Abstract
The association between elevated circulating levels of GP73 (and fucosylated GP73 in particular) and hepatocellular carcinoma suggests that a thorough analysis of the extent of GP73 glycosylation is warranted. Detailed analysis of the glycosylation patterns of such low abundance proteins are hampered by technical difficulties. Using conventional lectin affinity chromatography, we have established that three quarters of the GP73 secreted from a cell line derived from HCC is fucosylated. Using mass spectrometry, we have established that at least two of three potential sites of N-linked glycosylation are occupied on most molecules of GP73 secreted from cultured hepatoma cells. Furthermore, the oligosaccharides added to recombinant GP73 resemble those present in the bulk of secreted protein, mostly bi-antennary with core fucose, with a smaller fraction of tri- and tetra-antennary structures. The frequency of fucosylation observed on the recombinant protein agrees well with the pattern of lectin binding of the endogenous secreted protein. Finally, we have developed a method to interrogate the glycans added to either the near full length protein or at a particular sequon, providing proof of concept that a small peptide embedded in a heterologous context can preserve both fucosylation and a high level of branching of oligosaccharides added.
Keywords: hepatocellular carcinoma, fucosylation, Golgi protein 73
Hepatocellular carcinoma (HCC) is the 5th most frequent cancer in the world, with an estimate of more than 500,000 incidences in 2000. Although HCC is less common in the US, it is increasing in incidence, and in some “at risk” populations (such as Pacific Asians in California), it is among the three leading causes of death due to cancer [El-Serag and Mason, 1999]. HCC is usually associated with a high mortality with a 5-year survival rate of less than 5% without therapeutic intervention [Di Bisceglie et al., 1998]. The high mortality may be in part because the non-capsular part of the liver is lacking in sensory fibers and symptoms of HCC often occur late-making the case for serological early detection all the more compelling.
The major etiology of primary HCC is chronic HBV and HCV infection [Anzola, 2004]. Those chronically infected with HBV have a lifetime risk of death to HCC of between 10% and 25% [El-Serag and Mason, 1999]. The epidemiology and natural history of HCV is somewhat less understood, but it appears that the lifetime risk of HCC in those chronically infected with HCV will be between 2% and 7% [Lok and McMahon, 2001; Block et al., 2003]. In hepatitis virus carriers, the onset of serious disease usually presents after several decades of infection. The long latency between infection and disease onset in this high-risk population provides an opportunity for early detection [Block et al., 2005]. Currently, disease status is usually monitored by physical assessment, ultra sound imaging of the liver and/or analysis of serum for a panel of markers [Lok et al., 2001]. However, many of the constituents of the serum panel, for example, alanine amino-transferase (ALT) levels, vary throughout the course of chronic hepatitis and others are of limited use in early detection of HCC [Sherman, 2001]. Ultra-sound detection requires a tumor mass of at least a 3 cm, by which stage the prognosis may be very poor [Di Bisceglie et al., 1998]. Since early surgical and chemo-therapeutic intervention is the best hope for patient survival [Lok et al., 2001], accurate detection of HCC is necessary to identify the need for intervention.
To this end, multiple approaches have been used to identify genetic and protein-based biomarkers, with an emphasis on those that will permit early detection of disease. There is a correlation between the level of alpha-fetoprotein (AFP) and a diagnosis of HCC [Aoyagi et al., 1993; Aoyagi et al., 1998; Lok and McMahon, 2001]. However, AFP as a sole indicator of HCC is of limited value, as it is often elevated in the absence of serious disease [Sherman, 2001]. Nevertheless, the limited correlation between AFP and HCC demonstrates the potential of serum as a source of protein biomarkers of liver disease.
A number of N- and O-linked glycosylation changes have been associated with carcinogenesis in general and HCC in particular [Hakomori, 1996; Kobata, 1998]. The appearance of core fucosylation on many glycoproteins present in the circulation correlates with HCC [Comunale et al., 2006]. More specifically, an increase in fucosylated oligosaccharides associated with secreted α-fetoprotein has been observed in HCC [Aoyagi, 1995; Aoyagi et al., 1998; Miyoshi et al., 1999]. Alterations in the core fucosylation of alpha-1-antitrypsin also have been noted [Sekine et al., 1987]. Increased fucosylation in HCC has been attributed to an increase in both the alpha1-6 fucosyltransferase (Fut8) and its substrate, GDP-fucose [Noda et al., 2003]. Fucosylated N-glycan also has been associated with various disease states [Breborowicz et al., 1981; Yamashita et al., 1989; Aoyagi, 1995; Miyoshi et al., 1999; Callewaert et al., 2004]. Thus, it is desirable to monitor the fucosylation status of individual glycoproteins, possibly even individual glycopeptides. Other glycosylation changes besides fucosylation also have been associated with HCC [Fujiwara et al., 2002]. More generally, glycosylation changes may prove diagnostic of, and possibly serve as biomarkers for, a number of cancers, as well as offering potential therapeutic targets.
We have been systematically comparing the serum proteomes from individuals chronically infected with HBV or HCV as a function of disease status [Steel et al., 2001; Comunale et al., 2004; Comunale et al., 2006]. This work has identified the glycoprotein GP73 as a potential biomarker; elevated levels in the serum correlate with HCC [Marrero et al., 2005]. The protein had previously been reported to be elevated within the liver as a consequence of acute hepatitis [Kladney et al., 2000; Kladney et al., 2002]. GP73 is normally resident in the cis-Golgi, anchored by a single membrane spanning domain near the short cytoplasmic amino terminus of the protein, with the bulk of the protein in the lumenal space [Kladney et al., 2000]. GP73 and the related protein GPP130 may under certain conditions also redistribute to the cell surface and endosomes [Puri et al., 2002]. Although the function of GP73 remains uncertain, and it is unclear which cells are the source of circulating GP73 and how it arrives in the circulation in vivo, the correlation with HCC is sufficiently high to warrant further investigation as a potential disease marker.
Based on the potential importance of GP73 as a biomarker for early detection of HCC, we have characterized the glycosylation potential of secreted GP73 in more detail. These studies have employed two liver tumor derived cell lines, one of which makes and secretes detectable GP73, and one that secretes no detectable GP73. The status of potential sites of N-glycosylation within the protein has been investigated and we have begun to characterize the nature of the glycans, including fucosylation, that are added to the polypeptide.
