Abstract
Evaluation of the genetic contribution to the development of recurrent acute otitis media (rAOM) remains challenging. This study aimed to evaluate the potential association between single nucleotide polymorphisms (SNPs) in selected genes and rAOM and to analyze whether genetic variations might predispose to the development of complicated recurrent cases, such as those with tympanic membrane perforation (TMP).
A total of 33 candidate genes and 47 SNPs were genotyped in 200 children with rAOM (116 with a history of TMP) and in 200 healthy controls.
INFγ rs 12369470CT was significantly less common in the children with rAOM than in healthy controls (odds ratio [OR] 0.5, 95% confidence interval [CI] 0.25–1, P = 0.04). Although not significant, interleukin (IL)-1β rs 1143627G and toll-like receptor (TLR)-4 rs2737191AG were less frequently detected in the children with rAOM than in controls. The opposite was true for IL-8 rs2227306CT, which was found more frequently in the children with rAOM than in healthy controls. The IL-10 rs1800896TC SNP and the IL-1α rs6746923A and AG SNPs were significantly more and less common, respectively, among children without a history of TMP than among those who suffered from this complication (OR 2.17, 95% CI 1.09–4.41, P = 0.02, and OR 0.42, 95% CI 0.21–0.84, P = 0.01).
This study is the first report suggesting an association between variants in genes encoding for factors of innate or adaptive immunity and the occurrence of rAOM with or without TMP, which confirms the role of genetics in conditioning susceptibility to AOM.
INTRODUCTION
Acute otitis media (AOM) is a very common disease. More than 90% of children suffer from AOM in the first 5 years of life.1 Moreover, in 20% to 30% of these children, AOM tends to recur, ultimately causing significant medical, social, and economic problems.1,2 Several factors, including young age, day care attendance, and passive smoke exposure, have been associated with an increased risk of recurrent AOM (rAOM). However, it has been shown that host genetic factors significantly influence the risk of developing AOM.3 It has been calculated that AOM susceptibility is 40% to 60% heritable,4 although the exact mechanisms for this heritability have not been precisely described. Because AOM is an infectious disease and because the innate and adaptive immune systems play a fundamental role in the defense from infectious agents, most studies that were specifically designed to evaluate the potential role of genetics in conditioning rAOM have evaluated the association between variants of genes that encode factors of innate and adaptive immunity and susceptibility to AOM. With regard to innate immunity, genes encoding toll-like receptors (TLRs), CD14, mannose-binding lectins, and surfactants have been the most extensively studied.5 Regarding adaptive immunity, studies have primarily focused on genes encoding various cytokines, such as interleukin-1, (IL-1), IL-6, IL-10, transforming growth factor-β (TGF-β), tumor necrosis factor α (TNF-α), and interferon-γ (IFN-γ).5 Additionally, some genome-wide association studies of rAOM susceptibility have been performed.6–8 A set of known genetic polymorphisms that seem to lead to a predisposition to AOM have been identified, and genome-wide linkage scans have suggested multiple candidate regions of the genome that could be associated with rAOM.5,9
However, evaluating the genetic contribution to the development of a disease such as AOM, which is multifactorial in etiology, remains challenging. Most of the data collected to date could be debated. Their relevance varies significantly from study to study and according to the ethnicity and characteristics of the children. Finally, all of these studies have evaluated rAOM as a whole, without considering that in this complex condition, complicated and uncomplicated cases are included and genetics might play a role in conditioning the different clinical pictures. This study was designed to evaluate the potential association between single nucleotide polymorphisms (SNPs) in selected genes and rAOM and to analyze whether genetic variations might predispose to the development of complicated recurrent cases, such as those with tympanic membrane perforation (TMP).
METHODS
Study Population and Recruitment
The study was carried out between November 1, 2014, and January 31, 2015, and it involved children in the age group 1 to 5 years who had a history of rAOM (defined as at least 3 episodes in the preceding 6 months or at least 4 episodes in the preceding 12 months, with the most recent episode in the previous 2–8 weeks) and were regularly followed by the Outpatient Clinic of the Pediatric High Intensive Care Unit at Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, University of Milan, Milan, Italy. The minimum number of episodes of AOM required to include patients in the otitis-prone group must have been diagnosed by pneumatic otoscopy at the Outpatient Clinic of the Pediatric High Intensive Care Unit and documented by medical records, with at least 2 of these episodes also supported by tympanometric findings. The exclusion criteria were all of the factors that can, per se, favor the development of AOM, including severe atopy, acquired or congenital immunodeficiency, cleft palate, a chronically ruptured eardrum, craniofacial abnormalities or obstructive adenoids, sleep apnea syndrome, or the placement of tympanostomy tubes.
