Abstract
Both bioethics and law have governed human genomics by distinguishing research from clinical practice. Yet the rise of translational genomics now makes this traditional dichotomy inadequate. This paper pioneers a new approach to the ethics of translational genomics. It maps the full range of ethical approaches needed, proposes a “layered” approach to determining the ethics framework for projects combining research and clinical care, and clarifies the key role that return of results can play in advancing translation.
Keywords: translational, genomics, return of results, ethics, family
Introduction
Research on the use of genome and exome sequencing for diagnosis, identification of potential therapies, precision prescribing of pharmaceuticals, and identification of disease risk is progressing rapidly. Research projects now commonly yield findings of potential health importance for the individuals sequenced as well as their relatives, raising difficult questions about investigator responsibilities to offer those research findings for potential clinical work-up. As sequencing is moving into clinical application, the ethical questions are further multiplying. The ethical quandaries will only proliferate as use of sequencing in screening to achieve public health goals is debated more widely.
As in other fields of translational research, ethical issues will arise at every stage in the progression of translational genomics. Yet we currently lack a vision of the full scope of ethical approaches applicable to each stage in the translational pathway as genomics progresses, a means of integrating these approaches to cope with the dynamics of fast-moving translational science, and the place of the current narrower debates in this bigger picture. This article takes the ambitious step of trying to articulate the full roster of relevant ethics approaches, their role in the dynamics of translational genomics, and the pivotal role that return of results practices can play. Without this kind of larger vision, debates persist over return of results and incidental findings, how to reconcile research ethics with the ethics of clinical care as research findings are considered for return based on potential clinical importance, how to govern sequencing projects that combine research and clinical elements, and how to think about the ethics of genomic sequencing used to accomplish public health goals. In order to move forward, we propose a vision of the ethics of translational genomics that connects these disparate debates, moves beyond reliance on the traditional dichotomy between the ethics of research and clinical care, and offers a starting point for fully developing the ethical frameworks needed. We take genomics as our focus, but the fundamental approach we develop -- articulating the array of ethics frameworks that apply across the translational process and a method for resolving conflict between ethical frameworks -- can be applied beyond genomics to other fields of translational science.
The need for fundamental progress in conceptualizing the ethics of translational genomics is exemplified by the debates to date on return of results and how to think about such practices as well as entire projects that straddle the traditional research/clinical divide. The past decade has seen an explosion of interest in offering research participants – and now family members -- those individual research results and incidental findings that warrant clinicial evaluation and pursuit. Beginning in 2005,1 the National Human Genome Research Institute (NHGRI) and other institutes at the National Institutes of Health (NIH) have funded a growing portfolio of projects to study these practices and formulate guidelines. As genome and exome sequencing have moved into clinical application, concern over return of results and incidental findings has surfaced in the realm of clinical sequencing as well, with recommendations from the American College of Medical Genetics and Genomics (ACMG) published in 2013,2 vigorous ensuing debate,3 and revision of those recommendations in 2014.4 In 2013, the Presidential Commission on Bioethics published a report on return of results and incidental findings, divided into research return, clinical return, and return in direct-to-consumer genomics.5
The debate over return of results (which we will use here inclusively to refer to return of the primary results motivating the analysis, as well as return of additional incidental or secondary findings) has built on the traditional dichotomy between research and clinical care that grounds bioethics and health law.6 That dichotomy is historically foundational to 20th century bioethics and health law. Modern bioethics was born of a series of scandals in experimentation on human beings, leading to the formulation of the Nuremberg Code, the Belmont Report, and ultimately regulations from the U.S. Department of Health and Human Services (DHHS) that became the basis for the Common Rule.7 Both U.S. bioethics and law have sharply divergent approaches to human subjects research versus clinical care.8 In the research domain, investigators do not serve the individual participant but seek generalized knowledge to confer aggregate benefit, the IRB system reviews protocols to prevent harm, failures of research oversight are addressed primarily through administrative sanctions, and research participants have limited means of holding investigators and their institutions directly accountable. In the clinical domain, clinicians’ primary duty is to serve the patient’s well-being, clinicians are responsible for acting in conformance with professional standards, and patients can sue to hold clinicians and their institutions directly accountable for care that breaches those standards, causing compensable harm.
What has made return of research results controversial has been the fact that it straddles the domains of research and clinical care. The core question raised is whether some individual research findings raise sufficient concern that investigators should consider communicating those findings so that the individual has the option of pursuing the findings in the clinical domain. If research is on one side of the river (so to speak) with its own standards, and clinical care is on the other side with its different standards, the question is whether the researchers should somehow ferry information across to the clinicians, primarily by offering the findings to the individual research participant. An emerging consensus suggests the answer is “yes”; a number of policy groups and commentators have argued that investigators and others involved in the research enterprise bear obligations to research participants that include offering such findings in order to ferry the problem to professionals in the clinical domain.9 However, objections have focused on the research/clinical care dichotomy, with some commentators arguing that investigators should generally stay on the research side of the river and avoid returning information for pursuit on the clinical side. They have objected to refocusing research effort away from creating generalized knowledge to conferring individual benefit, devoting scarce research dollars to indentifying individual problems for clinical pursuit, and inviting participant confusion between research and clinical care.10
Even commentators who have supported return of results have thus far drawn a sharp line between return of results in research genomics and return of results in clinical genomics. Jarvik and colleagues, for example, argue that clinicians have robust obligations to serve the good of their individual patients, leading to strong obligations to offer results to patients, including incidental findings that may have important health implications.11 They contrast researchers, who do not shoulder the same obligations to individual participants, and thus have more degrees of freedom in designing research protocols with a greater or lesser scope of return.12 This comports with the analysis of the Presidential Commission on Bioethics, which also drew a sharp contrast between research return and clinical return. The PHG Foundation in England convened a workshop on the interdigitation of research and clinical components in genomics, reporting in 2014 that “There was support for a model of practice that enabled both clinical care and research to be done in parallel, but not for a distinct category or hybrid activity in which clinical and research elements were indistinguishable from an ethical and regulatory perspective.”13 They thus supported continued use of the research/clinical dichotomy.
This bifurcated analysis has served an important purpose by reminding investigators and policy makers of the goals of research in contrast to clinical care, while advancing thinking on investigator responsibilities to share research findings of potential health importance.14 Yet the research/clinical dichotomy, a creature of mid-20th century, post-Nuremberg bioethics, is increasingly under strain.15 The rise of translational science and particularly the rapid progress of translational genomics, has led to a deeper understanding of the multi-stage pathway from bench to bedside, to clinical integration and implementation, to population benefit. This multi-stage understanding makes the old dichotomy appear increasingly outmoded.
Translational genomics is epitomized by the NIH-funded Clinical Sequencing Exploratory Research (CSER) Consortium,16 eMERGE (Electronic Medical Records & Genomics) Network,17 and PGRN (Pharmacogenomics Research Network)18 projects. All of these projects aim to research and develop best practices for translating genomics into clinical application. This kind of research to develop an evidence-based foundation for clinical genomics requires a bioethics approach that transcends the 20th century research/clinical dichotomy and instead embraces a more nuanced 21st century translational vision. We propose the components and dynamics of that translational ethics below. That vision identifies the pivotal role that return of results can play in paving the translational pathway by impelling investigators to consider the subset of their findings that calls for going the next step in the translational process.
I. The Rise of Translational Genomics
Beginning in 2005, NIH articulated an explicit commitment to translational science.19 That commitment has since shaped significant intramural restructuring and extramural funding.20 Journals have been established devoted to translational science and a substantial literature has emerged.21 While the definition of “translational science” has been debated, Woolf characterizes translational research as embracing both “the ‘bench-to-bedside’ enterprise of harnessing knowledge from basic sciences to produce new drugs, devices, and treatment options for patients,” and “translating research into practice; ie, ensuring that new treatments and research knowledge actually reach the patients or populations for whom they are intended and are implemented correctly.”22
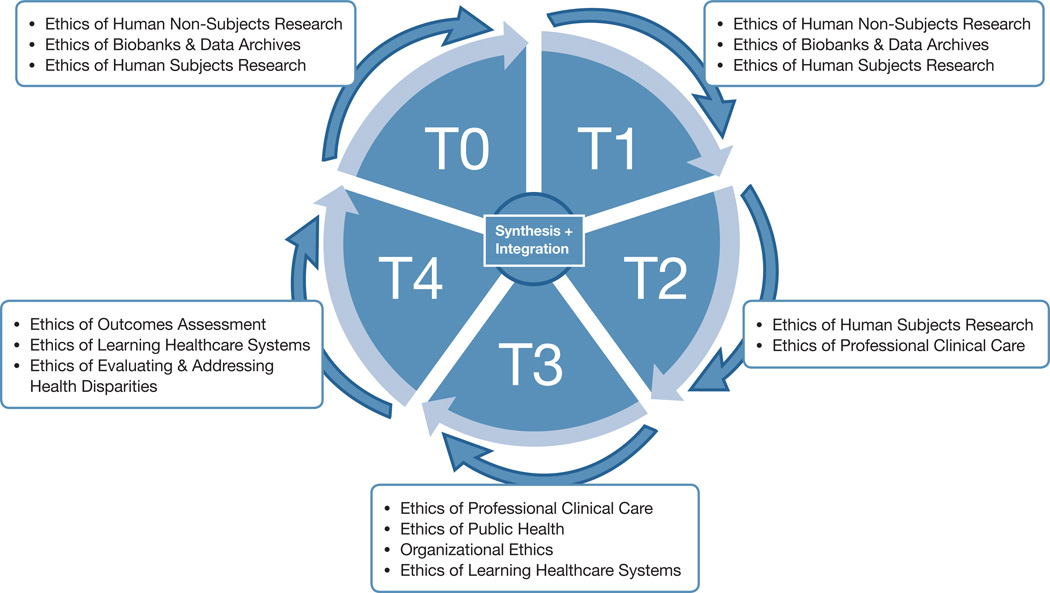
In 2011, NHGRI articulated a “vision for genomics…organized around five domains extending from basic research to health applications.”23 This translational “base pairs to bedside” vision has been further elaborated by scholars from the Centers for Disease Control and Prevention (CDC) Office of Public Health Genomics. Khoury and colleagues articulated four stages in the translational research in genetics: T1 “Discovery to candidate health application”; T2 “Health application to evidence-based practice guidelines”; T3 “Practice guidelines to health practice”; and T4 “Practice to population health impact.”24 They have since elaborated a more dynamic framework incorporationg T0, basic science, as well as feedback loops so that implementation stages can generate further research questions for basic science (T0) and bedside application (T1), and all stages contribute to knowledge synthesis.25 While not all commentators agree on the number and definition of stages,26 the basic idea of cycling through phases to reach a goal of evidence-based delivery of benefit to patients and populations pervades the various efforts.27 Thus, Goering and colleagues offer a model (further developed by Kelley and colleagues) depicting four overlapping stages (or phases) to comprise the translational cycle.28 These stages begin with “Discovery,” progress through “Development” and “Delivery,” and culminate in “Outcomes,” with all phases contributing to “Assessment and Priority Setting.” In both the T0–T4 depiction and the 4-stage model, research in one stage can lead to the next stage, but can also generate new questions to be pursued in a prior one. Thus, both models are “cyclical and iterative.”29
Translational genomics is already having a major impact on the structure of research projects. We have cited three of many funded research programs: CSER, eMERGE, and PGRN. These consist of multiple projects aimed at generating basic science insights through genome and exome sequencing, applying those insights in the care of patients, creating structures and practices for implementing genomic medicine, and studying process and outcomes in order to generate best practices and benefit.