MATERIALS AND METHODS
Cells and Reagents
HepG2.2.15 cells are a sub-line of HepG2 cells that contain a stable HBV genome that support continuous virus replication [Sells et al., 1987]. These cells were grown in RPMI containing 10% fetal calf serum. However, prior to harvest of supernatant for glycan analysis, cells were washed and then cultured for 24 h in serum free medium. Huh7 cells are a human hepatoma derived cell line and were normally maintained in DMEM plus 10% fetal bovine serum, but were incubated overnight in serum-free medium prior to collection of supernatants for analysis.
Lectin Extraction
These procedures were performed essentially as described previously [Comunale et al., 2006]. Culture supernatants were dialyzed extensively against low salt buffer solution and then concentrated prior to analysis. Enzymatic treatments were as indicated. For detection of fucosylated protein, HepG2.2.15 cell supernatant was collected in serum free medium and dialyzed against a lectin binding solution bringing the final concentration of the sample to 20 mM Tris buffered saline (TBS), 1 mM calcium chloride, 1 mM magnesium chloride, and 1 mM manganese chloride (pH 7.0). The samples were incubated for 16 h at 4°C with agarose beads bearing the fucose binding lectin Aleuria aurantia (AAL, Vector Laboratories). Unbound material was collected, then the lectin beads were washed thoroughly with lectin binding solution before the bound fraction was eluted by heating the beads in electrophoresis sample buffer. Equivalent fractions of bound and unbound material were loaded.
Glycan Analysis
Sample preparation for glycan analysis was performed essentially as described previously [Comunale et al., 2006]. HPLC analysis was performed using the Waters Alliance HPLC System, complemented with a Waters fluorescence detector, and quantified using the Millennium Chromatography Manager (Waters Corporation, Milford, MA). Glycan structures were identified by comparison to known standards as described previously [Rudd et al., 1999; Royle et al., 2002; Comunale et al., 2006].
Western Blotting and Antibodies
Proteins were resolved by SDS–PAGE, using commercially prepared gels in either Tris-glycine (Invitrogen, Carlsbad, CA) or Tris-HEPES buffers (Pierce). Proteins were transferred to PVDF, and GP73 was detected by incubation of the membrane with anti-GP73 as described previously [Marrero et al., 2005]. In some cases, proteins were detected using either horseradish peroxidase-conjugated goat anti-rabbit secondary antibody and enhanced chemiluminescence as described previously. Alternatively, protein was detected using IR-dye-conjugated goat anti-rabbit secondary antibody (Licor, Lincoln, NE) and infrared imaging on a Licor Odessey instrument.
Immunoprecipitation of GP73
GP73 was immunoprecipitated using protein A/G (Pierce). HepG2.215 lysate or human serum with a known high level of GP73 was precleared by incubation with protein A/G beads [100 μl beads (~600–700 μg IgG binding capacity)/1.0 ml lysate or sera]. The samples were incubated at room temperature rocking for 1 h. After incubation, beads were pelleted and supernatants were collected. Immunoprecipitation of GP73 from the supernatant was then performed with the addition of anti-GP73 antibody that is conjugated to protein A/G beads. The mixture was incubated at 4°C with rocking over night. The supernatant was collected and the beads were then washed with three washes of PBS. Once washed, the antibody-antigen complexes were released from the beads using a 3× volume of 0.1% TFA. The resulting antibody-antigen solution was stored at 4°C until further processing. The supernatant collected after overnight binding was subjected to second round of immunoprecipitation as described above.
Mass Spectrometer Analysis
Immunopreciptated GP73 starting from 2L of HepG2.215 media was dried down to near dryness then reconstituted in 200 μl “A” Buffer (2% ACN+0.1% TFA). The sample was purified using an ABI 140B HPLC (ABI, Foster city, CA) with a 1 mm×250 mm PLRP-S 100 A° column (Polymer labs, Amherst, MA) at a flowrate of 50 μl/min. “A” buffer was 2% ACN+0.1% TFA and “B” buffer was 90% ACN+0.1% TFA. A 1% B/min gradient was used, and fractions were collected every minute. Fractions 42 thru 48 were pooled as previous experiments had shown that GP73 elutes in this area (data not shown). The pooled fraction was evaporated to near dryness, and then resuspended in 100 μl of 25 mM NH4CO3+10% ACN. Two micrograms Glu-C or 1 μg trypsin protease (Promega, Madison, WI) was added and the sample was incubated at 37°C O/N. The sample was left at RT for an additional day, after which 20 μl of 5X PNGaseF buffer was added along with ~1 mU PNGaseF (Sigma, St. Louis, MO). The sample was incubated at 37°C O/N. The sample was loaded onto a QTRAP mass spectrometer (Applied Biosystems, Foster city, CA) which was fitted with a microspray unit with an integrated on-line desalting unit (Homebuilt). A 75 μm column with integrated frit (New objective, Woburn, MA) was packed with 10 cm of Reliasil C18 resin (Column Engineering, Ontario, Canada) and upstream was a desalting precolumn (Upchurch, Oak Harbor, WA) packed with the same resin. An micro pro pump (Eldex, Napa, CA) provide a flow rate of ~200 nl/min with a 0.5% B/min gradient. “A” buffer was 2% ACN+1% AcOH and “B” buffer was 90% ACN+1% AcOH. The mass spectrometer was set to scan from 450 to 1,400 m/z and the top 2 ions from each scan were automatically selected for sequential MS/MS scans. Data was analyzed manually using the Analyst software package (Applied Biosystems).