Upon enrollment, the demographic characteristics and medical history of the children were systematically recorded using standardized written questionnaires, paying particular attention to the characteristics of AOM and the occurrence in each episode of TMP. Finally, a 3 mL whole blood sample was obtained for genetic studies.
As the control group, a similar number of age- and gender-matched children without rAOM was enrolled, and a blood sample was drawn for genetic analyses.
The protocol was approved by the Ethics Committee of the Fondazione IRCCS Ca’ Granda, Ospedale Maggiore Policlinico, Milan, Italy, and written informed consent was obtained from the parents or legal guardian of each subject before enrollment.
Genetic Studies
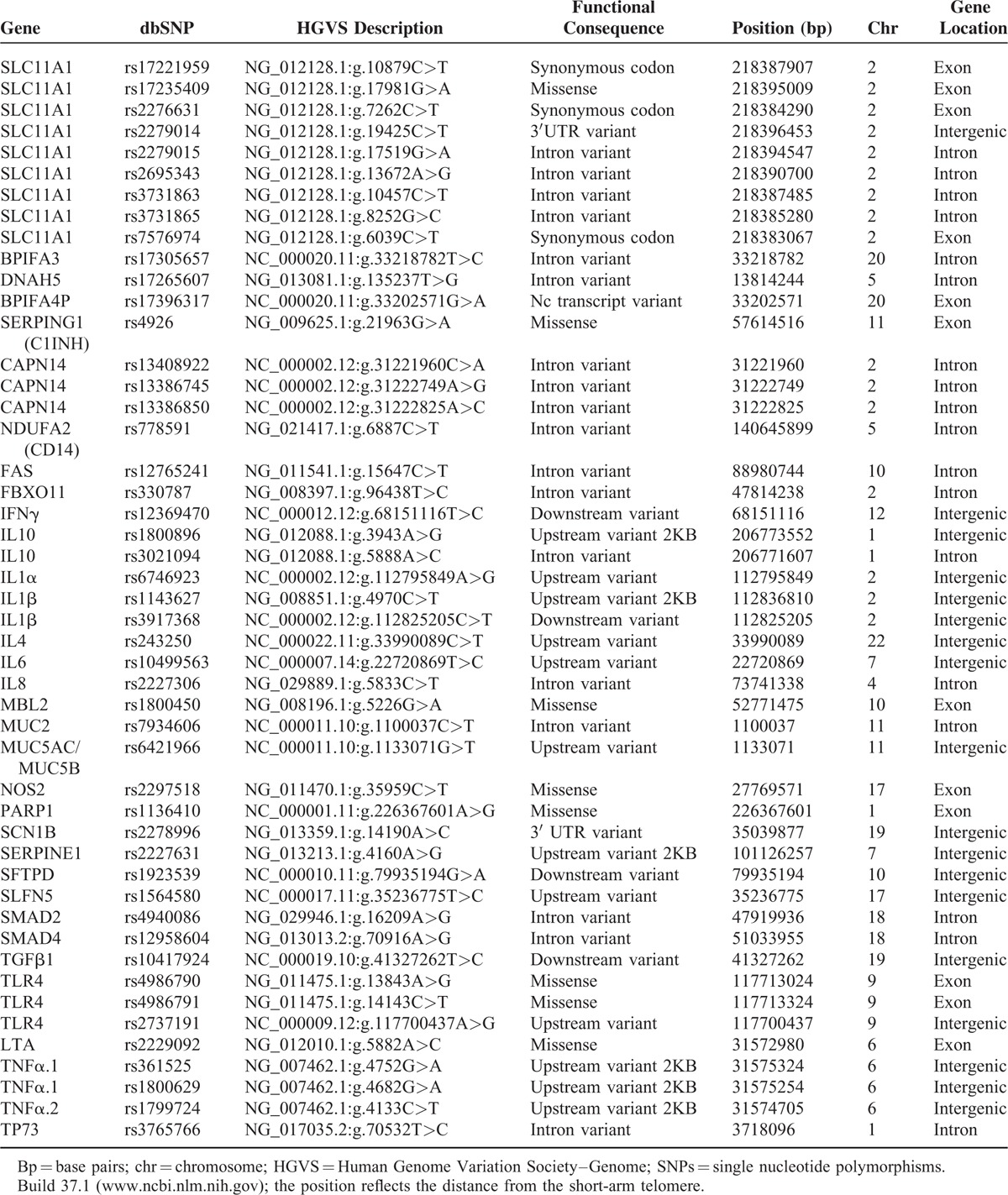
Thirty-three candidate genes and 47 single nucleotide polymorphisms (SNPs) were selected for analysis, including genes that are involved in immune regulation, the pathogenesis of inflammation, and the regulation of cell metabolism and function. Candidate genes included TLR-4, IL-1α, IL-1β, IL-4, IL-6, IL-8, IL-10, IFN-γ, TNF-α1, TNF-α2, lymphotoxin α (LTA), mannose-binding lectin 2 (MBL2), transforming growth factor β1 (TGFB1), nitric oxide synthase 2 (NOS2), poly(ADP-Ribose) polymerase-1 (PARP1), proton-coupled divalent metal ion transporter member 1 (SLC11A1), calpain 14 (CAPN14), mucin 2 (MUC2), mucin 5AC/AB (MUC5AC/AB), surfactant protein D (SFTPD), tumor protein 73 (TP73), serine protease inhibitors (serpine1), receptor-regulated SMAD2 (SMAD2), receptor-regulated SMAD4 (SMAD4), sodium channel voltage-gated type I beta subunit (SCN1β), dynein axonemal heavy chain 5(DNAH5), BPI fold containing family A member 3 (BPIFA3) and 4P (BFIFA4P), Fas cell surface death receptor (FAS), CD14, F-box protein 11 (FBXO11), and schlafen family member 5 (SLFN5). These SNPs are involved in causing or determining the severity or outcome of infectious or chronic immune-mediated diseases in experimental animals or humans or were previously supposed or found to be associated with an increased risk of developing AOM or otitis media with effusion (OME) or an abnormal immune response.5–8,10–20 The investigated genes and SNPs are listed in Table 1.
TABLE 1.
Gene and Single Nucleotide Polymorphisms (SNPs)
