Abstract
Background
Specific members of the plant Mildew Locus O (MLO) protein family act as susceptibility factors towards powdery mildew (PM), a worldwide-spread fungal disease threatening many cultivated species. Previous studies indicated that monocot and dicot MLO susceptibility proteins are phylogenetically divergent.
Methods
A bioinformatic approach was followed to study the type of evolution of Angiosperm MLO susceptibility proteins. Transgenic complementation tests were performed for functional analysis.
Results
Our results show that monocot and dicot MLO susceptibility proteins evolved class-specific conservation patterns. Many of them appear to be the result of negative selection and thus are likely to provide an adaptive value. We also tested whether different molecular features between monocot and dicot MLO proteins are specifically required by PM fungal species to cause pathogenesis. To this aim, we transformed a tomato mutant impaired for the endogenous SlMLO1 gene, and therefore resistant to the tomato PM species Oidium neolycopersici, with heterologous MLO susceptibility genes from the monocot barley and the dicot pea. In both cases, we observed restoration of PM symptoms. Finally, through histological observations, we demonstrate that both monocot and dicot susceptibility alleles of the MLO genes predispose to penetration of a non-adapted PM fungal species in plant epidermal cells.
Conclusions
With this study, we provide insights on the evolution and function of MLO genes involved in the interaction with PM fungi. With respect to breeding research, we show that transgenic complementation assays involving phylogenetically distant plant species can be used for the characterization of novel MLO susceptibility genes. Moreover, we provide an overview of MLO protein molecular features predicted to play a major role in PM susceptibility. These represent ideal targets for future approaches of reverse genetics, addressed to the selection of loss-of-function resistant mutants in cultivated species.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-015-0639-6) contains supplementary material, which is available to authorized users.
Keywords: MLO, Powdery mildew, Angiosperms, Evolution, Plant breeding
Background
The plant Mildew Locus O (MLO) gene family codes for proteins harboring seven transmembrane domains and a calmodulin-binding site, topologically reminiscent of metazoan and fungal G-protein coupled receptors (GPCRs) [1]. Following the completion of plant genome sequencing projects, a number of homologs varying from 12 to 19 has been identified in the MLO gene families of diploid species, namely Arabidopsis, rice, grapevine, cucumber, peach, woodland strawberry and sorghum [1–6].
Specific homologs of the MLO gene family act as susceptibility factors towards fungi causing the powdery mildew (PM) disease, worldwide spread and causing severe losses in agricultural settings. Inactivation of these genes, through loss-of function mutations or silencing, indeed results in resistance (referred to as mlo-based resistance) in several plant species [7]. The first MLO gene described as required for PM pathogenesis was barley HvMLO [8, 9]. Since then, MLO susceptibility genes have been functionally characterized in rice (OsMLO3), wheat (TaMLO_A1 and TaMLO_B1), Arabidopsis (AtMLO2, AtMLO6 and AtMLO12), tomato (SlMLO1), pepper (CaMLO2), tobacco (NtMLO1), pea (PsMLO1), lotus (LjMLO1) and barrel clover (MtMLO1) [10–17].
Defense mechanisms involved in mlo-based resistance prevent fungal penetration in epidermal cells and are associated with the formation of cell wall appositions, referred to as papillae [11]. Similar pre-penetration defense measures also take place in non-host resistance, following the interaction between PM fungal species and plant species beyond their host range. Consistent with the hypothesis of involvement of MLO genes in non-host resistance, loss of function of HvMLO in the interaction between barley and the wheat PM fungus Blumeria graminis f. sp. tritici is associated with decreased rate of penetration and lower incidence of epidermal cell death, the latter being a post-penetration defense mechanism [18, 19].
Several studies have been addressed to the characterization of regions of relevance for the functionality of MLO proteins. Multiple alignments have pointed out the occurrence of residues highly conserved within the whole MLO family, which were therefore predicted to provide a common protein structural scaffold [12, 20]. In addition, the occurrence of residues and motifs specifically conserved in putative orthologs of barley HvMLO has been reported [9]. Finally, functionally important residues for MLO susceptibility proteins have been inferred by the association of naturally occurring and induced mutations with partial or complete PM resistance [11, 12, 21–25].
In our previous studies, we showed that phylogenetically related dicot MLO genes of the same botanic family are conserved for their function as a susceptibility gene to PM [6, 16]. Notably, monocot and dicot MLO proteins involved in PM susceptibility group in clearly separated phylogenetic clades (e.g. [2, 9]). Here, we show that the evolution of Angiosperm PM susceptibility genes led to the fixation of class-specific molecular traits. Many of them appear to be the result of negative selection. By means of transgenic complementation assays, we demonstrate that, despite having different conservation patterns, monocot and dicot MLO susceptibility genes are essentially conserved with respect to functional features having a role in interactions with PM fungi. Consequences of our findings for plant breeding research are discussed.