Constructs for the Expression of GP73 Species
All plasmids used are summarized in Table I. A bacterial expression construct encoding the luminal portion of GP73 (amino acids 41–400, [Kladney et al., 2000]), lacking only the N-terminal membrane anchor region, was obtained from Dr. C. Fimmel. A BamHI-ApoI fragment encoding most of GP73 (amino acids 41–387) was removed and ligated to two complementary synthetic oligonucleotides that replaced the final 13 amino acids at the C-terminus, without the stop codon. (5′-AATTTACTTGATCAGCGTGAAAAGCGGAATCATACACTCA and 5′-GATCTGAGTGTATGATTCCGCTTTTCACGCTGATCAAGTA). This fragment was inserted into a BglII site within a mutated version of eukaryotic protein expression vector pFUSE-hFc2(IL2ss) (Invivogen, San Diego, CA), termed pFUSE-hFc2mut. The mutation removed the single N-glycan site within the Fc region by converting the acceptor asparagine residue to an alanine. (The protein produced by construct pFUSE-hFc2mut was expressed at levels comparable to that produced by the parental pFUSE-hFc2(IL2ss), and was confirmed to lack glycan based on both gel mobility change and lack of staining for glycan; data not shown.) The resulting fusion protein GP73-IgFc contains GP73 in-frame with an N-terminal signal sequence and a C-terminal human immunoglobulin heavy chain Fc region.
TABLE I.
Plasmids Used
| Construct | Description | Source |
|---|---|---|
| pQE9-GP73 | Bacterial expression vector for His tagged GP73 amino acids 41–400 | Kladney et al. [2000] |
| pFUSE-hFc2(IL2ss) | Eukaryotic expression construct with signal sequence and Ig Fc tag | Invivogen |
| pFUSE-hFc2mut | Same as above, but with the N-glycosylation site in Fc region mutated | This work |
| pFUSE-GP73.41-400 | The GP73 fragment from pQE9-GP73 inserted into pFUSE-hFc2mut | This work |
| pFUSE-GP73.106-115 | GP73 amino acids 106-115 inserted into pFUSE-hFc2(IL2ss) | This work |
To examine the modification present at a single site of N-linked glycosylation, synthetic oligonucleotides with the sequences that encode the GP73 peptide encompassing aa109 were inserted into expression vector pFUSE-hFc2(IL2ss). The inserts also carried a 6-His tag and a Flag tag. The resulting construct, pFUSE-GP73.106-115, has GP73 sequences fused in frame with the signal sequence of IL2 (at the N-terminus) and the human immunoglobulin heavy chain Fc region (at the C-terminus).
Preparation of Recombinant Proteins and Glycoproteins
Recombinant GP73 protein was expressed from pQE9-GP73 in E. coli with an N-terminal 6-His tag, and purified by metal affinity chromatography, essentially as described [Kladney et al., 2000]. To prepare recombinant glycoproteins, Huh7 hepatoma cells were transfected with either pFUSE-GP73.41-400 of pFUSE-GP73.106-115 using Lipofectamine 2000 essentially as described previously (Norton). Cells were transferred to serum free DMEM 2 days post-transfection and incubated for an additional 16 h. Secreted recombinant proteins were affinity purified via the Fc portion on Protein G-agarose beads (vendor) in PBS, eluted in 100 mM glycine-HCl, pH 2.5. A portion of material so enriched was separated by electrophoresis on a 12% SDS–PAGE gel and visualized by staining with Coomassie blue.
RESULTS
Glycan Content of GP73 as Determined by Lectin and Enzymatic Sensitivity
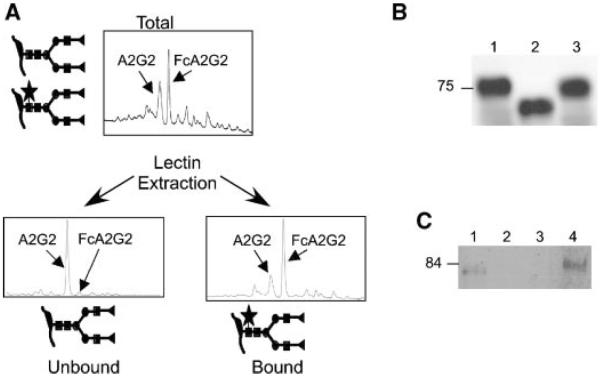
As the presence of core fucosylation on serum glycoproteins has been associated with liver disease in vivo, we have begun cataloguing the fucosylated proteome of liver-derived cell lines. A number of these transformed cells in culture preserve the phenotype associated with HCC, including secretion of alpha-fetoprotein. Of particular interest, HepG2.2.15 cells are derived from HepG2 hepatoblastoma cells and contain a stably transfected HBV genome [Sells et al., 1987]. Little GP73 is produced by HepG2 cells (Kladney et al. [2000] and data not shown), but HepG2.2.15 secretes immunologically detectable levels of GP73 (Kladney et al. [2000] and Fig. 1B). Previous studies of the hepatoma lines HepG2 and Huh-7 reported that the most abundant oligosaccharide present on the secreted alpha-fetoprotein is a fucosylated biantennary structure [Ohno et al., 1992]. Total secreted proteins concentrated from conditioned medium were subjected to PNGaseF treatment, labeled with 2-aminobenzene and separated by neutral phase HPLC. Identification of the released glycans is performed by normalization to the elution pattern of a “ladder” of glucose oligomers. Analysis of the oligosaccharides released from the total secreted glycoproteins derived from HepG2.2.15 cells reveals that core fucosylated FcA2G2 is the most abundant form (Fig. 1A; note that terminal sialic acid residues were enzymatically removed to simplify the analysis), and that the glycan content is indistinguishable from that secreted from the parental HepG2 cells (data not shown). Unfucosylated A2G2 was present at 90% of the level of FcA2G2, with smaller amounts of various larger oligosaccharide structures (peaks migrating to the right of the FcA2G2); these latter include tri- and tetra-antennary structures.
Fig. 1.
All GP73 produced by HepG2.2.15 cells contains N-linked glycans, with the majority bearing core fucose. A: Total glycan profiling of proteins secreted by HepG2.2.15 cells. The X-axes of the HPLC traces indicates relative elution time; the Y-axes indicate relative fluoresence intensity. Slightly more than half of the glycans contain core fucose, FcA2G2, as indicated by the star, with the remainder being largely A2G2. Terminology is based on that of Royle et al. [2002]. B: Supernatant collected and concentrated from HepG2.2.15 cells was either left untreated (lane 1) or was treated with PNGase F (lane 2) or EndoH (lane 3). GP73 was detected by immunoblotting. C: HepG2.2.15 cell supernatant was incubated with immobilized Aleuria aurantia lectin, and equal fractions of the unbound material (lane 1), two washed in binding buffer (lanes 2 and 3) or the eluted bound material (lane 4) were analyzed for the presence of GP73 by immunoblotting. Note that the apparent mobility of GP73 differs in panels B and C due to the use of different gel buffer systems.