DNA was extracted using the Masterpure DNA Purification kit (Epicentre, Madison, FL) in accordance with the manufacturer's instructions, and a 50 μL final elution volume was obtained after purification. Single nucleotide polymorphisms in the 33 genes were genotyped using the Custom TaqMan Array Microfluidic Cards genotyping system on an ABI 7900HT (Applied Biosystems, Foster City, CA). After PCR amplification, the alleles were detected by means of end-point analysis using SDS and the TaqMan Genotyper software (Applied Biosystems, Foster City, CA). The data were entered into a Progeny database (Progeny Software, LLC, South Bend, IN) to generate datasets for analysis.
Statistical Analysis
The categorical data were compared between the groups using contingency table analysis with Fisher's exact test. The continuous data were analyzed using a 2-sided Wilcoxon rank-sum test after ensuring that they were not normally distributed (by means of the Shapiro–Wilk statistic).
Genotype frequencies were determined by direct counting. To investigate Hardy–Weinberg equilibrium (HWE), the expected number of each genotype was compared with the observed number, and potential deviations were assessed using Fisher's exact test. Univariate odds ratios (OR), their 95% confidence intervals (CI), and pertinent P values obtained by Fisher's exact test were calculated to measure the associations between selected SNPs and (1) susceptibility to AOM by comparing all the children with rAOM and controls and (2) susceptibility to rAOM without TMP with respect to rAOM and at least 1 episode with TMP. The data were controlled for multiple testing using the false discovery rate method (with the Benjamini–Hochberg procedure). All of the statistical analyses were made using the R software package, version 3.1.1, with library genetics and epitools added.
RESULTS
A total of 200 children with rAOM (129 men; median age, 31 months) and 200 healthy controls (129 men; median age, 31 months) were enrolled. Among the children with rAOM, 84 (42.0%) had never experienced TMP, and 116 had a history of rAOM with TMP. Their demographic and clinical characteristics are reported in Table 2. The 2 groups were similar for all of the studied variables. None of the healthy controls had a previous history of rAOM.
TABLE 2.
