Significance
Sex chromosomes have repeatedly evolved in animals and plants, but the evolutionary forces driving this process are not entirely understood. Nonrecombining Y chromosomes undergo rapid loss of functional genes in animals; however, it remains unclear whether this holds true in plants. We report the first genome sequence-based analysis of sex chromosomes in white campion, to our knowledge, which evolved large sex chromosomes only 10 million years ago. We demonstrate that the Y chromosome has lost nearly half its functional genes, at a rate of Y degeneration comparable to that of animal Y chromosomes. This degeneration is accommodated for by highly variable dosage compensation. Our results resolve the puzzling discrepancy in evolutionary trajectories of sex chromosomes between the plant and animal kingdoms.
Keywords: sex chromosome evolution, Y degeneration, gene expression, dosage compensation, plants
Abstract
The nonrecombining regions of animal Y chromosomes are known to undergo genetic degeneration, but previous work has failed to reveal large-scale gene degeneration on plant Y chromosomes. Here, we uncover rapid and extensive degeneration of Y-linked genes in a plant species, Silene latifolia, that evolved sex chromosomes de novo in the last 10 million years. Previous transcriptome-based studies of this species missed unexpressed, degenerate Y-linked genes. To identify sex-linked genes, regardless of their expression, we sequenced male and female genomes of S. latifolia and integrated the genomic contigs with a high-density genetic map. This revealed that 45% of Y-linked genes are not expressed, and 23% are interrupted by premature stop codons. This contrasts with X-linked genes, in which only 1.3% of genes contained stop codons and 4.3% of genes were not expressed in males. Loss of functional Y-linked genes is partly compensated for by gene-specific up-regulation of X-linked genes. Our results demonstrate that the rate of genetic degeneration of Y-linked genes in S. latifolia is as fast as in animals, and that the evolutionary trajectories of sex chromosomes are similar in the two kingdoms.
In birds and mammals, sex is determined by sex chromosomes that arose more than 100 and 150 million years ago, respectively (1, 2). In contrast, separate sexes (dioecy) and sex chromosomes have evolved more recently in plants on multiple occasions (3, 4). Few of these dioecious species are known to have cytologically distinguishable (heteromorphic) sex chromosomes [e.g., Silene latifolia or Rumex hastatulus (5)]. Other dioecious plants, such as papaya (6) and persimmon (7), have very small nonrecombining sex-determining regions on homomorphic sex chromosomes. Recombination on the oldest section of the papaya Y chromosome ceased circa 7 million years ago (8). The large heteromorphic sex chromosomes of S. latifolia or R. hastatulus are slightly older, with the oldest parts being circa 10 and 15–16 million years old, respectively (9, 10). Because of their recent origin, plant sex chromosomes are particularly informative about the early stages of sex chromosome evolution.
A distinguishing feature shared by independently evolved sex chromosomes is the presence of a region of suppressed recombination on a sex-specific chromosome in the heterogametic sex (11). These nonrecombining regions tend to expand over evolutionary time (12), resulting in the formation of “evolutionary strata.” Greater levels of sequence divergence between homologous X- and Y-linked sequences are expected in older strata (1, 13, 14). Genes in nonrecombining regions are expected to gradually degenerate as complete linkage of multiple genes renders purifying selection less effective, leading to an accumulation of deleterious mutations and gene loss (15). This genetic degeneration has been observed in many animal species (11), whereas studies on plants have revealed less genetic degeneration (8, 16–19). Slower degeneration of plant Y-linked genes may be a result of stronger purifying selection acting at the haploid stage of the plant life cycle (18, 20, 21). Alternatively, it may be a result of undersampling of degenerate, weakly expressed genes in previous studies that were based on transcriptome sequencing (16–19, 22).
