SUMMARY
FACT, a heterodimer of Spt16 and Pob3, is an essential histone chaperone. We show that the H2A-H2B binding activity that is central to FACT function resides in short acidic regions near the C-termini of each subunit. Mutations throughout these regions impact binding and cause correlated phenotypes that range from mild to lethal, with the largest individual contributions unexpectedly coming from an aromatic residue and a nearby carboxylate residue within each domain. Spt16 and Pob3 bind overlapping sites on H2A-H2B, and Spt16-Pob3 heterodimers simultaneously bind two H2A-H2B dimers, the same stoichiometry as the components of a nucleosome. An Spt16:H2A-H2B crystal structure explains the biochemical and genetic data, provides a model for Pob3 binding, and implies a mechanism for FACT reorganization that we confirm biochemically. Moreover, unexpected similarity to binding of ANP32E and Swr1 with H2A.Z-H2B reveals that diverse H2A-H2B chaperones use common mechanisms of histone binding and regulating nucleosome functions.
Keywords: histone chaperone, nucleosome reorganization, histone binding, Spt16-Pob3, crystal structure
INTRODUCTION
Histone chaperones bind the highly charged and highly hydrophobic surfaces of free histone proteins to block non-productive interactions and promote the appropriate interactions required for nucleosome assembly (Gurard-Levin et al., 2014; Park and Luger, 2008; Ransom et al., 2010). Chaperones can also assist the disassembly of nucleosomes to facilitate access to DNA and/or coordinate assembly to maintain a stable chromatin barrier. FACT (FAcilitates Chromatin Transactions) is a highly conserved, essential histone chaperone that enhances both DNA accessibility and chromatin stability (Belotserkovskaya et al., 2004; Formosa, 2012; Hondele and Ladurner, 2013; Winkler and Luger, 2011). FACT has been proposed to act by binding a reorganized nucleosome in which the histones are dissociated from one another and from the DNA, but in which the components remain tethered together. FACT therefore enhances DNA availability without dispersing the histones, thereby promoting efficient reassembly of the original nucleosome (Formosa, 2012; Jamai et al., 2009).
FACT comprises multiple flexibly-associated structural domains, including one aminopeptidase-like motif, and several pleckstrin-homology (PH) motifs, mostly organized into distinctive double PH modules (Hondele et al., 2013; Kemble et al., 2013; Stuwe et al., 2008; VanDemark et al., 2006; VanDemark et al., 2008). Both subunits of FACT, Spt16 and Pob3, also contain 80-100 residue regions at their C-termini that are highly acidic, a feature that is found in many histone chaperones (Hondele and Ladurner, 2011; Park and Luger, 2008). The SSRP1 subunit, which is found in place of Pob3 in most eukaryotes other than yeast, has the same organization, but with the acidic region followed by an additional domain related to the HMGB family of DNA binding proteins (Belotserkovskaya et al., 2003). Yeast FACT also makes use of a member of this family, Nhp6, but as an independent protein (Stillman, 2010). The modular FACT structure is therefore highly conserved and is consistent with the reorganization/tethering model. Further understanding of FACT activity requires knowing how FACT domains contact histones during nucleosome reorganization.
The report of a structure of Chaetomium thermophilum Spt16-M expressed as a translational fusion with Xenopus laevis H2B in complex with H2A (Hondele et al., 2013) had the potential to provide new insight. However, we were unable to detect this interaction using independently expressed cognate proteins (Kemble et al., 2013), raising questions about the relevance of the Spt16-M:H2A-H2B interaction and prompting a search for authentic H2A-H2B binding sites in FACT. Our analysis now reveals primary H2A-H2B binding sites within the acidic Spt16-C and Pob3-C domains. These results confirm an expected role for dispersed non-specific charge neutralization (Belotserkovskaya et al., 2003; Belotserkovskaya et al., 2004), but also reveal that the biochemical binding and physiological function of each of these domains is even more strongly dependent on single aromatic residues, with specific nearby acidic residues also making major contributions. The Spt16-C and Pob3-C peptides bind competitively to the same site on H2A-H2B, such that FACT can bind two H2A-H2B dimers simultaneously, consistent with the ability to tether all of the components of a reorganized nucleosome (Formosa, 2012). Remarkably, our crystal structure of an Spt16-C:H2A-H2B complex shares close similarity with recently reported complexes of two unrelated H2A.Z-H2B binding proteins, ANP32E and Swr1, thereby disclosing a common binding mode that may be conserved broadly across highly diverse H2A-H2B chaperones.
RESULTS
Spt16-C and Pob3-C contain the H2A-H2B Binding Determinants of FACT
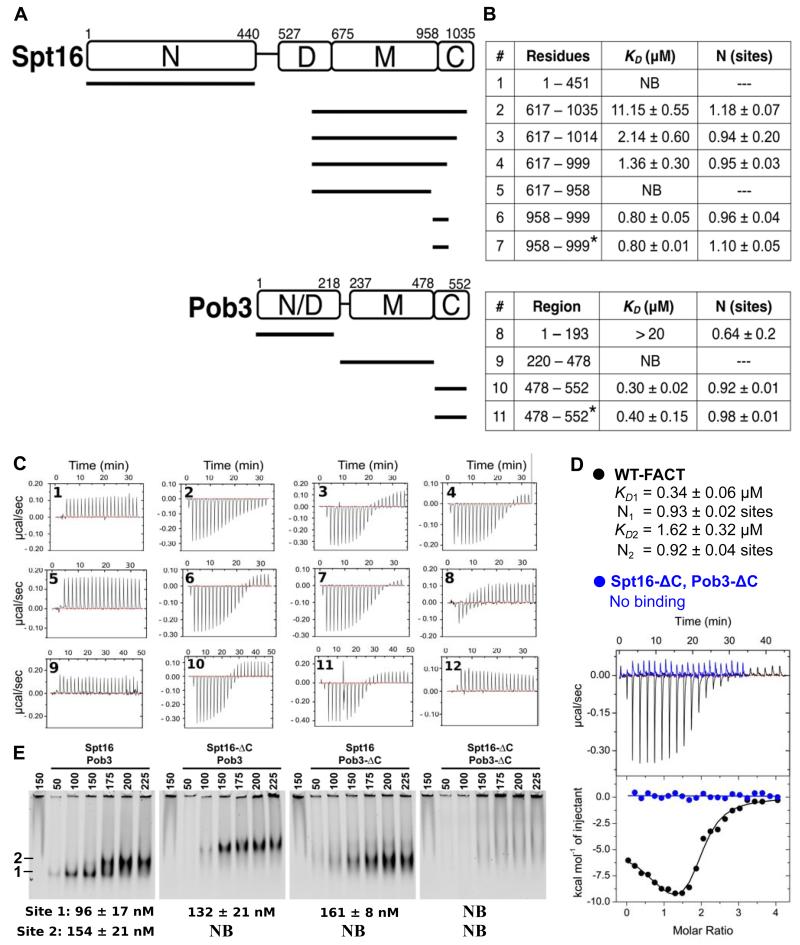
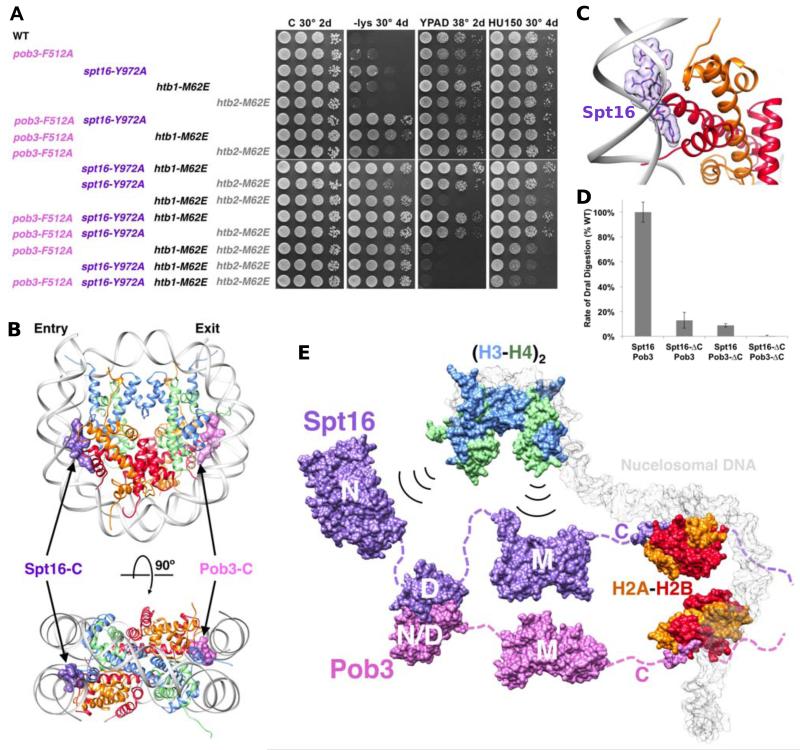
Working with cognate Saccharomyces cerevisiae proteins, we used Isothermal Titration Calorimetry (ITC) to map the regions of FACT that bind H2A-H2B (Figure 1). No binding was observed for Spt16-N or Spt16-M, whose titrations were superimposable with the heat of dilution for histones alone. In contrast, Spt16-C bound H2A-H2B with 1:1 stoichiometry and a 0.8 μM dissociation constant. Increased affinity of Spt16617-1014 relative to Spt16617-1035 indicates ~5-fold inhibition of binding by the C-terminal 21 residues. These residues include the W1017-G1030 sequence, which is predicted to adopt a helical conformation and is conserved from yeast to human (Figure S1), suggesting a physiological function. To test the importance of basic residues in H2A-H2B for this interaction we expressed H2A-H2B30-130, which lacks 8 basic residues, and found that binding was unaffected (Figure 1B). We conclude that Spt16-C958-999 recapitulates the full H2A-H2B binding activity of Spt16, and that this interaction is not dependent on the basic N-terminal tail of H2B.