Demographic, Clinical, and Familial Characteristics of Subjects With Recurrent Acute Otitis Media (rAOM) by Disease Characteristics
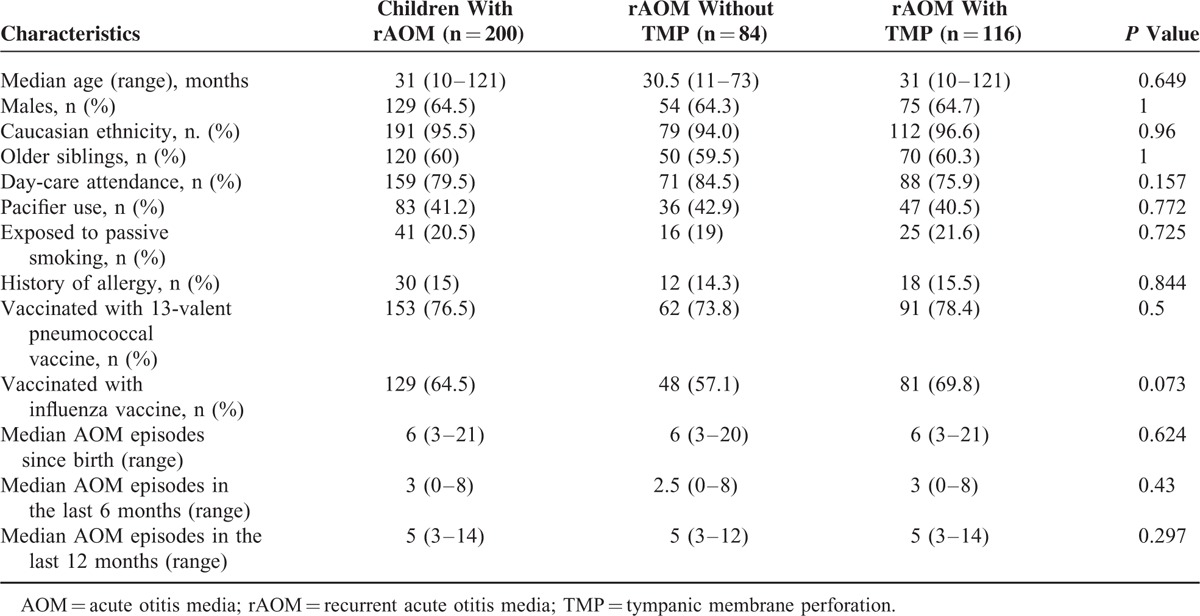
Table 3 lists the genotype frequencies with differences in the selected SNPs between the children with rAOM and otherwise healthy controls. As shown, INFγ rs 12369470CT was significantly less common in the children with rAOM than in healthy controls (OR 0.5, 95% CI 0.25–1, P = 0.04). Similarly, although not significant at the conventional 5% level, IL-1β rs 1143627G and TLR-4 rs2737191AG were less frequently detected in the children with rAOM than in controls (OR 0.57, 95% CI 0.31–1.05, P = 0.06; OR 0.68, 95% CI 0.43–1.05, P = 0.07, respectively). The opposite was observed for IL-8 rs2227306CT, which was found more frequently among the children with rAOM than in the healthy controls, although the difference was not statistically significant in this case (OR 1.52, 95% CI 0.97–2.04, P = 0.06). No other relevant genetic variation association was found between the children with rAOM and those without.
TABLE 3.
Genotype Frequencies With Differences in the Selected SNPs Between Controls and Children With Recurrent Acute Otitis Media (rAOM)
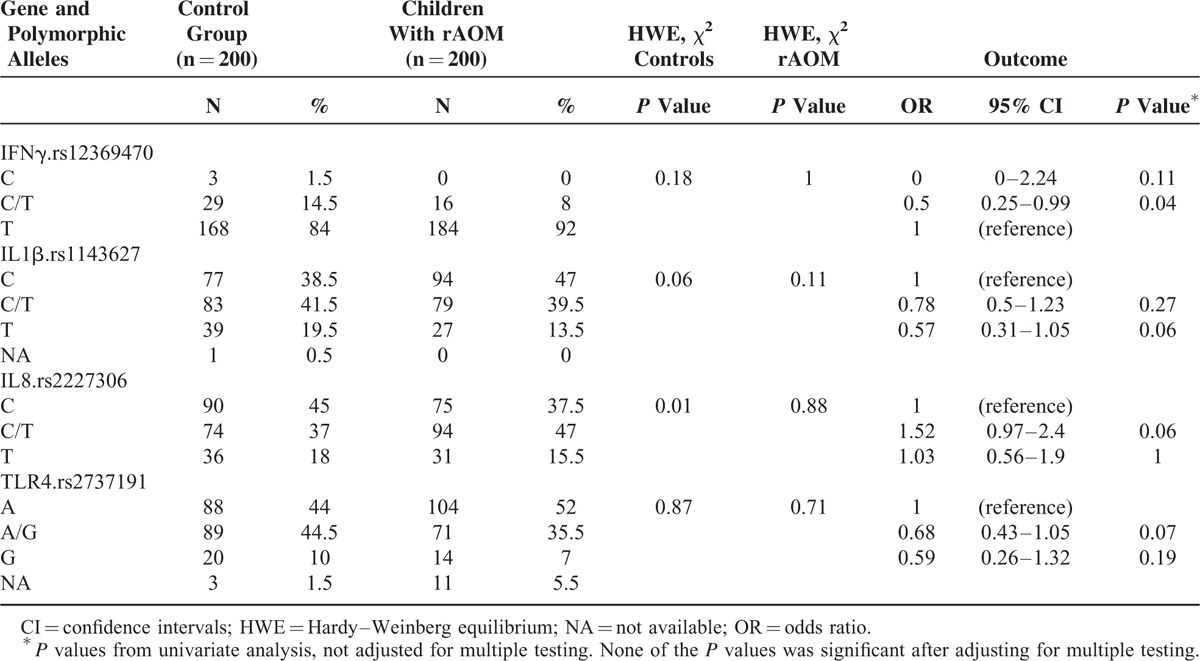
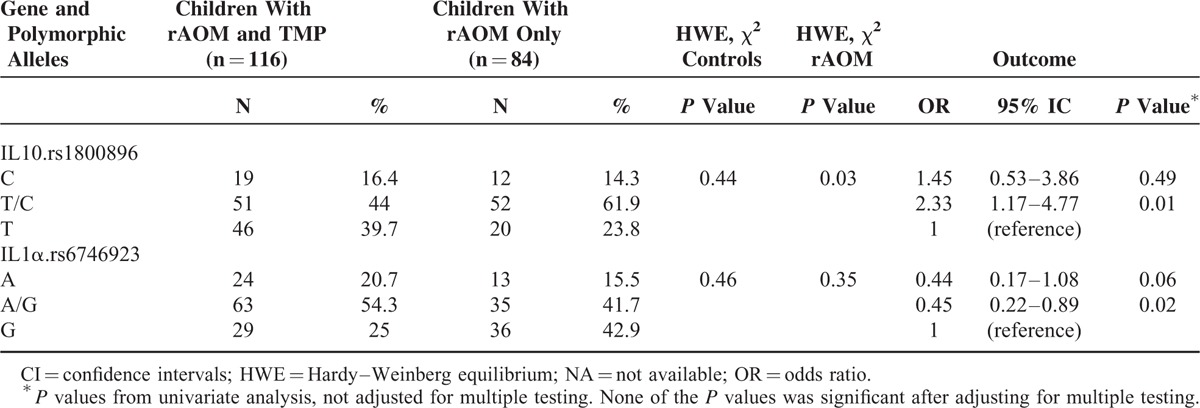
Table 4 summarizes the genotype frequencies with differences in the selected SNPs between the children with rAOM with TMP and those who did not experience TMP. The SNP IL-10 rs1800896TC and the IL-1α rs6746923A and AG SNPs were significantly more and less common, respectively, among children without a history of TMP