Abstract
Background: Preclinical and epidemiologic studies suggest that garlic intake is inversely associated with the progression of cancer and cardiovascular disease.
Objective: We designed a study to probe the mechanisms of garlic action in humans.
Methods: We conducted a randomized crossover feeding trial in which 17 volunteers consumed a garlic-containing meal (100 g white bread, 15 g butter, and 5 g raw, crushed garlic) or a garlic-free control meal (100 g white bread and 15 g butter) after 10 d of consuming a controlled, garlic-free diet. Blood was collected before and 3 h after test meal consumption for gene expression analysis in whole blood. Illumina BeadArray was used to screen for genes of interest, followed by real-time quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) on selected genes. To augment human study findings, Mono Mac 6 cells were treated with a purified garlic extract (0.5 μL/mL), and mRNA was measured by qRT-PCR at 0, 3, 6, and 24 h.
Results: The following 7 genes were found to be upregulated by garlic intake: aryl hydrocarbon receptor (AHR), aryl hydrocarbon receptor nuclear translocator (ARNT), hypoxia-inducible factor 1α (HIF1A), proto-oncogene c-Jun (JUN), nuclear factor of activated T cells (NFAT) activating protein with immunoreceptor tyrosine-based activation motif 1 (NFAM1), oncostatin M (OSM), and V-rel avian reticuloendotheliosis viral oncogene homolog (REL). Fold-increases in mRNA transcripts ranged from 1.6 (HIF1A) to 3.0 (NFAM1) (P < 0.05). The mRNA levels of 5 of the 7 genes that were upregulated in the human trial were also upregulated in cell culture at 3 and 6 h: AHR, HIF1A, JUN, OSM, and REL. Fold-increases in mRNA transcripts in cell culture ranged from 1.7 (HIF1A) to 12.1 (JUN) (P < 0.01). OSM protein was measured by ELISA and was significantly higher than the control at 3, 6, and 24 h (24 h: 19.5 ± 1.4 and 74.8 ± 1.4 pg/mL for control and garlic, respectively). OSM is a pleiotropic cytokine that inhibits several tumor cell lines in culture.
Conclusion: These data indicate that the bioactivity of garlic is multifaceted and includes activation of genes related to immunity, apoptosis, and xenobiotic metabolism in humans and Mono Mac 6 cells. This trial is registered at clinicaltrials.gov as NCT01293591.
Keywords: garlic, cancer, immunity, gene expression, Mono Mac 6
Introduction
Consumer demand for garlic (Allium sativum L.) has surged in recent years, with worldwide production almost doubling from 2002 to 2012 (1). This interest in garlic is partly driven by reports attributing various health benefits to garlic consumption. Given the prevalence of chronic diseases such as cancer and cardiovascular disease and their substantial personal, social, and financial impacts, there may be a role for garlic as part of a diet to promote and sustain human health (2, 3).
Preclinical studies have primarily focused on cardiovascular health and on prevention and treatment of cancer. Animal studies suggest that factors related to cardiovascular function such as plasma lipids, blood pressure, and platelet aggregation may be favorably modified by garlic or garlic-derived compounds (4–7). The inhibition of cholesterol synthesis has also been demonstrated in rat hepatoma cells. Of 9 garlic-derived compounds tested, diallyl disulfide, diallyl trisulfide, and allyl mercaptan were inhibitory, probably by suppression of 4α-methyl oxidase (8). Human clinical trials have produced mixed results with regard to lipid variables. LDL oxidation has been shown in cell studies and a small-scale human study to be suppressed by aged garlic extract (GE)7 (9, 10), which is high in S-allylcysteine in contrast to fresh crushed garlic, which is high in allicin (diallyl thiosulfinate) and γ-glutamyl-S-alkylcysteines (11, 12). A recent meta-analysis of 39 randomized, placebo-controlled clinical studies indicated that garlic intake over at least a 2-mo period significantly reduced total serum and LDL cholesterol in humans with very high baseline values (13). However, in humans with moderately high baseline LDL cholesterol, raw garlic, dried garlic powder, and aged GE did not influence total cholesterol, LDL cholesterol, HDL cholesterol, total- to HDL-cholesterol ratio, or TGs during the 6-mo intervention period (14).
The effects of garlic and garlic-derived compounds on the prevention and treatment of cancer have also been reported in a large number of preclinical studies and involve modulation of xenobiotic metabolism, cell cycle arrest, induction of apoptosis, inhibition of angiogenesis, and histone modification (15–23). Results from epidemiologic and clinical intervention studies are inconsistent. In the Iowa Women’s Health Study of 41,837 women over 5 y, an RR of 0.68 was reported for colon cancer for the uppermost compared with the lowermost consumption levels of garlic (24). The EPIC (European Prospective Investigation into Cancer and Nutrition) study of >500,000 individuals showed a weak inverse association between garlic and onion intake and risk of intestinal gastric cancer after an average 6.5-y follow-up, but when 477,312 of these participants were assessed after 11 y, this association was no longer present (25, 26). In a randomized double-blind study, supplementation with a combination of steam-distilled garlic oil and aged GE did not influence the incidence of precancerous gastric lesions or gastric cancer incidence after a 7.3-y intervention (27). However, in a double-blind intervention study of participants at high risk of gastric cancer, men (but not women) consuming 200 mg synthetic allitridum (diallyl trisulfide) with 100 μg selenium had a decreased cancer risk after 5 y (28).
There is substantial evidence for a role of garlic in reducing cancer risk and cardiovascular disease, but the inconsistent findings prevent a clear understanding of the role of garlic in health promotion. One reason for the inconsistent findings may be the need for additional biomarkers of biological activity. Gene expression in vivo is a biomarker at the most basic level of biological response and may provide important clues to the biological activity of bioactive food components. Therefore, we conducted a human clinical trial and follow-up in vitro studies to measure the influence of garlic intake on mRNA gene expression in whole blood. Gene expression in the leukocyte population of whole blood is responsive to dietary interventions and may correlate with tissue-specific gene expression (29–31). Whole blood is relatively accessible compared with other human tissues and thus is advantageous for obtaining and measuring mRNA. We chose to focus on genes related to immunity and cancer and studied their response to a single meal containing raw, crushed garlic (RCG).
Methods
Study design, diet, and treatments.