In males, reduced expression or complete loss of Y-linked genes is expected to result in an imbalance of transcripts (23); this may have negative consequences, as in aneuploids (24, 25). For some genes, lower expression in males may be tolerated, or the X-linked allele (gametolog) may be up-regulated to restore the balance (dosage compensation) (26). In organisms with old sex chromosomes, different types of chromosome-wide dosage compensation systems have evolved to rebalance gene expression in the heterogametic sex (27). Flies overexpress X-linked genes in males to match the X to autosome ratio in females (28). Mammals overexpress X-linked genes in both sexes and shut down expression from one of the X chromosomes in females (29, 30). In younger plant sex chromosomes, the pattern is much less clear. In S. latifolia, two studies reported no dosage compensation for X-linked genes that lacked expressed Y gametologs (18, 22), and a third study of expressed X- and Y-linked genes suggested that dosage compensation has evolved (16).
Large-scale analyses of plant sex chromosomes have generally focused on transcriptome sequencing (16–19). A notable exception to this is papaya, for which sequencing of the hermaphroditic, nonrecombining Yh-chromosome revealed faster accumulation of repetitive sequences than on the X chromosome, but similar rates of gene loss (8, 31). The rate of genetic degeneration is dependent on the number of genes linked together in nonrecombining regions (15, 32). With a larger number of functional genes linked in a nonrecombining region, more mutations can occur in that region that contribute to Hill-Robertson interference and background selection (11, 33). This leads to a reduction in the effective population size and more rapid accumulation of deleterious mutations via Muller’s ratchet (11, 15). Small nonrecombining regions, such as those found on papaya’s homomorphic Y chromosome, may contain too few genes for genetic degeneration to reach an appreciable rate. Thus, it is essential to study the genomes of plant species with larger heteromorphic sex chromosomes, such as those found in S. latifolia (34).
The sex chromosomes of white campion (S. latifolia) have long been the subject of study (5, 35). Most species in Silene are not dioecious, indicating that separate sexes and sex chromosomes have evolved de novo in Silene section Elisanthe (which includes S. latifolia) (36). Comparative genetic-mapping studies of S. latifolia and the nondioecious Silene vulgaris demonstrated that the sex chromosomes are derived from a single pair of autosomes (37, 38). Population genetic studies have demonstrated that genetic degeneration of S. latifolia Y-linked genes has already begun (39, 40), but recent transcriptome sequencing work indicated that this degeneration is relatively slow (16–18). With no genome sequence available, these transcriptome-based studies were limited to the analysis of actively expressed genes, potentially underestimating the extent of genetic degeneration. The current study overcomes this limitation by using a combination of genome and transcriptome sequencing of S. latifolia males and females. This revealed an extensive loss of Y-linked genes, resolving the apparent discrepancy between animal and plant sex chromosomes reported in the previous studies. Furthermore, the current study resolves the debate over the presence of dosage compensation in S. latifolia, revealing the extent to which loss of Y-expression is compensated for by X-gametolog up-regulation.
Results
Genetic Mapping.
To identify sex-linked genes and construct a high-density genetic map, we generated 126 Gb of paired-end transcriptome sequence data for the parents and 52 F2 progeny (SI Appendix, Table S1) and analyzed segregation of single-nucleotide polymorphisms (SNPs) in that genetic cross (see SI Appendix, Materials and Methods and Fig. S1 for details of segregation patterns used). The genetic map included 2,113 genes and was 1,009 cM long (Dataset S1). As expected, the map contained 12 linkage groups, corresponding to 11 autosomes and the sex chromosomes (SI Appendix, Figs. S2 and S3). The X chromosome was the longest (114 cM) and had the largest number of mapped genes (326 genes). Anchoring the genomic scaffolds to the genetic map (see following) brought the total number of mapped X-linked genes to 412 (total coding sequence length, 536.8kb). A further 629 genes identified as sex-linked by their segregation pattern could not be mapped to specific positions on the X chromosome either because of more than 20% missing data in the F2 progeny (45 genes) or because only segregation analysis in the male map was possible with SNPs heterozygous in the F1 male and homozygous in the F1 female (584 genes). Analysis of recombination between the 412 SNPs that were heterozygous in the male F1 parent revealed that 126 genes at one end of the X chromosome showed recombination during male meiosis, and hence were pseudoautosomal. The remaining 286 genes that mapped to the X chromosome were completely linked in male meiosis, but displayed considerable recombination (64 cM) in female meioses.