We were particularly interested in characterizing the glycan content of GP73. Heterogeneity in the apparent molecular size of GP73 present in the circulation is observed when the protein is detected by SDS–PAGE and immunoblotting [Marrero et al., 2005]. As previously noted, the predicted molecular mass of GP73 based on DNA sequence of cDNAs was 45 kDa, but the mobility of the protein on SDS–PAGE was consistent with a much larger protein [Kladney et al., 2000]. This discrepancy was due at least in part to the presence of one or more N-linked glycans, as a PNGaseF sensitive upshift was observed when in vitro translation was performed in the presence of microsomes. To investigate the number and nature of N-linked glycans present on secreted GP73, culture supernatant from HepG2.2.15 was treated with PNGase or EndoH and compared with untreated material by immunoblotting. All immunoreactive material was observed to migrate more rapidly with enzymatic deglycosylation (Fig. 1B, lane 2), indicating that all GP73 molecules are modified by N-linked glycosylation. The loss of approximately 13 kDa with deglycosylation is consistent with the presence of multiple side chains. Glycosylated GP73 was completely resistant to EndoH digestion indicating that all side chains are complex in structure (Fig. 1B).
The extent of fucosylation of GP73 was investigated by using lectin extraction. Supernatant from HepG2.2.15 cells was dialyzed against lectin binding buffer, then Aleuria lectin immobilized on agarose beads was added. Following incubation, beads were washed. Equivalent portions of bound, washes and unbound fractions were examined by immunoblotting for the presence of GP73 (Fig. 1C). Quantitation revealed that 72% of the GP73 secreted from HepG2.2.15 cells bound to lectin, indicating that this fraction was core fucosylated on at least one position. Thus, the extent of fucosylation of GP73 may be slightly higher than the bulk of the N-linked glycan in the total secreted protein analysis as shown in Figure 1A, with respect to core fucosylation.
N-glycan Addition Site Utilization
Based on sequence considerations, there are three potential sites of N-linked glycan addition within GP73, as indicated in Figure 2. In order to assess whether the peptides that are potentially N-glycosylated can be detected by mass spectrometry, recombinant GP73 was expressed in E. coli and subjected to MS/MS analysis. Two of the three peptides generated by digestion with either trypsin or endopeptidase Glu-c were detected; all the peptides detected following the latter digestion condition are indicated in Figure 2. The third potential site, which lies very near the C-terminus of the protein, was not identifiable following either proteolytic digestion, and thus its glycosylation status could not be investigated further.
Fig. 2.

Sequence of GP73. The conceptual translation of human GP73 based on cDNA cloning is shown, with potential sites for N-glycan addition indicated in green. The sequences shown in red represent peptides that were detected by mass spectrometry analysis of recombinant GP73 produced in E. coli, which lacked the N-terminal cytoplasmic domain and transmembrane domain (amino acids 1–40).
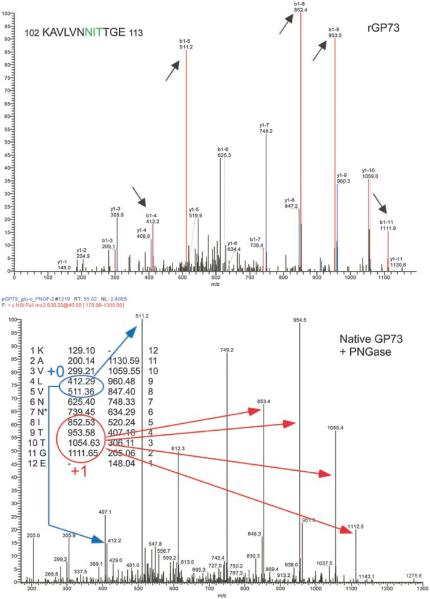
In order to determine whether either of the sequons within the two identified peptides were indeed used, GP73 was purified from HepG2.2.15 cells by immunoprecipitation with polyclonal antibody to GP73 and analyzed by mass spectrometry using a strategy similar to that described [Kaji et al., 2003]. Briefly, immunoprecipitated GP73 was digested with endopeptidase Glu-c, then de-N-glycosylated with PNGaseF. The enzymatic deglycosylation procedure results in deamidation of formerly glycosylated asparagines, and their conversion to aspartic acid. Thus, peptides that have undergone de-N-glycosylation will be shifted by one mass unit relative to the corresponding never-modified peptides, such as is obtained from recombinant protein produced in bacteria. The peptides produced from HepG2.2.15 cells and in bacteria were then identified by MS/MS analysis in parallel.
The spectra obtained (Figs. 3 and 4) comparing PNGaseF treated GP73 collected from HepG2.2.15 cells with recombinant GP73 prepared from E. coli suggest that both sites 1 and 2 are glycosylated. Smaller ions that do not extend to the N-glycan attachment site (denoted as +0 in the lower panels and highlighted in blue) have similar mass to the cognate ions observed with recombinant protein. In contrast, larger ions that do include the N-glycan attachment site (denoted as +1 in the lower panel of each figure, highlighted in red) are shifted up by approximately one mass unit relative to the cognate ion produced from the recombinant GP73. No information is available regarding the glycosylation status of site 3 at this time; peptide fragments derived from this region of the protein have not been detected from either native or recombinant GP73.
Fig. 3.
MS/MS analysis of GP73 amino acids 102–113. Top panel, MS/MS spectrum of fragment derived from recombinant GP73 purified from E. coli. Bottom panel, spectrum of same peptide derived from HepG2.2.15 cells. Long arrows in bottom panel indicate peaks representing fragments of identical mass in the two preparations (+0, blue) or shifted up one mass unit in the lower panel (+1, red), as expected for deamination of the templated asparagine (asterisk) to aspartic acid. The numbers shown next to the peptide sequence indicate the predicted species from the recombinant, unglycosylated protein. Short arrows indicating the peaks in the upper spectrum highlight some of the corresponding fragments that include the nonglycosylated residue derived from recombinant material.
Fig. 4.
MS/MS analysis of GP73 amino acids 137–148. See legend to Figure 3 for description of annotations.