Figure 1. Spt16-C and Pob3-C contain the H2A-H2B Binding Determinants of FACT.
(A) Domain organization of S. cerevisiae Spt16 and Pob3 showing constructs used for H2A-H2B binding assays. (B) Binding affinities (mean KD and standard deviations) and stoichiometry (N, sites) determined by ITC. Affinities are for full-length H2A-H2B or a variant (*) that lacks the first 29 residues of H2B (NB is no binding). (C) Isotherms for the constructs numbered in panel B. Isotherm 12 represents the heat of dilution for full-length H2A-H2B. (D) Overlay of raw (top panel) and integrated (bottom panel) ITC data for full-length H2A-H2B binding to full-length FACT (black) and Spt16-ΔC, Pob3-ΔC (blue). FACT binding was fit to a two-site binding model with stoichiometry and affinities shown for both sites. The data for full-length FACT also appears in Figures 3C and 4E. (E) EMSA of FACT (200 nM) titrated with H2A-H2B shows that binding requires the Spt16-C and Pob3-C domains and displays shifts that are consistent with a stoichiometry of 2:1 (H2A-H2B: FACT). See also Figure S1 for residue conservation, Table S1 for strains, and Table S2 for plasmids.
We followed an equivalent approach with Pob3, and discovered a second binding site for H2A-H2B in Pob3-C (Figure 1). (The weak interaction with the relatively insoluble Pob3-N domain is likely to be non-specific as suggested by the sub-stoichiometric binding). Pob3-C bound H2A-H2B with 1:1 stoichiometry and an affinity of 0.3 μM that, like Spt16, was not affected by deletion of the H2B N-terminal tail.
To determine if intact FACT heterodimers recapitulate these peptide-binding studies, we used ITC to quantify binding of full-length FACT to full-length H2A-H2B. Consistent with the finding that Spt16-C and Pob3-C each contained H2A-H2B binding sites, one full-length Spt16-Pob3 heterodimer bound two H2A-H2B dimers (Figure 1D, black). Moreover, the KD values obtained from fitting the data to a model with two independent binding sites (0.34 μM and 1.62 μM) were similar to those obtained with the isolated Spt16-C and Pob3-C peptides, while reasonable fits could not be obtained for alternative models such as those with one or three binding sites. Confirming that the two binding sites reside exclusively in the Spt16-C and Pob3-C domains of FACT, a mutant lacking these domains in otherwise full-length FACT (Spt16-ΔC, Pob3-ΔC) did not bind H2A-H2B (Figure 1D, blue). The complexity of our ITC data for full-length FACT prompted us to verify the stoichiometry by an independent method. An Electrophoretic Mobility Shift Assay (EMSA) was therefore performed using full-length FACT (Spt16-Pob3), FACT with the C-terminus of Spt16 deleted (Spt16-ΔC, Pob3), FACT with the C-terminus of Pob3 deleted (Spt16, Pob3-ΔC), and FACT with both C-termini deleted (Spt16-ΔC, Pob3-ΔC; Figure 1E). The EMSA results showed that full-length FACT complex binds to H2A-H2B with two distinct mobility shifts, thereby confirming the conclusion from the ITC data that FACT contains two distinct binding sites for H2A-H2B. The apparent affinities (KD) for these two EMSA binding events are ~100 nM and ~150 nM, which are slightly tighter than the values obtained by ITC, possibly due to restricted dissociation within the gel environment of this assay (see below). In comparison, similar but tighter (30 nM) binding has been reported for the non-cognate binding of human FACT with Xenopus H2A-H2B (Winkler et al., 2011). As expected, only one shift at ~150 nM was observed with each of the single deletions (Spt16-ΔC, Pob3 and Spt16, Pob3-ΔC) and binding was eliminated for the double deletion (Spt16-ΔC, Pob3-ΔC). Each FACT heterodimer therefore binds two H2A-H2B dimers and the two binding determinants are located in the Spt16-C and Pob3-C domains.
The shape of the isotherm obtained for full-length FACT binding to full-length H2A-H2B (Figure 1D) suggests a more complex interaction than expected for two completely independent binding sites. Because the C-terminal 21 residues of Spt16 appear to be inhibitory for H2A-H2B binding, one attractive possibility is that communication occurs between the two binding sites, which might allow Spt16-C and Pob3-C to coordinate with one another while depositing H2A-H2B during nucleosome assembly or while tethering components during nucleosome reorganization.
Binding is Mediated by Short Peptides of Spt16-C and Pob3-C
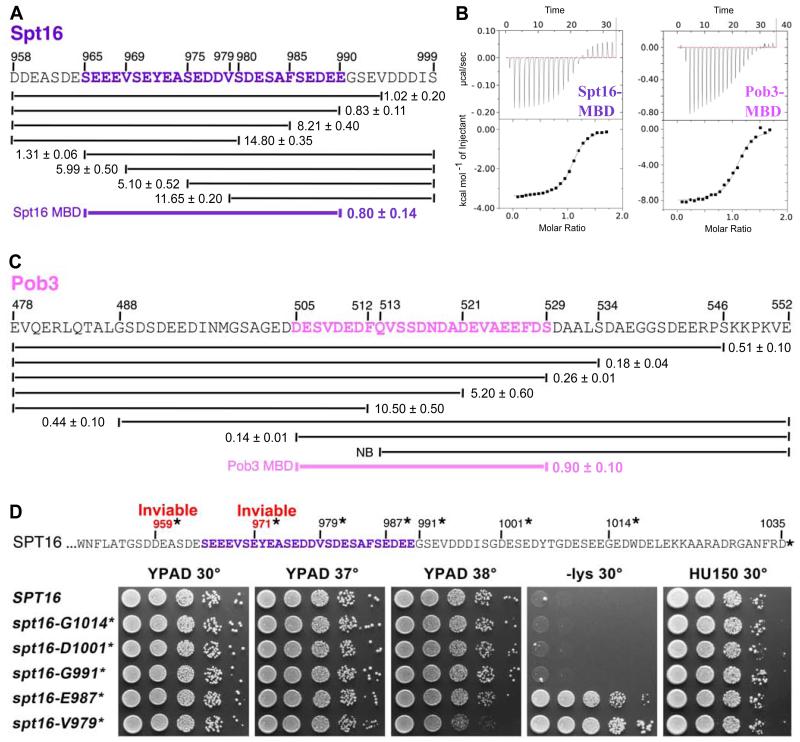
To further map the binding site in Spt16-C, we purified a series of peptides containing subsets of residues 958-999 and quantified binding to H2A-H2B30-130 using ITC (Figure 2A). Deleting residues from each end indicated that the minimal H2A-H2B binding domain (MBD) is between S965 and E990, and a peptide representing just this sequence retained essentially full affinity (Figure 2B, Spt16-MBD). An equivalent approach with Pob3 indicated that D505-S529 contains the minimal binding domain for this subunit (Figure 2C), and a peptide with just this sequence also retained nearly full binding affinity (Figure 2B, Pob3-MBD). These two short peptides therefore represent the primary binding sites for H2A-H2B in FACT.
Figure 2. Spt16 and Pob3 minimal binding domains.