Genome Sequencing.
As no genome sequence was available for S. latifolia or its close relatives, we generated 165 Gb of genomic sequence data (∼60-fold coverage) from a highly inbred line of this species (41) (SI Appendix, Table S1). To avoid problems with X–Y chimeric assemblies, which are possible for a genome with low divergence between these chromosomes, only female sequence data were used for a draft S. latifolia genome. The assembly comprised 307,782 scaffolds (range, 0.2–200 kb; with half of the assembly in scaffolds no shorter than 3,519 bp), with a total length of 665 Mb (SI Appendix, Table S2), or 25% of the S. latifolia genome [2,641 Mb (42)]. Annotation of the draft genome resulted in the identification of 13,711 protein coding genes with a total length of 52.3 Mb. A test of completeness of the genome annotation (43) revealed that 71% of the 248 ultraconserved core eukaryotic genes were present. Genetically mapped genes found on the genomic contigs allowed us to construct a hybrid physical/genetic map including 1,301 genomic scaffolds with total length of 26.4 Mb (Dataset S2). Of these scaffolds, 182 were anchored to the X chromosome, 131 of which were located in the nonrecombining portion and 51 in the pseudoautosomal region (PAR; total length, 2.5 and 1.1 Mb, respectively). The average length of the anchored scaffolds was considerably larger than the genome average (20.3 and 2.2 kb, respectively), as longer scaffolds are more likely to contain markers used in genetic mapping (SI Appendix, Table S2).
Reconstruction of Homologous X- and Y-Linked Gene Pairs.
Computational “subtraction” of the female genome from the sequences of the male genome allowed us to reconstruct 140.94 Mb of male-specific, putatively Y-linked contigs. Using the sequences of X-linked genes, we searched for Y-linked gametologs in male-specific contigs, which allowed us to reconstruct the X- and Y-linked gametolog pairs for 931 genes (total coding sequence length, 1.07 Mb), including 201 genes with known positions in the genetic map. The accuracy of this approach was confirmed by the close correspondence of previously published (10, 18) and newly reconstructed Y-linked genes (average number of differences per nucleotide, π = 0.0038). The gene ontology annotations for these genes reveal a range of functions (Dataset S3), with no significant enrichment for any specific gene ontology terms on the sex chromosomes [false discovery rate (FDR) < 0.1]. No sex-linked genes showed homology to transposable elements. Given the size of the sex chromosomes (Y is the largest and X is the second largest in the genome), our set of sex-linked genes is probably far from complete. Nevertheless, analysis of this gene set is highly informative about the evolution of sex chromosomes.
Divergence Between X and Y Chromosomes.
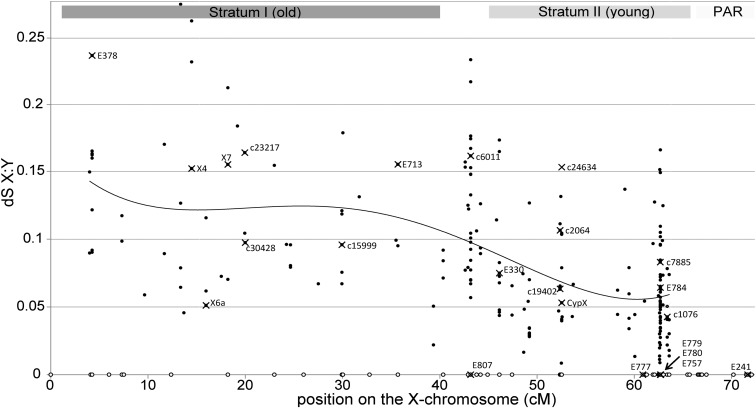
Synonymous divergence between X and Y gametologs ranged from 0 to 0.32 (median dS = 0.061), whereas nonsynonymous divergence ranged from 0 to 0.095 (median dN = 0.017) (SI Appendix, Fig. S4). To investigate the existence of “evolutionary strata” in S. latifolia, we used the set of 201 mapped X-linked genes and their reconstructed Y gametologs. Consistent with the strata hypothesis, silent X:Y divergence (dSxy) in the region of the X chromosome proximal to the PAR was indeed lower than in the distal region (Fig. 1). However, variation in synonymous X:Y-divergence is extensive all over the X chromosome, which makes it difficult to unambiguously delimit distinct strata. For consistency with the previously published lower-density map of the X chromosome (38), we marked the approximate positions of strata I and II, using the location of genes E713 and E330 (Fig. 1). In our map, the older stratum I is estimated to span 0–40 cM, and the younger stratum II is estimated to span 45–64 cM. To allow for uncertainty in the exact boundary position, genes located between 40 and 45 cM were not included in either stratum.