Within these research networks, application of the traditional research/clinical dichotomy has become problematic. For example, Henderson and colleagues analyzed consent forms from CSER studies investigating whole genome sequencing (WGS) and whole exome sequencing (WES).30 The studies ranged from one that placed no results in the participant’s medical record, to one that placed validated results relevant to patient care in the medical record, to one that gave participant’s a choice, to another that placed all results in the medical record. This runs the gamut from procedures one would expect in a research project (no results routinely incorporated into the clinical record) to procedures one would expect in clinical care (all results in the medical record). CSER thus seems to straddle translational stages T1 and T2.
This translational character is explicitly reflected in descriptions of the CSER Consortium: “CSER is a national consortium of projects…to develop and share innovations and best practices in the integration of genomic sequencing into clinical care”31 and “the CSER consortium supports…projects…to research the challenges of utilizing genomic sequence data in the clinic in the routine practice of medicine.”32 While the CSER projects are research funded by NIH and approved by IRBs, many of the projects enroll affected participants seeking health benefit from sequencing, the projects aim to determine how best to deliver such benefit, and seek to develop best practices. In this context, simply asking whether the ethics of research or clinical care should apply does not admit of an easy answer or do justice to the complexity of the issues. Those issues range from consent to sequencing, to what variants should be analyzed and reported, what results should be offered to participants and perhaps their family members, and how and where those result should be recorded.
II. The Need for a Full Ethics of Translational Genomics
While the rise of translational science has engendered a literature on ethical issues raised, much of that literature has focused not on developing bioethics to meet the challenge of translational analysis, but rather on critiquing the translational ambition to move drugs and other interventions to market. Maienschein and colleagues argue that translational research “is bringing a new social contract for the way science works in society. Instead of implicit promissory notes about eventual results, scientists must promise specific results up front…. [and] there is now far more guidance from public investors.”33 Indeed, Sofaer and Eyal maintain that “taken collectively, the literature on the ethics of translational research may leave readers with the impression that such research is particularly morally problematic and requires special scrutiny.”34 They argue, instead, that the ethical issues are focused in the early research stages (T0 and T1); in effect, they reinstantiate the research/clinical dichotomy. Kelley and colleagues as well as Shapiro and Layde go further, articulating values questions that arise at each stage in the pathway of translational science,35 yet neither offers a vision of the full range of ethics frameworks needed to guide the translational process.
More specific to translational genomics, Burke and colleagues caution against overselling the benefit of genomic analysis before it is rigorously established.36 Lin and colleeagues “describe a framework for assessing the evidence base for genomic tests (from discovery to clinical adoption),” by specifying six phases (0 to 5) from “Marker Identification & Assay Development” to “Initial Test Performance and Assay Refinement,” “Test Validation & Generalizability,” “Clinical Test Performance & Health Impacts,” “Comparison with Existing Tests,” and “Population Impacts.”37 While not expressly an ethics analysis, their framework has ethical implications: “Our framework…allows different stakeholders to specify different thresholds for decision-making, depending on their perspective and particular needs (e.g., for exploration, further development or discontinuation, regulation, clinical uptake, insurance coverage, dissemination, practice guideline development, or marketing).”38
What remains to be developed is an ethics framework that addresses the full scope of translational genomics and the dynamics of moving through translational stages, while providing normative guidance to researchers such as those in the CSER projects who face confusion about the admixture of research and clinical components in their projects. Although most funded research in genomics is currently at early translational stages,39 one can imagine later research (for example) on clinical sequencing in protocols that involve reporting variants of unknown significance (VUSs) to the individuals sequenced, though discerning the meaning of those VUSs will require further research. Whether to report VUSs is already an active debate, pitting those who question the utility and ethics of communicating potentially anxiety-provoking information of uncertain significance against others who maintain that patients are entitled to know their results and have the greatest stake in following the ensuing research to ascertain the meaning of these variants over time.40 Faced with this kind of controversy, simply invoking the ethics of clinical care will not provide adequate guidance, as sequencing will produce variants ranging from those that are well understood to those that are not, a circumstance likely to persist for a long time to come.
III. Developing an Ethics of Translational Genomics
There are three core problems with the past piecemeal approach to the ethics of genomics and traditional reliance on the research/clinical dichotomy. First, this approach fails to address the full spectrum of translational research. Second, it considers in isolation the question of how to approach projects mixing research and clinical elements, rather than placing that question in the context of translational dynamics. Third, it offers little concrete guidance when elements of research and clinical practice are interdigitated, as in CSER studies. Developing an ethics of translational genomics requires addressing all three.
A. Addressing the full translational cycle
The ethics frameworks relevant to each translational stage go considerably beyond the traditional duo of research and clinical ethics. In Table 1, we propose the range of ethical frameworks that should apply across the translational cycle. At the T0 stage of basic genomic research (corresponding to the early part of the Discovery phase in the model offered by Goering and colleagues as well as by Kelley and co-authors41), discovery using human data and specimens may or may not qualify as “human subjects research” (HSR) under the Common Rule as it currently stands, as federal guidance to date has treated research on deidentified samples and data that were not collected by the investigator for research as beyond the bounds of HSR.42 Brothers and Clayton have thus labeled this “human non-subjects research.”43 While the 2011 Advance Notice of Proposed Rulemaking (ANPRM) from DHHS to amend the Common Rule floated the idea of requiring at least rudimentary consent for such research44 and a 2014 federal statute directs NIH to treat research on dried bloodspots from newborn screening as HSR,45 much basic genomic research is currently not considered HSR. Serious ethical issues arise nonetheless, though they are not yet well addressed by the body of research ethics developed to cover HSR. For example, sequencing of the HeLa genome has raised serious issues of invasion of privacy as well as justice, given that the original cells were collected from the source individual without her knowledge and consent.46 Thus, even “human non-subjects research” raises issues including determining what notice and consent are needed, protecting the privacy of source individuals and their genetic relatives, considering re-identification of source individuals and return of results, and data-sharing. In addition, T0 genomics research often involves creation of biobanks and archives collecting specimens and data. A substantial literature addresses obligations of stewardship, protecting the privacy of source individuals, and other duties devolving on these research resources.47 This stage of translational research thus implicates both the ethics of “human non-subjects research” and the ethics of biobanks and data archives, as well as the ethics of human subjects research when data or specimens are collected by the investigator for research or identifiers are maintained.
Table 1. Identifying the ethics most applicable to each stage of translational genomics.
The definitions used for stages T0–T4 are based on, but modify the stages suggested by Khoury and colleagues.
| Translational Stage | Core questions include | Guiding Ethics |
|---|---|---|
| T0—basic genomic research (Early Discovery) |
|
|
| T1—early clinical research (Later Discovery) |
|
|
| T2—late clinical research to early implementation (Development) |
|
|
| T3—dissemination and implementation, in clinical care and in screening for public health purposes (Delivery) |
|
|
| T4—securing health benefit for patients & populations (Outcomes) |
|
|
The additional parenthetical label noted for each stage is based on, but slightly modifies the labels offered by Kelley and colleagues.
As this Table indicates, our approach works regardless of which labeling system is used for the stages of translational genomics.
M. J. Khoury et al., “Knowledge Integration at the Center of Genomic Medicine,” Genetics in Medicine 14, no. 7 (2012): 6432013;647.
M. Kelley et al., “Values in Translation: How Asking the Right Questions Can Move Translational Science Toward Greater Health Impact,” Clinical and Translational Science 5, no. 6 (2012): 445–451.
At the T1 stage of human clinical research (corresponding to the later part of the Discovery phase), the transition from bench science to early clinical trials occurs and HSR issues are abundant. These include issues not well handled by the Common Rule, though more fully addressed in Food and Drug Administration (FDA) guidance and policy, such as the question of what pre-clinical research provides an adequate basis for ethically moving into clinical trials, the ethics of first-in-human trials, and the application of Phase 0 versus Phase I trials. At this stage both the ethics of “human non-subjects research” (for research conducted on data and specimens collected in clinical care and then de-identified for research use) and the ethics of human subjects research may apply. In addition, the ethics of biobanks and data archives remain central, given that genomic research is increasingly fueled by these research assemblages.
