Summary
In 2010, Coxiella burnetii was identified at a high prevalence in the placentas of Northern fur seals (Callorhinus ursinus) collected at a single rookery on St. Paul Island Alaska; an area of the United States where the agent was not known to be present. As contamination was hypothesized as a potential cause of false positives, but nothing was known about environmental C. burnetii in the region, an environmental survey was conducted to look for the prevalence and distribution of the organism on the island. While environmental prevalence was low, two strains of the organism were identified using PCR targeting the COM1 and IS1111 genes. The two strains are consistent with the organism that has been increasingly identified in marine mammals as well as a strain type more commonly found in terrestrial environments and associated with disease in humans and terrestrial animals. Further work is needed to elucidate information regarding the ecology of this organism in this region, particularly in association with the coastal environment.
Introduction
Coxiella burnetii, the causative agent of human Q fever, is a gram negative intracellular bacteria that is found worldwide9. The organism causes a broad range of clinical disease in humans and animals, ranging from asymptomatic infection to reproductive loss and systemic infection10. Inhalation of contaminated aerosols are the most significant route of infection for humans, but ingestion, tick vectors and sexual transmission are possible1. In domestic animals, the organism is shed in abundance during parturition9 and can persist in the environment for protracted periods of time2 resulting in opportunities for spread. In the Netherlands, contaminated soil downwind from endemically infected goat farms was identified as a risk factor for human Q fever3. Environmental C. burnetii was identified in almost one quarter of the 1,622 samples collected from six regions in the USA8 however no samples from Alaska were included in that investigation.
Northern fur seals (NFS, Callorhinus ursinus) are a highly migratory, colonial pinniped which breed primarily on the Pribilof Islands (St. Paul, St. George) and other islands in the Bering Sea and North Pacific Ocean. They have a polygynous breeding system that is highly synchronous. About 95% of pregnant adult female NFS give birth within a six week period, with the midpoint of pupping around July 10 on St. Paul rookeries6. The seals are seasonal residents on their breeding islands and remain pelagic during the winter and spring before returning to their breeding islands. These animals show strong philopatry to their terrestrial6 sites and site fidelity to broad marine foraging areas11,14. The NFS population on the Pribilof Islands has declined severely over the past decade16, and are listed as ‘depleted’ under the Marine Mammal Protection Act. NMFS estimates approximately 93,627 pups were born on the 14 St. Paul Island breeding areas (rookeries) in 201015.
In 2010, C. burnetii was identified in 75% of the placentas collected from a single NFS rookery on St. Paul Island Alaska4. A striking feature of the C. burnetii DNA in the NFS was that PCR assays targeting the IS1111 insertion sequence were negative for these strains, even when C. burnetii was easily detected by PCR targeting the com1 gene. The feature of having a poorly amplifiable IS1111 has been found in C. burnetii derived from other marine mammals7, but in C. burnetii derived from all other animals and the environment, the IS1111 gene target is multi-copy resulting in a highly sensitive PCR. Thus, a lack of IS1111 amplification can potentially be used to distinguish marine mammal-derived C. burnetii from strains derived from terrestrial animals. As NFS have high site fidelity it was hypothesized that some of the PCR positive placentas may have been contaminated by soil on the rookery. Despite the high prevalence of C. burnetii in NFS placentas by PCR, the organism was only visualized by IHC in 3% of the tissues4. The discrepancy between PCR and IHC could be partly explained by the greater sensitivity of PCR, but contamination of placentas with C. burnetii from the environment could also play a role. The objective of this study was to look for the distribution of environmental C. burnetii on St. Paul Island in an attempt to identify potential sources of NFS infection and whether any non-marine mammal strains are on the island.
Results
Twenty-seven environmental samples were collected on Reef rookery prior to pupping season in April, allowing bulk samples to be taken directly from the breeding areas. Eighteen (67%) of these samples were positive by COM1, and no samples were positive by IS1111. The genome equivalents per gram were variable, ranging from 59-1,863. Positive samples were from both the center of the rookery as well as non-birthing habitat immediately behind the rookery and there was no apparent pattern in the positive samples.