Fig. 1.
Synonymous divergence between homologous X- and Y-linked genes plotted against position in the genetic map of the X chromosome. The PAR extends to the right of this plot, from 64 to 114 cM. The curve is a fourth-order polynomial fitted to the data. Mapped X-linked genes with no homologous reconstructed Y-linked copy are shown as open circles on the horizontal axis. Data points corresponding to genes that were mapped in previous studies (22, 38) are represented by crossed circles and labeled with gene names used previously. Gray rectangles at the top signify approximate locations of the evolutionary strata, as defined in ref. 38.
Genetic Degeneration of Y-Linked Genes.
A significantly higher number of ORFs were broken by stop codons on the Y compared with the X chromosome (210 and 12, respectively; G-test, P < 0.00001; SI Appendix, Table S3). The same holds true for the subset of sex-linked genes included in the genetic map (SI Appendix, Table S4). The proportion of Y-linked genes interrupted by stop codons increased with silent X:Y divergence (i.e., time), going from 13% in the 100 genes with the least X:Y divergence (dSxy < 3%) to 31% in the 100 most-diverged genes (dSxy > 12%). However, there was no significant difference in the number of stop codons accumulated in Y-linked gametologs (27 and 45 stop codons) between the two strata after correction for the total number of codons in the genes of each stratum (24,121 and 52,711 codons). In contrast to degeneration observed on the Y, genetically mapped X-linked genes contained only one premature stop codon in the older stratum, and no stop codons in the younger stratum. There was no significant difference between the strata in the number of X-linked genes that lacked Y gametologs (i.e., X-only genes), with 22% and 30% of such genes present in the old (I) and young (II) strata, respectively (contingency χ2, P > 0.05).
To compare the rates of nonsynonymous (dN) and synonymous (dS) substitutions on the X and Y chromosomes, we used S. vulgaris (Sv), a nondioecious relative of S. latifolia, as an outgroup for decomposing X:Y divergence into divergence accumulated in the X- and Y-linked gene copies. Analysis of 841,383 gap-free positions in tripartite alignments (X:Y:Sv) revealed significantly (P < 10−5) more nonsynonymous changes in Y-linked over X-linked genes (8,853 and 3,118, respectively), after correction for the number of synonymous substitutions (7,991 and 6,439, respectively; SI Appendix, Table S3). This is consistent with a reduced efficacy of purifying selection on the Y chromosome. Correspondingly, median dN/dS was much higher for the Y-linked genes compared with all other classes (SI Appendix, Fig. S5A). Significantly higher dN/dS for Y-linked genes held true in both the older and the younger strata (SI Appendix, Fig. S5B). Neither the X-linked nor the Y-linked genes showed significant differences in dN/dS between the two strata.
Expression of S. latifolia Sex-Linked Genes.