than among those who suffered from this complication (OR 2.17, 95% CI 1.09–4.41, P = 0.02, and OR 0.42, 95% CI 0.21–0.84, P = 0.01, respectively). The other SNPs were similar in both groups of children with rAOM, regardless of the history of TMP.
TABLE 4.
Genotype Frequencies With Differences in the Selected SNPs Between Children With Recurrent Acute Otitis Media (rAOM) With Tympanic Membrane Perforation (TMP) or rAOM Without TMP
DISCUSSION
The results of this study contribute to the knowledge about the relationship between genetics and rAOM. Together with data regarding the potential role of SNPs in conditioning rAOM, this study suggests for the first time that genetics might be implicated in the determination of AOM with TMP. Concerning rAOM, this study shows that the SNP rs 12369470CT of the IFN-γ gene is less common among children with rAOM than in healthy controls, which suggests that this genetic variant might protect children from repeated AOM episodes. Although they are not supported by a statistically significant difference at the conventional 5% level, the same conclusions could be drawn for the SNP rs 1143627G of the IL-1β gene and the SNP rs2737191AG of the TLR4 gene. Conversely, a negative effect with an increased risk of rAOM seems to be associated with the IL-8 rs2227306CT SNP, which was more common among children with rAOM than in healthy controls, although the difference between the groups did not reach statistical significance in this case. For the further study, it is suggested that increasing the number of patients with rAOM might increase the statistical power of a similar study.