The study protocol was approved by the MedStar Health Research Institute (Hyattsville, Maryland). Written, informed consent was obtained from each study participant. A randomized crossover design with two 11-d treatment periods separated by a 17-d washout period was used. Subjects consumed a basal garlic-free diet for 10 d. On the 11th day, participants receiving the control treatment ate a breakfast consisting of 100 g white bread with 15 g butter. Participants receiving the garlic treatment consumed 100 g white bread with 15 g butter and 5 g RCG. The basal diet consisted of adequate protein,∼35% of calories from fat, and 3 servings of fruits and vegetables daily to be in accord with average intakes in the United States (32, 33). Participants were instructed to consume all foods and only foods that were provided by the Beltsville Human Nutrition Research Center. Breakfast and dinner on weekdays were consumed at the Beltsville Human Nutrition Research Center, and lunches and weekend meals were packed for carry-out. Study participants were asked to abstain from vitamin and mineral supplements beginning 2 wk before the study and continuing throughout the study.
One week before the garlic meal of the first period, raw cloves of California Early garlic were minced and homogenized. Five-gram portions in sealed plastic containers were stored at −20°C until used. A subsample was reserved for the measurement of organosulfur compounds by using previously published HPLC methods (Silliker) (34).
Study participants.
Participants were recruited from Beltsville, Maryland. Eighteen participants began the study. One participant left the study due to a scheduling conflict and 17 participants completed the study. Participant characteristics are reported in Supplemental Table 1.
Whole-blood collection and mRNA gene expression.
Whole blood was collected into PAXgene blood RNA tubes (Qiagen) immediately before the day 11 breakfast (consisting of a control or garlic meal) and at 3 h after breakfast. Total RNA was isolated according to the manufacturer’s instructions. Global mRNA gene expression by Illumina HumanHT-12 v4 BeadChip was used to screen for genes of interest in 12 randomly selected participants (Expression Analysis). Differentially expressed genes (P < 0.01) were considered for analysis by qRT-PCR. We selected genes related to immunity and genes potentially involved in cancer-related processes (Table 1). All probes and primers for qRT-PCR were designed by using the Primer Express (Applied Biosystems) software package and nucleotide sequences obtained from GenBank.
TABLE 1.
Genes measured in human whole blood by qRT-PCR1
| Gene | Symbol |
| AHR pathway genes | |
| Aryl hydrocarbon receptor | AHR |
| Aryl hydrocarbon receptor nuclear translocator | ARNT |
| Aryl hydrocarbon receptor nuclear translocator 2 | ARNT2 |
| Hypoxia-inducible factor 1, α subunit | HIF1A |
| Cancer-related genes | |
| Excision repair cross-complementation group 1 | ERCC1 |
| IL 6 | IL6 |
| Proto-oncogene c-Jun | JUN |
| Leukemia inhibitory factor | LIF |
| Oncostatin M | OSM |
| V-rel avian reticuloendotheliosis viral oncogene homolog | REL |
| Tumor necrosis factor receptor superfamily, member 21 | TNFRSF21 |
| Immunity-related genes | |
| Calcium/calmodulin-dependent protein kinase IIγ | CAMK2G |
| Chemokine (C-X-C motif) ligand 14 | CXCL14 |
| IL 2 | IL2 |
| Nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 3 | NFATC3 |
| NFAT activating protein with immunoreceptor tyrosine-based activation motif 1 | NFAM1 |
| Housekeeping gene | |
| Cyclophilin A | PPIA |
Several genes could be assigned to more than one classification, but for ease of reference are placed within only one group.
The RNA of the 12 participants used to assess global mRNA gene expression was also used to synthesize cDNA for qRT-PCR. Total RNA from the whole blood of the remaining 5 participants was extracted from PAXgene blood RNA tubes by using the BioRobot Universal system (Qiagen) according to the manufacturer’s instructions. RNA quality was assessed by using Experion RNA gel electrophoresis analysis chips (Bio-Rad Laboratories), and the concentration was determined by using a Nanodrop spectrophotometer (Thermo Scientific). cDNA was prepared from 1.35 μg total RNA by using SuperScript II reverse transcriptase according to the manufacturer’s protocol (Life Technologies). Quantitative real-time PCR was conducted by using iQ Supermix and a CFX96 real-time PCR system (both, Bio-Rad Laboratories). Data were adjusted for the housekeeping gene cyclophilin A (PPIA). Quantitative mRNA fold-changes were derived by using the ΔΔCt (threshold cycle) method (35) and are presented as the fold-change due to the garlic treatment relative to that of the control = 2−∆C′t, where ∆C′t = (Ctgarlic, hour 3 – Ctgarlic, hour 0) – (Ctcontrol, hour 3 – Ctcontrol, hour 0).
Preparation of GE for in vitro studies.
Garlic from a local grocery store was processed with a juicer to produce a crude garlic homogenate, which was centrifuged at 4500 g for 10 min at 20°C. The supernatant was filtered through a 0.45-μm syringe filter (Millipore) followed by filtration with a 0.22-μm centrifugal filter (Millipore) and centrifuged at 4500 g for 5 min at 20°C. The GE was placed into 1.5-mL microcentrifuge tubes and frozen at −80°C. The endotoxin concentration in the extract was below detection (0.01 ng endotoxin/mL) according to the Limulus amebocyte lysate assay (ThermoScientific). Organosulfur compounds were measured by HPLC.
Cell culture and sample analysis of Mono Mac 6 cells.
Human study gene expression findings were confirmed in vitro by using the Mono Mac 6 cell line. The Mono Mac 6 cell line was selected because it expresses all of the genes that are significantly expressed in humans. In addition, Mono Mac 6 cells are monocytic and therefore model an important class of leukocytes. Cells were obtained from the Leibniz-Institut Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany) and were cultured according to the supplier’s instructions. Cells were maintained at a density of 1.0 × 106 cells/mL.
At the beginning of each experiment, 1 mL medium was transferred to the wells of a cell culture plate and combined with 1 mL of cell-free medium with or without GE, resulting in concentrations of 0.0 or 0.5 μL GE/mL medium (n = 3). Viability was determined by flow cytometric analysis on an Attune flow cytometer (Life Technologies) by using an Annexin-fluorescein isothiocyanate/propidium iodide staining kit (Trevigen). Cell gating was performed by using forward and side scatter, and the percentage of live, apoptotic, and necrotic cells was determined from 10,000 cells. The percentage of viable, early apoptotic, late apoptotic, and necrotic cells averaged 93.3%, 3.6%, 1.9%, and 1.1%, respectively, in control cells, and averaged 91.7%, 4.8%, 2.2%, and 1.2%, respectively, in garlic-treated cells.
Samples were collected at 0, 3, 6, and 24 h after adding the GE. Total RNA was isolated from cells by using the RNeasy Plus Mini Kit (Qiagen). RNA quality and concentration were determined as for the human samples. One microgram of total RNA was transcribed to cDNA by using the Quantitect reverse transcription kit (Qiagen). Genes that were significant (P < 0.05) in the qRT-PCR analysis of the human samples were selected for measurement in Mono Mac 6 cells. qRT-PCR was performed by using iQ Supermix and a CFX96 real-time PCR system, with ribosomal protein L32 (RPL32) used as the housekeeping gene. Ct data were subtracted from 45 (total number of PCR cycles) so that increasing levels of mRNA are represented by increasing values of the difference between 45 and Ct.