(A) ITC-derived binding affinities (as in Figure 1) for H2A-H2B30-130 with Spt16-C peptides. The Minimal Binding Domain (MBD) is indicated. (B) Isotherms and integrated heat responses for Spt16-MBD and Pob3-MBD peptides with H2A-H2B30-130. (C) Binding affinities for full-length H2A-H2B and the indicated Pob3 peptides. (D) Sequence of the Spt16 acidic domain indicating the sites of truncations assayed in vivo. YPAD is rich medium, -lys is synthetic medium lacking lysine (growth indicates the Spt− phenotype resulting from incomplete repression of the lys2-128∂ reporter allele in these strains; (Rando and Winston, 2012), and HU150 is YPAD with 150 mM hydroxyurea.
To evaluate the relevance of these results in vivo, we introduced a series of C-terminal truncations (stop codons, indicated as “*” in Figure 2D) into the native SPT16 gene at the normal genomic locus (Table S1). spt16-D959* and spt16-E971* did not support viability (not shown), but spt16-V979* did. This demonstrates that yeast tolerate deletion of almost half of the Spt16-C MBD, but loss of more of this sequence is lethal. Previous reports concluded that Spt16-C is essential, but used a construct that also lacked a significant portion of Spt16-M (Belotserkovskaya et al., 2003) or attributed inviability to loss of a nuclear localization signal at the extreme C-terminus (Hondele et al., 2013). Our data show that the essential function of Spt16-C maps to the region between E971 and V979, and that the extreme C-terminus is not essential. Notably, the viable truncation mutants that lacked portions of Spt16-C MBD displayed the Spt− phenotype (spt16-E987* and spt16-V979*), revealing a defect in maintaining chromatin-based repression (Rando and Winston, 2012) (Figure 2D). The essential region of Spt16-C therefore coincides with the binding site for H2A-H2B, and partial disruption of this region leads to loss of efficient transcriptional repression.
We were unable to perform an equivalent in vivo deletion analysis of Pob3-C because pob3-K548* causes a severe growth defect (Schlesinger and Formosa, 2000), and even some single residue substitutions near the C-terminus were lethal (not shown). Because these residues do not contribute to binding H2A-H2B in our in vitro assays, we conclude that Pob3 residues 448-552 have an additional essential activity that is distinct from the interaction with H2A-H2B.
Single aromatic residues are important for binding of Spt16-C and Pob3-C
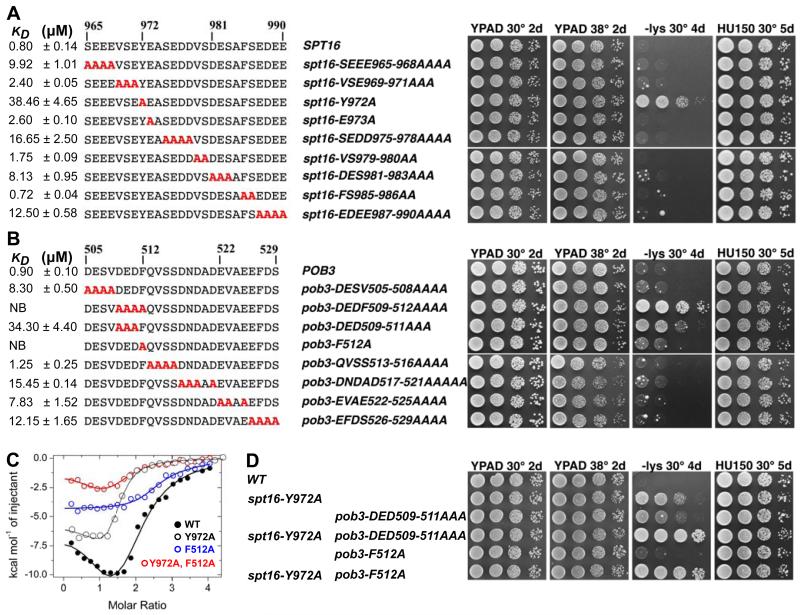
To determine the importance of specific residues in binding H2A-H2B, we introduced alanine substitutions into peptides corresponding to Spt16 residues 965-990 (Figure 3A, left panel). Substantial decreases in affinity were observed with quadruple mutations SEEE965-968AAAA (12-fold) and EDEE987-990AAAA (15-fold), which affect both flanks of the minimal binding domain. Two other substitutions that removed multiple negative charges also affected binding substantially: SEDD975-978AAAA (20-fold) and DES981-983AAA (10-fold). This indicates that negatively charged residues throughout the targeted region contribute to H2A-H2B binding, with effects that are loosely proportional to the number of charges, supporting the expectation that charge neutralization is an important component of histone binding by Spt16-C. However, the greatest effect we observed was for the single residue substitution Y972A, which diminished binding 48-fold without altering the charge of the peptide. Thus, while distributed electrostatic effects make an important contribution, the most dramatic reduction in binding resulted from loss of a single specific hydrophobic contact.
Figure 3. Specific Tyr/Phe residues make the largest contributions to binding for Spt16 and Pob3.
(A) Alanine mutations (red) in the Spt16-MBD peptide with affinity estimated from ITC for H2A-H2B30-130 (left) and phenotype when introduced into the SPT16 locus (right). Mean KD is given in μM with the standard deviation indicated. (B) Equivalent analysis for Pob3. (C) Integrated ITC data for H2A-H2B binding to full-length wild type and mutant FACT. The data for full-length FACT also appears in Figures 1D and 4E. (D) Selected Spt16-C and Pob3-C-MBD mutants display synthetic phenotypes. See also Figure S2.
Consistent with the biochemical data obtained using short peptides, substituting negatively charged residues with alanines in full-length Spt16 expressed from its normal locus had no noticeable effects in vivo, but the single Y972A point mutation caused a moderate Spt− phenotype (Figure 3A, right panel). More sensitive tests conducted in a background in which Pob3 function was also impaired confirmed the importance of some blocks of acidic residues in FACT function, especially those nearest to Spt16 Y972 (Figure S2) as expected from the biochemical analysis. Once again, however, the strongest defects, including severely impaired growth, mapped to the same single point mutation that had the most severe effect on H2A-H2B binding, spt16-Y972A. We conclude that acidic residues within the minimal binding domain contribute to a distributed H2A-H2B interaction locus, but unexpectedly a single hydrophobic residue also has a crucial role.
An equivalent approach was used to analyze Pob3-MBD (Figure 3B). Most mutations removing negative charges again caused moderately decreased H2A-H2B binding, with loss of three negative charges causing the largest effects (17-fold with DNDAD517-521AAAAA and 38-fold with DED509-511AAA), whereas the substitution of multiple neutral residues in QVSS513-516AAAA had no effect. Once more, however, the largest defect was caused by the single substitution of an aromatic residue, F512A, which reduced binding at least 100-fold to an undetectable level by this assay. Consistent with these observations, integration of these substitutions into the native POB3 gene in vivo caused significant phenotypes only for pob3-DED509-511AAA and pob3-F512A, which displayed moderate or mild Spt− phenotypes, respectively (Figure 3B, right panel). Collaboration between Spt16-Y972 and Pob3-F512 was revealed in vitro (Figure 3C) and in vivo (Figure 3D) by the finding that double mutant combinations displayed enhanced loss of H2A-H2B binding relative to single mutants and also enhanced loss of transcriptional repression. Pob3-C and Spt16-C are therefore similar in that each uses a combination of acidic residues and a singularly important aromatic residue to bind H2A-H2B, these hydrophobic contributions are important for maintaining transcriptional repression by FACT in vivo, and impairing both binding sites is more detrimental than disturbing either one alone. H2A-H2B binding sites centered on Spt16 Y972 and Pob3 F512 therefore collaborate in mediating a physiologically important function of FACT.