The impact of polymorphisms of the IFNγ, IL-1β, and TLR4 genes has been well studied in several clinical conditions, including AOM, with conflicting results. The protective effect of the IFNγ rs 12369470CT SNP found in this study is new information that might be useful for differentiating otitis-prone children from those for whom rAOM is less likely. Gentile et al found that genetic variations of the IFNγ gene were associated with an increased frequency of AOM.21 Ilia et al reported that the same SNP could be considered a predictor of progress to AOM following upper respiratory infection (URI).22 By contrast, results consistent with our study were published by Alper et al, who did not find any association between this SNP and the development of AOM in children with URIs.23
The data describing the IL-1β rs 1143627G and TLR4 rs2737191AG SNPs merit further evaluation because the protective effect associated with these genetic variations is not substantiated by the statistical analysis and was not found in previously published studies in which the same or other IL-1β SNPs were examined. Nokso-Koivisto et al studied the occurrence of AOM following URI and concluded that the presence of the IL-1β rs1143627G SNP did not increase susceptibility to AOM.24 By contrast, the IL-1β rs1143634 SNP was associated with a higher risk of severe inflammation after AOM.25 Similarly, conflicting data were reported for the TLR4 SNPs. The TLR4 rs4986790 and TLR4 rs49867912 SNPs, 2 of the SNPs evaluated in this study for which no association with rAOM was found, were reported to be more common in otitis-prone children by Emonts et al13 but were considered independent from AOM by Carroll et al.26
The IL-8 rs2227306CT SNP seems to be associated with an increased risk of rAOM because it is more common among children with this condition than controls. Although the difference between groups did not reach statistical significance, this finding merits attention and further studies to confirm the data because IL-8 has been repeatedly reported as a factor that increases susceptibility to ear disease and chronic ear inflammation both in vitro and in vivo.27,28
The IL-10 rs1800896TC SNP and the IL-1α rs6746923A and AG SNPs were associated with a reduced or an increased risk, respectively, of rAOM with TMP. Differences in genetic characteristics between subjects with and without TMP are not surprising because AOM complicated by TMP significantly differs from AOM without this complication in several factors. AOM with TMP is frequently caused by Streptococcus pyogenes, a bacterial pathogen that is not common in AOM without TMP, and it frequently has a complicated course.29 Moreover, the administration of vitamin D 30 or influenza vaccine,31 which can reduce the incidence of new episodes of AOM in children who have never had TMP, is not effective in patients with rAOM that is occasionally complicated by TMP. The importance of SNPs in IL-10 and IL-1α in conditioning susceptibility to respiratory infections has been reported by others. Nokso-Koivisto et al showed that the IL-10 rs1800896TC SNP was more common in subjects with a reduced risk of URI and an occurrence of AOM during URI episodes,24 whereas Joki-Erkkila et al found that SNPs in the IL-1α gene were associated with an increased risk of rAOM.32 Conversely, the role of IL-1α in predisposing to ear diseases is supported by the demonstration that the expression of IL-1α is higher in cases of chronic otitis media, with a strong positive correlation between the cytokine level and the degree of bone destruction.33 However, the findings of this study extend previous knowledge and seem to indicate that genetic variants of the IL-1α genes might be associated with complicated AOM at risk of negative evolution.
In this study, several associations between variants in genes encoding for factors of innate or adaptive immunity and the occurrence of rAOM were identified, which confirmed the role of genetics in conditioning susceptibility to AOM. Moreover, for the first time, an association between genetic variants of IL-10 and IL-1α and the risk of development of rAOM complicated by TMP was indicated. However, before these data can be used in clinical practice, linkage studies and genome-wide association studies might be useful to definitively solve the problem of the real role of genetic variants in conditioning susceptibility to rAOM and in the development of complicated cases.