Oncostatin M (OSM) protein was measured in cell culture supernatant by ELISA (R&D Biosystems) following the manufacturer’s instructions. Samples were centrifuged at 14,000 g for 15 min at 4°C before being added to the plate. In vitro experiments were performed twice, with similar results.
Statistical analysis.
ANOVAs of the gene expression data from the human study were performed by using the MIXED procedure in SAS (version 9.3; SAS Institute). Data were tested for normality with the Shapiro-Wilk statistic and by inspection of stem-leaf plots and normal probability plots of residuals. The data for nuclear factor of activated T cells (NFAT) activating protein with immunoreceptor tyrosine-based activation motif 1 (NFAM1) and OSM were skewed and therefore were ln-transformed. To account for the serial correlation of repeated measures on the same experimental unit (participant), covariance structures were fit to the data by using Akaike’s and Bayesian information criteria. The models for aryl hydrocarbon receptor (AHR), proto-oncogene c-Jun (JUN), leukemia inhibitory factor (LIF), and OSM were fit with the compound symmetry covariance structure, and the models for hypoxia-inducible factor 1α (HIF1A), NFAM1, and V-rel avian reticuloendotheliosis viral oncogene homolog (REL) were fit with the variance components structure. Study period, sequence, and sex were treated as fixed effects, whereas study participant was treated as a random effect. Model effects are reported as least squares means (LSmeans; P < 0.05). LSmeans for NFAM1 and OSM were inverse transformed to their original scale. The statistical model for the in vitro gene expression data was a treatment × hour 2-way factorial mixed-effects model, with repeated measures observed on each treatment replication at 0, 3, 6, and 24 h. Fisher’s protected least significant difference tests were applied to compare LSmeans at each hour in the in vitro experiment. Data are reported as LSmeans and SEMs.
Results
Composition of the garlic products
The concentrations of the organosulfur compounds in RCG and GE are given in Supplemental Table 2. The concentrations of the compounds in RCG may be calculated on a dry weight basis by multiplying the values by 2.42 (mg/g dry wt). The RCG contained >3 times the concentration of γ-glutamyl-S-alkylcysteines (14.5 mg/g) compared with thiosulfinates (4.5 mg/g), in contrast to the GE in which the concentrations of γ-glutamyl-S-alkylcysteines (3.7 mg/g) and thiosulfinates (3.5 mg/g) were similar. In both products, the concentrations of allyl sulfides were much lower than those of the other organosulfur compounds.
Human clinical trial
Gene expression in whole blood.
Sixteen genes were selected for mRNA gene expression measurement. Of these genes, AHR, aryl hydrocarbon receptor nuclear translocator (ARNT), HIF1A, JUN, NFAM1, OSM, and REL were significantly upregulated 3 h after the RCG intervention (Table 2). NFAM1 had the largest fold-increase (3.0) and HIF1A had the smallest increase (1.6). For both NFAM1 and OSM, there was a significant treatment × sex interaction (P < 0.05) in which the gene expression in men did not change, whereas that in women increased significantly. In women, the NFAM1 fold-change was 5.6 (P < 0.001) and the OSM fold-change was 2.5 (P < 0.001)
TABLE 2.
Measurement by qRT-PCR of mRNA gene expression in human whole blood 3 h after garlic consumption relative to the control meal1
| Gene | Fold of control | P |
| P ≤ 0.05 | ||
| AHR | 2.6 | 0.017 |
| ARNT | 1.8 | 0.020 |
| HIF1A | 1.6 | 0.027 |
| JUN | 1.7 | 0.045 |
| NFAM12 | 3.0 | <0.001 |
| OSM3 | 1.8 | 0.001 |
| REL | 1.7 | 0.016 |
| P > 0.05 | ||
| ARNT2 | 1.0 | 0.97 |
| CAMK2G | 1.2 | 0.35 |
| CXCL14 | 1.3 | 0.74 |
| ERCC1 | 1.3 | 0.11 |
| IL2 | 1.1 | 0.61 |
| IL6 | 1.3 | 0.14 |
| LIF | 1.4 | 0.08 |
| NFATC3 | 1.4 | 0.11 |
| TNFRSF21 | 1.6 | 0.09 |
Fold of control = 2−∆C′t, where ∆C′t = (Ctgarlic, hour 3 – Ctgarlic, hour 0) – (Ctcontrol, hour 3 – Ctcontrol, hour 0). AHR, aryl hydrocarbon receptor; ARNT, aryl hydrocarbon receptor nuclear translocator; ARNT2, aryl hydrocarbon receptor nuclear translocator 2; CAMK2G, calcium/calmodulin-dependent protein kinase IIγ Ct, threshold cycle; CXCL14, chemokine (C-X-C motif) ligand 14; ERCC1, excision repair cross-complementation group 1; HIF1A, hypoxia-inducible factor 1α JUN, proto-oncogene c-Jun; LIF, leukemia inhibitory factor; NFAM1, NFAT activating protein with immunoreceptor tyrosine-based activation motif 1; NFATC3, nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 3; OSM, oncostatin M; REL, V-rel avian reticuloendotheliosis viral oncogene homolog; TNFRSF21, tumor necrosis factor receptor superfamily, member 21.
Significant sex × treatment interaction, P = 0.010. Fold-change for women = 5.6 (P < 0.001) and for men = 1.5 (P = 0.32).
Significant sex × treatment interaction, P = 0.007. Fold-change for women = 2.5 (P < 0.001) and for men = 1.2 (P = 0.29).
In vitro experiments with Mono Mac 6 cells
Gene expression in response to GE.