Analysis of N-linked Glycans Attached to GP73 Sequons
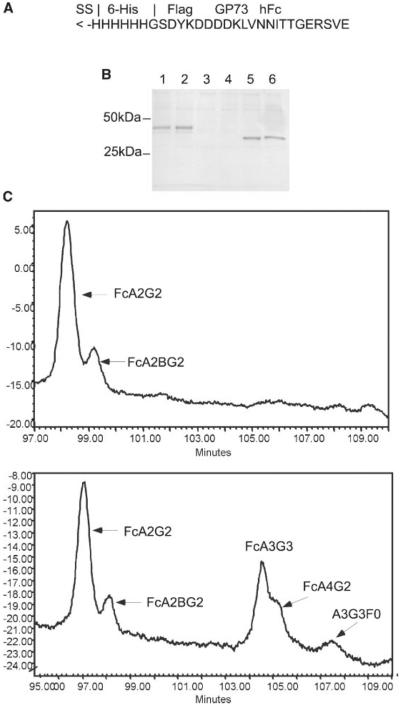
Secreted GP73 is not an abundant protein, making direct sequence analysis of its glycan content technically challenging. Repeated attempts to isolate sufficient protein by immunoprecipitation from HepG2.2.15 cell supernatants to perform direct glycan analysis were unsuccessful. Thus, a strategy to generate recombinant protein was pursued. The luminal domain of GP73 was subcloned into eukaryotic expression construct pFUSE-hFc2mut so as to be in frame with a signal sequence at the N-terminus and an immunoglobulin Fc domain at the C-terminus to permit protein recovery. Note that this version of the expression vector was modified to remove the N-glycan sequon normally present in the immunoglobulin domain (the asparagine residue was mutagenized to alanine). Huh7 hepatoma cells (which do not secrete endogenous GP73 or immunoglobulin) were transfected with this construct, pFUSE-GP73.41-400, and secreted protein captured on Protein G beads. Bound GP73-IgFc fusion protein was resolved on a 10% polyacrylamide gel and detected by staining with Coomassie Blue (Fig. 5A). The GP73-IgFc protein was excised, and oligosaccharides were released as described above. This analysis indicated that predominant species present were non-fucosylated A2G2, FcA2G2 (and the bisect variant) and a large fucosylated structure, FcA4G2 (Fig. 5B). Quantiation revealed that when these four major species are considered, the core fucosylated forms represent 72% of the total, in very good agreement with the fraction of secreted GP73 observed to bind Aleuria lectin. Because the vector used lacked the N-glycan sequon site normally present in the Fc region, the profile is entirely based on oligosaccharides added to the GP73 region.
Fig. 5.

Isolation and glycan sequence analysis of secreted GP73-IgFc. A: Coomassie stained gel showing the isolated fusion protein of expected size of ca. 90 kDa, with the contributions of the two portions diagrammed below. B: HPLC profile of released oligosaccharides, identified based on mobility relative to a dextrose ladder.
Analysis of N-linked Glycans Added at a Single GP73 Sequon
Analysis of the glycans released from the total protein does not address the nature of the glycans added at each individual site. To explore this issue, a recombinant protein was generated in which a short peptide encompassing glycan site at amino acid 109 (LVNNITTGER) was fused in frame to the human immunoglobulin Fc domain in pFUSE-hFc2(IL2ss). The recombinant fusion protein was recovered from the medium of Huh7 hepatoma cells that had been transfected with pFUSE-GP73.106-115 and captured on Protein G agarose. Because the IgFc region is itself glycosylated at a single site, recombinant protein was isolated from cells that were transfected with the pFUSE-hFc2(IL2ss) empty vector (Fig. 6A). Based on the appearance of the proteins on the Coomassie stained gel, the majority of each recombinant protein is present as a single species, suggesting that glycan site utilization is efficient, with no unglycosylated material. Analysis of the glycans present on the two proteins established that the glycan added to the IgFc region is core fucosylated FcA2G2, with a small amount of this oligosaccharide with a bisect (Fig. 6B). In contrast, the glycoprotein protein produced by the pFUSE-GP73.106-115 construct bears the same structures, but also contains additional triand tetra-antennary fucosylated structures, including the FcA4G2 oligosaccharide observed when the near full length recombinant GP73 was analyzed (Fig. 5). It seems likely that the latter oligosaccharides are added exclusively to the GP73 peptide sequon, the only other site present. Thus, the short peptide encompassing the N-glycan site at 109 is core fucosylated less than 100% of the time, as are both native secreted and recombinant GP73; in contrast, the IgFc site present within the same molecule is completely fucosylated. These results provide initial proof of concept for this analytical strategy to identify the glycans added at individual sequons.
Fig. 6.
Isolation and glycan sequence analysis of secreted peptide-IgFc. A: Structure of insert containing GP73 peptide cloned into pFUSE-hFc2(IL2ss). B: Coomassie stained gel showing the isolated fusion protein derived from supernatants of cells transfected with pFUSE-GP73.106–115, with the approximate expected size (28 kDa plus 2 glycan side chains; lanes 1 and 2), relative to the protein produced by transfection of empty vector (25 kDa plus one glycan side chain; lanes 5 and 6). Lanes 3 and 4 were prepared from supernatant from non-transfected Huh7 cells. C: HPLC profiles of oligosaccharides released from protein with no insert (upper panel) or the GP73 amino acids 106–115 insert (lower panel), with identities based on mobility relative to a dextrose standard. Note that although only a part of the spectrum is shown, there was no peak corresponding to unfucosylated A2G2 in either profile (not shown).
DISCUSSION
Elevated circulating levels of GP73 overall [Marrero et al., 2005], and fucosylated GP73 in particular [Drake et al., 2006], have been associated with hepatocellular carcinoma, suggesting that a more detailed analysis of GP73 glycosylation is warranted. Detailed analysis of the glycosylation patterns of such low abundance proteins is hampered by technical difficulties. Using lectin affinity chromatography, we have established that much of the GP73 secreted from a cell line derived from HCC is fucosylated. In addition, the results indicated that all GP73 molecules secreted from these cells are modified by addition of N-linked glycan at one or more sites. However, the PNGase treatment did not reduce the molecule to the apparent size predicted from the primary sequence, which would represent a reduction of ca. 30 kDa instead of the 13 kDa shift observed. This could reflect anomalous migration of the protein, or the presence of additional post-translational modifications. No change in mobility was observed with EndoH treatment, indicating that GP73-linked glycans are complex, not high mannose. These results also indicate that the GP73 secreted from the cells has transited through the Golgi. In addition, it has been postulated that cells of the liver other than hepatocytes might be the source for circulating GP73 [Iftikhar et al., 2004]. Our results do not rule out the latter possibility, but do demonstrate that hepatocytic cells are capable of secreting fucosylated GP73.