The T2 stage of later clinical trials and post-approval Phase IV trials48 (corresponding to the Development phase) begins to strain the research/clinical dichotomy. While later clinical trials typically raise issues in HSR, post-approval Phase IV trials and “post-market surveillance” involve collecting data in the context of clinical application and use. CSER projects that sequence affected patients to aid diagnosis and treatment while developing an evidence-base for best practices in clinical sequencing epitomize the integration of research and clinical care that raises questions about whether research ethics or clinical ethics applies, or whether some combination is needed. Thus, both the ethics of human subjects research and the ethics of professional clinical care are germane. Determining how they apply to a given project and how they interdigitate can be difficult.
The T3 stage involves dissemination and implementation science, moving interventions into practice.49 Goering, Kelley, and colleagues regard this as the Delivery phase.50 It is not at all clear what ethics undergirds this stage of translation. Certainly the ethics of professional clinical care would apply, but so would organizational ethics.51 Indeed, as organizations have increasingly recognized the goal and responsibility to become learning healthcare systems engaged in evidence-based quality assessment, quality improvement, and innovation,52 bioethicists have begun to address the challenge of evolving an ethics for learning healthcare systems. Faden and colleagues propose an ethics framework that prioritizes seven obligations, including “[c]onduct continuous learning activities that improve the quality of clinical care and health care systems.”53
A further complexity at the T3 stage is that delivery may involve implementation in individual patient care as well as implementation in screening for public health purposes. This is particularly true in the application of genomic sequencing, which may allow individual diagnosis or identification of therapeutic targets, but may also permit opportunistic screening54 or even population screening.55 Thus, the ethics of public health comes to the fore. There is a robust history of criteria for appropriate application of public health screening measures, beginning in modern times with the criteria advanced by Wilson and Jungner.56 More recently, Burke and colleagues have developed a set of six recommendations for global public health practice, including continuing “[e]fforts to integrate genomics into public health research and practice,” building and sustaining needed “infrastructure for generating an evidence-base for genomic medicine,” and developing an ethics that includes “responsible stewardship of resources.”57 Despite tremendous leadership from the CDC’s Office of Public Health Genomics and others, we still have only a sketch of what a full ethics of public health genomics would look like.58
The T4 stage (corresponding to the Outcomes phase) finally addresses patient and population health impact. Khoury and colleagues characterize this stage as involving outcomes research--“‘research that describes, interprets and predicts the impact of various influences, especially (but not exclusively) interventions on “final” endpoints that matter to decision makers. The decision makers may include patients, families, individuals at risk, providers, private and public payers, [and others].’”59 This kind of research will be essential to ensure patient and population benefit from genomics applications and avert non-beneficial and even harmful applications of genome and exome sequencing. Here, the ethics of outcomes assessment becomes relevant, including comparative effectiveness research.60 This means that the ethics of learning healthcare systems remains germane, as “Quality improvement and comparative effectiveness research are emblematic of the kinds of ongoing learning activities that a learning health care system is designed to promote.”61 In addition, a core issue that outcomes assessment must address is whether the intervention alleviates or exacerbates health disparities.62 This makes central the ethics of evaluating and addressing health disparities, including justice, stakeholder engagement, and inclusive governance of healthcare research, delivery, and evaluation.
The traditional research/clinical dichotomy thus fails to capture the full range of ethics frameworks essential to cope with the full cycle of translational research. Table 1 depicts the far greater range of ethical approaches applicable to the cycle of translational genomics research.
B. Placing the interdigitation of research and clinical care in a dynamic translational context
Seen in the context of the full range of ethics approaches needed to address the entire cycle of translational research, the problem of how to approach domains in which research and clinical care are interdigitated is actually a subset of a larger problem—how to approach stages and projects that intermix different ethical domains. If T2 is a stage likely to see the collision and confusion of human subjects research and clinical care, T3 is a stage likely to see tension and possible confusion between the ethics of individual health care professionals (committed to serving their patients) and that of their organizations (committed to serving their population, and usually requiring constraints on individual patient care), as well as the imperative to build learning healthcare systems (that may deliberately aim to alter individual professional practice, including a clinician’s discharge of ethical duties to individual patients). And when use of genomics in stage T3 foregrounds the ethics of public health, application of genomic screening will often (though not always) be conducted in the context of clinician-patient relationships and in healthcare organizations. Thus, the ethics of public health, requiring (for example) demonstrated positive predictive value and net population benefit as a predicate to instituting genomic screening, may war with the clinicians’ felt imperative to use the genomic tools available to serve their patients’ well-being.63 In stage T4, outcomes assessment will involve evaluating both patient and population impacts, calling for analysis at all levels. What individual patients and clinicians value may not yield positive benefits at the population level and may worsen rather than relieve health disparities.
All of these tensions are born of the dynamic nature of the translational research process, as well as path-breaking projects mixing approaches to advance genomics to serve patients and the public. Yet at present, with most funded genomics research in early translational stages, the confusion between research and clinical ethics in stage T2 projects such as the CSER and eMERGE projects looms especially large. This was why the PHG Foundation in England convened a 2013 project on whether some kind of hybrid research/clinical ethics was needed.64
The answer to how to negotiate this tension, however, is to recognize translational research as a process in motion. The reason why elements of research and clinical care mix in uneasy proximity in stage T2 is because genomics aims to progress—the goal is to move from the domain of human subjects research to the domain of clinical application. Thus we see some projects recording all results in research records, for example, while other projects record all in medical records, and still other projects are in between, offering participants the choice.
Moving genomics forward responsibly is, after all, the goal. Unpacking what “responsibly” entails at every step is the job of investigators, clinicians, their institutions, oversight authorities, funders, participants, patients, and the public. In short, it is the work of research, clinical care, public health, and ethics. Advancing entails reconciling the imperative to progress with responsibly ensuring an adequate evidence base (including validity and quality), protection for and accountability to participants, benefit to patients, and gains for public health.65 Our collective vision of the translational process must capture that dynamic motion.
There are a number of different visual models of the translational process, but few capture the tension between the imperative to make progress and the need to progress responsibly. As noted above, Khoury and colleagues have offered an influential model that can serve as a starting point.66 But their multi-sided polygon—a helpful early rendering of communication among stages—does not fully capture the idea of forward movement. The model offered by Goering, Kelley, and colleagues goes a step further, developing a 4-stage picture of the translational cycle.67 We suggest progressing to the model of a wheel, where the hub continues to depict synthesis and knowledge integration (or assessment and priority setting, in the Goering et al. model). The wheel itself depicts stages T0 to T4 (though it would work equally well depicting the four stages that Goering, Kelley et al. advocate). However, that wheel is framed by standards and ethics—a missing element of prior models. (See Figure 1) An important caveat is that all schematic depictions of translational research (and translational genomics) are a simplification. A given project may mix T2 and T3 elements, for example. Moreover, although we have tried to identify the primary ethical approaches associated with each translational stage, the actual elements of a T3 project, for example, may make the ethics of human subjects research (for instance) germane.
Figure 1. The translational research process, framed by the ethical domains most relevant to each stage.
This re-visualization allows us to understand the problem of research practices mixing with clinical practices, as genomics moves through stages toward increased clinical integration, as a problem of what standards and ethics should apply. In the T2 context of CSER projects, this question has arisen in multiple ways. We have already noted the question of where results should be recorded (research record, medical record, or both). Another example is the debate concerning return of results from laboratories without CLIA certification; a powerful argument has been made that such return should be allowed, as long as those results are clearly communicated as research results requiring further confirmation in a CLIA-certified lab and clinical work-up.68 This argument rests on concluding that some projects remain dominantly human subjects research rather than clinical care, even if some research results trigger concern and an alert to the participant with the recommendation of clinical confirmation and consultation.