Point mutations in the C-terminal domains of full-length FACT had large effects on binding when measured by ITC (Figure 3C), but more subtle effects in EMSA experiments (not shown). Similar relative differences between ITC and EMSA data were seen in Figure 1, and are consistent with ITC measuring changes in enthalpy under equilibrium conditions in solution and being inherently more sensitive than EMSA, which depends upon retention of complex formation in a crowded polyacrylamide gel environment. Together, these data indicate that the H2A-H2B binding site is distributed over the minimal binding domains in Spt16-C and Pob3-C, and that Spt16 Y972 and Pob3 F512 make the single largest contributions to binding. Phenotypic analysis supports this model (Figure 3D), with some loss of repression observed with single mutants, more severe effects in the double mutant, and loss of viability resulting from complete removal of the entire binding site (Figures 1 and 2)
Structural Basis for the Spt16-C:H2A-H2B interaction
To further understand the mechanism of FACT binding, we determined the structure of Spt16-C in complex with H2A-H2B. We obtained crystals of Spt16958-990 in complex with H2A-H2B30-130, determined the structure by molecular replacement using the coordinates of yeast H2A-H2B from the yeast nucleosome (PDB code 1ID3) (White et al., 2001), and refined the model against 1.80 Å data to an R free value of 22.5% (Table 1). The crystal contains four H2A-H2B dimers per asymmetric unit and a single copy of the Spt16 peptide, for which 6 residues are visible in electron density. The ordered model comprises residues H2A Q16-V101, H2B M29-S126 and Spt16 E967-Y972. The ordered Spt16 residues are clearly defined in an unbiased omit map (Figure 4A). The equivalent Spt16-binding sites in the other three H2A-H2B dimers in the asymmetric unit are blocked by lattice contacts.
Table 1. Crystallographic data collection and refinement.
| Data Collection | |
|---|---|
| Beamline / Detector | SSRL, 7-1 / Q315 |
| Wavelength (Å) | 1.12709 |
| No. Reflections: observed / unique | 287,848 / 77,661 |
| Space group | R3(H3) |
| Cell dimensions: a, b, c (Å) | 109.67, 109.67, 187.47 |
| Resolution (Å) | 40.0 – 1.80 (1.86 – 1.80) |
| I/σI | 9 (1.8) |
| Mosaicity (°) | 0.3 |
| Completeness (%) | 98.7 (95.6) |
| Rsym (%) | 0.086 (0.928) |
| Refinement | |
| Resolution (high) | 40.0 – 1.8 (1.82 – 1.80) |
| Rwork / Rfree (%) | 0.181 / 0.225 (0.283 / 0.341) |
| Number of protein atoms | 5,768 |
| Number of water molecules | 388 |
| Number of phosphates | 2 |
| RMSD bond length (Å) / angles(°) | 0.007 / 0.952 |
| ϕ/ψ most favored / allowed (%) | 98.3 / 1.3 |
| <B> (Å2): protein / solvent | 50 / 53 |
Values in parentheses refer to the highest resolution shell.
Rsym = (|(ΣI − <I>)|)/(ΣI), where <I> is the average intensity.
Rwork = Σ||Fo| − |Fc||/Σ|Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively, summed over structure factors used in refinement calculations.
Rfree = R factor calculated using a random set of reflections (5% of total) that were not used in refinement calculations.
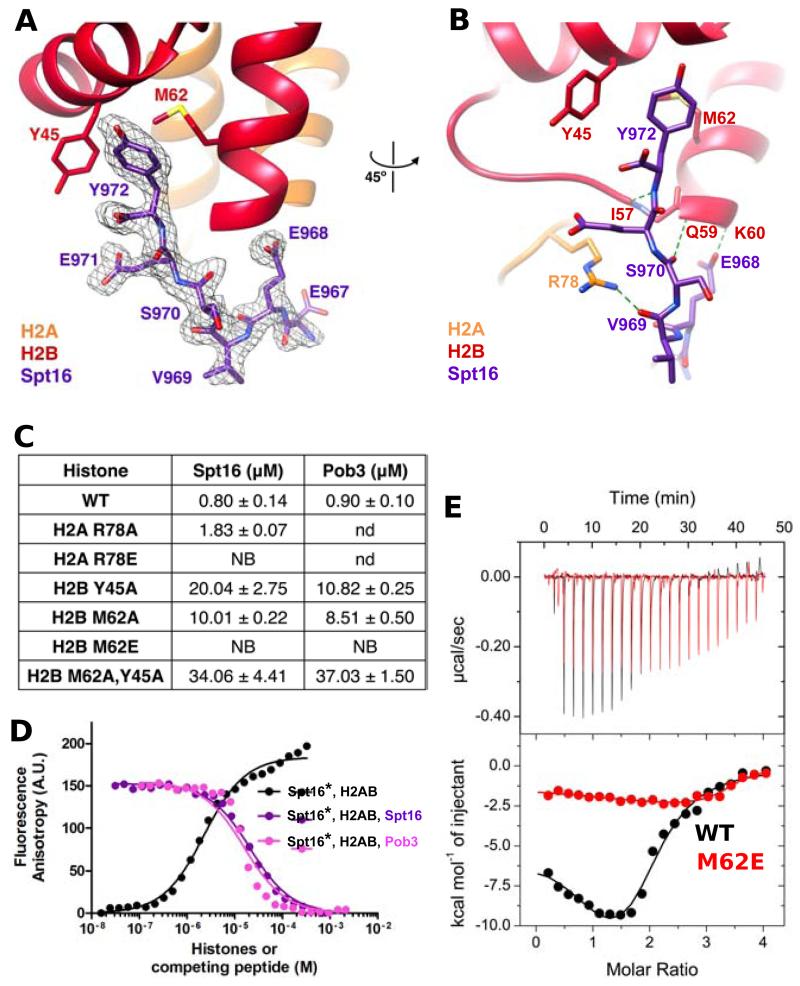
Figure 4. Spt16MBD:H2A-H2B structure and biochemical validation of binding to Spt16 and Pob3.
(A) Fo-Fc omit map (3× RMSD) around Spt16 E967-Y972. The map was phased on an H2A-H2B model that was refined in the absence of the Spt16 peptide. (B) Hydrogen bonding interactions (green dashed lines). (C) ITC-derived binding affinities of Spt16 and Pob3-MBDs to H2A-H2B30-130. NB is no binding, nd is not determined. (D) Fluorescence anisotropy binding assays. Fluorescein-Spt16 MBD and unlabeled H2A-H2B (black line), competition of Fluorescein-Spt16 bound to H2A-H2B by unlabeled Spt16-MBD (purple) and unlabeled Pob3-MBD (pink). (E) Raw (top) and integrated (bottom) ITC binding data for full-length FACT and H2A-H2B (black) or full-length FACT and H2A-H2B M62E (red). The data for full-length FACT also appears in Figures 1D and 3C. See also Figure S1 and Table S2.
Binding of the ordered Spt16 residues against H2A-H2B buries a total of 821 Å2 of surface area. Importantly, the last visible Spt16 residue, Y972, which is the side chain whose substitution had the most pronounced consequence for binding affinity and phenotype (above), nestles into a hydrophobic pocket formed by the conserved H2B residues Y45 and M62 (Figures 4A, 4B and S1). The crystallographic interface was further validated by measuring Spt16958-990 binding to H2A-H2B30-130 bearing H2B Y45A, M62A, or both mutations. The single residue substitutions diminished binding 25-fold (Y45A) and 12-fold (M62A) and the effects were additive (42-fold) in the double mutant, indicating that each of these residues contributes to the interface in solution (Figure 4C). A more severe single mutant, H2B M62E, reduced binding to undetectable levels by this assay, as expected for the introduction of a negative charge into a hydrophobic interaction pocket. Our structure and the associated binding data therefore demonstrate that this is a specific interaction and explain the importance of Spt16 Y972 for the binding of H2A-H2B.
The Spt16 interface also includes capping of the N-terminus of H2B helix 2 by a main chain-to-main chain hydrogen bond and the side chain of E968 (Figure 4B). The E968 carboxylate interaction has the potential to be especially favored by the partial positive charge associated with the N-terminus of a helix (Shoemaker et al., 1987). The importance of this interaction was further validated in vitro by ITC experiments that demonstrated that the Spt16 E968A mutation reduced affinity by 6-fold to a KD of 5 μM (not shown).
H2A R78 is the only residue of H2A that contacts Spt16. This interaction, which buries just 107 Å2 of surface area, primarily comprises a hydrogen bond between the R78 guanidinium and the main chain carbonyl oxygen of Spt16 V969 (Figure 4B). The H2A R78A mutation had little effect on binding, but affinity was completely lost with the more severe H2A R78E charge-reversal mutation (Figure 4C). These results further support the validity of the crystallographic interface; H2A R78 makes little contribution to either the interaction surface or to binding affinity, but insertion of an inappropriate residue at this position disrupts the interaction.
Spt16 and Pob3 bind competitively to H2A-H2B
Given the similar use of a single dominant aromatic residue in the binding of both Spt16 and Pob3, we sought to determine if they bind to the same site on H2B. This was accomplished by asking if the H2B Y45A and M62A substitutions that impair binding of Spt16958-990 also diminish binding of Pob3505-529. Quantification by ITC demonstrated that these histone mutations reduced Pob3 binding affinity by 12-fold and 9-fold, respectively, and the double mutant displayed an additive effect with a 41-fold reduction in KD (Figure 4C). These observations indicated that Spt16-C and Pob3-C have overlapping binding sites on H2A-H2B and that their binding may be competitive.