In July, during peak NFS pupping, 149 environmental samples were collected; 122 soil samples from various coastal and interior locations on the island and 27 swabs were collected from Reef rookery. The majority (105) of the samples were classified as ‘seal associated’ as they were collected from non-birthing, but seal resting areas behind NFS rookeries. Thirty-five samples were classified as ‘human associated’ with 17 of these being from areas of perceived high human usage, such as public locations in town, and 18 from areas of low usage, such as roadsides and remote sites away from the town. The remaining 9 samples were classified as ‘non-seal, non-human associated’ and represented soil samples collected from sites with little or no human or marine mammal presence not accessible by road.
The distribution of positive and negative samples from the NFS pupping season sampling is shown in figure 1. Overall, 16 of the 149 samples (10.7%) were positive by one or both PCRs. Four of the samples were positive for only com1, 11 samples were only positive for IS1111, and 1 sample was positive for both. There was significant PCR inhibition present in 59 of the samples despite multiple sample cleaning attempts using techniques previously found to be very effective at cleaning up environmental DNA5. As inhibition was thought to only result in false negatives, inhibited samples in which no organism was detected were excluded from further analysis and the adjusted total sample size was 95, representing 72 soil samples and 23 swabs collecting during the pupping season. The quantity of DNA within the environmental samples was low, ranging from 4-4502 genome equivalents per gram of soil or per swab extract. The highest genomic equivalents were com1 positive samples from Northeast Point and Lukanin rookeries.
Figure 1.
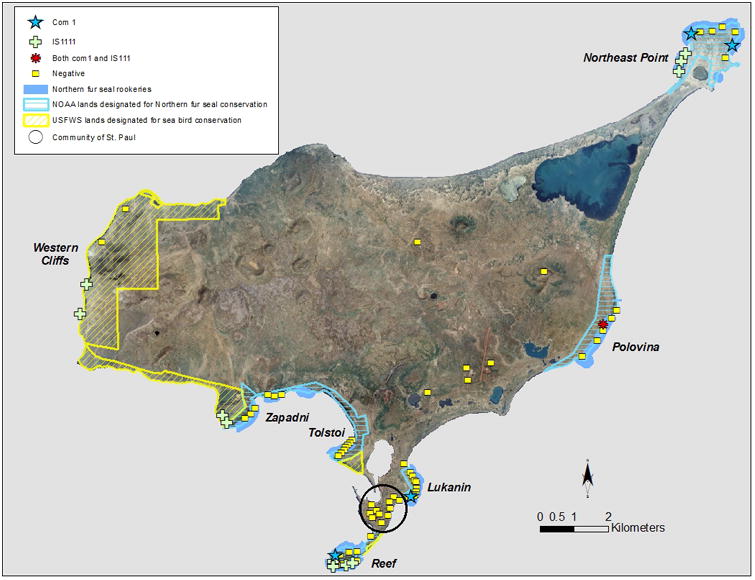
Distribution and C. burnetii PCR results for environmental samples on St. Paul Island, Alaska
Twelve (75%) of the positive environmental samples were classified as ‘seal associated’; 7 were positive for IS1111 only, 4 were positive for COM1 only and 1 was positive by both PCRs. Four of the positive samples (25%) were classified as ‘non-seal non-human associated’; all of these samples were positive for IS1111 and negative for COM1 and all had very low organism counts. No positives were found in either the low or high intensity human-associated (town or in inland, non-seal-associated) samples.
The number of positive samples by NFS rookery is listed in Table 1. On Northeast Point, 3 of the 5 positive samples (for IS1111) were clustered in the south western section of the area (Figure 1). Two of 27 (7.4%) swab samples collected from the walkways on Reef rookery were positive, both for IS1111 and negative for COM1. The COM1 positive samples were from Reef periphery, Northeast Point, Polovina and Lukanin rookeries. Of the ‘non-seal, non-human associated’ samples, 2 of 6 (33.3%) of the samples above the High Bluffs bird colonies and 2 of 3 (66.6%) samples from above the Zapadni bird colonies were positive by IS1111 but negative on COM1; these areas are referred to as the Western Cliffs sea bird conservation area.
Table 1.