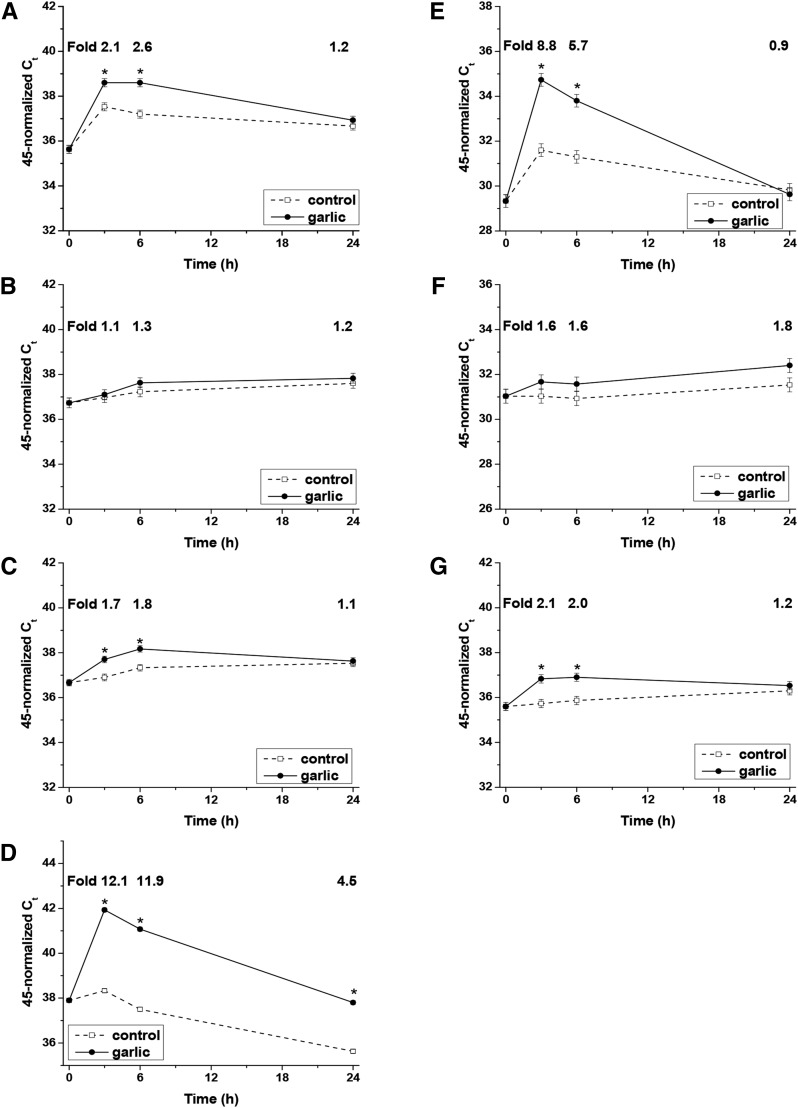
Five of the 7 genes that were significantly upregulated in the human clinical trial also were upregulated in Mono Mac 6 cells treated with GE: AHR, HIF1A, JUN, OSM, and REL (Figure 1). The expression of these genes was higher in GE-treated cells than in control cells at 3 and 6 h, and with the exception of JUN, expression did not differ between GE-treated and control cells at 24 h. The largest fold-increases occurred for JUN, with a fold-increase of 12.1 at 3 h and 11.9 at 6 h, followed by OSM with fold increases of 8.8 at 3 h and 5.7 at 6 h. The smallest fold-change that was significant was for HIF1A, which increased 1.7-fold at 3 h.
FIGURE 1.
Expression of AHR (A), ARNT (B), HIF1A (C), JUN (D), OSM (E), NFAM1 (F), and REL (G) in Mono Mac 6 cells in response to garlic extract (0.5 μL/mL). Values are least squares means ± SEs; n = 3. *Different from control at that time, P < 0.01 (Fisher’s protected least significant different test). The Ct data were normalized to RPL32 and are subtracted from 45 (total number of PCR cycles), so that increasing levels of mRNA are represented by increasing values on the y-axis. AHR, aryl hydrocarbon receptor; ARNT, aryl hydrocarbon receptor nuclear translocator; Ct, threshold cycle; HIF1A, hypoxia-inducible factor 1α JUN, proto-oncogene c-Jun; RPL32, ribosomal protein L32.
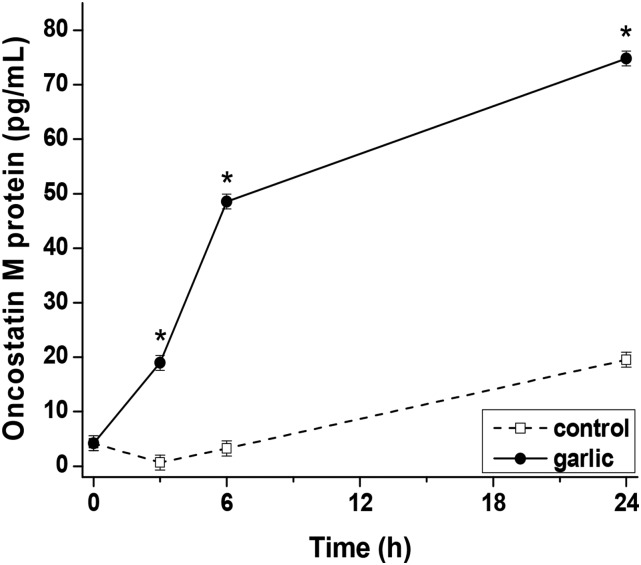
Concentration of OSM protein in response to GE.
The concentration of OSM was significantly higher in GE-treated cells than in control cells at 3, 6, and 24 h (Figure 2). The most rapid accumulation of OSM occurred from 3 to 6 h, when OSM concentration increased by 157% from 19.0 ± 1.4 to 48.6 ± 1.4 pg/mL. The beginning of this high rate of OSM protein synthesis coincided with the peak concentration of the OSM transcript, which occurred at 3 h. The concentration of OSM at 24 h in the GE-treated medium was 74.8 ± 1.4 pg/mL, which was 3.8-fold that of the control cell medium.
FIGURE 2.
Concentration of oncostatin M protein produced by Mono Mac 6 cells treated with garlic extract (0.5 μL/mL). Values are least squares means ± SEs; n = 3. *Different from control at that time, P < 0.01.
Discussion
We chose to use RCG because it is less processed than cooked garlic and other garlic formulations and thus represents garlic in one of its most basic forms. The organosulfur composition of the RCG was similar to that reported in previous studies (34). Concentrations of the organosulfur compounds tended to be lower in GE than in RCG. This difference may be a result of the fact that the garlic from which the GE was produced was purchased several months after harvest and that storage causes degradation of γ-glutamyl-S-alkylcysteines by means of γ-glutamyltranspeptidase (36). Also of note is that the GE used in the present study is chemically distinct from the similarly named aged GE, in which thiosulfinates are present at very low concentrations (11).
Genes affected by garlic.