Results
Class-specific molecular features of Angiosperm MLO homologs required for PM susceptibility
Previous studies indicated that dicot and monocot MLO proteins with a putative or ascertained role in susceptibility to PM fungi group in two different phylogenetic clades (e.g. [2, 9]). This was confirmed by performing a new UPGMA-based phylogenetic analysis involving all the 12 MLO homologs which have been until recently functionally related to PM susceptibility (Fig. 1). Aiming to detect molecular features responsible for such phylogenetic divergence, the same MLO homologs were used as dataset for protein multiple alignment (Fig. 2). Notably, this led to the identification of 41 alignment positions in which residues invariable throughout dicots are absent in monocots, and 84 alignment positions in which residues invariable throughout monocots are absent in dicots. In 44 alignment positions, class-specific residues are replaced in the other class with residues having different properties, according to the chemical features of their side-chain group (hydrophobic, polar basic, polar acidic and polar uncharged).
Fig. 1.
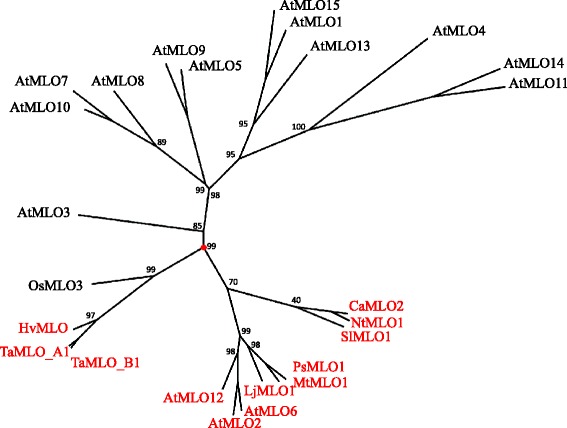
Unrooted radial phylogenetic tree of MLO powdery mildew susceptibility proteins. The tree includes, in red, all the monocot and dicot MLO homologs shown to be required for powdery mildew susceptibility (Arabidopsis AtMLO2, AtMLO6 and AtMLO12, tomato SlMLO1, pepper CaMLO2, tobacco NtMLO1, pea PsMLO1, lotus LjMLO1, barrel clover MtMLO1, barley HvMLO, wheat TaMLO_B1 and TaMLO_A1b and rice OsMLO3), and the remaining homologs of the Arabidopsis AtMLO family. Numbers at each node represent bootstrap support values (out of 100 replicates)
Fig. 2.
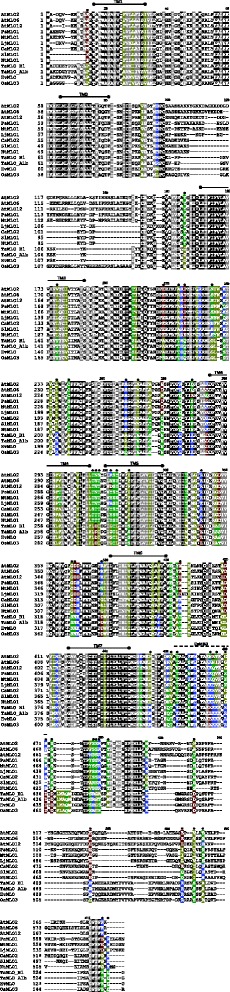
Multiple alignment of MLO powdery mildew susceptibility proteins. The dataset is composed of all the monocot (barley HvMLO, rice OsMLO3, wheat TaMLO_B1 and TaMLO_A1b), and dicot (Arabidopsis AtMLO2, AtMLO6 and AtMLO12, tomato SlMLO1, pepper CaMLO2, tobacco NtMLO1, pea PsMLO1, lotus LjMLO1 and barrel clover MtMLO1) MLO homologs shown to act as powdery mildew susceptibility factors. The positions of the seven MLO transmembrane domains (TM1-TM7) and the calmodulin binding domain (CaMBD) are identical to the ones reported by Feechan et al. [2], Functional Plant Biology, 35: 1255–1266. Black color indicates alignment positions in which invariable residues are present. Grey color indicates alignment positions which do not contain class-specific residues and are conserved with respect to biochemical properties. Other colors indicate alignment positions in which there are class-specific residues in monocots, dicots, or both: yellow indicates hydrophobic residues (G, A, V, L, I, F, W, M, P); blue indicates polar basic residues (K,R,H); red indicates polar acidic residues (D, E); green indicates polar uncharged residues (S, T, C, Y, N, Q). Black dots highlight 44 alignment positions in which class-specific residues are substituted in the other class by residue(s) having different biochemical properties
Adaptive relevance of class-specific molecular features supported by evolutionary analysis
In order to make inference on the evolutionary events leading to the above mentioned class-specific molecular features, we performed a codon-based Single-Likelihood Ancestor Counting (SLAC) analysis on the difference of nonsynonymous to synonymous substitutions per nonsynonymous and synonymous sites (dN-dS). Tests were conducted to predict the evolution of each codon: neutral/dN = dS or negative (purifying)/dN < dS. We decided to restrict the analysis to a panel of nine dicot MLO susceptibility genes, as only four monocot MLO homologs have been so far associated with PM pathogenesis and the dN-dS analysis can provide significant results only when using a sequence dataset which is not too small. We found 130 codons under significant negative selection, coding for amino acids scattered throughout MLO protein domains. Among the 130 codons, 27 are translated into class-specific residues, which are therefore predicted to provide an adaptive value (Additional file 1).