C. burnetii positive and inhibited PCR samples taken near or on NFS rookeries on St. Paul Island, Alaska.
| Rookery | Positive samples | Samples collected | Percent positive | Inhibited samples |
|---|---|---|---|---|
| Reef on rookery, pre pupping season (soil) | 18 | 27 | 66.7 | 0 |
| Reef on rookery, pupping season (swabs) | 2 | 27 | 7.4 | 6 |
| Reef periphery | 3 | 12 | 25.0 | 1 |
| Lukanin | 1 | 12 | 8.3 | 1 |
| Tolstoi | 0 | 8 | 0 | 1 |
| Zapadni | 0 | 15 | 0 | 9 |
| Polovina | 1 | 12 | 8.3 | 6 |
| Northeast Point | 5 | 19 | 26.3 | 13 |
NFS placentas were also collected for this study. Of 119 placentas collected at Reef rookery, 91 (76.5%) were positive on PCR for COM1. Genomes per gram was variable, ranging from 42-270,929,000 with a mean of 4,154,309 ± 28,722,289; 12 of 119 (10%) placentas had organism counts greater or equal to 3×105/g, the cutoff in which organism was observed histologically in the previous study4. Eight (6.7%) of the COM1-positive placentas were also positive for IS1111 on PCR, but the Ct values of the IS1111 PCR were much lower than would be expected based on the Ct values obtained by COM1 PCR. This supports that NFS-associated C. burnetii has an altered form of IS1111 and can be distinguished from terrestrial strains based on poor or absent IS1111 amplification.
Discussion
Results of this study suggest that multiple strains of Coxiella burnetii are present on St. Paul Island, Alaska. The distribution of the organism on land is irregular and the numbers of organism are low, but the bacterium is present in all coastal sections of the island tested. All of the positive samples came from coastal areas and no environmental Coxiella was identified in areas identified as high or low human usage. This pattern suggests that irregular distribution of the organism may be associated with breeding and marine-foraging wild animal distribution.
Almost 40% of the environmental samples had significant inhibition, even after multiple cleaning attempts. This is surprising given that the techniques used here for DNA isolation have previously been shown to be quite effective at removal of PCR inhibitors5. The cause of the high prevalence of inhibition is not known, but the high concentrations of animal waste deposited at the sampling sites where large numbers of seals return every year could be a factor. Molecular diagnostics can be inhibited by numerous environmental compounds; however, this phenomenon is most likely to result in false negatives but not false positives18. In this study, inhibited samples that were positive on PCR likely had falsely low organism counts, the cause of the inhibition in this group of samples is unclear as they were taken from multiple areas and represented both seal and non-seal associated samples. It is unknown how many inhibited samples may have been positive; however, given the exclusion of inhibited negative samples from this analysis, the true prevalence of environmental C. burnetii on St. Paul Island is likely higher than identified in the present study.
On Reef rookery, the site in which a high prevalence of infection was identified in 20104, placental testing confirmed an equally high infection prevalence in 2011. In contrast, the prevalence of organisms within the habitat on the rookery during the pupping season was low, less than 10% of the non-inhibited on-rookery samples were positive. When these results are compared to the pre-pupping samples there was a significant difference as 66.7% of the pre-pupping environmental samples were positive. This discrepancy is thought to be due to differences in sample collection protocol. The seal rookeries are predominantly rocky beaches and there is minimal ‘soil’ to be sampled. Before animals have returned to the rookery it was possible to collect samples by hand in a 15ml falcon tube while during the pupping season only swabs could be safely be collected from the overhead walkway system without disturbing the animals. The use of swabs at this location is thought to have resulted in different recovery opportunities compared to soil samples collected elsewhere on the island.
To date, C. burnetii isolated from marine mammals has a genetic pattern that differs from the more common terrestrial strains; marine mammal samples are positive for COM1 with low to absent IS1111 signal4,7. Northern fur seal placentas have been tested for two seasons and only 3% have been positive for IS1111. This data suggests that the IS1111 environmental samples may have arisen from another source. The ‘non-seal non-human associated’ sites at Zapadni and High Bluffs (along the west coast) are found above large seabird breading colonies and, although cliff tops where sampling occurred are not typically occupied by seabirds, the molecular signature and distance of some samples from NFS rookeries suggests that birds may play a role in the regional ecology this the bacteria. In recent years, Coxiella has been increasingly reported in psittacines where it is associated with multi-systemic clinical disease12,17 but asymptomatic infection of birds found shedding the organism are also reported13. Birds can also be mechanical vector for organisms and many of the piscivous or planktivorous birds nesting on St. Paul travel great distances during the year resulting in potential opportunities for pathogen movement from endemic areas. Terrestrial strains bearing multiple copies of IS1111 have been routinely identified in the environment of mainland USA8 although the source of the organism is often unknown.