A single meal with RCG induced the expression of 7 genes involved in immunity and cancer-related processes 3 h after consumption. Remarkably, 5 of these genes were also upregulated in GE-treated Mono Mac 6 cells. AHR is a ligand-activated member of the basic helix-loop-helix/Per-ARNT-single-minded (SIM) superfamily of transcription factors (37). When activated by ligand binding, AHR protein moves from the cytoplasm to the nucleus where it dimerizes with ARNT and binds to xenobiotic response elements (XREs) within the promoters of target genes (38). AHR has been classically associated with its activation by halogenated aromatic hydrocarbons and nonhalogenated polycyclic aromatic hydrocarbons and the subsequent induction of xenobiotic metabolizing enzymes such as cytochrome P450, family 1, subfamily A, polypeptide 1 (CYP1A1), cytochrome P450, family 1, subfamily A, polypeptide 2 (CYP1A2), uridine diphosphate glucuronosyltransferase 1 family, polypeptide A1 (UGT1A1), uridine diphosphate glucuronosyltransferase 1 family, polypeptide A6 (UGT1A6), and glutathione S-transferase α 1 (GSTA1) (39–41). In addition, AHR suppressed intestinal carcinogenesis in adenomatosis polyposis coli (Apc)multiple intestinal neoplasia/+ mice by its participation in a ligand-dependent E3 ubiquitin ligase that degrades β-catenin, a protein whose overexpression is associated with cancers of the colon, skin, liver, ovaries, and prostate (42, 43). AHR was activated by natural ligands derived from dietary tryptophan and glucosinolates, a class of secondary plant metabolites associated with protection from colon cancer (44). These results, together with our finding that the consumption of RCG and GE induced AHR expression, suggest a mechanism by which garlic may inhibit intestinal cancer in humans. AHR also has a role in the development and function of effector and regulatory T cells, a subpopulation of the blood cells collected in our human clinical trial, supporting a role for RCG in the modulation of immune function (45).
Similar to AHR, HIF1A is a dimeric partner with ARNT, forming the transcription factor hypoxia-inducible factor 1 (HIF-1). Genes activated by HIF-1 increase glucose transport, glycolysis, angiogenesis, and erythropoietin (46). In hypoxic environments such as in solid tumors, the increase in HIF-1 leads to increased oxygen transport and mediates adaptive responses to oxygen deprivation, which may support tumor development (47). In ischemic disorders, upregulation of HIF-1 can counteract the pathologic effects of hypoxic conditions and promote reparative neovascularization (48). Therefore, the RCG-induced expression of HIF1A may have different implications depending on the health status of the individual, with benefits for those with ischemic conditions and risks for those with existing tumors.
JUN codes for c-JUN, a protein that partners with the proto-oncogene FBJ murine osteosarcoma viral oncogene homolog (c-FOS) to form activator protein 1 (AP-1), an early response transcription factor. Although c-JUN is required for normal development, many studies suggest that c-JUN is associated with cancer development (49, 50). In contrast, other studies indicate that c-JUN interferes with tumorigenesis. In c-JUN–deficient breakpoint cluster region-Abelson+ (BCR-ABL+) tumor cell lines, mRNA and protein of the tumor suppressor and cell cycle inhibitor p16inhibitor of cyclin-dependent kinase 4a (INK4a) are downregulated, suggesting that c-JUN inhibits the tumorigenic silencing of p16INK4a (51). In tylophorine-treated carcinoma cells, altered binding of c-JUN to the regulatory regions of the cyclin A2 promoter resulted in decreased expression of cyclin A2 and increased G1 phase arrest (52). These results represent mechanisms by which the induction of JUN after RCG consumption may inhibit tumorigenesis.
NFAM1 is expressed in B cells, T cells, and monocytes and is involved in B cell signaling and development (53). Given that NFAM1 was upregulated in whole blood from the human study but not in Mono Mac 6 cells, which are monocytic, garlic intake may primarily influence B and/or T cells. NFAM1 activates the NFAT signaling pathway, leading to activation of tumor necrosis factor (TNF) and IL13 promoters in the human mast cell line 1 (HMC-1) (54). The upregulation of NFAM1 points to an immunomodulating effect of RCG intake, an effect that has been hypothesized, but not proven, to reduce cancer risk (55).
REL codes for V-Rel avian reticuloendotheliosis viral oncogene homolog (c-REL), which exists as homodimers or heterodimers in the NF-κB family of transcription factors. These transcription factors have many roles in both normal and pathologic processes and activate genes related to apoptosis, development, and immune and inflammatory responses (56). Overexpression of c-REL has been linked with mammary tumorigenesis in Michigan Cancer Foundation-7 (MCF7) cells and in primary human breast cancer tissue samples; in addition, the proliferation of B cell lymphoma cell lines was stimulated by c-REL (57–59). In contrast, 3 of 6 REL−/− mice developed lymphoproliferative lesions after 12 mo of infection with Helicobacter pylori, suggesting that c-REL–mediated signaling may reduce the risk of lymphomagenesis in gastric mucosa–associated lymphoid tissue (60). REL has many functions, and the effect of its upregulation by RCG may be equally diverse and depend on an individual’s health status.
OSM encodes a pleiotropic cytokine belonging to the IL-6 family of cytokines, which is characterized by a common signal transducing receptor component, glycoprotein 130. OSM protein is produced by activated T cells, monocytes, dendritic cells, and neutrophils (61–64). The induction of OSM after RCG consumption seems not to be a general inflammatory response because IL6 and LIF (also an IL-6 family gene) did not change. Note that OSM has been shown to inhibit proliferation of Human Tumor Bank 10 (HTB10) neuroblastoma cells, A549 lung carcinoma cells, and A375 and Sloan Kettering Melanoma 28 (SK-MEL-28) melanoma cells and in 4 of 5 chondrosarcoma cell lines, because this indicates that OSM may have a role in reducing cancer risk (65, 66). It may also be significant that OSM upregulated AHR and HIF1A in hepatoma G2 (HepG2) cells and JUN in human fibroblasts and M1 leukemic cells, because these results suggest that the increased expression of AHR, HIF1A, and JUN measured in our human clinical trial and in vitro study may have occurred in response to increased expression of OSM (67–69).
Although gene expression is an important determinant of protein abundance, gene expression is not perfectly correlated to amounts of protein present. The abundance of mRNA is correlated to cellular protein concentration, but only approximately half of the variation in cellular protein concentration can be attributed to mRNA levels (70–72). The other half is the result of post-transcriptional modification and protein degradation (71, 72). Therefore, although our results highlight potential pathways that may be influenced by garlic, they should not be considered definitive proof of alterations in metabolism.
Conclusions.
Seven genes related to immunity and/or cancer were upregulated in whole blood 3 h after RCG consumption, and 5 of these genes were also upregulated in the monocytic cell line Mono Mac 6 when treated with garlic extract. The upregulated genes have a variety of functions, including roles in xenobiotic metabolism, inflammation, B cell and T cell development, apoptosis, and tumorigenesis. The measurement of gene expression allows insight into early events initiated by RCG intake. It has been used infrequently in preclinical studies, and to our knowledge, this is the first clinical human trial to assess gene expression in response to the consumption of garlic.