The mechanism by which GP73 is secreted from these cells (or how it appears in the circulation in vivo) is unclear. The membrane anchored GP73 protein might be cleaved in the Golgi and then secreted. This result would be consistent with the possibility that hepatocytes in the diseased tissue are the source of circulating GP73. However, GP73 can cycle to the cell surface and to late endosomes, at least under some circumstances [Puri et al., 2002], and perhaps the protein is shed from the surface. It is also a possibility that another start codon, such as Met46 can be used as an alternative site of translation initiation, producing a protein with no membrane anchor (Fig. 2). Although not the focus of the present study, we recognized that the mass spectrometric analysis might provide useful information regarding the termini of the secreted form of the GP73 protein. A peptide lying very close to the C-terminus was identified (Fig. 2), suggesting that this end of the protein may be intact in the secreted version. However, as the most N-terminal peptide detected from the recombinant protein begins at amino acid 79 (Fig. 2), it is difficult to interpret the absence of peptides from the region proximal to the transmembrane domain from secreted GP73. Thus, the amino terminus of secreted GP7 remains ill-defined.
Using an MS/MS approach, it was demonstrated that GP73 is N-glycosylated on at least two out of a possible three sites (Figs. 3 and 4). The Glu-C peptide fragment that includes the third site, which lies very close to the C-terminus of the protein, was never detected from either the native or the recombinant protein, and thus we remain unsure of its usage. Analysis of the primary sequence of GP73 using the NetNGlyc server (bs.dtu.dk/services/NetNGlyc/) suggests that only the most N-terminal sequon is favored for N-glycosylation, with a score of 0.5474 exceeding the threshold of 0.50. The C-terminal site is the least favorable for N-glycan addition, with a score of 0.3603; however, the score for the middle sequon also does not meet the threshold (0.4604). Thus, the failure to meet the prediction threshold does not preclude use of the site.
Although we have been unable to investigate directly the glycan side chain structures of GP73 secreted endogenously from HepG2.2.15 cells, the glycans added to a recombinant version of GP73 which contains all three glycosylation sequons were analyzed directly. This analysis suggested that the majority of the glycans added displayed core fucosylation, consistent with observations with the native protein (Fig. 1). In addition, some larger species were present, indicating some heterogeneity of the oligosaccharide side chain maturation. However, the fraction of GP73 that includes binds to Aleuria aurantia lectin, and thus presumably contains core fucose (72%, Fig. 1C), corresponds exactly with the fraction of the recombinant GP73-IgFc that contains core fucose (72%, Fig. 5).
To begin to address side chain heterogeneity, a small GP73 peptide that includes the first glycosylation sequon was fused to the IgFc region and protein secreted from hepatoma cells was analyzed for the glycans added. This analysis demonstrated that this site is still incompletely fucosylated, albeit to a greater extent than near full length GP73, but that the structures vary and include large tri- and tetra-antennary structures. In contrast, the glycans added at the immunoglobulin Fc region site appear to be restricted to a single glycan, FcA2G2, which may be further modified by addition of a bisect. Thus, we can conclude that the single GP73 N-glycan site can accommodate a range of oligosaccharide modifications, and that features of the surrounding peptide sequence might contribute the efficiency of core fucosylation. Variation in the extent of core fucosylation on different sites within a protein has been reported previously [James et al., 1995; Tajiri et al., 2005], but relatively little is known about what regulates these differences. The expression system described here will allow us to investigate further how local sequences may influence the addition of core fucose. Further characterization of the range of N-glycans that can be added at individual sequons might reveal additional structures that correlate with disease.
Changes in glycan structures have been correlated with both developmental stage and disease status in a number of situations [Hakomori, 1996; Kobata, 1998]. Specifically, glycosylation changes that have been associated with liver disease include increased fucosylation and an increase in oligosaccharide branching [Naitoh et al., 1999; Comunale et al., 2006; Morelle et al., 2006]. However, these changes do not appear to be associated with a specific underlying disease state, but instead appear to relate to any liver dysfunction. In addition to changes in the bulk glycan composition of serum or plasma, modifications can be detected on individual liver-derived proteins such as alpha-fetoprotein or alpha-1-acid glycoprotein [Aoyagi et al., 1993; Aoyagi et al., 1998; Anderson et al., 2002; Higai et al., 2005]. These glycoforms have been proposed as potential markers for HCC and cirrhosis. We recently reported that a total of 19 circulating proteins exhibit increased fucosylation in patients with HCC [Comunale et al., 2006]. While total serum GP73 levels correlate well with HCC [Marrero et al., 2005], it appears that fucosylated GP73 has both a higher sensitivity (90% vs. 65%) and specificity (100% vs. 90%) in a recently published smaller study [Drake et al., 2006]. Studies of larger cohorts will be needed to fully establish the utility of fucosylated GP73 as a biomarker for HCC.
Understanding the biological relevance of glycosylation changes has been somewhat more elusive. It has recently been reported that fucosylation appears to preferentially direct hepatocyte-derived proteins to be secreted into the bile ducts as opposed to the circulation [Nakagawa et al., 2006]. Derangements in the secretory system of hepatocytes involved in HCC might account for the appearance of increased fucosylation seen on many proteins in the sera of patients with HCC [Comunale et al., 2006]. Secretory system defects might also be responsible for the increased secretion of GP73 seen with liver disease [Marrero et al., 2005]. Whether the differentially modified species of GP73 are in any way functionally distinct remains unexplored.
ACKNOWLEDGMENTS
Danielle Loughlin and Angela Evans are thanked for their help in preparing native and recombinant GP73. This work was supported by the NIH NCI Early Detection Research Network, by a grant from the Department of Education and by an appropriation from the Commonwealth of Pennsylvania, through the Hepatitis B Foundation of America's Institute for Hepatitis and Virus Research.