Functional conservation of monocot and dicot MLO susceptibility genes
We tested whether different molecular features between monocot and dicot MLO proteins are specifically required by PM fungal species infecting either one or the other class of Angiosperms. To this aim, we developed two constructs for the transgenic expression of a monocot (barley HvMLO) and a dicot (pea PsMLO1) MLO gene in the tomato Slmlo1 line, which is homozygous for a loss-of-function mutation in the endogenous gene SlMLO1 and therefore resistant to the tomato PM fungus Oidium neolycopersici. We reasoned that complementation and restoration of PM symptoms would have occurred only in case of functional conservation between SlMLO1 and any of the two tested transgenes. In total, nineteen 35S::PsMLO1 and 20 35S::HvMLO transformants were obtained. In both cases, 18 individuals were obtained showing variable transgene expression levels. For each construct, three T1 plants displaying high transgene expression (35S::PsMLO1-4,−6 and−7 and 35S::HvMLO-9,−10 and−15) were self-pollinated to generate T2 families (Additional file 2 ). Ten individuals from each T2 family were tested for the presence or the absence of the transgene and challenged with O. neolycopersici. Transgenic individuals of the three T2 families overexpressing PsMLO1 (35S::PsMLO1_(+)) displayed PM symptoms with an average D.I. (disease index) score ranging from 2.87 to 2.92. Transgenic individuals of the three T2 families overexpressing HvMLO (35S::HvMLO_(+)) showed an average D.I. score ranging from 1.8 to 2.4. In contrast, all non-transgenic 35S::PsMLO1_(−) and 35S::HvMLO_(−) T2 individuals displayed, similar to the Slmlo1 plants, hardly any fungal growth (Fig. 3 and Additional file 3). For transgenic plants of the three 35S::HvMLO T2 families, positive correlation was found between average D.I. and transgene expression level of corresponding T1 plants (Fig. 3 and Additional file 2 and 3). Together, these results indicate that monocot and dicot MLO susceptibility genes are functionally conserved with respect to molecular features required for PM pathogenesis.
Fig. 3.
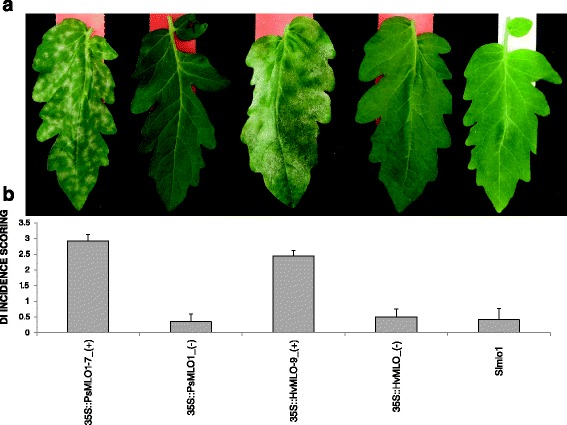
Transgenic overexpression of pea PsMLO1 and barley HvMLO in the tomato mutant line Slmlo1. Panel a shows the phenotypes of two selected individuals of the T2 family 35S::PsMLO1-7, segregating for the presence (first from the left) or the absence (second from the left) of the transgene, two selected individuals of the T2 family 35S::HvMLO-9, segregating for the presence (third from the left) or the absence (second from the right) of the transgene, and one individual of the Slmlo1 line (first from the right), in response to the tomato powdery mildew fungus Oidium neolycopersici. Panel b from left to right shows average disease index (DI) values relative to transgenic plants (+) of the 35S::PsMLO1-7 T2 family, non-transgenic plants (−) of three T2 families segregating for the 35S::PsMLO1 construct, transgenic plants of the 35S::HvMLO-9 T2 family, non-transgenic plants of three T2 families segregating for the 35S::HvMLO construct and the Slmlo1 line. Standard deviation bars refer to six 35S::PsMLO1_(+) individuals, nine 35S::HvMLO_(+) individuals, 7 PsMLO1_(−) individuals, 7 HvMLO_(−) individuals and 10 Slmlo1 individuals
Functional conservation of monocot and dicot MLO susceptibility genes in non-host interactions
We next investigated whether functional conservation between monocot and dicot MLO homologs also holds true in non-host plant-PM interactions. To this aim, we used the PM species B. graminis f.sp. hordei (Bgh) to inoculate plants of the Slmlo1 mutant line, the cultivar Moneymaker (MM), carrying wild-type SlMLO1, and two of the 35S::HvMLO T2 families (35S::HvMLO-9 and −10, previously described in Fig. 3, Additional file 2 and 3). Bgh is an adapted PM on barley and a non-adapted PM to tomato. In the Slmlo1 line, 75.4 % of infection units were associated with papilla formation and 24.6 % with cell death response (Fig. 4). Compared with the Slmlo1 line, transgenic 35S::HvMLO-9 T2 plants displayed a lower level of papilla formation (31.3 %) and a higher level of cell death response (68.7 %). In MM, papilla formation and cell death occurred at a rate similar to the one in 35S::HvMLO-9 plants (14.6 % and 85.4 %, respectively). Taken together, this body of evidence indicates that both HvMLO and SlMLO1 predispose to the penetration of a non-host pathogen.
Fig. 4.