Footnotes
Abbreviations: AOM = acute otitis media, BFIFA4P = BPI fold containing family A member 3 (BPIFA3) and 4P, CAPN14 = calpain 14, CI = confidence interval, DNAH5 = dynein axonemal heavy chain 5, FAS = Fas cell surface death receptor, FBXO11 = F-box protein 11, IFN = interferon, IL = interleukin, LTA = lymphotoxin α, MBL2 = mannose-binding lectin 2, MUC2 = mucin 2, MUC5AC/AB = mucin 5AC/AB, NOS2 = nitric oxide synthase 2, OME = otitis media with effusion, OR = odds ratio, PARP1 = poly(ADP-Ribose) polymerase-1, rAOM = recurrent acute otitis media, SCN1β = sodium channel voltage-gated type I beta subunit, serpine1 = serine protease inhibitors, SFTPD = surfactant protein D, SLC11A1 = proton-coupled divalent metal ion transporter member 1, SLFN5 = schlafen family member 5), SMAD2 = receptor-regulated SMAD2, SMAD4 = receptor-regulated SMAD4, SNPs = single nucleotide polymorphisms, TGF = transforming growth factor, TLRs = toll-like receptors, TMP = tympanic membrane perforation, TNF = tumor necrosis factor interferon-γ (IFN-γ), TP73 = tumor protein 73, TGFB1 = transforming growth factor β1.
The authors have no conflicts of interest to disclose.
This study was supported by a grant from the Italian Ministry of Health (Bando Giovani Ricercatori 2009 and Ricerca Corrente Grant 2014850/01).
REFERENCES
- 1.Daly KA, Rovers MM, Hoffman HJ, et al. Recent advances in otitis media. 1. Epidemiology, natural history, and risk factors. Ann Otol Rhinol Laryngol Suppl 2005; 194:8–15. [DOI] [PubMed] [Google Scholar]
- 2.Daly KA, Hoffman HJ, Kvaerner KJ, et al. Epidemiology, natural history, and risk factors: panel report from the Ninth International Research Conference on Otitis Media. Int J Pediatr Otorhinolaryngol 2010; 74:231–240. [DOI] [PubMed] [Google Scholar]
- 3.Casselbrant ML, Mandel EM, Fall PA, et al. The heritability of otitis media: a twin and triplet study. JAMA 1999; 282:2125–2130. [DOI] [PubMed] [Google Scholar]
- 4.Casselbrant ML, Mandel EM. Genetic susceptibility to otitis media. Curr Opin Allergy Clin Immunol 2005; 5:1–4. [DOI] [PubMed] [Google Scholar]
- 5.Mittal R, Robalino G, Gerring R, et al. Immunity genes and susceptibility to otitis media: a comprehensive review. J Genet Genomics 2014; 41:567–581. [DOI] [PubMed] [Google Scholar]
- 6.Casselbrant ML, Mandel EM, Jung J, et al. Otitis media: a genome-wide linkage scan with evidence of susceptibility loci within the 17q12 and 10q22.3 regions. BMC Med Genet 2009; 10:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rye MS, Warrington NM, Scaman ESH, et al. Genome-wide association study to identify the genetic determinants of otitis media susceptibility in childhood. PloS One 2012; 7:e48215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen EK, Chen WM, Weeks DE, et al. A Genome-wide association study of chronic otitis media with effusion and recurrent otitis media identifies a novel susceptibility locus on chromosome 2. J Assoc Res Otolaryngol 2013; 14:791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen EK, Manichaikul A, Sale MM. Genetic contributors to otitis media: agnostic discovery approaches. Curr Allergy Asthma Rep 2014; 14:411. [DOI] [PubMed] [Google Scholar]
- 10.Lee HY, Chung JH, Lee SK, et al. Toll-like receptors, cytokines & nitric oxide synthase in patients with otitis media with effusion. Indian J Med Res 2013; 138:523–530. [PMC free article] [PubMed] [Google Scholar]
- 11.Revai K, Patel JA, Grady JJ, et al. Association between cytokine gene polymorphisms and risk for upper respiratory tract infection and acute otitis media. Clin Infect Dis 2009; 49:257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rye MS, Wiertsema SP, Scaman ES, et al. Genetic and functional evidence for a role for SLC11A1 in susceptibility to otitis media in early childhood in a Western Australian population. Infect Genet Evol 2013; 16:411–418. [DOI] [PubMed] [Google Scholar]
- 13.Emonts M, Veenhoven RH, Wiertsema SP, et al. Genetic polymorphisms in immunoresponse genes TNFA, IL6, IL10, and TLR4 are associated with recurrent acute otitis media. Pediatrics 2007; 120:814–823. [DOI] [PubMed] [Google Scholar]