Interestingly, the two swab samples that were PCR positive on Reef rookery during the pupping season, were positive only for IS1111 and negative for COM1. In contrast, only a small number of NFS placentas from Reef rookery were positive for IS1111 and all of these were also positive on COM1. This discordant result suggests that both strains of Coxiella may be present in some areas of the island. Arctic foxes and gulls are known to scavenge marine mammal placentas and their presence at the rookery during the pupping season may represent the source of the second strain.
The COM1 positive samples were collected from resting areas near Lukanin, Northeast Point and Reef periphery. The association of these COM1 positive with nearby NFS rookeries further supports the hypothesis that NFS are the most likely source for the IS1111-negative organism in the coastal environment. No placentas or animals have been tested for C. burnetii in these locations however the presence of the bacteria in the environment in these locations suggesting that seals in this location may be similarly infected to those at Reef.
The high pre-season prevalence of organism within the environment on Reef rookery supports the possibility of placental contamination by environmental organism; however there was marked discrepancy in the quantity of organism in the respective samples. Only two of 119 NFS placentas had genomic equivalents less than 1000/gram of tissue while only a single environmental sample had over 1000 genomic equivalents per gram of soil. It seems highly unlikely that the low concentration of environmental organism could result in such high numbers of organism per gram of placental tissue. This finding emphasizes the benefit of quantitative assays to differentiate true positivity from environmental contamination.
In conclusion, given the high prevalence of COM1 positive NFS placentas, many with extremely high genome equivalents per gram of tissue, the presence of only 5 environmental samples that fit the marine mammal profile suggests that the spread of NFS associated C. burnetii into the environment is low. Investigation into other sources of environmental organism may be warranted.
Experimental procedures
Environmental sampling was conducted at two time points; prior to the animals arrival to reef rookery and at the time of peak pupping in the 2011 season. The pre-arrival samples were all collected at Reef rookery on April 27 2011. Approximately 5 grams of soil was collected in a 15ml Falcon tube from 27 locations in the rookery area; 18 from the walkway area and 9 samples approximately 2 meters behind the walkway system in the direction of the haul out area. The second sampling period was from July 20-26th 2011. Targeted sampling techniques were employed in attempt to best represent regions of the island that with differing usage. Sample areas were classified as ‘seal associated’ if they were collected within a known haul out or rookery area. To minimize animal disturbance, samples were collected from the haul out area behind the rookery at most sites. From these areas 3-6 grams of soil was collected in a 15ml falcon tube and a GPS waypoint was collected at each site. On Reef rookery where a NOAA walkway system is in place, 27 additional swab samples were collected on the rookery. In these areas a sterile, cotton tipped swab was attached to a bamboo pole and lowered to the ground from the walkway. The swab was dragged along the rookery substrate, which consisted largely of rocks, retrieved and placed in a sterile 15ml falcon tube. Placentas were collected from the walkway system on Reef rookery consistent with previous reports4.
DNA was purified from the environmental samples using methods previously described5. Briefly, microorganisms in the soil were extracted with PBS, and a high-speed pellet of the PBS extract was subjected to the Qiagen QIAmp DNA stool mini-kit followed by the Qiagen QIAmp DNA mini kit tissue protocol (Qiagen, Valencia, CA). DNA was eluted in 200 μl of the elution buffer supplied with the kits. DNA was tested for PCR inhibition by spiking a PCR reaction containing 200 genome equivalents of C. burnetii Nine Mile Phase 1 DNA with 1 μl of the environmental sample DNA eluate. IS1111 PCR was run on these samples and compared to samples spiked with 1 μl of water. Inhibition was considered present if the DNA sample caused an increase of ≥1 C(t) value. IS1111 and COM1 quantitative PCR was performed as described previously7.
Acknowledgments
Sincere thanks to individuals who helped collect placental and environmental samples for use in this study: Dr. Wendi Roe, Sophie Peirszalowski, Kirsten Dullen and Dustin Carl. All tissue samples were collected under authority of U.S. Marine Mammal Permit No. 782-1708 issued to the National Marine Mammal Lab, Seattle. A.V.K. was supported by an appointment to the Emerging Infectious Diseases (EID) Fellowship program administered by the Association of Public Health Laboratories (APHL) and funded by the Centers for Disease Control and Prevention (CDC).
Footnotes
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the CDC or the Department of Health and Human Services.