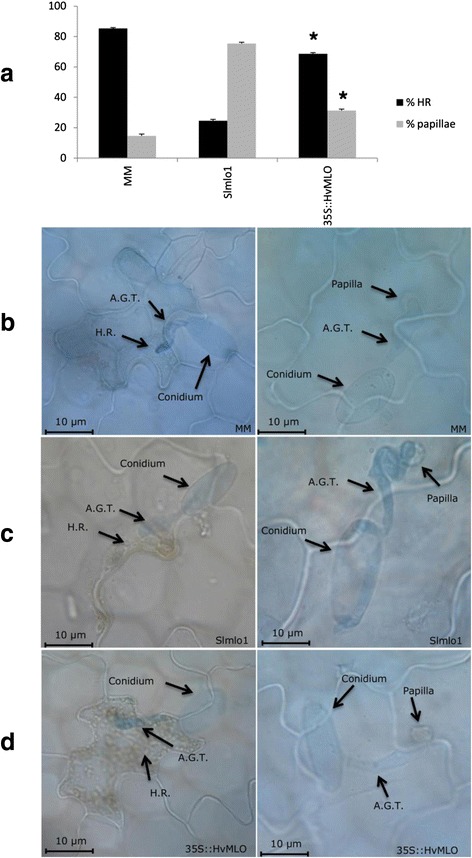
Functional conservation of SlMLO1 and HvMLO in the tomato/Blumeria graminis f.sp. hordei (Bgh) interaction. Panel a shows the ratio of penetrated and non-penetrated epidermal cells, assessed in function of infection units showing hypersensitive response (H.R.) and papillae, respectively, in the following genotypes: the mlo mutant line Slmlo1; the cultivar MM, with a similar genetic background and carrying wild-type SlMLO1; transgenic plants of a T2 family overexpressing barley HvMLO in the Slmlo1 genetic background (35S::HvMLO-9). Panel b, c and d show, in the same genotypes, fungal structures (conidiospore and appressorium germination tube -A.G.T.-) and cellular events (the formation of papillae and H.R.) arresting fungal growth before and after penetration, respectively
Discussion
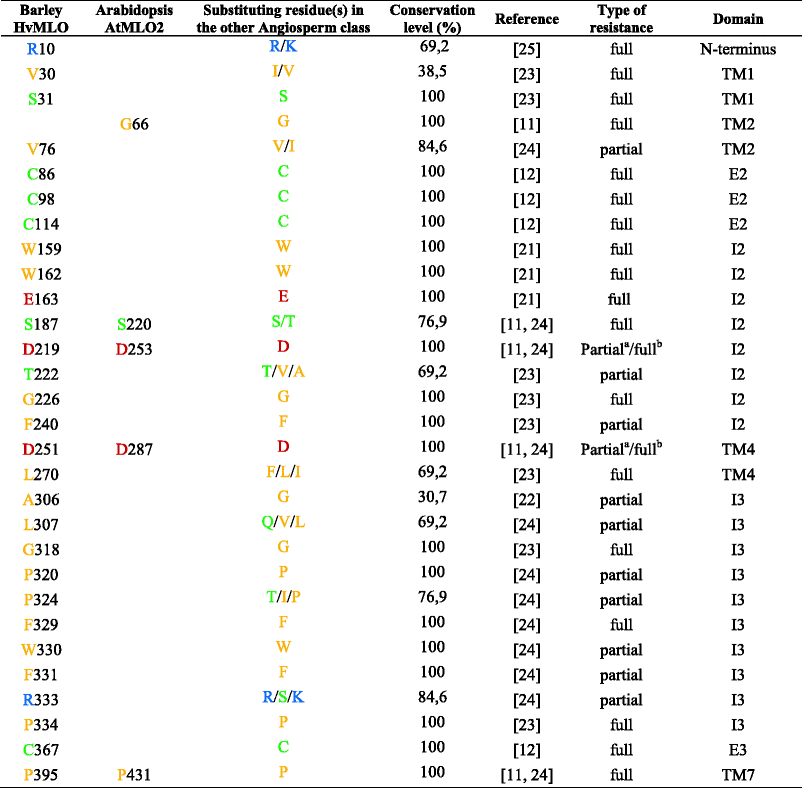
The functional characterization of MLO homologs involved in PM susceptibility is of great interest for basic research on plant-microbe interactions as well as for plant breeding, as loss-of-function genotypes could be conveniently used to introduce durable and broad-spectrum resistance in cultivated species [7]. Results of previous investigations indicated that mlo-based resistance in a certain plant species can be lost by the heterologous expression of MLO susceptibility genes from related species of the same botanical family. Indeed, restored susceptibility has been observed in barley HvMLO mutants transformed with wheat TaMLO_B1 and rice OsMLO3, as well as in pea PsMLO1 mutants expressing lotus LjMLO1 or barrel clover MtMLO1[12, 13]. Recently, similar evidence was shown on tomato SlMLO1 mutants transformed with pepper CaMLO2 or tobacco NtMLO1 [16, 17]. Here, we investigated whether complementation can also occur by transferring MLO genes from more evolutionary divergent plant species. We found that, in a tomato mlo mutant background, transgenic expression of a MLO susceptibility gene from pea (a distantly related dicot species) and barley (a monocot species) is sufficient to re-establish PM susceptibility (Fig. 3 and Additional file 3). This finding indicates that, despite phylogenetic distance and the evolution of peculiar molecular traits (Fig. 1 and 2), monocot and dicot MLO proteins are essentially conserved with respect to features involved in the interaction with PM pathogens. In support of this conclusion, we show that the monocot gene HvMLO and the dicot gene SlMLO1 both enhance penetration of the non-adapted pathogen B. graminis f.sp. hordei compared to a tomato mlo-mutant (Fig. 4). Moreover, after reviewing scientific literature, we found that only one out of thirty MLO protein substitutions so far associated with PM resistance involves a class-specific residue (a monocot-specific alanine residue in position 350 of the alignment in Fig. 2) (Table 1) [22]. The same residue is replaced in dicots by a glycine (sharing similar non-polar chemical properties of alanine, Table 1), indicating that, in this case, class-specific conservations are not associated with important changes in protein structure or function.