Grant sponsor: NCI; Grant number: 5U01 CA084951; Grant sponsor: Commonwealth of PA.
REFERENCES
- Anderson N, Pollacchi A, Hayes P, Therapondos G, Newsome P, Boyter A, Smith K. A preliminary evaluation of the differences in the glycosylation of alpha-1-acid glycoprotein between individual liver diseases. Biomed Chromatogr. 2002;16:365–372. doi: 10.1002/bmc.167. [DOI] [PubMed] [Google Scholar]
- Anzola M. Hepatocellular carcinoma: Role of hepatitis B and hepatitis C viruses proteins in hepatocarcinogenesis. J Viral Hepat. 2004;11:383–393. doi: 10.1111/j.1365-2893.2004.00521.x. [DOI] [PubMed] [Google Scholar]
- Aoyagi Y. Carbohydrate-based measurements on alpha-fetoprotein in the early diagnosis of hepatocellular carcinoma. Glycoconj J. 1995;12:194–199. doi: 10.1007/BF00731319. [DOI] [PubMed] [Google Scholar]
- Aoyagi Y, Suzuki Y, Igarashi K, Yokota T, Mori S, Suda T, Naitoh A, Isemura M, Asakura H. Highly enhanced fucosylation of alpha-fetoprotein in patients with germ cell tumor. Cancer. 1993;72:615–618. doi: 10.1002/1097-0142(19930715)72:2<615::aid-cncr2820720246>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Aoyagi Y, Isokawa O, Suda T, Watanabe M, Suzuki Y, Asakura H. The fucosylation index of alpha-fetoprotein as a possible prognostic indicator for patients with hepatocellular carcinoma. Cancer. 1998;83:2076–2082. [PubMed] [Google Scholar]
- Block TM, Mehta AS, Fimmel CJ, Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22:5093–5107. doi: 10.1038/sj.onc.1206557. [DOI] [PubMed] [Google Scholar]
- Block TM, Comunale MA, Lowman MA, Steel LF, Romano PR, Fimmel CJ, Tennant BC, London WT, Evans AA, Blumberg BS, Dwek RA, Mattu TS, Mehta AS. Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Proc Natl Acad Sci USA. 2005;102:779–784. doi: 10.1073/pnas.0408928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breborowicz J, Mackiewicz A, Breborowicz D. Micro-heterogeneity of alpha-fetoprotein in patient serum as demonstrated by lectin affino-electrophoresis. Scand J Immunol. 1981;14:15–20. doi: 10.1111/j.1365-3083.1981.tb00179.x. [DOI] [PubMed] [Google Scholar]
- Callewaert N, Van Vlierberghe H, Van Hecke A, Laroy W, Delanghe J, Contreras R. Noninvasive diagnosis of liver cirrhosis using DNA sequencer based total serum protein glycomics. Nat Med. 2004;10:429–434. doi: 10.1038/nm1006. [DOI] [PubMed] [Google Scholar]
- Comunale MA, Mattu TS, Lowman MA, Evans AA, London WT, Semmes OJ, Ward M, Drake R, Romano PR, Steel LF, Block TM, Mehta A. Comparative proteomic analysis of de-N-glycosylated serum from hepatitis B carriers reveals polypeptides that correlate with disease status. Proteomics. 2004;4:826–838. doi: 10.1002/pmic.200300625. [DOI] [PubMed] [Google Scholar]
- Comunale MA, Lowman MA, Long RE, Krakover J, Philip R, Seeholzer S, Evans AA, Hann HW, Block TM, Mehta AS. Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma. J Proteome Res. 2006;5:308–315. doi: 10.1021/pr050328x. [DOI] [PubMed] [Google Scholar]
- Di Bisceglie AM, Carithers RL, Jr., Gores GJ. Hepatocellular carcinoma. Hepatology. 1998;28:1161–1165. doi: 10.1002/hep.510280436. [DOI] [PubMed] [Google Scholar]
- Drake RR, Schwegler EE, Malik G, Diaz J, Block TM, Mehta A, Semmes OJ. Lectin capture strategies combined with mass spectrometry for the discovery of serum glycoprotein biomarkers. Mol Cell Proteomics. 2006;5:1957–1967. doi: 10.1074/mcp.M600176-MCP200. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340:745–750. doi: 10.1056/NEJM199903113401001. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Shimada M, Takenaka K, Kajiyama K, Shirabe K, Sugimachi K. The Sialyl Lewis X expression in hepatocarcinogenesis: Potential predictor for the emergence of hepatocellular carcinoma. Hepatogastroenterology. 2002;49:213–217. [PubMed] [Google Scholar]
- Hakomori S. Tumor malignancy defined by aberrant glycosylation and sphingo(glyco)lipid metabolism. Cancer Res. 1996;56:5309–5318. [PubMed] [Google Scholar]
- Higai K, Aoki Y, Azuma Y, Matsumoto K. Glycosylation of site-specific glycans of alpha1-acid glycoprotein and alterations in acute and chronic inflammation. Biochimica et Biophysica Acta. 2005;1725:128–135. doi: 10.1016/j.bbagen.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Iftikhar R, Kladney RD, Havlioglu N, Scmitt-Graff A, Gusmirovic I, Solomon H, Luxon BA, Bacon BR, Fimmel CJ. Disease- and cell-specific expression of GP73 in human liver disease. Am J Gastroenterol. 2004;99:1087–1095. doi: 10.1111/j.1572-0241.2004.30572.x. [DOI] [PubMed] [Google Scholar]
- James DC, Freedman RB, Hoare M, Ogonah OW, Rooney BC, Larionov OA, Dobrovolsky VN, Lagutin OV, Jenkins N. N-glycosylation of recombinant human inter-feron-gamma produced in different animal expression systems. Biotechnology. 1995;13:592–596. doi: 10.1038/nbt0695-592. [DOI] [PubMed] [Google Scholar]
- Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K-I, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- Kladney RD, Bulla GA, Guo L, Mason AL, Tollefson AE, Simon DJ, Koutoubi Z, Fimmel CJ. GP73, a novel Golgi-localized protein upregulated by viral infection. Gene. 2000;249:53–65. doi: 10.1016/S0378-1119(00)00136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladney RD, Cui X, Bulla GA, Brunt EM, Fimmel CJ. Expression of GP73, a resident Golgi membrane protein, in viral and nonviral liver disease. Hepatology. 2002;35:1431–1440. doi: 10.1053/jhep.2002.32525. [DOI] [PubMed] [Google Scholar]
- Kobata A. A retrospective and prospective view of glycopathology. Glycoconj J. 1998;15:323–331. doi: 10.1023/a:1006961532182. [DOI] [PubMed] [Google Scholar]
- Lok A, McMahon B. Chronic hepatitis B. Hepatology. 2001;34:1225–1241. doi: 10.1053/jhep.2001.29401. [DOI] [PubMed] [Google Scholar]
- Lok AS, Heathcote EJ, Hoofnagle JH. Management of hepatitis B: 2000—Summary of a workshop. Gastroenterology. 2001;120:1828–1853. doi: 10.1053/gast.2001.24839. [DOI] [PubMed] [Google Scholar]
- Marrero JA, Romano PR, Nikolaeva O, Steel LF, Mehta AS, Fimmel CJ, Comunale MA, D'Amelio A, Lok AS, Block TM. GP73, a resident Golgi glycoprotein, is a novel serum marker for Hepatocellular Carcinoma. Hepatology. 2005;43:1007–1012. doi: 10.1016/j.jhep.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Miyoshi E, Noda K, Yamaguchi Y, Inoue S, Ikeda Y, Wang W, Ko JH, Uozumi N, Li W, Taniguchi N. Altered glycosylation of serum transferrin of patients with hepatocellular carcinoma. J Biol Chem. 1999;264:2415–2423. [PubMed] [Google Scholar]
- Morelle W, Flahaut C, Michalski J-C, Louvet A, Mathurin P, Klein A. Mass spectrometric approach for screening modifications of total serum N-glycome in human diseases: Application to cirrhosis. Glycobiology. 2006;16:281–293. doi: 10.1093/glycob/cwj067. [DOI] [PubMed] [Google Scholar]
- Naitoh A, Aoyagi Y, Asakura H. Highly enhanced fucosylation of serum glycoproteins in patients with hepatocellular carcinoma. J Gastroenterol Hepatol. 1999;14:436–445. doi: 10.1046/j.1440-1746.1999.01882.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Uozumi N, Nakano M, Mizuno-Horikawa Y, Okuyama N, Taguchi T, Gu J, Kondo A, Taniguchi N, Miyoshi E. Fucosylation of N-glycans regulates the secretion of hepatic glycoproteins into bile ducts. J Biol Chem. 2006;281:29797–29806. doi: 10.1074/jbc.M605697200. [DOI] [PubMed] [Google Scholar]
- Noda K, Miyoshi E, Gu J, Gao CX, Nakahara S, Kitada T, Honke K, Suzuki K, Yoshihara H, Yoshikawa K, Kawano K, Tonetti M, Kasahara A, Hori M, Hayashi N, Taniguchi N. Relationship between elevated FX expression and increased production of GDP-L-fucose, a common donor substrate for fucosylation in human hepatocellular carcinoma and hepatoma cell lines. Cancer Res. 2003;63:6282–6289. [PubMed] [Google Scholar]
- Ohno M, Nishikawa A, Koketsu M, Taga H, Endo Y, Higashino K, Taniguchi N. Enzymatic basis of sugar structures of alpha-fetoprotein in hepatoma and hepatoblastoma cell lines: Correlation with activities of alpha 1-6fucosyltransferase and N-acetylglucosaminyltransferases III and V. Int J Cancer. 1992;51:315–317. doi: 10.1002/ijc.2910510223. [DOI] [PubMed] [Google Scholar]
- Puri S, Bachert C, Fimmel CJ, Linstedt AD. Cycling of early Golgi proteins via the cell surface and endosomes upon lumenal pH disruption. Traffic. 2002;3:641–653. doi: 10.1034/j.1600-0854.2002.30906.x. [DOI] [PubMed] [Google Scholar]
- Royle L, Mattu TS, Hart E, Langridge JI, Merry AH, Murphy N, Harvey DJ, Dwek RA, Rudd PM. An analytical and structural database provides a strategy for sequencing O-glycans from microgram quantities of glycoproteins. Anal Biochem. 2002;304:70–90. doi: 10.1006/abio.2002.5619. [DOI] [PubMed] [Google Scholar]
- Rudd PM, Mattu TS, Zitzmann N, Mehta AS, Colominas C, Hart E, Opdenakker G, Dwek RA. Glycoproteins: Rapid sequencing technology for N-linked and GPI anchor glycans. Biotechnol Genet Eng Rev. 1999;16:1–21. doi: 10.1080/02648725.1999.10647969. [DOI] [PubMed] [Google Scholar]
- Sekine C, Aoyagi Y, Suzuki Y, Ichida F. The reactivity of a-1-antitrypsin with Lens culinaris agglutinin and its usefulness in the diagnosis of neoplastic diseases of the liver. Br J Cancer. 1987;56:371–375. doi: 10.1038/bjc.1987.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M. Alphafetoprotein: An obituary. J Hepatol. 2001;34:603–605. doi: 10.1016/s0168-8278(01)00025-3. [DOI] [PubMed] [Google Scholar]
- Steel LF, Mattu TS, Mehta A, Hebestreit H, Dwek R, Block /SNM> T A proteomics approach for the discovery of early detection markers of hepatocellular carcinoma. Dis Markers. 2001;17:179–183. doi: 10.1155/2001/963023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajiri M, Yoshida S, Wada Y. Differential analysis of site-specific glycans on plasma and cellular fibronectins: Application of a hydrophilic affinity method for glycopeptide enrichment. Glycobiology. 2005;15:1332–1340. doi: 10.1093/glycob/cwj019. [DOI] [PubMed] [Google Scholar]
- Yamashita K, Koide N, Endo T, Iwaki Y, Kobata A. Altered glycosylation of serum transferrin of patients with hepatocellular carcinoma. J Biol Chem. 1989;264:2415–2423. [PubMed] [Google Scholar]