We directly tested the possibility of competitive binding by Spt16 and Pob3 using a fluorescence anisotropy binding assay. A fluorescently labeled Spt16965-990 peptide bound unlabeled H2A-H2B with an estimated KD of 2.3 μM (Figure 4D), in good agreement with ITC. The assay was further validated by titration of unlabeled Spt16 peptide into reactions containing labeled Spt16 peptide at 80% saturation of 10 μM H2A-H2B (Figure 4D). The unlabeled Spt16 peptide effectively competed the labeled peptide, with an IC50 of 21.1 +/− 0.1 μM and a Ki of 2.3 +/− 0.1 μM. An equivalent titration experiment with unlabeled Pob3505-529 also showed displacement of labeled Spt16 peptide, with an IC50 of 15.7 +/− 0.1 μM and a Ki of 1.3 +/− 0.1 μM. These results demonstrate that Pob3 can displace Spt16 from H2A-H2B, and support the model that Spt16 and Pob3 have overlapping binding sites on H2A-H2B.
To confirm that an intact FACT heterodimer binds to the same histone interface identified in our peptide studies, we used ITC to demonstrate a severe reduction in the binding of full-length FACT to the histone mutant H2A-H2B M62E (Figure 4E). The H2A-H2B M62E mutation also diminished binding to full-length FACT detected by EMSA, but the effect was less acute (not shown). Thus, once again, ITC suggested a more severe binding defect than EMSA, but both revealed impaired binding, further supporting the conclusions that Spt16 Y972 and Pob3 F512 are the single most important residues for H2A-H2B binding, that the crystal structure accurately depicts the binding geometry, and that other residues in the minimal binding domains also contribute to binding in vitro and in vivo.
The H2B binding site is important for FACT function in vivo
We verified the functional relevance of the histone residues contacted by Spt16 and Pob3 by mutating these residues in vivo. The H2B mutations Y45A, M62A, and M62E were integrated into both of the two endogenous genomic loci encoding H2B (HTB1 and HTB2) as described previously (McCullough et al., 2011). Single mutations caused weak or mild Spt− phenotypes, and combination with FACT mutations revealed overlapping functions through strong loss of repression (Figure 5A). Combination of either single FACT mutation with mutation of one histone gene, htb1-M62E, caused a strong loss of repression (for example, compare rows 3 and 4 with row 9 on the -lys plate). Mutating both copies of the H2B gene also caused loss of repression as well as additional phenotypes including temperature sensitivity and sensitivity to the replication toxin hydroxyurea. Similar, but weaker effects were observed with H2B M62A and H2B Y45A (not shown). Together, these observations recapitulate the in vitro binding data, and show that the H2B Y45 and M62 residues that mediate binding to Spt16-C and Pob3-C are important for functional interactions with FACT in vivo.
Figure 5. Spt16-C and Pob3-C are required for nucleosome reorganization.
(A) Phenotypes of selected Pob3 (pink) or Spt16 (purple) mutations alone or combined with htb1-M62E (black) or htb2-M62E (gray), all integrated at their native loci. (B) The Spt16-C complex superimposed onto H2A-H2B dimers of the canonical nucleosome (PDB 1ID3; (White et al., 2001). One of these sites is labeled Pob3 in accord with the model that Spt16 and Pob3 bind equivalently to the same reorganized nucleosome. (C) Closeup demonstrating the conflict between Spt16 and DNA binding to H2A-H2B. (D) The rate of digestion by DraI was determined for nucleosomes reconstituted with recombinant S. cerevisiae histones in the presence of Nhp6 and the Spt16-Pob3 heterodimers indicated (Xin et al., 2009); see Figure 1). The rates for Nhp6 alone and with full-length Spt16-Pob3 were set to 0% and 100%, respectively. Each construct was tested at least in triplicate, with the standard deviation shown. (E) Model of FACT binding the components of a nucleosome including two H2A-H2B dimers bound by Spt16-C and Pob3-C and H3-H4 histones bound by the Spt16-N and Spt16-M domains (Kemble et al., 2013; Stuwe et al., 2008) and DJK, TF, CPH unpublished). See also Figure S3.
FACT interactions with H2A-H2B facilitate nucleosome reorganization
Inspection of the nucleosome structure (White et al., 2001) shows that binding of Spt16 to H2B is incompatible with the DNA:H2A-H2B interaction that comprises the second contact point between DNA and the histone octamer core (Figure 5B, 5C). Because of the nucleosome’s dyad symmetry, this second contact is equivalent to the penultimate of the fourteen DNA:H2A-H2B contacts (Luger et al., 1997), suggesting a model in which Spt16 disrupts one of these DNA contacts while the other is disrupted by Pob3. This is consistent with a report that FACT competes with DNA for binding to H2A-H2B (Hsieh et al., 2013), and indicates that FACT binding to H2A-H2B may induce or trap an open, reorganized nucleosome conformation with enhanced DNA accessibility (Formosa, 2012).
To test the importance of H2A-H2B binding for nucleosome reorganization by FACT, we asked whether Spt16-Pob3 heterodimers lacking C-terminal domains could produce the increased rate of endonuclease digestion characteristic of this altered structure (Xin et al., 2009). Indeed, deleting one C-terminal tail from Spt16 or Pob3 caused a decrease in reorganization activity, and deleting both tails caused a complete loss of activity (Figure 5D). These data parallel the H2A-H2B binding defects (Figure 1E) and indicate that interactions with H2A-H2B are central for nucleosome reorganization by FACT, presumably because disruption of histone:DNA contacts is needed to achieve or maintain reorganization. We conclude that full-length FACT reorganizes nucleosome structure at least in part by using Spt16-C and Pob3-C to compete with DNA to bind H2A-H2B through the interface defined by our binding and crystallographic data, as illustrated in Figure 5E. In addition to two H2A-H2B dimers, FACT can also bind H3-H4 and thereby simultaneously tether of all of the components of a nucleosome. The H3-H4 interface with FACT is not known, but we used EMSA to show that its binding is compatible with, and does not significantly affect, the interaction of FACT with two H2A-H2B dimers (Figure S3).
Similarity of Spt16-C:H2A-H2B with ANP32E:H2A.Z-H2B and Swr1:H2A.Z-H2B
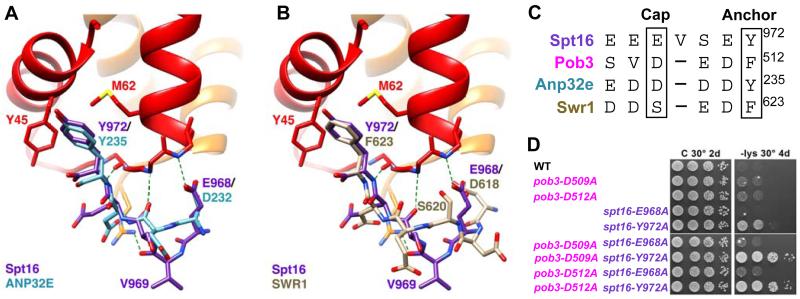
Our Spt16:H2A-H2B structure showed unexpected similarity with the structure of human ANP32E (acidic nuclear phosphoprotein 32 kilodalton e) in complex with the histone variant H2A.Z-H2B (Mao et al., 2014; Obri et al., 2014). ANP32E binds H2A.Z through its acidic H2A.Z interacting domain (ZID) and participates in the deposition of H2A.Z at promoters. Despite the absence of sequence similarity, ANP32E displays remarkable structural similarity over the visible Spt16-C residues (Figure 6A). After aligning on H2B, Spt16 residues E968, S970, E971 and Y972 overlap corresponding ANP32E Cα atoms with an RMSD of 1.17 Å. The one Spt16 residue that lacks an equivalent in ANP32E is V969, which adopts a bulged conformation that allows residues on either side to maintain structural equivalence. Strikingly, the two Spt16 side chains that make the primary contacts with H2B, E968 and Y972, superimpose closely with ANP32E D232 and Y235 (Figure 4A). Notably, Spt16 E968 and ANP32E D232 form equivalent hydrogen bonding contacts with the N-terminus of H2B helix 2, despite being 4 and 3 residues N-terminal to the conserved tyrosine, respectively. This is achieved by the bulging of Spt16 V969 and the longer side chain of E968 compared to ANP32E D232, resulting in the two carboxylates occupying the same relative position.