Table 1.
Amino acid residues in dicot AtMLO2 and monocot HvMLO whose mutation has been associated with PM resistance. For each amino-acid, localization in any of the MLO protein domains, including seven transmembrane (TM) regions, three extracellular loops (E), three intracellular (I) loops, the N-terminus and the C-terminus, is indicated
|
ᅟ |
Numbers adjacent to each amino acid indicate their position in either HvMLO or AtMLO2 proteins
Barley and Arabidopsis residues in the same row correspond to each other in HvMLO/AtMLO2 protein alignment
Percentage of conservation is calculated based on the alignment of 13 MLO proteins functionally associated with powdery mildew susceptibility (AtMLO2, AtMLO6, AtMLO12, SlMLO1, CaMLO2, NtMLO1, PsMLO1, LjMLO1, MtMLO1, TaMLO_A1b, TaMLO_B1, OsMLO3 and HvMLO)
Amino acid color is according to its chemical properties: non-polar (yellow), polar, uncharged (green), polar, acidic (red), polar, basic (blue)
apartial resistance observed in barley, bfull resistance observed in Arabidopsis
We cannot exclude that class-specific traits might have minor effects on interactions with PM fungi. Indeed, by comparing three independent T2 families for each construct, we found that that overexpression of PsMLO1 results in higher D.I. index scores than the one of HvMLO (Fig. 3 and Additional file 3). Clearly, complementation tests with several other monocot and dicot transgenes could help to answer this question.
Through the analysis of the dN-dS difference, we provide evidence for negative selection acting on several class-specific residues, which are thus likely to play a major adaptive role (Additional file 1). However, as mentioned before, transgenic complementation tests indicate that these class-specific residues are not crucial for the outcome of the interaction between plants and PM pathogens. Possibly, some of the class-specific residues identified in this study might underlie roles which are not related with the interaction with PM fungi. The implication of MLO susceptibility proteins in other physiological processes would explain why, in spite of being required for pathogenesis, they have been not excluded by evolution. With this respect, it is worth to mention that PM resistance in Arabidopsis and barley mlo mutants has been associated with the induction of leaf senescence, a pleiotropic phenotype [11].
We show that MLO homologs required for PM pathogenesis can complement a mlo mutant background in transgenic assays, irrespective of the phylogenetic distance between the donor and the recipient species (Fig. 3). This would be of great advantage in order to test the function of candidate MLO susceptibility genes which are currently being identified by several authors across cultivated species [4, 5]. Moreover, we provide an overview of MLO protein regions which are under negative selection and thus are expected to be of functional relevance. These regions represent ideal targets to select loss-of-function mutants resistant to the PM disease. With this respect, breeders may apply diverse tools, such as conventional targeted mutagenesis approaches of TILLING (targeted induced local lesions in genomes) or advanced technologies of genome editing, based on zinc finger nucleases (ZFNs), clustered regularly interspaced short palindromic repeat (CRISPR) and transcription activator-like effector nucleases (TALEN) [26–28].
Conclusion
This work provides insights on the evolution and function of Angiosperm MLO susceptibility genes. We show that complementation assays similar to those carried out in this study are suitable for future activities aimed at the characterization of novel PM susceptibility factors across cultivated species. Moreover, we indicate a series of gene targets for the selection of loss-of-function mlo resistant mutants.
Methods
Bioinformatic analyses
The following MLO proteins, experimentally shown to be required for PM susceptibility, were used as dataset for CLUSTAL alignment using the CLC sequence viewer software (http://www.clcbio.com/products/clc-sequence-viewer/): Arabidopsis AtMLO2 [GenBank: NP172598], AtMLO6 [GeneBank: NP176350] and AtMLO12 [GeneBank: NP565902], tomato SlMLO1 [GeneBank: NP001234814], pea PsMLO1 [GeneBank: ACO07297], pepper CaMLO2 [GeneBank: AFH68055], lotus LjMLO1 [GeneBank: AAX77015], barrel clover MtMLO1 [GeneBank: ADV40949], barley HvMLO [GeneBank: P93766], rice OsMLO3 [GeneBank: AAK94907], wheat TaMLO_B1 [GeneBank: AAK94904] and TaMLO_A1b [GeneBank: AAK94905]. The alignment was given to Geneious v8 software (http://www.geneious.com, [29] ), to highlight amino acids with different polarity, and the online web service Phylogeny.fr (http://www.phylogeny.fr/) to construct an unrooted radial phylogenetic tree.