C. “Layering” ethics and standards when research is mixed with clinical care
When research is mixed with clinical care in translational genomics—as in many of the T2 CSER projects--there are different possible approaches to clarifying the ethics and standards that should apply. Theoretically, one could try to distinguish the elements of each protocol or intervention that constitute research versus those that constitute clinical care, applying the standards from the corresponding realm. However, this will raise difficulties, as participants are asked to consent to the entire process, investigators must decide in designing the protocol where results will be recorded (the research record or medical record), and investigators must set up a pipeline relative to research or clinical standards for ascertaining, interpreting, and reporting genomic sequence.69
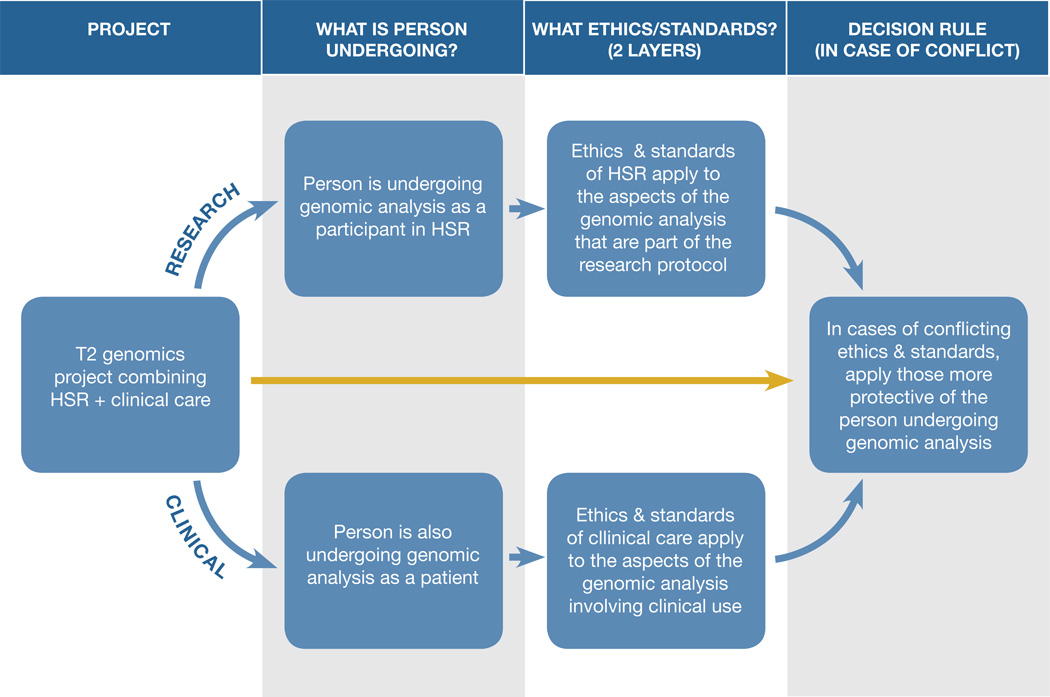
More realistic may be to ask first whether the protocol or intervention constitutes human subjects research, even if the research includes clinical components and is ultimately designed to help develop best practices for clinical application. We start with that question, because the history of research involving human beings and the late-20th century development of modern bioethics teaches the perils of failing to prioritize protection of human beings in research, as the search for generalized knowledge creates a common temptation to subordinate protection of participants to public priorities. Beginning with that question places the individual undergoing genomic analysis at the center of the ethics analysis. If the answer to the research question is “yes,” then the safeguards attending human subjects research should generally apply, including advance review and approval by an IRB, careful scrutiny of consent processes and privacy protections, more elaborate consent than in clinical care, ongoing IRB oversight, the opportunity to exit the research at any point, and the other protections for participants mandated by the Common Rule and FDA’s comparable (though not identical) regulations. This establishes the research layer of protections. (See Figure 2)
Figure 2. “Layering” ethical approaches to deal with T2 genomics projects combining human subjects research (HSR) with clinical care.
This method asks whether the person is undergoing genomic analysis as a participant in human subjects research (HSR) and as a patient (2nd column). This method then “layers” ethics by applying the ethics and standards of HSR to genomic analysis that is part of the research protocol, while applying the ethics and standards of clinical care to the aspects of the genomic analysis that involve clinical use (3rd column). If both ethics apply to some aspect of the genomic analysis and conflict, this method uses a decision rule, applying to that aspect the ethics and standards that are more protective of the person undergoing genomic analysis (4th column).
The next step of the inquiry would be to ask if the person is also undergoing genomic analysis as a patient and the project involves clinical components (such as recording the results in the medical record). If so, then the protections for patients that attend that clinical practice (such as federal privacy protections under HIPAA) should also apply, establishing a clinical layer of protection as well. For example, if some sequencing results are offered to participants for direct diagnostic and/or therapeutic application (instead of being offered as mere research results for subsequent validation in a clinical lab setting before diagnostic or therapeutic use), then they are being handled as clinical results and should meet clinical standards for quality and validation.70
The final step is to recognize that there will be scenarios in which this “layered” ethics approach to the complex realities of translational genomics will yield quandaries, because both research protections and clinical protections will seem to apply to the same aspect of the analysis, but they will conflict. In cases of conflict, the approach more protective of the individual undergoing the genomic analysis should apply.71 For example, if applicable clinical practice is to record all sequencing results in the medical record but research practice is to record results in the research record unless validated to clinical standards and used for clinical care, then the heightened privacy protections in the research context would take priority. There may be instances in which deciding which approach is more protective is challenging, but the goal would be to make that determination. This approach embraces a decision rule that prioritizes protection for the rights, interests, and well-being of the individual who is undergoing genomic analysis. The approach thus maps a way to navigate the confusion of interdigitated research and clinical care.
The core of this “layered” approach--retaining a role for research ethics and for clinical ethics--corresponds to the way clinical research combining human subjects research and clinical care is overseen. When an oncology patient is enrolled in a clinical trial assessing a new chemotherapeutic agent, for instance, the full protections and oversight that go along with human subjects research apply. Yet at the same time, the dimensions of the patient’s treatment that are not research are governed as clinical care.
The “layered” approach to ethics and standards also has the virtue of being generalizable. At stage T2 in translational genomics, the intermixing of research and clinical practices appears the main source of confusion. But at stage T3, the problem may be the intermixing of human subjects research with studies aimed to yield quality improvement in a learning healthcare system. Our “layered” approach would again start by asking whether the protocol or intervention consitutes human subjects research, even if elements of institutional or system-wide quality improvement are present as well. Once protections for human subjects are secured, layering on the ethics needed to appropriately design and oversee the learning healthcare system elements, would fill out the ethical picture.
Critics might object that this “layered” approach prioritizes protection of human subjects, when translational genomics ultimately aims to yield both patient benefit and public health benefit. The ethics of public health indeed requires balancing protections for individual autonomy and privacy against the public good.72 Yet translational genomics cannot advance without the trust and confidence of individuals willing to participate in research and partner with investigators in pioneering the genomics revolution in medicine.73 Protecting the interests of those individuals has to rank high in ethics priorities, to avoid the errors of the past, to respect the rights of those willing to participate, to shape genomic medicine in a way that is responsive, and simply to recruit enough participants to make progress. In later stages of the translational process, the answer to whether there is a clear element of human subjects research may be “no.” Instead, research and interventions may aim to evaluate aggregate trends and deidentified data to ascertain whether screening applications of genomics yield net population benefit.
IV. The Role of Return of Results in Translational Genomics—The Leading Edge
Where does return of results (including incidental or secondary findings) fit in this picture of translational genomics? Seen in the context of translational genomic research moving toward clinical integration, implementation, and eventually deployment for patient and population benefit, return of results can play a pivotal role. At early stages of human subjects research, recognition of some results as sufficiently validated, pathogenic, and actionable to be offered to participants—applying commonly recognized criteria for return74 —allows investigators to pioneer practices they will need to expand with increasing clinical integration. To undertake return of results, investigators need to anticipate the kinds of returnable results they may generate, create a pathway to evaluate the suitability of those results for return, seek participant consent for return, create a process for communicating results, and determine what reports to generate for communication to the participants’ clinicians and how to record the results to be returned. Return of results thus can act as the leading edge of a process expected to grow into a full-blown set of clinical genomics practices as genomics progresses along the translational pathway.
Return of results consequently helps pave the translational pathway by requiring investigators to think through, try out, and scrutinize the success or failure of the associated procedures. This is an important and unrecognized role. Yet on reflection, it stands to reason. Khoury and colleagues place knowledge synthesis and integration at the center of the translational process;75 each translational stage adds to accumulating knowledge about the capacity of genomics to generate information of value to individuals and the public health and allows refinement of next steps. Return of results is precisely about recognizing the capacity of genomics to generate information of value, sometimes unexpectedly, as in the case of unanticipated incidental findings. Studying the results generated, what results individuals wish to receive, what they and their clinicians do with those results, and what mix of benefits and burdens eventuate, generates invaluable knowledge whose synthesis can deeply inform next steps in the translational process.
Indeed, return of research results is by its nature a translational practice.76 It occurs when genomic information generated in research is recognized to hold such potential clinical importance that investigators offer it back to participants so that those individuals can take the information into the clinical context and address it with their clinicians. The return of results process is thus one that moves research-generated information into the domain of clinical care. But what has not been previously recognized is how important return of results can be for paving the translational pathway, advancing knowledge integration (both on the meaning of genomics and the “how” of evaluating results and offering them to individuals), and thus fueling translational progress.
Seen in this light, return of results beyond participants themselves, to family—the next step in the evolution of return of results practices77—takes the translational process one step further. Though the question of when, whether, and how to offer an individual’s genomic results to relatives is under debate, the idea of expanding the circle of those offered such results in order to confer wider benefit is consistent with the trajectory of T0–T4 genomics toward increasingly distributed benefit. The early approaches toward return of genomic results to relatives take differing stands on whether investigators should reach out to relatives to offer some subset of results in “active return” or generally limit communication to relatives by allowing them to initiate a request for results, in what is called “passive return.”78 Either mechanism (or some combination, as urged by Wolf and colleagues79) requires investigators to go a step further than in preparing for return to participants themselves. Anticipating return to relatives involves considering what consent may be needed from the participant him- or herself; what set of results has sufficient importance beyond participants that return to family members is appropriate to consider; how return to relatives should be conducted, given that the investigators may have no direct relationship with the relatives if they are not themselves enrolled in the study; and how such return should be recorded. Of equal importance, investigators should consider how to study the process of return of results to relatives, and its impacts.
All of this further paves the translational pathway. It expands the uses of genomic information and the scope of individuals who may benefit. It calls for innovation in consent and communication practices. And it drives further knowledge synthesis to inform next steps on the translational pathway.
Conclusion
This article reveals the deep connections between the developing vision of translational genomics, confusion over whether projects combining research and clinical genomics should be governed as research or clinical care, and the debates over return of results. By re-envisioning translational genomics as a dynamic process framed by a set of ethics approaches and standards, and by fully articulating the range of ethics involved in the full translational cycle, we have advanced the tools available to make progress in genomics in a way that comports with applicable ethical standards.