Figure 6. Superposition of Spt16 with ANP32E and Swr1.
(A) Spt16 and ANP32E (PDB 4cay; (Obri et al., 2014) following overlap on H2B molecules. (B) Spt16 and Swr1 (PDB 4m6b; (Hong et al., 2014). (C) Alignment of Spt16, Pob3, ANP32E and SWR1 sequences. (D) Ability to maintain transcriptional repression in strains bearing mutations in aromatic anchor and acidic helix-capping residues, integrated at the endogenous POB3 or SPT16 loci as in Figure 2. See also Figure S4.
Our Spt16:H2A-H2B structure also showed unexpected similarity with the structure of S. cerevisiae Swr1 in complex with H2A.Z-H2B (Hong et al., 2014). Swr1 is an ATP-dependent remodeler that deposits H2A.Z-H2B into the nucleosomes that flank nucleosome-depleted regions (Ranjan et al., 2013; Yen et al., 2013). Swr1 lacks sequence similarity with Spt16 or ANP32E, and the similarity in binding mode was not previously noted. Nevertheless, overlap on H2B Cα atoms reveals an RMSD of 0.89 Å for Spt16 E968, S970, E971 and Y972 with corresponding Swr1 Cα atoms (Figure 6B). Swr1 F623 superimposes with Spt16 Y972 and ANP32E Y235. Like ANP32E, Swr1 lacks a bulge residue equivalent to Spt16 V969, although it has a serine, S620, at the positions of Spt16 E968 and ANP32E D232. Like the Spt16 and ANP32E carboxylates, the Swr1 S620 side chain hydrogen bonds with a main chain amide at the N-terminus of H2B helix 2, although it necessarily lacks the stabilizing charge interaction expected for a carboxylate at this position. Instead, the Swr1 E618 carboxylate nestles adjacent to S620, thereby providing a variation of the H2B helix 2 capping motifs of Spt16 and ANP32E.
The inference that Spt16 Y972 and Pob3 F512 bind equivalent sites on H2B prompted us to consider the further possibility that Pob3 D509 might perform the same helix-capping role as Spt16 E968, ANP32E D232, and Swr1 S620/E618. As noted above, Spt16 E968A displayed 6-fold reduced affinity (KD = 5 μM), and we found that Pob3 D509A displayed an even stronger 39-fold reduction in binding affinity (KD = 35 μM; not shown), thereby supporting the sequence alignment shown in Figure 6C. We also found that these residues are important in vivo. Mutating them alone had minimal effects on transcriptional repression, but combining pob3-D509A with spt16-Y972A generated a robust phenotype similar to that of the pob3-F512A spt16-Y972A double mutant (Figure 6D). Consistent with the smaller effect on H2A-H2B binding in vitro, spt16-E968A pob3-F512A double mutants did not have a significant defect (Figure 6D). Thus, all four histone binding proteins (Spt16, Pob3, Anp32E, and Swr1) use a conserved bidentate binding motif, in which a carboxylate (or hydroxyl and carboxylate in Swr1) side chain caps H2B helix 2 and an aromatic side chain binds in the H2B Y45, M62 hydrophobic pocket. The relative importance of these elements varies between Spt16 and Pob3, but each displays strong correlation between the impacts on binding in vitro and phenotypic consequences in vivo.
These four proteins overlap in their binding with H2B, with ANP32E and Swr1 making additional H2A.Z-discriminating contacts that are not captured in our structure. Consistent with this, EMSA with full-length FACT and ITC with C-terminal peptides both demonstrate that FACT does not differentiate between H2A.Z-H2B and H2A-H2B (Figure S4). Thus, ANP32E and Swr1 make extensive contacts with H2A.Z that explain their specificity for this variant, whereas Spt16 and Pob3 use a more flexible network of nearby charged residues to enhance binding to any H2A family member. Residues outside the common binding region therefore contribute to affinity in both cases, with the details reflecting the biological function of each factor.
DISCUSSION
Binding of H2A-H2B is a core activity of the essential histone chaperone FACT. We have found that the Spt16 and Pob3 subunits of FACT can each bind H2A-H2B, and that ~1 μM binding affinity is mediated by short peptides within natively unstructured acidic domains in each of the C-terminal tails. These short peptides recapitulate the affinity of intact subunits, and FACT lacking just these domains fails to bind H2A-H2B within the detection limits of our ITC or EMSA experiments. H2A-H2B binding through the sites identified is physiologically relevant because mutations that impair binding in vitro cause defects in transcriptional repression in vivo, with complete removal of the binding site being lethal and impairment by a variety of point mutants generating proportional phenotypic responses. The mechanistic role of the FACT interaction with H2A-H2B is indicated by our crystal structure, which revealed incompatibility with DNA binding at the 2nd and 13th nucleosome contact sites, and by our finding that both H2A-H2B binding domains are important for FACT’s nucleosome reorganization activity.
Our crystal structure of the Spt16:H2A-H2B complex revealed that two Spt16 side chains make distinctive histone interactions, with E968 capping the N-terminus of H2B helix 2 and Y972, the aromatic anchor, filling a hydrophobic pocket formed by the highly conserved H2B residues Y45 and M62. Remarkably, the ordered residues of Spt16 superimpose closely with the previously reported structures of ANP32E (Obri et al., 2014) and Swr1 (Hong et al., 2014) bound to the histone variant H2A.Z-H2B, including both the capping residue and the aromatic anchor. Our analysis indicated that Pob3-C uses an equivalent binding mechanism, with D509 and F512 serving as the helix capping and aromatic anchor residues, respectively. We validated this model biochemically, and demonstrated the physiological importance of the histone-binding residues for both FACT subunits.
The similarities among Spt16, Pob3, ANP32E, and Swr1, and the conservation of the binding site residues in H2B, strongly suggest that this is a conserved mechanism for regulating the stability of interactions between DNA and H2A-H2B family members that is likely to be employed by other histone-binding proteins. The low complexity of the motif makes it challenging to assess its conservation even among known homologs or to search for it in other chaperones. However, the ConSurf algorithm (Ashkenazy et al., 2010; Celniker et al., 2013) revealed conservation of the crucial aromatic residues Spt16-Y972 and Pob3-F512 (Figure S1) and conservation of ANP32E Y235 was noted previously (Obri et al., 2014). Further, manual examination of the acidic domains that are common features of many histone chaperones revealed other potential matches to this motif, consistent with the possibility that this binding mechanism is not limited to the four factors noted here. The emergence of a common mode for H2A-H2B interaction parallels recent developments with H3-H4 chaperones in which targeting of the H3-H3’ intranucleosomal interface as a binding site has been observed (Elsasser et al., 2012; Gurard-Levin et al., 2014).
Our data indicate that the major FACT interactions are with H2B, and that any contribution to binding from H2A is modest. Nevertheless, we did find that the mutations SEDD975-978AAAA and Spt16 EDEE987-990AAAA, which are not visible in our structure and may contact H2A, disrupted binding by 20- and 15-fold, respectively. Our finding that the affinity of FACT was the same for H2A.Z-H2B and H2A-H2B indicates that any contact between FACT and H2A is likely to be limited to conserved regions. Consistent with limited contact interfaces in FACT, Spt16 and Pob3 have modest affinities for H2A-H2B (~1 μM) compared to Swr1 (7 nM; (Hong et al., 2014). This is consistent with the idea that FACT functions in a general way to modulate nucleosome structure, whereas the specific chaperones need to make tight, specific interactions with defined variant histones.
Acidic domains, such as those found in Spt16 and Pob3 and many other histone chaperones, are typically portrayed as unstructured charged “clouds” that associate with histones through non-specific mechanisms (Belotserkovskaya et al., 2003). Our analysis reveals that unstructured acidic residues are indeed important for binding, but also drives an important revision of this model by showing that the acidic domains of Spt16-C and Pob3-C bind H2A-H2B as specific 1:1 complexes, binding is not affected by deletion of a highly basic region of H2B, and the affinity is strongly dependent on single aromatic residues within the acidic domains. Thus, while negative charges are an important component of binding, specific recognition by an aromatic anchor residue and an associated helix-capping motif are central to the interaction of Spt16 and Pob3 with H2A-H2B.