Acknowledgments
We thank John A Milner (deceased) for his input into the design of the experiments. CSC, HDD, and JAN designed the research; CSC, HDD, GPA, PMS, GIS-A, AM, and JAN conducted the research; CSC, HDD, PMS, and BTV analyzed the data; CSC wrote the manuscript; and CSC and JAN had primary responsibility for final content. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: AHR, aryl hydrocarbon receptor; AP-1, activator protein 1; Apc, adenomatosis polyposis coli; ARNT, aryl hydrocarbon receptor nuclear translocator; c-FOS, FBJ murine osteosarcoma viral oncogene homolog; c-JUN, proto-oncogene c-Jun; c-REL, V-Rel avian reticuloendotheliosis viral oncogene homolog; Ct, threshold cycle; CYP1A1, cytochrome P450, family 1, subfamily A, polypeptide 1; CYP1A2, cytochrome P450, family 1, subfamily A, polypeptide 2; GE, garlic extract; GSTA1, glutathione S-transferase α 1; HIF-1, hypoxia-inducible factor 1; HIF1A, hypoxia-inducible factor 1; INK4a, inhibitor of cyclin-dependent kinase 4a; JUN, proto-oncogene c-Jun; LIF, leukemia inhibitory factor; LSmean, least squares mean; NFAM1, NFAT activating protein with immunoreceptor tyrosine-based activation motif 1; NFAT, nuclear factor of activated T cells; OSM, oncostatin M; PPIA, cyclophilin A; RCG, raw, crushed garlic; REL, V-rel avian reticuloendotheliosis viral oncogene homolog; RPL32, ribosomal protein L32; SIM, single-minded; TNF, tumor necrosis factor; UGT1A1, uridine diphosphate glucuronosyltransferase 1 family, polypeptide A1; UGT1A6, uridine diphosphate glucuronosyltransferase 1 family, polypeptide A6; XRE, xenobiotic response element.
References
- 1.FAOSTAT [Internet]. New York: Food and Agriculture Organization of the United Nations; c2002–2012 [cited 2014 Jan 13]. Available from: http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567#ancor.
- 2.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S, et al. Heart disease and stroke statistics—2014 update: a report from the American Heart Association. Circulation 2014;129:e28–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- 4.Mohammadi A, Bazrafshani MR, Oshaghi EA. Effect of garlic extract on some serum biochemical parameters and expression of Npc1l1, Abca1, Abcg5 and Abcg8 genes in the intestine of hypercholesterolemic mice. Indian J Biochem Biophys 2013;50:500–4. [PubMed] [Google Scholar]
- 5.Heidarian E, Jafari-Dehkordi E, Seidkhani-Nahal A. Effect of garlic on liver phosphatidate phosphohydrolase and plasma lipid levels in hyperlipidemic rats. Food Chem Toxicol 2011;49:1110–4. [DOI] [PubMed] [Google Scholar]
- 6.Elkayam A, Peleg E, Grossman E, Shabtay Z, Sharabi Y. Effects of allicin on cardiovascular risk factors in spontaneously hypertensive rats. Isr Med Assoc J 2013;15:170–3. [PubMed] [Google Scholar]
- 7.Chan KC, Yin M, Chao W. Effect of diallyl trisulfide-rich garlic oil on blood coagulation and plasma activity of anticoagulation factors in rats. Food Chem Toxicol 2007;45:502–7. [DOI] [PubMed] [Google Scholar]
- 8.Singh DK, Porter TD. Inhibition of sterol 4α-methyl oxidase is the principal mechanism by which garlic decreases cholesterol synthesis. J Nutr 2006;136:759S–64S. [DOI] [PubMed] [Google Scholar]
- 9.Lau BHS. Suppression of LDL oxidation by garlic compounds is a possible mechanism of cardiovascular health benefit. J Nutr 2006;136(Suppl):765S–8S. [DOI] [PubMed] [Google Scholar]
- 10.Ide N, Nelson AB, Lau BHS. Aged garlic extract and its constitutuents inhibit Cu2+-induced oxidative modification of low density lipoprotein. Planta Med 1997;63:263–4. [DOI] [PubMed] [Google Scholar]
- 11.Lawson LD, Wang Z-YJ, Hughes BG. Identification and HPLC quantitation of the sulfides and dialk(en)yl thiosulfinates in commercial garlic products. Planta Med 1991;57:363–70. [DOI] [PubMed] [Google Scholar]
- 12.Lawson LD. The composition and chemistry of garlic cloves and processed garlic. In: Koch HP, Lawson LD, editors. Garlic: the science and therapeutic application of Allium sativum L and related species. 2nd ed. Baltimore (MD): Williams & Waverly; 1996. p. 37–107.
- 13.Ried K, Toben C, Fakler P. Effect of garlic on serum lipids: an updated meta-analysis. Nutr Rev 2013;71:282–99. [DOI] [PubMed] [Google Scholar]
- 14.Gardner CD, Lawson LD, Block E, Chatterjee LM, Kiazand A, Balise RR, Kraemer HC. Effect of raw garlic vs commercial garlic supplements on plasma lipid concentrations in adults with moderate hypercholesterolemia: a randomized clinical trial. Arch Intern Med 2007;167:346–53. [DOI] [PubMed] [Google Scholar]
- 15.Hu X, Benson PJ, Srivastava SK, Xia H, Bleicher RJ, Zaren HA, Awasthi S, Awasthi YC, Singh SV. Induction of glutathione S-transferase π as a bioassay for the evaluation of potency of inhibitors of benzo(a)pyrene-induced cancer in a murine model. Int J Cancer 1997;73:897–902. [DOI] [PubMed] [Google Scholar]
- 16.Ban JO, Yuk DY, Woo KS, Kim TM, Lee US, Jeong HS, Kim DJ, Chung YB, Hwang BY, Oh KW, et al. Inhibition of cell growth and induction of apoptosis via inactivation of NF-kappaB by a sulfurcompound isolated from garlic in human colon cancer cells. J Pharmacol Sci 2007;104:374–83. [DOI] [PubMed] [Google Scholar]
- 17.Bose C, Guo J, Zimniak L, Srivastava SK, Singh SP, Zimniak P, Singh SV. Critical role of allyl groups and disulfide chain in induction of Pi class glutathione transferase in mouse tissues in vivo by diallyl disulfide, a naturally occurring chemopreventive agent in garlic. Carcinogenesis 2002;23:1661–5. [DOI] [PubMed] [Google Scholar]
- 18.Balasenthil S, Rao KS, Nagini S. Retinoic acid receptor-β mRNA expression during chemoprevention of hamster cheek pouch carcinogenesis by garlic. Asia Pac J Clin Nutr 2003;12:215–8. [PubMed] [Google Scholar]
- 19.Lee Y, Kim H, Lee J, Kim K. Anticancer activity of S-allylmercapto-L-cysteine on implanted tumor of human gastric cancer cell. Biol Pharm Bull 2011;34:677–81. [DOI] [PubMed] [Google Scholar]