At the same time, we have placed the dynamic nature of fast-moving translational genomics front and center, and have suggested a method for ascertaining the governing ethics when genomics research and interventions mix research and clinical care or mix other elements across translational stages. That method involves a decisional process that begins with a commitment to protecting the interests of those undergoing genomic analysis, while “layering” on the additional ethics needed to fully analyze and govern the range of research and clinical practices (or other practices) involved. In cases of conflict between applicable standards, this method embraces a decision rule that favors the standard more protective of the person undergoing genomic analysis.
Finally, we argue that return of results is itself a pivotal translational process. By prompting investigators to address the subset of their findings ready for communication and use, return of results drives those investigators to advance their thinking and practices in the direction of translation. This can make a significant early contribution to paving the translational pathway.
We offer a new vision of the ethics of translational genomics. This vision attempts to fully specify the range of ethics frameworks in play, to array them around the wheel of translational genomics, and to reveal the dynamics driving the translational process. Those dynamics will regularly interdigitate frameworks such as research and clinical care, while making return of results a leading-edge practice that helps propel progress. The vision we present offers new tools for analysis while showing the connections between previouly disparate debates--over translational genomics, the research-clinical divide, and return of results.
Making progress in translational genomics ultimately requires more than current schemas of T0 to T4 or Discovery to Outcomes. Progress requires an appreciation of the ethics and standards that frame and help power the translational process. We urge an ethics of translational genomics that prioritizes the interests of the partipants and patients crucial to genomic progress. The only way to pave the translational highway is in partnership with those willing to undergo genomic analysis and to help shape the future of genomics in patient care and population health.
Acknowledgments
Preparation of this article was supported by National Institutes of Health (NIH), National Cancer Institute (NCI) and National Human Genomic Research Institute (NHGRI) grant # 1-R01-CA154517 (Petersen, Koenig, Wolf, PIs); Robert Wood Johnson Foundation (RWJF) Investigator Award # 69763 (Wolf, PI); NIH, NHGRI University of Washington CEER grant # P50-HG003374 (Burke, PI); and NIH, NHGRI UCSF CEER grant # P20-HG007243 (Koenig, Somkin, PIs). The authors additionally participate in the Clinical Sequencing Exploratory Research (CSER) Consortium supported by NHGRI and NCI. All views expressed are those of the authors and do not necessarily reflect the views of NIH, NCI, NHGRI, RWJF, or the CSER Consortium. Thanks to Ellen Wright Clayton, James P. Evans, and Muin Khoury for helpful comments and to Rebecca Branum and Emily Scholtes for research assistance. All errors are our own.
Biographies
Susan M. Wolf, J.D., is McKnight Presidential Professor of Law, Medicine & Public Policy; Faegre Baker Daniels Professor of Law; Professor of Medicine; Faculty member, Center for Bioethics; and Chair, Consortium, on Law and Values in Health, Environment & the Life Sciences at the University of Minnesota. She is one of three Principal Investigators on NIH/NCI/NHGRI grant # 1-R01-CA154517 on return of genomic results to family members, including after the death of the proband.
Wylie Burke, M.D., Ph.D., is a Professor of Bioethics and Humanities at the University of Washington and Adjunct Professor of Medicine (Medical Genetics). She is Director of the University of Washington's Center for Genomics and Healthcare Equality, which has been funded by the National Human Genome Research Institute as a Center of Excellence in ELSI Research (CEER).
Barbara A. Koenig, Ph.D., is Professor of Bioethics and Medical Anthropology based at the Institute for Health & Aging, University of California, San Francisco. Currently, she co-directs a Center of Excellence in ELSI Research that focuses on translational genomics, co-leads an NCI/NHGRI R01 on return of results in genomic biobanks, and directs the ELSI component of a U19 award focused on newborn screening in an era of whole genome analysis.
Contributor Information
Susan M. Wolf, McKnight Presidential Professor of Law, Medicine & Public Policy, Faegre Baker Daniels Professor of Law, Professor of Medicine; Faculty Member, Center for Bioethics, Chair, Consortium on Law and Values in Health, Environment & the Life Sciences, University of Minnesota, 325 Johnston Hall, 101 Pleasant St. S.E., Minneapolis, MN 55455, Tel.: 612-301-1121, swolf@umn.edu
Wylie Burke, Department of Bioethics & Humanities, Adjunct Professor, Department of Medicine, Member, Fred Hutchinson Cancer Research Center, University of Washington School of Medicine, 1107 NE 45th St., Suite 305, Seattle, WA 98105-4690, Tel.: 206-221-5482, wburke@u.washington
Barbara A. Koenig, Department of Social & Behavioral Sciences, Institute for Health and Aging, University of California, San Francisco, 3333 California St., Suite 340, San Francisco, CA 94118, Tel.: 415-710-8217, Barbara.koenig@ucsf.edu
References
- 1.See Wolf SM, et al. Managing Incidental Findings in Human Subjects Research: Analysis and Recommendations. Journal of Law, Medicine & Ethics. 2008;36(2):219–248. doi: 10.1111/j.1748-720X.2008.00266.x.
- 2.Green RC, et al. ACMG Recommendations for Reporting of Incidental Findings in Clinical Exome and Genome Sequencing. Genetics in Medicine. 2013;15(7):565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]; American College of Medical Genetics and Genomics (ACMG) Incidental Findings in Clinical Genomics: A Clarification. Genetics in Medicine. 2013;15(8):664–666. doi: 10.1038/gim.2013.82. [DOI] [PubMed] [Google Scholar]
- 3.See, e.g., Burke W, et al. Recommendations for Returning Genomic Incidental Findings? We Need to Talk! Genetics in Medicine. 2013;15(11):854–859. doi: 10.1038/gim.2013.113. McGuire AL, et al. Ethics and Genomic Incidental Findings. Science. 2013;340(6136):1047–1048. doi: 10.1126/science.1240156. Wolf SM, Annas GJ, Elias S. Patient Autonomy and Incidental Findings in Clinical Genomics. Science. 2013;340(6136):1049–1050. doi: 10.1126/science.1239119.
- 4.ACMG Board of Directors. ACMG Policy Statement: Updated Recommendations Regarding Analysis and Reporting of Secondary Findings in Clinical Genome-Scale Sequencing. Genetics in Medicine. 2015;17(1):68–69. doi: 10.1038/gim.2014.151. [DOI] [PubMed] [Google Scholar]
- 5.Anticipate and Communicate: Ethical Management of Incidental and Secondary Findings in the Clinical, Research, and Direct-to-Consumer Contexts. Washington, D.C.: Presidential Commission for the Study of Bioethical Issues; 2013. [last visited January 13, 2015]. Presidential Commission for the Study of Bioethical Issues. at < http://bioethics.gov/node/3169>. [DOI] [PubMed] [Google Scholar]
- 6.See, Wolf SM. Return of Individual Research Results and Incidental Findings: Facing the Challenges of Translational Science. Annual Review of Genomics and Human Genetics. 2013;14:557–577. doi: 10.1146/annurev-genom-091212-153506. Wolf SM. The Role of Law in the Debate over Return of Research Results and Incidental Findings: The Challenge of Developing Law for Translational Science. Minnesota Journal of Law, Science & Technology. 2012;13(2):435–448. doi: 10.2139/ssrn.2117289. Wolf SM. Incidental Findings in Neuroscience Research: A Fundamental Challenge to the Structure of Bioethics and Health Law. In: Illes J, Sahakian BJ, editors. The Oxford Handbook of Neuroethics. New York: Oxford University Press; 2011. pp. 623–634.
- 7.For the Common Rule, see Department of Health and Human Services (DHHS) Protection of Human Subjects 45 C.F.R. Part 46. [last visited January 18, 2015]; available at < http://www.hhs.gov/ohrp/humansubjects/guidance/45cfr46.html>. DHHS, Federal Policy for the Protection of Human Subjects. Common Rule. [last visited January 18, 2015]; at < http://www.hhs.gov/ohrp/humansubjects/commonrule/index.html>.
- 8.See references cited in note 6m supra.
- 9.See, e.g., Wolf SM, et al. Managing Incidental Findings and Research Results in Genomic Research Involving Biobanks & Archived Datasets. Genetics in Medicine. 2012;14(4):361–384. doi: 10.1038/gim.2012.23. Fabsitz RR, et al. Ethical and Practical Guidelines for Reporting Genetic Research Results to Study Participants: Updated Guidelines from a National Heart, Lung and Blood Institute Working Group. Circulation: Cardiovascular Genetics. 2010;3(6):574–580. doi: 10.1161/CIRCGENETICS.110.958827. Wolf SM, et al. Managing Incidental Findings in Human Subjects Research: Analysis and Recommendations. Journal of Law, Medicine & Ethics. 2008;36(2):219–248. doi: 10.1111/j.1748-720X.2008.00266.x.
- 10.See, e.g., Bledsoe MJ, et al. Return of Research Results from Genomic Biobanks: Cost Matters. Genetics in Medicine. 2013;15(2):159–160. doi: 10.1038/gim.2012.105. Clayton EW, McGuire AL. The Legal Risks of Returning Results of Genomic Research. Genetics in Medicine. 2012;14(4):473–477. doi: 10.1038/gim.2012.10.
- 11.Jarvik GP, et al. Return of Results to Research Participants: The Floor, the Ceiling, and the Choices In Between. American Journal of Human Genetics. 2014;94(6):818–826. doi: 10.1016/j.ajhg.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Presidential Commission supra note 5.
- 13.Hall A, et al. PHG Foundation. Genomics and the Boundary Between Research and Clinical Care and Treatment. [last visited January 4, 2015];2014 Apr; available at < http://www.phgfoundation.org/briefing_notes/303/>. [Google Scholar]
- 14.For an influential argument that researchers bear obligations of ancillary care, see Richardson HS. Moral Entanglements: The Ancillary-Care Obligations of Medical Researchers. New York: Oxford University Press; 2013.