Our finding that binding of Spt16-C is incompatible with binding of H2A-H2B to DNA in canonical nucleosomes explains the observed competition between FACT and DNA for binding to H2A-H2B (Hsieh et al., 2013). DNA wrapping around the nucleosome core is stabilized by 14 sets of contacts (Luger et al., 1997). As DNA enters the nucleosome, the first set of contacts primarily involves H3-H4, and the second set primarily H2A-H2B. This second set is incompatible with Spt16-C binding because the hydrophobic pocket is occluded by DNA and because Spt16-C and DNA make mutually exclusive contacts with H2A R78. The equivalent H2A.Z R87 residue also makes a similar contact with ANP32E, Swr1, and DNA, so this is a conserved feature of the binding motif (Hong et al., 2014; Obri et al., 2014). Given the similarity of Pob3-C binding to H2A-H2B, we strongly anticipate that binding of Pob3-C would also disrupt this interaction.
Thus, binding of FACT to a nucleosome presumably involves equivalent binding of Spt16-C and Pob3-C to the two symmetry-related H2A-H2B dimers, competing for binding to the DNA at each of the second points of contact internal to the entry/exit sites. We therefore propose that FACT promotes reorganization of nucleosomes by competing with DNA for these H2A-H2B interactions as they are transiently broken (Tims et al., 2011). Moreover, by maintaining contact with both H2A-H2B dimers and also with H3-H4 and DNA (Winkler et al., 2011) FACT could additionally tether the components together to prevent their dispersal (Formosa, 2012; Jamai et al., 2009) and promote reassembly with the same histone molecules. This model is further supported by our observation that loss of Spt16 and Pob3 C-terminal domains rendered FACT unable to induce reorganization of nucleosomes in vitro. Although our data explain how FACT contacts H2A-H2B to promote reorganization, we do not propose that these moderate ~1 μM interactions function to recruit FACT to specific chromatin locations. Instead, recruitment more likely results from interactions with more specialized cellular machinery, such as DNA Pol1 (Wittmeyer et al., 1999), PAF1C (Krogan et al., 2002), and Swi6 (Takahata et al., 2009), while the H2A-H2B interaction functions to alter the structure of the nucleosome to which FACT has been recruited.
Our results contrast with a previous report, in which a different domain, Spt16-M, was identified as the H2A-H2B binding site in FACT (Hondele et al., 2013). That study used a translational fusion of Spt16-M from the thermophilic yeast C. thermophilum with X. laevis H2B in a complex with H2A to define the binding interface crystallographically. This ensured that crystals would contain both Spt16-M and histones, but increased the risk of forming a physiologically irrelevant interaction. Residues important for the interaction were not highly conserved in either Spt16 or H2B, and the largely hydrophobic interface was only disrupted by substituting multiple residues simultaneously, invoking the possibility that non-specific interactions were stabilized by the fusion construct. This same study also reported binding of free C. thermophilum Spt16-M and Spt16-M,C with X. laevis H2A-H2B in solution, with Spt16-C providing the major contribution (Hondele et al., 2013). Our results with cognate proteins recapitulate the importance of Spt16-C, but do not reveal interaction of H2A-H2B with Spt16-M or any other FACT domain apart from Pob3-C. Indeed, structural overlap indicates extensive clashes that would preclude simultaneous formation of the published Spt16-M interaction and the interaction seen in our structure. We therefore conclude that Spt16-M does not contribute to the physiological interaction of FACT with H2A-H2B.
A previous study of the metazoan homolog of Pob3, SSRP1, did not detect binding of H2A-H2B (Winkler et al., 2011). While SSRP1 contains an acidic domain that contains a match to the consensus motif identified here, it also has other features at its C-terminus, including an HMGB DNA binding domain and an additional serine-rich, mixed acidic and basic region. Given our observation that the presence of other features, such as the C-terminal region of Spt16, can mask H2A-H2B binding, it will be necessary to reexamine SSRP1 and other chaperones with this signature in a more focused way. One attractive possibility is that the SSRP1 interaction with H2A-H2B might be dependent upon prior association of the C-terminal domain with DNA.
In conclusion, our biochemical, structural, and genetic observations support the nucleosome reorganization model, in which FACT maintains nucleosomes in an altered, loosened structure at least in part by competing with DNA for binding both H2A-H2B dimers in a nucleosome. Our data also indicate that the Spt16-C and Pob3-C interactions with H2A-H2B are an essential, core function of FACT in vivo. The demonstration that specific interactions are central to H2A-H2B binding forces a revision of the model that acidic domains mediate non-specific, cloud-like binding. Finally, the conservation of a binding motif between Spt16, Pob3, ANP32E and SWR1 suggests that equivalent mechanisms of binding and nucleosome modulation might be used by a wide range of histone chaperones and chromatin remodelers.
EXPERIMENTAL PROCEDURES
Protein expression and purification
Saccharomyces cerevisiae FACT constructs are described in Table S2. Heterodimers were expressed in yeast, subdomains (inlcuding peptides) were expressed in bacteria, and purifications were performed as described and verified by mass spectrometry (Biswas et al., 2005; Kemble et al., 2013; Ruone et al., 2003). Yeast H2A-H2B was purified as a dimer expressed from a plasmid kindly provided by Dr. Robert Dutnall (Anderson et al., 2010). Yeast H2A.Z-H2B and yeast H3-H4 were purified as described (Dyer et al., 2004). Nucleosomes were assembled from recombinant yeast histones and 5S rDNA as described (Xin et al., 2009).
Isothermal Titration Calorimetry
ITC experiments were performed at 25° C using a MicroCal iTC200 MicroCalorimeter. For ITC, the final purification step was gel filtration chromatography in 25 mM Na-K phosphate pH 7.4, 200 mM NaCl, followed by dialysis against several changes of the same solution over 48 hours. Titrations included an initial injection volume of 0.4 μL (omitted from analysis) and 25 injections of 1.8 μL spaced at intervals of 90 s (120 s for FACT constructs). Data were analyzed using Origin 7 software wherein the heat of dilution was subtracted from the raw values, and the stoichiometry (N), association constant (KA), and change in enthalpy (ΔH) were calculated by fitting the isotherm. All reactions were performed at least in triplicate.
EMSA
Affinities were estimated from half-maximal complex formation on native polyacrylamide gels, as described previously (Ruone et al., 2003; Xin et al., 2009). Titrations were performed at least in triplicate.
Genetic analysis
Mutations were introduced into the genomic SPT16, POB3, HTB1, and HTB2 loci in diploids along with selectable markers downstream of each gene by PCR-mediated site-directed mutagenesis (McCullough et al., 2011), and confirmed by sequencing. Haploids were derived and strains with combinations of mutations were constructed using standard methods. Strains (Table S1) were grown to saturation in rich medium, then aliquots of 10-fold serial dilutions were tested as noted in each experiment.
Structure determination
S. cerevisiae Spt16958-990 and H2A-H2B30-130 were purified as described above and eluted from Superdex SD200 in 20 mM Tris pH 7.5, 200 mM NaCl. Proteins were mixed at stoichiometric amounts in the presence of 0.6 M NDSB-256 (Hampton, Dimethylbenzylammonium propane sulfonate) and concentrated to 10 mg/ml (Bradford). Crystals of ~60 × 60 μm formed within 10 days in drops of 2 μL protein and 2 μL reservoir solution [0.1M SPG (succinic acid:sodium dihydrogen phosphate:glycine) pH 7, 25% PEG 1500] (A4 of the PACT screen; Qiagen). Crystals were transferred from mother liquor to a coating of mineral oil, suspended in a rayon loop, and cooled by plunging into liquid nitrogen. Diffraction data were collected at SSRL remotely with Blu-ice (McPhillips et al., 2002; Soltis et al., 2008). Data processing and scaling were performed using the HKL-2000 package (Otwinowski, 1997). The crystal structure was determined by molecular replacement with Phaser (CCP4 program suite) using the coordinates of yeast H2A-H2B (PDB code 1ID3; (White et al., 2001) as the search model. Model building used COOT (Emsley et al., 2010). Refinement used Phenix (Adams et al., 2010). Figures of atomic structures were prepared using Chimera (Pettersen et al., 2004).
Fluorescence Anisotropy
Spt16965-990 was labeled at the N-terminus with fluorescein during synthesis and purified by anion exchange (Q HiTrap) and gel filtration (Superdex SD200). Protein concentration was determined by infrared spectrometry (Direct Detect Spectrometer, EMD Millipore). For direct binding assays, yeast H2A-H2B was titrated with a fixed concentration of Spt16965–990 using a Tecan Infinite200 spectrometer. Raw anisotropy values were analyzed by nonlinear regression in GraphPad Prism 6 to obtain the average KD value for 3 independent experiments. For competition experiments, unlabeled Spt16965–990 or unlabeled Pob3505–529 were added to a fixed concentration of labeled Spt16965–990 at 80% saturation of the H2A-H2B. IC50’s were used to calculate the apparent Ki (http://sw16.im.med.umich.edu/software/calc_ki/).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Michael Kay and Michael Jacobson for synthesis of fluorescein-labeled peptides, Zaily Connell for technical assistance, and Robert Dutnall for the H2A-H2B expression plasmids. Mass spectrometry was performed by the University of Utah Mass Spectrometry and Proteomics Core Facility, which is supported by an award from the National Cancer Institute (P30CA042014). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). This work was supported by National Institutes of Health Grants R01 GM59135 (to CPH and TF), P50 GM082545 (to CPH), and GM064649 (to TF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information comprises 4 figures and 2 tables.