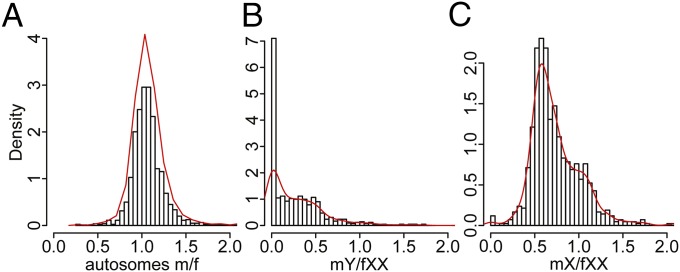
We used transcriptome sequence data from 22 male and 35 female S. latifolia individuals (SI Appendix, Table S1) to analyze the expression of X- and Y-linked genes (SI Appendix, Materials and Methods). Overall, expression is highly correlated between males and females for both the autosomal (r = 0.986) and X-linked (r = 0.961) genes. For autosomal genes, the distribution of male to female expression ratios is centered on 1 (mode = 1.03; median = 1.06), denoting near-equal transcript abundance in the two sexes (Fig. 2A). In contrast, Y-linked genes show drastically reduced expression, with many genes having zero or nearly zero expression (Fig. 2B; mode = 0.03; median = 0.19). For X-linked genes, the distribution is centered close to mX/fXX = 0.5 expected from the dosage difference between males (X) and females (XX) (Fig. 2C; mode = 0.58; median = 0.67). However, a “shoulder” is apparent in the distribution, with a substantial proportion of genes showing equal or nearly equal expression of X-linked genes in the two sexes (mX/fXX ∼1).
Fig. 2.
Distribution of relative transcript abundance in S. latifolia males and females. (A) Relative male (m) versus female (f) expression for autosomal genes with known positions in the genetic map (1,657 genes). (B) Expression of Y-linked genes in males normalized by expression of X-linked gametologs in females (866 genes). (C) Expression of X-linked genes in males relative to females (848 genes). The curve shows the kernel-smoothed density function.
To analyze gene expression in X- and Y-linked genes, we used expression in the nondioecious S. vulgaris that has no sex chromosomes, providing a proxy for ancestral expression of sex-linked genes in S. latifolia. Although overall gene expression was very similar between S. vulgaris and both S. latifolia sexes (SI Appendix, Fig. S6), sex-linked genes showed reduced expression in S. latifolia males compared with females, and loss of expression from Y-linked genes appears to account for most of this reduction (SI Appendix, Fig. S7). Degeneration of gene expression of Y-linked genes is clearly apparent in both evolutionary strata (SI Appendix, Fig. S8A) and in the genes from the lower and upper quartiles of synonymous divergence between the sex chromosomes (SI Appendix, Fig. S8B). Expression of the sex-linked compared with autosomal genes within a sex is arguably more biologically relevant than expression differences between the sexes (e.g., see refs. 23 and 26). The X/A expression ratio in females is 0.984, indicating similar levels of X and A expression in S. latifolia. If Y degeneration and X dosage compensation are absent, we expect both Y/A and X/A ratios to be 0.5 in males. However, in males, Y/A = 0.183 and X/A = 0.656, an indication of both Y degeneration and slightly higher expression of X-linked genes in males than is expected in the absence of dosage compensation.
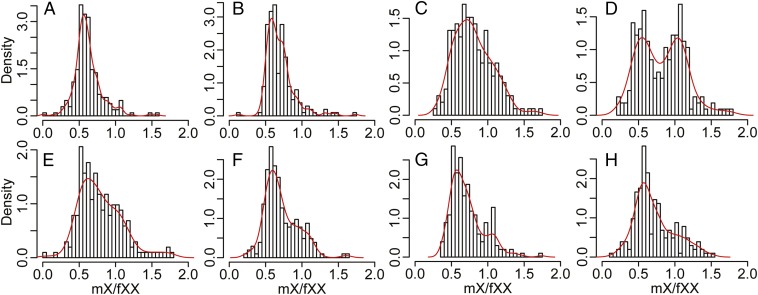
To assess whether dosage compensation is operating, X expression data were partitioned on the basis of the loss of male Y (mY) expression compared with the ancestral expression inferred from Sv (Fig. 3 A–D). For genes where the Y gametolog was still actively expressed (mY/Sv > 0.256), expression of X-linked genes in males was around half that in females (Fig. 3 A and B), which is consistent with twofold larger gene dosage for X-linked genes in females. With decreasing mY expression compared with the inferred ancestral level (mY/Sv ≤ 0.256), the distribution of relative male to female expression of X-linked genes (mX/fXX) becomes wider (Fig. 3C). A bimodal distribution is apparent for gametologs for which no mY expression was detected (Fig. 3D; dip test for unimodality; D = 0.0421, P = 0.007106). Thus, a subset of X-linked genes showed little to no dosage compensation (mX/fXX peak centered at ∼0.5; Fig. 3D), whereas the other subset showed near-complete dosage compensation (mX/fXX peak centered at ∼1; Fig. 3D).