- 15.See Angrist M, Jamal L. Living Laboratory: Whole-Genome Sequencing as a Learning Healthcare Enterprise. Clinical Genetics. 2015;87(4):311–318. doi: 10.1111/cge.12461. Berkman BE, Hull S, Eckstein LG. The Unintended Implications of Blurring the Line between Research and Clinical Care in a Genomic Age. Personalized Medicine. 2014;11(3):1–18. doi: 10.2217/pme.14.3. Brody H, Miller FG. The Research-Clinical Practice Distinction, Learning Health Systems, and Relationships. Hastings Center Report. 2013;43(5):41–47. doi: 10.1002/hast.199. at 45 (discussing “reasons to hang onto the distinction, even in the face of examples like learning health systems that seem at first to challenge its utility”); Faden RR, et al. An Ethics Framework for a Learning Health Care System: A Departure from Traditional Research Ethics and Clinical Ethics. Hastings Center Report. 2013;43(1):S16–S27. doi: 10.1002/hast.134. Lyon GJ, Segal JP. Practical, Ethical and Regulatory Considerations for the Evolving Medical and Research Genomics Landscape. Applied & Translational Genomics. 2013;2(1):34–40. doi: 10.1016/j.atg.2013.02.001. Phimister EG, Feero WG, Guttmacher AE. Realizing Genomic Medicine. New England Journal of Medicine. 2012;366(8):757–759. doi: 10.1056/NEJMe1200749. at 757 (“The potential consequences of blurring clinical and research infrastructures are considerable, and such a merger should not be undertaken without extensive public debate”); Largent EA, Joffe S, Miller FG. Can Research and Care Be Ethically Integrated? Hastings Center Report. 2011;41(4):37–46. doi: 10.1002/j.1552-146x.2011.tb00123.x. at 38 (“Thirty years after Belmont, the sharp distinction between research and care is becoming increasingly blurred”).
- 16.CSER. Clinical Sequencing Exploratory Research. [last visited January 13, 2015]; at < https://cser-consortium.org/>. [Google Scholar]; National Human Genome Research Institute (NHGRI) Clinical Sequencing Exploratory Research (CSER) [last visited January 13, 2015]; at < http://www.genome.gov/27546194>.
- 17.eMERGE Network: Electronic Medical Records & Genomics. [last visited December 31, 2014]; at < http://emerge.mc.vanderbilt.edu/>. [Google Scholar]; NIH, Electronic Medical Records and Genomics (eMERGE) Network. [last visited January 18, 2015]; at < http://www.genome.gov/27540473>.
- 18.Pharmacogenomics Research Network (PGRN) [last visited January 18, 2015]; at < http://pgrn.org/display/pgrnwebsite/PGRN+Home>. [Google Scholar]; NIH, National Institute of General Medical Sciences (NIGMS) NIH Pharmacogenomics Research Network. [last visited January 18, 2015]; at < http://www.nigms.nih.gov/Research/SpecificAreas/PGRN/Pages/default.aspx>.; NIH, National Heart, Lung and Blood Institute (NHLBI) PGRN: Pharmacogenomics Research Network. [last visited January 18, 2015]; at < http://www.nhlbi.nih.gov/research/resources/genetics-genomics/pgrn>.
- 19.See Zerhouni EA. Translational and Clinical Science: Time for a New Vision. New England Journal of Medicine. 2005;353(15):1621–1623. doi: 10.1056/NEJMsb053723. See also Zerhouni EA. Clinical Research at a Crossroads: The NIH Roadmap. Journal of Investigative Medicine. 2006;54(4):171–173. doi: 10.2310/6650.2006.X0016. For prior NIH history, see NIH, Intramural Research Program, Advancing Translational Science. [last visited January 20, 2015]; at < http://irp.nih.gov/nih-clinical-center/advancing-translational-science>.
- 20.See Woolf SH. The Meaning of Translational Research and Why It Matters. [last visited January 18, 2015];JAMA. 2008 299(2):211–213. doi: 10.1001/jama.2007.26. NIH, National Center for Advancing Translational Sciences (NCATS) at < http://www.ncats.nih.gov/>. NIH. NCATS, Clinical and Translational Science Awards. [last visited January 18, 2015]; at < http://www.ncats.nih.gov/research/cts/ctsa/ctsa.html>. See also Institute of Medicine Leshner AI, et al. The CTSA Program at NIH: Opportunities for Advancing Clinical and Translational Research. Washington, D.C.: National Academies Press; 2013. [last visited January 6, 2015]. available at < http://www.iom.edu/Reports/2013/The-CTSA-Program-at-NIH-Opportunities-for-Advancing-Clinical-and-Translational-Research.aspx>.
- 21.See, e.g., [last visited January 18, 2015];Clinical and Translational Science. at < http://onlinelibrary.wiley.com/journal/10.1111/%28ISSN%291752-8062>. [last visited January 18, 2015];Applied & Translational Genomics. at < http://www.journals.elsevier.com/applied-and-translational-genomics/>.
- 22.Woolf supra note 20, at 211.
- 23.Green ED, Guyer MS. National Human Genome Research Institute: Charting a Course for Genomic Medicine from Base Pairs to Bedside. Nature. 2011;470(7333):204–213. doi: 10.1038/nature09764. at 204. [DOI] [PubMed] [Google Scholar]
- 24.Khoury MJ, et al. The Continuum of Translational Research in Genomic Medicine: How Can We Accelerate the Appropriate Integration of Human Genome Discoveries into Health Care and Disease Prevention? Genetics in Medicine. 2007;9(10):665–674. doi: 10.1097/GIM.0b013e31815699d0. at 666. [DOI] [PubMed] [Google Scholar]
- 25.Khoury MJ, et al. Beyond Base Pairs to Bedside: A Population Perspective on How Genomics Can Improve Health. American Journal of Public Health. 2012;102(1):34–37. doi: 10.2105/AJPH.2011.300299. [DOI] [PMC free article] [PubMed] [Google Scholar]; Khoury MJ, et al. Knowledge Integration at the Center of Genomic Medicine. Genetics in Medicine. 2012;14(7):643–647. doi: 10.1038/gim.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.On the multiple models proposed, see Trochim W, et al. Evaluating Translational Research: A Process Marker Model. Clinical and Translational Science. 2011;4(3):153–162. doi: 10.1111/j.1752-8062.2011.00291.x.
- 27.See, e.g., Evans JP, Khoury MJ. The Arrival of Genomic Medicine to the Clinic Is Only the Beginning of the Journey. Genetics in Medicine. 2013;15(4):268–269. doi: 10.1038/gim.2012.133. Manolio TA, et al. Implementing Genomic Medicine in the Clinic: The Future Is Here. Genetics in Medicine. 2013;15(4):258–267. doi: 10.1038/gim.2012.157.
- 28.See Goering S, Holland S, Edwards K. Making Good on the Promise of Genetics: Justice in Translational Science. In: Burke W, et al., editors. Achieving Justice in Genomic Translation: Rethinking the Pathway to Benefit. New York: Oxford University Press; 2011. pp. 3–21. Kelley M, et al. Values in Translation: How Asking the Right Questions Can Move Translational Science Toward Greater Health Impact. Clinical and Translational Science. 2012;5(6):445–451. doi: 10.1111/j.1752-8062.2012.00441.x.
- 29.Goering et al. supra note 28, at 7.
- 30.Henderson GE, et al. The Challenge of Informed Consent and Return of Results in Translational Genomics: Empirical Analysis and Recommendations. Journal of Law, Medicine & Ethics. 2014;42(3):344–355. doi: 10.1111/jlme.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.CSER: Clinical Sequencing Exploratory Research. [last visited January 13, 2015]; at < https://cser-consortium.org/> (emphasis added). [Google Scholar]
- 32.National Human Genome Research Institute (NHGRI) Clinical Sequencing Exploratory Research (CSER) [last visited January 13, 2015]; at < http://www.genome.gov/27546194> (emphasis added).
- 33.Maienschein J, et al. The Ethos and Ethics of Translational Research. AJOB. 2008;8(3):43–51. doi: 10.1080/15265160802109314. at 43. [DOI] [PubMed] [Google Scholar]
- 34. Sofaer N, Eyal N. The Diverse Ethics of Translational Research. AJOB. 2010;10(8):19–30. doi: 10.1080/15265161.2010.494214. at 20. Kimmelman and London respond to the concerns over translational research by offering a model of the drug pipeline that envisions information about the safe and effective use of products as the main output, rather than the products themselves. Kimmelman J, London AJ. The Stucture of Clinical Translation: Efficiency, Information, and Ethics. Hastings Center Report. 2015;45(2):27–39. doi: 10.1002/hast.433.
- 35.Kelley et al. supra note 28; Shapiro RS, Layde PM. Integrating Bioethics into Clinical and Translational Science Research: A Roadmap. Clinical and Translational Science. 2008;1(1):67–70. doi: 10.1111/j.1752-8062.2008.00005.x.
- 36.Burke W, et al. Translational Genomics: Seeking a Shared Vision of Benefit. AJOB. 2008;8(3):54–56. doi: 10.1080/15265160802109488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin JS, et al. Evaluating Genomic Tests from Bench to Bedside: A Practical Framework. BMC Medical Informatics and Decision Making. 2012;12(1):117–125. doi: 10.1186/1472-6947-12-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Id., at 123.