In order to make predictions on the type of evolution (negative or neutral) of class-specific molecular features, all the above mentioned dicot MLO susceptibility genes were used as dataset for a codon-based evolutionary analysis based on the difference of nonsynonymous-to-synonymous substitutions per nonsynonymous and synonymous sites (dN/dS). This was performed by using the Single-likelihood Ancestor Counting (SLAC) method implemented by the Datamonkey web server (www.datamonkey.org). The default p-value of 0.1 was taken as threshold to call codons under significant negative selection.
Isolation and cloning of full-length PsMLO1 and HvMLO
Total RNAs from pea (cultivar Sprinter) and barley (cultivar Maythorpe) were isolated by using the RNeasy plant mini kit (Qiagen), and corresponding cDNAs were synthesized by using the SuperScript III first-strand synthesis kit (Invitrogen) and the oligo(dT)20 primer. Specific primer pairs, named PsMLO1-Fw/PsMLO1-Rev and HvMLO-Fw/HvMLO-Rev (Additional file 4: Table S2) were manually designed in order to amplify the PsMLO1 and HvMLO full-length coding sequences, respectively. PCR reactions were performed by using the high-fidelity Phusion DNA polymerase (New England Biolabs) and an annealing temperature of 55 °C. Amplicons were ligated into the Gateway-compatible vector pENTR D-TOPO (Invitrogen) and cloned into the E. coli One Shot® TOP10 cells (Invitrogen), according to the manufacturer’s instructions. After selecting positive colonies by colony PCR, using the two gene-specific primer pairs above mentioned, recombinant plasmids were extracted and their inserts were sequenced. A single colony for each construct was selected, in which the inserts resulted to have sequences identical to those of HvMLO and PsMLO1 deposited in the NCBI database.
Generation and functional characterization of transgenic SlMLO1 mutant tomato plants expressing PsMLO1 and HvMLO
Following the manufacturer instructions (Invitrogen), cloned HvMLO and PsMLO1 gene sequences were inserted by LR recombination into the binary plasmid vector pK7WG2, which harbors the 35S Cauliflower Mosaic Virus (CaMV) promoter and the marker gene nptII for kanamycin resistance selection. Plasmids were then transferred to E. coli and positive colonies were screened by colony PCR and sequencing, as previously mentioned. Finally, recombinant vectors were extracted and transferred to the AGL1-virG strain of A. tumefaciens by electroporation.
The transformation of the tomato ol-2 mutant line, carrying a loss-of-function mutation of the PM susceptibility gene SlMLO1, was performed according to the methods described by [6] and [16]. The evaluation of the expression levels of PsMLO1 and HvMLO in T1 plants was carried out by real-time qPCR using the primer pairs qPsMLO1-Fw/qPsMLO1-Rev and qHvMLO-Fw/qHvMLO-Rev (Additional file 4). A primer pair designed on the elongation factor 1α gene (qEF-Fw/qEF-Rev) was used for relative quantification (Additional file 4).
Functional characterization of host and non-host interactions
For each of the two transgenes above mentioned, three T1 individuals showing the highest expression levels were allowed to self-pollinate, resulting in a total of six T2 families. Individuals of each family were assayed for the presence/absence of the overexpression construct by means of PCR, using the primer pairs NPTII_Fw/ NPTII_Rev and 35S-Fw/35S-Rev designed on the nptII marker gene and the 35S promoter, respectively (Additional file 4). Ten resistant Slmlo1 plants carrying the loss-of-function SlMLO1 allele and ten individuals of each family were challenged with an isolate of the tomato PM fungus O. neolycopersici maintained at the Plant Breeding Department of the University of Wageningen, The Netherlands. Inoculation was performed as described by [30], spraying 4 weeks-old plants with a suspension of conidiospores obtained from freshly sporulating leaves of heavily infected plants and adjusted to a final concentration of 4 × 104 spores/ml. Inoculated plants were grown in a greenhouse compartment at 20 ± 2 °C with 70 ± 15 % relative humidity. Disease evaluation was visually carried out 15 days after inoculation, based on the presence of disease signs on the third and fourth leaf, according to the scale from 0 to 3 reported by [10].
For the functional characterization of a non-host interaction, seeds from one of the three 35S::HvMLO T2 families previously tested were surface-sterilized and sown on half-strength Murashige and Skoog (MS) agar supplemented with 50 mg/ml kanamycin for selection of transgenic plants. Seeds were left for 2 days at 4 °C and then transferred to a growing chamber for 10 days. Five transgenic seedlings were transplanted in pots and transferred to a greenhouse compartment. Three barley plants of the PM susceptible cultivar Manchuria, five Slmlo1 plants and five MoneyMaker plants were used as controls. An isolate of B. graminis f. sp. hordei (Bgh) collected in Wageningen (Wag.04) was used for the inoculation. This was performed by rubbing Manchuria leaves heavily infected with Bgh on the third tomato leaf. After 72 h, in which inoculated plants were kept in a climate chamber at 20 °C, 16 h of light/day and 70 % RH, a 4 cm2 segment was cut from the inoculated leaves (third leaf). Three samples were taken from 3 plants of each genotype.
Each leaf segment was bleached is a 1:3 (v/v) acetic-acid/ethanol solution and 48 h later stained in 0.005 % Trypan Blue as described by [31]. The rate of fungal penetration was estimated by the frequency of infection units showing epidermal cell death. For each genotype, three biological replicates were considered, considering at least 100 infection units.