References
- 1.Angelakis E, Raoult D. Q fever. Veterinary Microbiology. 2010;140:297–309. doi: 10.1016/j.vetmic.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 2.Arricau-Bouvery N, Rodolakis A. Is Q fever an emerging or re-emerging zoonosis? Vet Res. 2005;36:327–349. doi: 10.1051/vetres:2005010. [DOI] [PubMed] [Google Scholar]
- 3.Dijkstra F, van der Hoek W, Wijers N, Schimmer B, Rietveld A, Wijkmans CJ, Vellema P, Schneeberger PM. FEMS Immunology & Medical Microbiology. 2011. The 2007-2010 Q fever epidemic in the Netherlands: characteristics of notified acute Q fever patients and the association with dairy goat farming; pp. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 4.Duncan C, Kersh G, Spraker T, Patyk K, Fitzpatrick K, Massung R, Gelatt T. Coxiella burnetii in northern fur seal (Callorhinus ursinus) placentas from St.Paul Island, Alaska. Vector Borne Zoonotic Dis. 2011 doi: 10.1089/vbz.2011.0715. Online Ahead of Print: October, 21, 2011. [DOI] [PubMed] [Google Scholar]
- 5.Fitzpatrick KA, Kersh GJ, Massung RF. Practical method for extraction of PCR-quality DNA from environmental soil samples. Appl Environ Microbiol. 2010;76:4571–4573. doi: 10.1128/AEM.02825-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gentry RL. Behavior and ecology of the northern fur seal. Princeton University Press; Prinston, NJ: 1998. p. 392. [Google Scholar]
- 7.Kersh GJ, Lambourn DM, Self JS, Akmajian AM, Stanton JB, Baszler TV, Raverty SA, Massung RF. Coxiella burnetii infection of a Steller sea lion (Eumetopias jubatus) found in Washington State. J Clin Microbiol. 2010;48:3428–3431. doi: 10.1128/JCM.00758-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kersh GJ, Wolfe TM, Fitzpatrick KA, Candee AJ, Oliver LD, Patterson NE, Self JS, Priestley RA, Loftis AD, Massung RF. Presence of Coxiella burnetii DNA in the environment of the United States, 2006 to 2008. Appl Environ Microbiol. 2010;76:4469–4475. doi: 10.1128/AEM.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maurin M, Raoult D. Q fever. Clinical microbiology reviews. 1999;12:518–553. doi: 10.1128/cmr.12.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McQuiston JH, Childs JE. Q fever in humans and animals in the United States. Vector Borne Zoonotic Dis. 2002;2:179–191. doi: 10.1089/15303660260613747. [DOI] [PubMed] [Google Scholar]
- 11.Robson BW, Goebel ME, Baker JD, Ream RR, Loughlin TR, Francis RC, Antonelis GA, Costa DP. Separation of foraging habitat among breeding sites of a colonial marine predator, the northern fur seal (Callorhinus ursinus) Canadian Journal of Zoology. 2004;82:20–29. [Google Scholar]
- 12.Shivaprasad HL, Cadenas MB, Diab SS, Nordhausen R, Bradway D, Crespo R, Breitschwerdt EB. Coxiella-Like Infection in Psittacines and a Toucan. Avian Diseases. 2008;52:426–432. doi: 10.1637/8192-120707-Reg. [DOI] [PubMed] [Google Scholar]
- 13.Stein A, Raoult D. Pigeon Pneumonia in Provence: A Bird-Borne Q Fever Outbreak. Clinical Infectious Diseases. 1999;29:617–620. doi: 10.1086/598643. [DOI] [PubMed] [Google Scholar]
- 14.Sterling JT, Ream RR. At-sea behavior of juvenile male northern fur seals (Callorhinus ursinus) Canadian Journal of Zoology. 2004;82:1621–1637. [Google Scholar]
- 15.Towell R, Ream R, Bengtson J, Sterling J. 2010 northern fur seal pup production and adult male counts on the Pribil of Islands, Alaska. 2011 [Google Scholar]
- 16.Towell RG, Ream RR, York AE. Decline in northern fur seal (Callorhinus ursinus) pup production on the Pribil of Islands. Marine Mammal Science. 2006;22:486–491. [Google Scholar]
- 17.Vapniarsky N, Barr BC, Murphy B. Systemic Coxiella-like Infection With Myocarditis and Hepatitis in an Eclectus Parrot (Eclectus roratus) Veterinary Pathology Online. 2011 doi: 10.1177/0300985811409251. [DOI] [PubMed] [Google Scholar]
- 18.Wilson IG. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