- 14.Sale MM, Chen WM, Weeks DE, et al. Evaluation of 15 functional candidate genes for association with chronic otitis media with effusion and/or recurrent otitis media (COME/ROM). PLoS One 2011; 6:e22297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bueno MT, Reyes D, Valdes L, et al. Poly(ADP-ribose) polymerase 1 promotes transcriptional repression of integrated retroviruses. J Virol 2013; 87:2496–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segade F, Daly KA, Allred D, et al. Association of the FBXO11 gene with chronic otitis media with effusion and recurrent otitis media: the Minnesota COME/ROM Family Study. Arch Otolaryngol Head Neck Surg 2006; 132:729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fliegauf M, Olbrich H, Horvath J, et al. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am J Respir Crit Care Med 2005; 171:1343–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bossaller L, Chiang PI, Schmidt-Lauber C, et al. FAS mediates non-canonical IL-1β and IL-18 maturation via caspase-8 in a Rip3-independent manner. J Immunol 2012; 189:5508–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harslund J, Frees D, Leifsson PS, et al. The role of Serpine-1 and tissue inhibitor of metalloproteinase type-1 in early host responses to Staphylococcus aureus intracutaneous infection of mice. Pathog Dis 2013; 68:96–104. [DOI] [PubMed] [Google Scholar]
- 20.Tateossian H, Morse S, Parker A, et al. Otitis media in the Tgif knockout mouse implicates TGFβ signalling in chronic middle ear inflammatory disease. Hum Mol Genet 2013; 22:2553–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentile DA, Doyle WJ, Zeevi A, et al. Cytokine gene polymorphisms moderate illness severity in infants with respiratory syncytial virus infection. Hum Immunol 2003; 64:338–344. [DOI] [PubMed] [Google Scholar]
- 22.Ilia S, Goulielmos GN, Samonis G, et al. Polymorphisms in IL-6, IL-10, TNF-α, IFN-γ and TGF-β1 genes and susceptibility to acute otitis media in early infancy. Pediatr Infect Dis J 2014; 33:518–521. [DOI] [PubMed] [Google Scholar]
- 23.Alper CM, Winther B, Hendley JO, et al. Cytokine polymorphisms predict the frequency of otitis media as a complication of rhinovirus and RSV infections in children. Eur Arch Otorhinolaryngol 2009; 266:199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nokso-Koivisto J, Chonmaitree T, Jennings K, et al. Polymorphisms of immunity genes and susceptibility to otitis media in children. PLoS One 2014; 9:e93930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormick DP, Grady JJ, Diego A, et al. Acute otitis media severity: association with cytokine gene polymorphisms and other risk factors. Int J Pediatr Otorhinolaryngol 2011; 75:708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carroll SR, Zald PB, Soler ZM, et al. Innate immunity gene single nucleotide polymorphisms and otitis media. Int J Pediatr Otorhinolaryngol 2012; 76:976–979. [DOI] [PubMed] [Google Scholar]
- 27.Smirnova MG, Birchall JP, Pearson JP. In vitro study of IL-8 and goblet cells: possible role of IL-8 in the aetiology of otitis media with effusion. Acta Otolaryngol 2002; 122:146–152. [DOI] [PubMed] [Google Scholar]
- 28.Kaur R, Casey J, Pichichero M. Cytokine, chemokine, and toll-like receptor expression in middle ear fluids of children with acute otitis media. Laryngoscope 2015; 125:E39–E44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segal N, Givon-Lavi N, Leibovitz E, et al. Acute otitis media caused by Streptococcus pyogenes in children. Clin Infect Dis 2005; 41:35–41. [DOI] [PubMed] [Google Scholar]
- 30.Marchisio P, Consonni D, Baggi E, et al. Vitamin D supplementation reduces the risk of acute otitis media in otitis-prone children. Pediatr Infect Dis J 2013; 32:1055–1060. [DOI] [PubMed] [Google Scholar]
- 31.Marchisio P, Esposito S, Bianchini S, et al. Efficacy of injectable trivalent virosomal-adjuvanted inactivated influenza vaccine in preventing acute otitis media in children with recurrent complicated or noncomplicated acute otitis media. Pediatr Infect Dis J 2009; 28:855–859. [DOI] [PubMed] [Google Scholar]
- 32.Joki-Erkkilä VP, Puhakka H, Hurme M. Cytokine gene polymorphism in recurrent acute otitis media. Arch Otolaryngol Head Neck Surg 2002; 128:17–20. [DOI] [PubMed] [Google Scholar]
- 33.Kaur R, Casey J, Pichichero M. Cytokine, chemokine, and Toll-like receptor expression in middle ear fluids of children with acute otitis media. Laryngoscope 2015; 125:E39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]