Fig. 3.
Probability density histograms of relative transcript abundance for X-linked genes in S. latifolia males and females (mX/fXX). The data are partitioned according to transcript abundance for Y-linked genes in S. latifolia males (mY) compared with S. vulgaris (Sv) (A–D) and quartiles for synonymous divergence (dS) between homologous X- and Y-linked genes (E–H). (A) Genes with mY/Sv ≥ 0.667 (176 genes); (B) 0.667 > mY/Sv ≥ 0.256 (201 genes); (C): 0.256 > mY/Sv ≥ 0.0001 (250 genes); and (D): mY = 0 (233 genes). (E) Includes the genes with dS ≤ 0.038 (225 genes); (F) 0.038 < dS ≤ 0.0602 (218 genes); (G) 0.0602 < dS ≤ 0.0975 (212 genes); and (H) dS > 0.0975 (216 genes). The curve shows the kernel-smoothed density function.
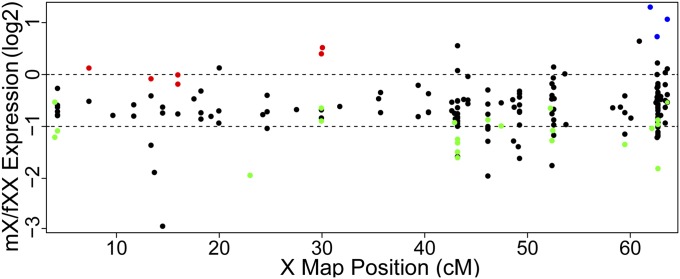
We performed univariate Gaussian mixed model analyses of mX/fXX expression ratios to determine whether distinct populations of dosage-compensated and noncompensated genes were present among the sex-linked genes. Three distinct clusters were detected among X-linked genes without Y-gametolog expression (SI Appendix, Fig. S9). One cluster, signifying genes with no dosage compensation, was centered at an mX/fXX ratio of 0.54 (n = 99; σ2 = 0.017). Another cluster, comprising putatively dosage-compensated genes, was centered at 1.03 (n = 108; σ2 = 0.026). The remaining 12 genes, with considerably higher mX compared with fXX expression, formed the third cluster (SI Appendix, Fig. S9). Genes in these clusters belong to a range of functions (Dataset S3) and show no significantly over- or under-represented gene ontology annotations. Interestingly, all of the dosage-compensated genes with known map locations are present only in the older stratum, whereas genes with high mX/fXX expression are all next to the PAR boundary, although the number of mapped genes in these categories is very small (Fig. 4). Three groups were also detected among the X-linked genes with Y-gametolog expression (SI Appendix, Fig. S10). However, it is less clear whether these correspond to clusters of dosage compensated and noncompensated genes (mX/fXX peaks center at 0.60, 0.83, and 2.57), and the uncertainty in assigning genes to clusters is high (SI Appendix, Fig. S10D).
Fig. 4.
Relative transcript abundance in males and females for genes with known genetic positions on S. latifolia X chromosome. Expression ratios are shown for sex-linked genes with detectable expression (>0) (black points) or no expression (colored points) of the Y gametolog in males. Colors denote Gaussian mixed-model clusters based on mX/fXX expression ratios. Red and green points correspond to putatively dosage compensated and noncompensated genes, respectively. Blue points denote genes falling in the small cluster with high mX/fXX expression ratio. Dotted lines mark the null expectation for complete dosage compensation [log2(mX/fXX) = 0] and no compensation [log2(mX/fXX) = −1].
