10.1242/dev.02749. [DOI] [PubMed] [Google Scholar]
- Nomura R, Kamei E, Hotta Y, Konishi M, Miyake A, Itoh N. Fgf16 is essential for pectoral fin bud formation in zebrafish. Biochem Biophys Res Commun. 2006;347 (1):340–6. doi: 10.1016/j.bbrc.2006.06.108. [DOI] [PubMed] [Google Scholar]
- Ny A, Koch M, Schneider M, Neven E, Tong RT, Maity S, Fischer C, Plaisance S, Lambrechts D, Heligon C, Terclavers S, Ciesiolka M, Kalin R, Man WY, Senn I, Wyns S, Lupu F, Brandli A, Vleminckx K, Collen D, Dewerchin M, Conway EM, Moons L, Jain RK, Carmeliet P. A genetic Xenopus laevis tadpole model to study lymphangiogenesis. Nat Med. 2005;11 (9):998–1004. doi: 10.1038/nm1285. [DOI] [PubMed] [Google Scholar]
- Olivera-Martinez I, Thélu J, Dhouailly D. Molecular mechanisms controlling dorsal dermis generation from the somitic dermomyotome. Int J Dev Biol. 2004;48 (2–3):93–101. doi: 10.1387/ijdb.15272374. [DOI] [PubMed] [Google Scholar]
- Osterfield M, Du X, Schüpbach T, Wieschaus E, Shvartsman SY. Three-dimensional epithelial morphogenesis in the developing Drosophila egg. Dev Cell. 2013;24 (4):400–10. doi: 10.1016/j.devcel.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn. 2009;238 (12):2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon F, Ghysen A. Evolution of posterior lateral line development in fish and amphibians. Evol Dev. 2004;6 (3):187–93. doi: 10.1111/j.1525-142X.2004.04024.x. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, Baker CV. The development of lateral line placodes: taking a broader view. Dev Biol. 2014;389 (1):68–81. doi: 10.1016/j.ydbio.2014.02.016. [DOI] [PubMed] [Google Scholar]
- Pouyet J, Beaumont A. Ultrastructure du pancréas larvaire d’un Amphibien Anoure, Alytes obstetricians L. en culture organotypique. Compt Rend Soc Biol. 1975:846–850. [PubMed] [Google Scholar]
- Quint E, Smith A, Avaron F, Laforest L, Miles J, Gaffield W, Akimenko MA. Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc Natl Acad Sci U S A. 2002;99 (13):8713–8. doi: 10.1073/pnas.122571799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders JW. The proximo-distal sequence of origin of the parts of the chick wing and the role of the ectoderm. J Exp Zool. 1948;108 (3):363–403. doi: 10.1002/jez.1401080304. [DOI] [PubMed] [Google Scholar]
- Sennett R, Rendl M. Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Semin Cell Dev Biol. 2012;23 (8):917–27. doi: 10.1016/j.semcdb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleman C, Holtzman Na. Growth and Maturation in the Zebrafish, Danio Rerio: A Staging Tool for Teaching and Research. Zebrafish. 2014;11(4):396–406. doi: 10.1089/zeb.2014.0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck TS, Miyoshi H. The role of stromal stem cells in tissue regeneration and wound repair. Science. 2009;324 (5935):1666–9. doi: 10.1126/science.1172687. [DOI] [PubMed] [Google Scholar]
- Sugimura R, Li L. Noncanonical Wnt signaling in vertebrate development, stem cells, and diseases. Birth Defects Res C Embryo Today. 2010;90 (4):243–56. doi: 10.1002/bdrc.20195. [DOI] [PubMed] [Google Scholar]
- Sugiyama M, Sakaue-Sawano A, Iimura T, Fukami K, Kitaguchi T, Kawakami K, Okamoto H, Higashijima S, Miyawaki A. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc Natl Acad Sci U S A. 2009;106 (49):20812–7. doi: 10.1073/pnas.0906464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TH, Jarrell A, Zentner GE, Welsh A, Brownell I, Scacheri PC, Atit R. Role of canonical Wnt signaling/ β-catenin via Dermo1 in cranial dermal cell development. Development. 2010;137 (23):3973–84. doi: 10.1242/dev.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN. Genome evolution and biodiversity in teleost fish. Heredity (Edinb) 2005;94 (3):280–94. doi: 10.1038/sj.hdy.6800635. [DOI] [PubMed] [Google Scholar]
- Warren M, Puskarczyk K, Chapman SC. Chick embryo proliferation studies using EdU labeling. Dev Dyn. 2009;238 (4):944–9. doi: 10.1002/dvdy.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio) Eugene, OR: Univ. of Oregon Press; 2000. Ch. 1 General Methods for Zebrafish Care. [Google Scholar]
- Widelitz R. Wnt signaling in skin organogenesis. Organogenesis. 2008;4 (2):123–33. doi: 10.4161/org.4.2.5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widelitz R, Chuong C. Early events in skin appendage formation: induction of epithelial placodes and condensation of dermal mesenchyme. J Investig Dermatol Symp Proc. 1999;4 (3):302–6. doi: 10.1038/sj.jidsp.5640234. [DOI] [PubMed] [Google Scholar]
- Widelitz R, Jiang T, Noveen A, Ting-Berreth S, Yin E, Jung H, Chuong C. Molecular histology in skin appendage morphogenesis. Microsc Res Tech. 1997;38 (4):452–65. doi: 10.1002/(SICI)1097-0029(19970815)38:4<452::AID-JEMT13>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Wu P, Hou L, Plikus M, Hughes M, Scehnet J, Suksaweang S, Widelitz R, Jiang TX, Chuong CM. Evo-Devo of amniote integuments and appendages. Int J Dev Biol. 2004;48 (2–3):249–70. doi: 10.1387/ijdb.041825pw. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Abe G, Yokoyama H, Kawakami K, Tamura K. Mechanism of pectoral fin outgrowth in zebrafish development. Development. 2012;139 (16):2916–25. doi: 10.1242/dev.075572. [DOI] [PubMed] [Google Scholar]
- Zeller R, López-Ríos J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet. 2009;10 (12):845–58. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1. 3D rotation of DAPI-stained barbel bud epithelium
S2. 3D rotation of TCFsiam:mCherry+ maxillary barbel core cells.