Acknowledgements
We acknowledge Dr. Henk Schouten, Dr. Anne-Marie Wolters and Dr. Rients Niks for critical reading and valuable suggestions during the preparation of the manuscript. We also acknowledge Romero Cynara for her help for inoculation with Blumeria graminis f.sp. hordei. The work of MA, CL, LR and SP were supported by the Italian Ministry of University and Research through the GenHort PON R&C project.
Additional files
Codons under significant negative selection in PM susceptibility genes. Codon numbers refer to positions in the alignment of nine dicot MLO genes (AtMLO2, AtMLO6, AtMLO12, PsMLO1, MtMLO1, LjMLO1, CaMLO2, SlMLO1, NtMLO1) experimentally shown to act as powdery mildew susceptibility genes. Amino acid residues corresponding to each codon in barley HvMLO and pea PsMLO1 are indicated. For each residue, localization in any of the MLO protein domains, including seven transmembrane (TM) regions, three extracellular loops (E), three intracellular (I) loops, the N-terminus and the C-terminus, is indicated. Codons marked in bold are translated into class-specific residues. The threshold p-value was 0.1, representing the default value for Single-likelihood Ancestor Counting (SLAC) analysis implemented by the Datamonkey web server. (DOCX 30 kb)
Expression levels of PsMLO1 and HvMLO after transformation. Panel A) and panel B) show the expression of PsMLO1 and HvMLO in 19 and 20 T1 individuals, respectively, which were obtained by the transformation of the tomato mutant line Slmlo1, harboring a loss-of-function mutation of the endogenous SlMLO1 gene. Asterisks indicate T1 individuals selected for self-pollination and the development of T2 families. (PDF 173 kb)
Effects of transgenic overexpression of pea PsMLO1 and barley HvMLO in the tomato mutant line Slmlo1. Average disease index (DI) values and phenotypes are referred to transgenic plants of two additional T2 families segregating for PsMLO1 [35S::PsMLO1-4 and 35S::PsMLO1-6, panel a) and b)] and two additional T2 families segregating for HvMLO [35S::HvMLO-10 and 35S::HvMLO-15), panel c) and d)]. Data relative to the Slmlo1 mutant line, used as genetic background for transformation, and non-transgenic plants of three T2 families for each overexpression construct (35S::PsMLO1_(−) and 35S::HvMLO_(−)) are also shown. (PDF 304 kb)
Primer pairs used in this study. (DOCX 14 kb)
Footnotes
Competing interest
The authors declare no competing interest.
Authors’ contributions
MA developed transgenic plants, performed disease tests and was involved in experimental design, interpretation of results and manuscript drafting. MISM performed histological analyses. ZZ prepared overexpression constructs for transgenic complementation assays. DC, CL, RGFV, LR and YB were involved in experimental design and critical revision of the manuscript. SP performed evolutionary analyses and was involved in experimental design, interpretation of results and manuscript drafting. All authors read and approved the final manuscript.
Contributor Information
Yuling Bai, Phone: +31 (0)317 482136, Email: bai.yuling@wur.nl.
Stefano Pavan, Phone: +39 080 5442994, Email: stefano.pavan@uniba.it.
References
- 1.Devoto A, Hartmann HA, Piffanelli P, Elliott C, Simmons C, Taramino G, et al. Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J Mol Evol. 2003;56(1):77–88. doi: 10.1007/s00239-002-2382-5. [DOI] [PubMed] [Google Scholar]
- 2.Feechan A, Jermakow AM, Torregrosa L, Panstruga R, Dry IB. Identification of grapevine MLO gene candidates involved in susceptibility to powdery mildew. Funct Plant Biol. 2008;35(12):1255–1266. doi: 10.1071/FP08173. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q, Zhu H. Molecular evolution of the MLO gene family in Oryza sativa and their functional divergence. Gene. 2008;409(1–2):1–10. doi: 10.1016/j.gene.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 4.Pessina S, Pavan S, Catalano D, Gallotta A, Visser R, Bai Y, et al. Characterization of the MLO gene family in Rosaceae and gene expression analysis in Malus domestica. BMC Genomics. 2014;15(1):618. doi: 10.1186/1471-2164-15-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou SJ, Jing Z, Shi JL. Genome-wide identification, characterization, and expression analysis of the MLO gene family in Cucumis sativus. Genet Mol Res. 2013;12(4):6565–6578. doi: 10.4238/2013.December.11.8. [DOI] [PubMed] [Google Scholar]
- 6.Singh VK, Singh AK, Chand R, Singh BD. Genome wide analysis of disease resistance mlo gene family in sorghum [Sorghum bicolor (l.) Moench] J Plant Genom. 2012;2(1):18–27. [Google Scholar]
- 7.Pavan S, Jacobsen E, Visser RGF, Bai Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol Breed. 2009;25(1):1–12. doi: 10.1007/s11032-009-9323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell. 1997;88(5):695–705. doi: 10.1016/S0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 9.Panstruga R. Discovery of novel conserved peptide domains by ortholog comparison within plant multi-protein families. Plant Mol Biol. 2005;59(3):485–500. doi: 10.1007/s11103-005-0353-0. [DOI] [PubMed] [Google Scholar]