AUTHOR CONTRIBUTIONS
DJK, TF, and CPH designed the experiments. DJK and LLM conducted the experiments. FGW made important contributions to the X-ray crystallography analysis. DJK, TF, and CPH wrote the paper.
ACCESSION NUMBERS
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under the accession code 4WNN.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M, Huh JH, Ngo T, Lee A, Hernandez G, Pang J, Perkins J, Dutnall RN. Co-expression as a convenient method for the production and purification of core histones in bacteria. Protein Expr Purif. 2010;72:194–204. doi: 10.1016/j.pep.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Saunders A, Lis JT, Reinberg D. Transcription through chromatin: understanding a complex FACT. Biochim Biophys Acta. 2004;1677:87–99. doi: 10.1016/j.bbaexp.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Biswas D, Yu Y, Prall M, Formosa T, Stillman DJ. The yeast FACT complex has a role in transcriptional initiation. Mol Cell Biol. 2005;25:5812–5822. doi: 10.1128/MCB.25.14.5812-5822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker G, Nimrod G, Ashkenazy H, Glaser F, Martz E, Mayrose I, Pupko T, Ben-Tal N. ConSurf: Using Evolutionary Data to Raise Testable Hypotheses about Protein Function. Israel Journal of Chemistry. 2013;53:199–206. [Google Scholar]
- Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- Elsasser SJ, Huang H, Lewis PW, Chin JW, Allis CD, Patel DJ. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature. 2012;491:560–565. doi: 10.1038/nature11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T. The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta. 2012;1819:247–255. doi: 10.1016/j.bbagrm.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurard-Levin ZA, Quivy JP, Almouzni G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu Rev Biochem. 2014;83:487–517. doi: 10.1146/annurev-biochem-060713-035536. [DOI] [PubMed] [Google Scholar]
- Hondele M, Ladurner AG. The chaperone-histone partnership: for the greater good of histone traffic and chromatin plasticity. Curr Opin Struct Biol. 2011;21:698–708. doi: 10.1016/j.sbi.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Hondele M, Ladurner AG. Catch me if you can: how the histone chaperone FACT capitalizes on nucleosome breathing. Nucleus. 2013;4:443–449. doi: 10.4161/nucl.27235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondele M, Stuwe T, Hassler M, Halbach F, Bowman A, Zhang ET, Nijmeijer B, Kotthoff C, Rybin V, Amlacher S, et al. Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature. 2013;499:111–114. doi: 10.1038/nature12242. [DOI] [PubMed] [Google Scholar]
- Hong J, Feng H, Wang F, Ranjan A, Chen J, Jiang J, Ghirlando R, Xiao TS, Wu C, Bai Y. The catalytic subunit of the SWR1 remodeler is a histone chaperone for the H2A.Z-H2B dimer. Molecular cell. 2014;53:498–505. doi: 10.1016/j.molcel.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh FK, Kulaeva OI, Patel SS, Dyer PN, Luger K, Reinberg D, Studitsky VM. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1222198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamai A, Puglisi A, Strubin M. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Molecular cell. 2009;35:377–383. doi: 10.1016/j.molcel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Kemble DJ, Whitby FG, Robinson H, McCullough LL, Formosa T, Hill CP. Structure of the Spt16 Middle Domain Reveals Functional Features of the Histone Chaperone FACT. J Biol Chem. 2013;288:10188–10194. doi: 10.1074/jbc.C113.451369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA Polymerase II Elongation Factors of Saccharomyces cerevisiae: a Targeted Proteomics Approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Mao Z, Pan L, Wang W, Sun J, Shan S, Dong Q, Liang X, Dai L, Ding X, Chen S, et al. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell research. 2014;24:389–399. doi: 10.1038/cr.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough L, Rawlins R, Olsen AE, Xin H, Stillman DJ, Formosa T. Insight into the Mechanism of Nucleosome Reorganization from Histone Mutants that Suppress Defects in the FACT Histone Chaperone. Genetics. 2011;188:835–846. doi: 10.1534/genetics.111.128769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, et al. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- Obri A, Ouararhni K, Papin C, Diebold ML, Padmanabhan K, Marek M, Stoll I, Roy L, Reilly PT, Mak TW, et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature. 2014;505:648–653. doi: 10.1038/nature12922. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z.a.M. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. W. [DOI] [PubMed] [Google Scholar]
- Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Winston F. Chromatin and transcription in yeast. Genetics. 2012;190:351–387. doi: 10.1534/genetics.111.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Mizuguchi G, FitzGerald PC, Wei D, Wang F, Huang Y, Luk E, Woodcock CL, Wu C. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell. 2013;154:1232–1245. doi: 10.1016/j.cell.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom M, Dennehey BK, Tyler JK. Chaperoning Histones during DNA Replication and Repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruone S, Rhoades AR, Formosa T. Multiple Nhp6 Molecules are Required to Recruit Spt16-Pob3 to Form yFACT Complexes and Reorganize Nucleosomes. J Biol Chem. 2003;278:45288–45295. doi: 10.1074/jbc.M307291200. [DOI] [PubMed] [Google Scholar]
- Schlesinger MB, Formosa T. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics. 2000;155:1593–1606. doi: 10.1093/genetics/155.4.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker KR, Kim PS, York EJ, Stewart JM, Baldwin RL. Tests of the helix dipole model for stabilization of alpha-helices. Nature. 1987;326:563–567. doi: 10.1038/326563a0. [DOI] [PubMed] [Google Scholar]
- Soltis SM, Cohen AE, Deacon A, Eriksson T, Gonzalez A, McPhillips S, Chui H, Dunten P, Hollenbeck M, Mathews I, et al. New paradigm for macromolecular crystallography experiments at SSRL: automated crystal screening and remote data collection. Acta Crystallogr D Biol Crystallogr. 2008;64:1210–1221. doi: 10.1107/S0907444908030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman DJ. Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim Biophys Acta. 2010;1799:175–180. doi: 10.1016/j.bbagrm.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T, Hothorn M, Lejeune E, Rybin V, Bortfeld M, Scheffzek K, Ladurner AG. The FACT Spt16 “peptidase” domain is a histone H3-H4 binding module. Proc Natl Acad Sci U S A. 2008;105:8884–8889. doi: 10.1073/pnas.0712293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S, Yu Y, Stillman DJ. The E2F functional analogue SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters. EMBO J. 2009;28:3378–3389. doi: 10.1038/emboj.2009.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tims HS, Gurunathan K, Levitus M, Widom J. Dynamics of nucleosome invasion by DNA binding proteins. J Mol Biol. 2011;411:430–448. doi: 10.1016/j.jmb.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDemark AP, Blanksma M, Ferris E, Heroux A, Hill CP, Formosa T. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Molecular cell. 2006;22:363–374. doi: 10.1016/j.molcel.2006.03.025. [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Xin H, McCullough L, Rawlins R, Bentley S, Heroux A, Stillman DJ, Hill CP, Formosa T. Structural and Functional Analysis of the Spt16p N-terminal Domain Reveals Overlapping Roles of yFACT Subunits. J Biol Chem. 2008;283:5058–5068. doi: 10.1074/jbc.M708682200. [DOI] [PubMed] [Google Scholar]
- White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. Embo J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler DD, Luger K. The histone chaperone Fact: Structural insights and mechanisms for nucleosome reorganization. J Biol Chem. 2011;286:18369–18374. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler DD, Muthurajan UM, Hieb AR, Luger K. The histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem. 2011;286:41883–41892. doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmeyer J, Joss L, Formosa T. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry. 1999;38:8961–8971. doi: 10.1021/bi982851d. [DOI] [PubMed] [Google Scholar]
- Xin H, Takahata S, Blanksma M, McCullough L, Stillman DJ, Formosa T. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Molecular cell. 2009;35:365–376. doi: 10.1016/j.molcel.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Pugh BF. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell. 2013;154:1246–1256. doi: 10.1016/j.cell.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.