- 20.Chu YL, Ho CT, Chung JG, Raghu R, Lo YC, Sheen LY. Allicin induces anti-human liver cancer cells through the p53 gene modulating apoptosis and autophagy. J Agric Food Chem 2013;61:9839–48. [DOI] [PubMed] [Google Scholar]
- 21.Powolny AA, Singh SV. Multitargeted prevention and therapy of cancer by diallyl trisulfide and related Allium vegetable-derived organosulfur compounds. Cancer Lett 2008;269:305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuura N, Miyamae Y, Yamane K, Nagao Y, Hamada Y, Kawaguchi N, Katsuki T, Hirata K, Sumi S-I, Ishikawa H. Aged garlic extract inhibits angiogenesis and proliferation of colorectal carcinoma cells. J Nutr 2006;136(Suppl):842S–6S. [DOI] [PubMed] [Google Scholar]
- 23.De Martino A, Filomeni G, Aquilano K, Ciriolo MR, Rotilio G. Effects of water garlic extracts on cell cycle and viability of HepG2 hepatoma cells. J Nutr Biochem 2006;17:742–9. [DOI] [PubMed] [Google Scholar]
- 24.Steinmetz KA, Kushi LH, Bostick RM, Folsom AR, Potter JD. Vegetables, fruit, and colon cancer in the Iowa Women’s Health Study. Am J Epidemiol 1994;139:1–15. [DOI] [PubMed] [Google Scholar]
- 25.González CA, Pera G, Agudo A, Bueno-De-Mesquita HB, Ceroti M, Boeing H, Schulz M, Del Giudice G, Plebani M, Carneiro F, et al. Fruit and vegetable intake and the risk of stomach and oesophagus adenocarcinoma in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST). Int J Cancer 2006;118:2559–66. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez CA, Lujan-Barroso L, Bueno-De-Mesquita HB, Jenab M, Duell EJ, Agudo A, Tjønneland A, Boutron-Ruault MC, Clavel-Chapelon F, Touillaud M, et al. Fruit and vegetable intake and the risk of gastric adenocarcinoma: a reanalysis of the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST) study after a longer follow-up. Int J Cancer 2012;131:2910–9. [DOI] [PubMed] [Google Scholar]
- 27.You WC, Brown LM, Zhang L, Li J-y, Jin M-l, Chang Y-s, Ma J-l, Pan K-f, Liu W-d, Hu Y, et al. Randomized double-blind factorial trial of three treatments to reduce the prevalence of precancerous gastric lesions. J Natl Cancer Inst 2006;98:974–83. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Li H-Q, Wang Y, Xu H-X, Fan W-T, Wang M-L, Sun P-H, Xie X-Y. An intervention study to prevent gastric cancer by micro-selenium and large dose of allitridum. Chin Med J (Engl) 2004;117:1155–60. [PubMed] [Google Scholar]
- 29.van Erk MJ, Blom WAM, van Ommen B, Hendriks HFJ. High-protein and high-carbohydrate breakfasts differentially change the transcriptome of human blood cells. Am J Clin Nutr 2006;84:1233–41. [DOI] [PubMed] [Google Scholar]
- 30.Liew C-C, Ma J, Tang H-C, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med 2006;147:126–32. [DOI] [PubMed] [Google Scholar]
- 31.de Mello VDF, Kolehmanien M, Schwab U, Pulkkinen L, Uusitupa M. Gene expression of peripheral blood mononuclear cells as a tool in dietary intervention studies: what do we know so far? Mol Nutr Food Res 2012;56:1160–72. [DOI] [PubMed] [Google Scholar]
- 32.Kennedy ET, Bowman SA, Powell R. Dietary-fat intake in the US population. J Am Coll Nutr 1999;18:207–12. [DOI] [PubMed] [Google Scholar]
- 33.Collins AR, Dobson VL, Dusinska M, Kennedy G, Stetina R. The comet assay: what can it really tell us? Mutat Res 1997;375:183–93. [DOI] [PubMed] [Google Scholar]
- 34.Lawson LD, Gardner CD. Composition, stability, and bioavailability of garlic products used in a clinical trial. J Agric Food Chem 2005;53:6254–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 36.Lawson LD, Wang Z-YJ, Hughes BG. γ-Glutamyl-S-alkylcysteines in garlic and other allium spp.: precursors of age-dependent trans-1-propenyl thiosulfinates. J Nat Prod 1991;54:436–44. [Google Scholar]
- 37.Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol 1995;35:307–40. [DOI] [PubMed] [Google Scholar]
- 38.Beischlag TV, Morales JL, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit Rev Eukaryot Gene Expr 2008;18:207–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aleksunes LM, Klaassen CD. Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARα-, and Nrf2-null mice. Drug Metab Dispos 2012;40:1366–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawajiri K, Fujii-Kuriyama Y. Cytochrome P450 gene regulation and physiological functions mediated by the aryl hydrocarbon receptor. Arch Biochem Biophys 2007;464:207–12. [DOI] [PubMed] [Google Scholar]
- 41.Rushmore TH, Pickett CB. Transcriptional regulation of the rat glutathione S-transferase Ya subunit gene: characterization of a xenobiotic-responsive element controlling inducible expression by phenolic antioxidants. J Biol Chem 1990;265:14648–53. [PubMed] [Google Scholar]
- 42.Kawajiri K, Kobayashi Y, Ohtake F, Ikuta T, Matsushima Y, Mimura J, Pettersson S, Pollenz RS, Sakaki T, Hirokawa T, et al. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in Apc Min/+ mice with natural ligands. Proc Natl Acad Sci USA 2009;106:13481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morin PJ. β-Catenin signaling and cancer. BioEssays 1999;21:1021–30. [DOI] [PubMed] [Google Scholar]
- 44.Arikawa AY, Gallaher DD. Cruciferous vegetables reduce morphological markers of colon cancer risk in dimethylhydrazine-treated rats. J Nutr 2008;138:526–32. [DOI] [PubMed] [Google Scholar]
- 45.Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, Weiner HL. Control of Treg and TH17 cell differentiation by the aryl hydrocarbon receptor. Nature 2008;453:65–71. [DOI] [PubMed] [Google Scholar]
- 46.Semenza GL. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol Med 2001;7:345–50. [DOI] [PubMed] [Google Scholar]
- 47.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Overexpression of hypoxia-inducible factor 1α in common human cancers and their metastases. Cancer Res 1999;59:5830–5. [PubMed] [Google Scholar]