- 10.Bai Y, Pavan S, Zheng Z, Zappel NF, Reinstädler A, Lotti C, et al. Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of Mlo function. Mol Plant-Microbe Interact. 2008;21(1):30–39. doi: 10.1094/MPMI-21-1-0030. [DOI] [PubMed] [Google Scholar]
- 11.Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet. 2006;38(6):716–720. doi: 10.1038/ng1806. [DOI] [PubMed] [Google Scholar]
- 12.Elliott C, Müller J, Miklis M, Bhat RA, Schulze-Lefert P, Panstruga R. Conserved extracellular cysteine residues and cytoplasmic loop-loop interplay are required for functionality of the heptahelical MLO protein. Biochem J. 2005;385(1):243–254. doi: 10.1042/BJ20040993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humphry M, Reinstädler A, Ivanov S, Bisseling T, Panstruga R. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol Plant Pathol. 2011;12(9):866–878. doi: 10.1111/j.1364-3703.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavan S, Schiavulli A, Appiano M, Marcotrigiano AR, Cillo F, Visser RGF, et al. Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Theor Appl Genet. 2011;123(8):1425–1431. doi: 10.1007/s00122-011-1677-6. [DOI] [PubMed] [Google Scholar]
- 15.Várallyay É, Giczey G, Burgyán J. Virus-induced gene silencing of Mlo genes induces powdery mildew resistance in Triticum aestivum. Arch Virol. 2012;157(7):1345–1350. doi: 10.1007/s00705-012-1286-y. [DOI] [PubMed] [Google Scholar]
- 16.Zheng Z, Nonomura T, Appiano M, Pavan S, Matsuda Y, Toyoda H, Wolters AMA, Visser RGF, Bai Y: Loss of Function in Mlo Orthologs Reduces Susceptibility of Pepper and Tomato to Powdery Mildew Disease Caused by Leveillula taurica. PLoS ONE 2013, 8(7). 10.1371/journal.pone.0070723 [DOI] [PMC free article] [PubMed]
- 17.Appiano M, Pavan S, Catalano D, Zheng Z, Bracuto V, Lotti C, et al. Identification of candidate MLO powdery mildew susceptibility genes in cultivated Solanaceae and functional characterization of tobacco NtMLO1. Transgenic Res. 2015;24:1–12. doi: 10.1007/s11248-015-9878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterhansel C, Freialdenhoven A, Kurth J, Kolsch R, Schulze-Lefert P. Interaction Analyses of Genes Required for Resistance Responses to Powdery Mildew in Barley Reveal Distinct Pathways Leading to Leaf Cell Death. Plant Cell. 1997;9(8):1397–1409. doi: 10.1105/tpc.9.8.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, et al. Pre- and Postinvasion Defenses Both Contribute to Nonhost Resistance in Arabidopsis. Science. 2005;310(5751):1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- 20.Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, Von Heijne G, et al. Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J Biol Chem. 1999;274(49):34993–35004. doi: 10.1074/jbc.274.49.34993. [DOI] [PubMed] [Google Scholar]
- 21.Müller J, Piffanelli P, Devoto A, Miklis M, Elliott C, Ortmann B, et al. Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell. 2005;17(1):149–163. doi: 10.1105/tpc.104.026625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panstruga R, Molina-Cano JL, Reinstädler A, Müller J. Molecular characterization of mlo mutants in North American two- and six-rowed malting barley cultivars. Mol Plant Pathol. 2005;6(3):315–320. doi: 10.1111/j.1364-3703.2005.00277.x. [DOI] [PubMed] [Google Scholar]
- 23.Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, et al. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002;129(3):1076–1085. doi: 10.1104/pp.010954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinstädler A, Müller J, Czembor JH, Piffanelli P, Panstruga R. Novel induced mlo mutant alleles in combination with site-directed mutagenesis reveal functionally important domains in the heptahelical barley Mlo protein. BMC Plant Biol. 2010;10:31. doi: 10.1186/1471-2229-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiberg A. Sources of resistance to powdery mildew in barley. Hereditas. 1974;78(1):1–40. doi: 10.1111/j.1601-5223.1974.tb01426.x. [DOI] [PubMed] [Google Scholar]
- 26.McCallum CM, Comai L, Greene EA, Henikoff S. Targeting Induced Local Lesions IN Genomes (TILLING) for Plant Functional Genomics. Plant Physiol. 2000;123(2):439–442. doi: 10.1104/pp.123.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaj T, Gersbach CA, Barbas CF., III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terns RM, Terns MP. CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends Genet. 2014;30(3):111–118. doi: 10.1016/j.tig.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavan S, Zheng Z, Borisova M, Van Den Berg P, Lotti C, De Giovanni C, et al. Map- vs. homology-based cloning for the recessive gene ol-2 conferring resistance to tomato powdery mildew. Euphytica. 2008;162(1):91–98. doi: 10.1007/s10681-007-9570-8. [DOI] [Google Scholar]
- 31.Anker C, Niks R. Prehaustorial resistance to the wheat leaf rust fungus, Puccinia triticina, in Triticum monococcum (s.s.) Euphytica. 2001;117(3):209–215. doi: 10.1023/A:1026577307163. [DOI] [Google Scholar]