- 39.Clyne M, et al. Horizon Scanning for Translational Genomic Research Beyond Bench to Bedside. Genetics in Medicine. 2014;16(7):535–538. doi: 10.1038/gim.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schully SD, Khoury MJ. What Is Translational Genomics? An Expanded Research Agenda for Improving Individual and Population Health. Applied & Translational Genomics. 2014;3(4):82–83. doi: 10.1016/j.atg.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Puggal MA, et al. Translation of Genetics Research to Clinical Medicine: The National Heart, Lung and Blood Institute Perspective. Circulation: Cardiovascular Genetics. 2013;6(6):634–639. doi: 10.1161/CIRCGENETICS.113.000227. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schully SD, Benedicto CB, Khoury MJ. How Can We Stimulate Translational Research in Cancer Genomics Beyond Bench to Bedside? Genetics in Medicine. 2012;14(1):169–170. doi: 10.1038/gim.2011.12. [DOI] [PubMed] [Google Scholar]; Schully SD, et al. Translational Research in Cancer Genetics: The Road Less Traveled. Public Health Genomics. 2011;14(1):1–8. doi: 10.1159/000272897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Compare, e.g. Berg JS, Khoury MJ, Evans JP. Deploying Whole Genome Sequencing in Clinical Practice and Public Health: Meeting the Challenge One Bin at a Time. Genetics in Medicine. 2011;13(6):499–504. doi: 10.1097/GIM.0b013e318220aaba. at 500 (arguing for “[t]he imperative to ignore variants of unknown significance”); McGuire AL, Lupski JR. Personal Genome Research: What Should the Participant Be Told? Trends in Genetics. 2010;26(5):199–201. doi: 10.1016/j.tig.2009.12.007. at 200 (“At this early stage of WGS research there should not be a moral or legal obligation to return results of unproven significance”); Richards CS, et al. ACMG Recommendations for Standards for Interpretation and Reporting of Sequence Variations: Revision 2007. Genetics in Medicine. 2008;10(4):294–300. doi: 10.1097/GIM.0b013e31816b5cae. at 296 (“Because this information may be used for medical decisions, such as surgery or pregnancy termination, the recommended conservative approach is to avoid speculation and simply classify the variant as one of unknown clinical significance, even if potentially more frustrating for the patient”). See also Institute of Medicine Beachy SH. Assessing Genomic Sequencing Information for Health Care Decision Making: Workshop Summary. Washington, DC: National Academies Press; 2014. [last visited January 19, 2015]. at 17 available at < http://www.iom.edu/reports/2014/assessing-genomic-sequencing-information-for-health-care-decision-making.aspx>. “The vast majority of genetic variants have no known clinical relevance. The challenge, Berg said, is therefore to parse through variants to determine which ones can be used to inform clinical decisions. This process requires setting a high bar for which variants from a genome-scale test to report; otherwise, reporting variants with unknown clinical validity…or unknown implications for the asymptomatic patient’s health could potentially have negative impacts, such as patient concern…or unnecessary medical costs for testing…”). For the experience of an informed patient dealing with variants of unknown significance (VUSs), see Couzin-Frankel J. Unknown Significance. Science. 2014;346(6214):1167–1170. doi: 10.1126/science.346.6214.1167. For data on participants’ interest in receiving VUSs, see, e.g., Facio FM, et al. Intentions To Receive Individual Results from Whole-Genome Sequencing Among Participants in the ClinSeq Study. European Journal of Human Genetics. 2013;21(3):261–265. doi: 10.1038/ejhg.2012.179.
- 41.See Goering et al. and Kelley et al. supra note 28.
- 42.See discussion in Wolf, et al. Managing Incidental Findings and Research Results in Genomic Research Involving Biobanks & Archived Datasets. supra. (9) doi: 10.1038/gim.2012.23. at 364.
- 43.Brothers KB. Biobanking in Pediatrics: The Human Nonsubjects Approach. Personalized Medicine. 2011;8(1):79. doi: 10.2217/pme.10.70. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brothers KB, Clayton EW. Human Non-Subjects Research”: Privacy and Compliance. AJOB. 2010;10(9):15–17. doi: 10.1080/15265161.2010.492891. [DOI] [PubMed] [Google Scholar]
- 44. U.S. Department of Health and Human Services (DHHS) Human Subjects Research Protections: Enhancing Protections for Research Subjects and Reducing Burden, Delay, and Ambiguity for Investigators. [last visited January 9, 2015];Federal Register. 2011 76:44,512–44,531. available at < http://www.gpo.gov/fdsys/pkg/FR-2011-07-26/html/2011-18792.htm>. A 2014 statute calls for taking the next step -- issuance of a notice of proposed rule-making -- by June 2015 and promulgation of the final rules by December 2016. See Newborn Screening Saves Lives Reauthorization Act of 2014, H. R. 1281, 113th Congress, Pub. L. [last visited March 30, 2015];:113–240. Section 12 available at < https://www.govtrack.us/congress/bills/113/hr1281>. Note that the NIH Genomic Data Sharing Policy that applies beginning. 2015 Jan 25; states that “For studies proposing to use genomic data from cell lines or clinical specimens that were created or collected after the effective date of the Policy, NIH expects that informed consent for future research use and broad data sharing will have been obtained even if the cell lines or clinical specimens are de-identified” National Institutes of Health Genomic Data Sharing Policy. [last visited March 30, 2015];2014 Aug 27; at 5 available at < http://gds.nih.gov/PDF/NIH_GDS_Policy.pdf> (citation omitted).
- 45.Newborn Screening Saves Lives Reauthorization Act of 2014 supra note 44.
- 46.Hudson KL, Collins FS. Biospecimen Policy: Family Matters. Nature. 2013;500(7461):141–142. doi: 10.1038/500141a. [DOI] [PMC free article] [PubMed] [Google Scholar]; National Institutes of Health (NIH) Advisory Committee to the Director, HeLa Genome Data Access Working Group, Background, last reviewed. [last visited January 13, 2015];2013 Aug 7; at < http://acd.od.nih.gov/hlgda.htm>.
- 47.See, e.g., Wolf, et al. Managing Incidental Findings and Research Results in Genomic Research Involving Biobanks & Archived Datasets. supra. 9 doi: 10.1038/gim.2012.23. at 377. National Cancer Institute (NCI) Office of Biorepositories and Biospecimen Research, NCI Best Practices for Biospecimen Resources, 2011. [last visited January 9, 2015]; at < http://biospecimens.cancer.gov/bestpractices/2011-NCIBestPractices.pdf>.
- 48.Trochim et al. supra note 26.
- 49.Khoury et al., “Beyond Base Pairs supra note 25; Trochim et al. supra note 26; Khoury et al., “The Continuum supra note 24.
- 50.See Goering et al. and Kelley et al. supra note 28.
- 51.On organizational ethics, see, e.g., Boyle PJ, et al. Organizational Ethics in Health Care: Principles, Cases, and Practical Solutions. San Francisco: John Wiley & Sons; 2001. literature cited and analyzed in Bishop LJ, Cherry MN, Darragh M National Reference Center for Bioethics Literature. Organizational Ethics and Health Care: Expanding Bioethics to the Institutional Arena,” Kennedy Institute of Ethics, Scope Note 36. [last visited January 13, 2015]; doi: 10.1353/ken.1999.0009. at < http://220.227.128.112/downloads/ProfessionalEthics/sn36.pdf>.
- 52.Institute of Medicine. Committee on the Learning Health Care System in America. Best Care at Lower Cost: The Path to Continuously Learning Health Care in America [Google Scholar]; Smith M, editor. Washington, D.C.: National Academies Press; 2012. [Google Scholar]; Grossmann C, et al. for the Institute of Medicine. Engineering a Learning Healthcare System: A Look at the Future, Workshop Summary. Washington, D.C.: National Academies Press; 2011. Committee on the Learning Health Care Systems in America. [Google Scholar]; Olsen L, Aisner D, McGinnis JM, editors. The Learning Healthcare System: Workshop Summary. Washington, D.C.: National Academies Press; 2007. Institute of Medicine, IOM Roundtable on Evidence-Based Medicine. [PubMed] [Google Scholar]
- 53.Faden et al. supra note 15, at S22.
- 54.See Green et al. supra note 2 (advocating opportunistic screening in individual sequencing); Burke et al. supra note 3 (arguing that the evidence base for such screening is not yet in place).
- 55.See e.g., Knoppers BM, et al. Whole-Genome Sequencing in Newborn Screening Programs. [last visited January 20, 2015];Science Translational Medicine. 2014 6(229):cm2–cm5. doi: 10.1126/scitranslmed.3008494. posted online at < http://stm.sciencemag.org/content/6/229/229cm2.full.pdf>. For controversy over genomic sequencing in newborn screening, see also Goldenberg AJ, Sharp RR. The Ethical Hazards and Programmatic Challenges of Genomic Newborn Screening. JAMA. 2012;307(5):461–462. doi: 10.1001/jama.2012.68. Tarini BA, Goldenberg AJ. Ethical Issues with Newborn Screening in the Genomics Era. Annual Review of Genomics and Human Genetics. 2012;13:381–393. doi: 10.1146/annurev-genom-090711-163741. Clayton EW. State Run Newborn Screening in the Genomic Era, or How To Avoid Drowning When Drinking from a Fire Hose. Journal of Law, Medicine & Ethics. 2010;38(3):697–700. doi: 10.1111/j.1748-720X.2010.00522.x.
- 56.Wilson JMG, Jungner YG. Principles and Practice of Screening for Disease. Geneva: World Health Organization; 1968. [last visited January 6, 2015]. available at < http://www.who.int/bulletin/volumes/86/4/07-050112bp.pdf>. [Google Scholar]
- 57.Burke W, et al. Extending the Reach of Public Health Genomics: What Should Be the Agenda for Public Health in an Era of Genome-Based and “Personalized” Medicine? Genetics in Medicine. 12(12):785–791. doi: 10.1097/GIM.0b013e3182011222. at 789. [DOI] [PubMed] [Google Scholar]
- 58. Goddard KAB, et al. Description and Pilot Results from a Novel Method for Evaluation Return of Incidental Findings from Next-Generation Sequencing Technologies. Genetics in Medicine. 2013;15(9):721–728. doi: 10.1038/gim.2013.37. Howard HC, et al. The Ethical Introduction of Genome-Based Information and Technologies into Public Health. Public Health Genomics. 2013;16(3):100–109. doi: 10.1159/000346474. Khoury et al., “Beyond Base Pairs,” supra note 25; Berg et al., supra note 38; Burke et al. (2010), supra note 54, at 789 (“Developments in public health genomics require that attention is focused on managing multiple ethical issues…. Where ethically informed practices do not already exist, they should be developed.”).