- 48.Bosch-Marce M, Okuyama H, Wesley JB, Sarkar K, Kimura H, Liu YV, Zhang H, Strazza M, Rey S, Savino L, et al. Effects of aging and hypoxia-inducible factor-1 activity on angiogenic cell mobilization and recovery of perfusion after limb ischemia. Circ Res 2007;101:1310–8. [DOI] [PubMed] [Google Scholar]
- 49.Takeda K, Kinoshita I, Shimizu Y, Ohba Y, Itoh T, Matsuno Y, Shichinohe T, Dosaka-Akita H. Clinicopathological significance of expression of p-c-Jun, TCF4 and beta-Catenin in colorectal tumors. BMC Cancer 2008;8:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tiniakos DG, Mitropoulos D, Kyroudi-Voulgari A, Soura K, Kittas C. Expression of c-jun oncogene in hyperplastic and carcinomatous human prostate. Urology 2006;67:204–8. [DOI] [PubMed] [Google Scholar]
- 51.Kollmann K, Heller G, Sexl V. c-JUN prevents methylation of p16 INK4a (and Cdk6): the villain turned bodyguard. Oncotarget 2011;2:422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang CW, Lee YZ, Hsu HY, Wu CM, Chang HY, Chao YS, Lee SJ. C-Jun-mediated anticancer mechanisms of tylophorine. Carcinogenesis 2013;34:1304–14. [DOI] [PubMed] [Google Scholar]
- 53.Ohtsuka M, Arase H, Takeuchi A, Yamasaki S, Shiina R, Suenaga T, Sakurai D, Yokosuka T, Arase N, Iwashima M, et al. NFAM1, an immunoreceptor tyrosine-based activation motif-bearing molecule that regulates B cell development and signaling. Proc Natl Acad Sci USA 2004;101:8126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J, Hu G, Wang SW, Li Y, Martin R, Li K, Yao Z. Calcineurin/nuclear factors of activated T cells (NFAT)-activating and immunoreceptor tyrosine-based activation motif (ITAM)-containing protein (CNAIP), a novel ITAM-containing protein that activates the calcineurin/NFAT-signaling pathway. J Biol Chem 2003;278:16797–801. [DOI] [PubMed] [Google Scholar]
- 55.Lamm DL, Riggs DR. Enhanced immunocompetence by garlic: role in bladder cancer and other malignancies. J Nutr 2001;131(Suppl):1067S–70S. [DOI] [PubMed] [Google Scholar]
- 56.Fullard N, Wilson CL, Oakley F. Roles of c-Rel signalling in inflammation and disease. Int J Biochem Cell Biol 2012;44:851–60. [DOI] [PubMed] [Google Scholar]
- 57.Wu M, Lee H, Bellas RE, Schauer SL, Arsura M, Katz D, Fitzgerald MJ, Rothstein TL, Sherr DH, Sonenshein GE. Inhibition of NF-κB/Rel induces apoptosis of murine B cells. EMBO J 1996;15:4682–90. [PMC free article] [PubMed] [Google Scholar]
- 58.Sovak MA, Bellas RE, Kim DW, Zanieski GJ, Rogers AE, Traish AM, Sonenshein GE. Aberrant nuclear factor-κB/Rel expression and the pathogenesis of breast cancer. J Clin Invest 1997;100:2952–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS Jr. Selective activation of NF-κB subunits in human breast cancer: potential roles for NF-κB2/p52 and for Bcl-3. Oncogene 2000;19:1123–31. [DOI] [PubMed] [Google Scholar]
- 60.Burkitt MD, Williams JM, Duckworth CA, O’Hara A, Hanedi A, Varro A, Caamaño JH, Pritchard DM. Signaling mediated by the NF-κB sub-units NF-κB1, NF-κB2 and c-Rel differentially regulate Helicobacter felis-induced gastric carcinogenesis in C57BL/6 mice. Oncogene 2013;32:5563–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown TJ, Lioubin MN, Marquardt H. Purification and characterization of cytostatic lymphokines produced by activated human T lymphocytes: synergistic antiproliferative activity of transforming growth factor β1, interferon-γ, and oncostatin M for human melanoma cells. J Immunol 1987;139:2977–83. [PubMed] [Google Scholar]
- 62.Malik N, Kallestad JC, Gunderson NL, Austin SD, Neubauer MG, Ochs V, Marquardt H, Zarling JM, Shoyab M, Wei CM, et al. Molecular cloning, sequence analysis, and functional expression of a novel growth regulator, oncostatin M. Mol Cell Biol 1989;9:2847–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hurst SM, McLoughlin RM, Monslow J, Owens S, Morgan L, Fuller GM, Topley N, Jones SA. Secretion of oncostatin M by infiltrating neutrophils: regulation of IL-6 and chemokine expression in human mesothelial cells. J Immunol 2002;169:5244–51. [DOI] [PubMed] [Google Scholar]
- 64.Suda T, Chida K, Todate A, Ide K, Asada K, Nakamura Y, Suzuki K, Kuwata H, Nakamura H. Oncostatin M production by human dendritic cells in response to bacterial products. Cytokine 2002;17:335–40. [DOI] [PubMed] [Google Scholar]
- 65.Zarling JM, Shoyab M, Marquardt H, Oncostatin M. A growth regulator produced by differentiated histiocytic lymphoma cells. Proc Natl Acad Sci USA 1986;83:9739–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.David E, Guihard P, Brounais B, Riet A, Charrier C, Battaglia S, Gouin F, Ponsolle S, Bot RL, Richards CD, et al. Direct anti-cancer effect of oncostatin M on chondrosarcoma. Int J Cancer 2011;128:1822–35. [DOI] [PubMed] [Google Scholar]
- 67.Vollmer S, Kappler V, Kaczor J, Flügel D, Rolvering C, Kato N, Kietzmann T, Behrmann I, Haan C. Hypoxia-inducible factor 1α is up-regulated by oncostatin M and participates in oncostatin M signaling. Hepatology 2009;50:253–60. [DOI] [PubMed] [Google Scholar]
- 68.Stobbe-Maicherski N, Wolff S, Wolff C, Abel J, Sydlik U, Frauenstein K, Haarmann-Stemmann T. The interleukin-6-type cytokine oncostatin M induces aryl hydrocarbon receptor expression in a STAT3-dependent manner in human HepG2 hepatoma cells. FEBS J 2013;280:6681–90. [DOI] [PubMed] [Google Scholar]
- 69.Liu J, Clegg C, Shoyab M. Regulation of EGR-1, c-jun, and c-myc gene expression by oncostatin M. Cell Growth Differ 1992;3:307–13. [PubMed] [Google Scholar]
- 70.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst 2009;5:1512–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012;13:227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett 2009;583:3966–73. [DOI] [PubMed] [Google Scholar]