- 59.Khoury et al., “The Continuum supra note 24, at 671 quoting from Lipscomb J, Donaldson MS, Hiatt RA. Cancer Outcomes Research and the Arenas of Application. Journal of the National Cancer Institute Monographs. 2004;33:1–7. doi: 10.1093/jncimonographs/lgh038.
- 60.On the ethical issues raised by outcomes assessment and comparative effectiveness research, see, e.g., Frank L, et al. Conceptual and Practical Foundations of Patient Engagement in Research at the Patient-Centered Outcomes Research Institute. [last visited January 19, 2015];Quality of Life Research. 2015 Jan 6; doi: 10.1007/s11136-014-0893-3. 2015, published online at < http://download.springer.com/static/pdf/142/art%253A10.1007%252Fs11136-014-0893-3.pdf?auth66=1421720914_420ea4c5e4967fb29363839619224674&ext=.pdf>. Gray EA, Thorpe JH. Comparative Effectiveness Research and Big Data: Balancing Potential with Legal and Ethical Considerations. Journal of Comparative Effectiveness Research. 2015;4(1):61–74. doi: 10.2217/cer.14.51. Lantos JD, Feudtner C. SUPPORT and the Ethics of Study Implementation: Lessons for Comparative Effectiveness Research from the Trial of Oxygen Therapy for Premature Babies. Hastings Center Report. 2015;45(1):30–40. doi: 10.1002/hast.407. Frank L, Basch E, Selby J. The PCORI Perspective on Patient-Centered Outcomes Research. JAMA. 2014;312(15):1513–1514. doi: 10.1001/jama.2014.11100. Lantos JD, Spertus JA. The Concept of Risk in Comparative-Effectiveness Research. New England Journal of Medicine. 2014;371(22):2129–2130. doi: 10.1056/NEJMhle1413301. Platt R, Kass NE, McGraw D. Ethics, Regulation, and Comparative Effectiveness Research: Time for a Change. JAMA. 2014;311(15):1497–1498. doi: 10.1001/jama.2014.2144. Kass N, Faden R, Tunis S. Addressing Low-Risk Comparative Effectiveness Research in Proposed Changes to US Federal Regulations Governing Research. JAMA. 2012;307(15):1589–1590. doi: 10.1001/jama.2012.491. See also Faden et al. supra note 15.
- 61.Faden et al. supra note 15, at S16.
- 62.See, e.g., IOM Assessing Genomic Sequencing supra note 37, at 25 more data need to be generated from different racial and ethnic groups Lynch J, et al. Race and Genomics in the Veterans Health Administration. American Journal of Public Health. 2014;104(suppl. 4):S522–S524. doi: 10.2105/AJPH.2014.302202. (“There are multiple barriers to genomic medicine for minorities…. Most large scale genome wide association studies have been conducted on populations of European ancestry”) (reference omitted); Kohane IS, Hsing M, Kong SW. Taxonomizing, Sizing, and Overcoming the Incidentalome. Genetics in Medicine. 2012;14(4):399–404. doi: 10.1038/gim.2011.68. at 401–403. “Fundamentally, the utility of the clinical annotation of a genomic variant is only as useful as its applicability to a patient. That is, if a variant were found to track with a disease in a specified group of patients, that annotation may in fact serve well if one belongs to that specific group of patients but serve rather poorly if one does not”). On a cautionary note, see Sankar P, et al. Genetic Research and Health Disparitie. JAMA. 2004;291(24):2985–2989. doi: 10.1001/jama.291.24.2985. at 2985. (“overemphasis on genetics as a major explanatory factor in health disparities could lead researchers to miss factors that contribute to disparities more substantially”). On disparities in health care more broadly, see IOM, Smedley BD, Stith AY, Nelson AR, editors. Unequal Treatment: Confronting Racial and Ethnic Disparities in Health Care. Washington, D.C.: National Academies Press; 2003. [last visited January 19, 2015]. Committee on Understanding and Eliminating Racial and Ethnic Disparities in Health Care. available at < https://www.iom.edu/Reports/2002/Unequal-Treatment-Confronting-Racial-and-Ethnic-Disparities-in-Health-Care.aspx>.
- 63.Compare, e.g., Green et al. supra note 2 (urging opportunistic genomic screening) with the critique by Burke et al. supra note 3 (arguing that the evidence base is not in place to justify such screening).
- 64.See Hall et al. supra note 13.
- 65.See, e.g., IOM Assessing Genomic Sequencing supra note 38, at 26–27 (on unresolved challenges in establishing the reproducibility of genomic data, eliminating inaccurate published associations, and agreeing on standards to prove a genotype-phenotype association).
- 66.Khoury at al., “Beyond Base Pairs supra note 25.
- 67.See Goering et al. and Kelley et al. supra note 28.
- 68. Burke W, Evans BJ, Jarvik GP. Return of Results: Ethical and Legal Distinctions Between Research and Clinical Care. American Journal of Medical Genetics. 2014;Part C 166C:105–111. doi: 10.1002/ajmg.c.31393. The Clinical Laboratory Improvement Amendments (CLIA) are explained at Centers for Medicare & Medicaid Services (CMS) Clinical Laboratory Improvement Amendments (CLIA) [last visited March 30, 2015]; at < http://www.cms.gov/Regulations-and-Guidance/Legislation/CLIA/index.html?redirect=/clia/>. See also Centers for Disease Control and Prevention (CDC) Clinical Laboratory Improvement Amendments (CLIA) [last visited March 30, 2015]; at < http://wwwn.cdc.gov/clia/regulatory/default.aspx>.
- 69.On research versus clinical standards for sequencing, see, e.g., Burke et al. supra note 68. FDA standards for sequencing are in transition. See. e.g, Lander ES. Cutting the Gordian Helix – Regulating Genomic Testing in the Era of Precision Medicine. New England Journal of Medicine. 2015;372(13):1185–1186. doi: 10.1056/NEJMp1501964. Food and Drug Administration (FDA) Optimizing FDA’s Regulatory Oversight of Next Generation Sequencing Diagnostic Tests – Preliminary Discussion Paper. [last visited April 1, 2015];2015 available at < http://www.fda.gov/downloads/MedicalDevices/NewsEvents/WorkshopsConferences/UCM427869.pdf>. FDA announcement of a new approach to regulating laboratory-developed tests (LDTs), including in genomics, has met with considerable controversy. Compare, e.g., Sharfstein J. FDA Regulation of Laboratory-Developed Diagnostic Tests: Protect the Public, Advance the Science. JAMA. 2015;313(7):667–668. doi: 10.1001/jama.2014.18135. with Evans JP, Watson MS. Genetic Testing and FDA Regulation: Overregulation Threatens the Emergence of Genomic Medicine. JAMA. 2015;313(7):669–670. doi: 10.1001/jama.2014.18145.
- 70.Compare IOM Assessing Genomic Sequencing supra note 40, at 7 (“Exome sequencing can be used for either clinical or research purposes, though recently the boundaries between the two have been blurring. In Hegde’s laboratory, exome data are divided according to why the sequencing is being done. For new disease presentations the diagnostic yield, or likelihood that the test will provide enough information to make an appropriate diagnosis, ranges roughly from 30 percent to 40 percent, depending on which laboratory is reporting and what kinds of cases are considered, Hegde said. When writing clinical reports, she said, it is critical to sorting the data into categories of what can be interpreted in the clinic and what is clinically actionable…”).
- 71.Author S. M. W. thanks Hank Greely for early conversation on this point.
- 72.See, e.g., Callahan D, Jennings B. Ethics and Public Health: Forging a Strong Relationship. American Journal of Public Health. 2002;92(2):169–176. doi: 10.2105/ajph.92.2.169. Kass NE. An Ethics Framework for Public Health. American Journal of Public Health. 2001;91(11):1776–1782. doi: 10.2105/ajph.91.11.1776.
- 73.See Trinidad SB, et al. Research Practice and Participant Preferences: The Growing Gulf. Science. 2011;331(6015):287–288. doi: 10.1126/science.1199000. at 288 (“We propose a shift from paternalistic protections to respectful engagement with individuals and groups whose conceptions of risk, benefit, and harm deserve consideration. Such an approach would treat participants as true stakeholders in research, who willingly take on risk because they see the potential benefits to society as outweighing potential harms”); Kohane IS, et al. Reestablishing the Researcher-Patient Compact. Science. 2007;316(5826):836–837. doi: 10.1126/science.1135489. at 837 (arguing that it is “ethically superior” to treat “patients as partners in research rather than passive, disenfranchised purveyors of biomaterials and data”).
- 74.See Wolf et al., “Managing Incidental Findings and Research Results in Genomic Research Involving Biobanks & Archived Datasets supra note 9; Fabsitz et al. supra note 9.
- 75.Khoury et al., “Knowledge Integration supra note 25.
- 76.See references cited in note 6 supra.
- 77.Wolf SM, et al. Returning a Research Participant’s Genomic Results to Relatives: Analysis and Recommendations. Journal of Law, Medicine & Ethics. doi: 10.1111/jlme.12288. __, no. __(2015): __-__. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.See sources cited and discussed in id.
- 79.Id.