To assess the rapidity by which dosage compensation can arise, the comparison of mX expression with fXX expression was repeated, but the data were partitioned according to levels of X:Y silent site divergence. For each of the partitions, the distribution of mX/fXX relative expression was similar (Fig. 3 E–H). Therefore, dosage compensation mechanisms appear to not be strongly dependent on the amount of X:Y divergence. At least partial dosage compensation could be achieved via preexisting transcriptional buffering in gene networks (26). However, if up-regulation of X-linked genes evolved to compensate for dysfunctional Y-linked copies, then X up-regulation should also be observed for actively expressed Y-linked genes broken by premature stop codons. We observed that the presence of premature stop codons in Y-linked genes does not significantly correlate with expression of X-linked gametologs (SI Appendix, Fig. S11).
Discussion
Our study combined high-throughput genome and transcriptome sequencing to uncover the evolution of heteromorphic plant sex chromosomes. Analysis of S. latifolia sex-linked genes revealed a massive accumulation of stop codons and widespread reduction in expression of Y-linked genes. The rate of genetic degeneration was comparable to that observed in bird (1), mammal (44), and Drosophila (45) sex chromosomes. Our genetic map demonstrates that degeneration is extensive even in the youngest parts of the Y chromosome. This study compared gene expression of S. latifolia sex-linked genes with ancestral expression in an outgroup species without sex chromosomes, revealing dosage compensation only in a fraction of X-linked genes.
Degeneration of Y-Linked Genes.
Only two cases of independent heteromorphic sex chromosome evolution in vascular plants have been studied so far: S. latifolia and R. hastatulus. Two further cases were analyzed in bryophytes (46, 47). The previous large-scale analyses of S. latifolia and R. hastatulus were based on transcriptome sequencing (16–19), which is likely to have resulted in analyses being biased toward sex-linked genes with actively expressed Y-linked copies. Using genome sequence data from both sexes in S. latifolia, we were able to analyze sex-linked genes, regardless of their expression level. This revealed many more severely degenerated Y-linked gametologs compared with that observed in previous transcriptome-only studies (16–18). In particular, 48% of Y-linked gametologs in the current genome-based analysis had 10-fold reduced expression, and 65% were expressed at least threefold lower compared with X-linked gametologs. In contrast, for genes reported in the previous transcriptome-based analyses in this species (17, 18), less than 2% of the Y-linked genes had a 10-fold reduction, and only about 6% had a threefold reduction. Furthermore, the weakly expressed or nonexpressed Y-linked genes in the current study were twice as likely to be interrupted by premature stop codons relative to the entire set of sex-linked genes. Most of these weakly expressed genes would have been missed in the previous transcriptome-based analyses. As a result of this difference, the number of Y-linked genes interrupted by stop codons was much higher in the current genome-based analysis, at 23%, compared with 8% in ref. 18 or 1% in ref. 17. X-linked and autosomal genes contained very few stop codons in both this and the previous studies. This contrast between the Y- and non-Y-linked genes is unlikely to be a result of the difference in assembly strategies or poor gene models, as newly assembled sequences for known genes closely match previously published sequences.
Differences and uncertainty in both contemporary and ancestral generation times complicate direct comparisons of the rate of Y degeneration between S. latifolia and other species. Assuming that S. latifolia sex chromosomes originated 10 million years ago (35), as well as generation times of 10, 1, and 0.2 per year for Drosophila, Silene, and primates, respectively, the sex (or neo-sex) chromosomes in these groups evolved (or youngest primate strata formed) 8–10 million generations ago. The extent of Y degeneration in Silene is similar to that found in the extensively studied neo-Y chromosome of Drosophila miranda (48), in which roughly 40% of genes have lost their function in the last million years. Even more extensive Y degeneration (∼75% gene loss) was reported for the youngest evolutionary strata (strata 4 and 5) of human sex chromosomes, which evolved 30–40 million years ago (44). The ∼40% faster rate of gene loss from the most recent strata in primates may stem from smaller population sizes, which speed up degenerative processes, such as Muller’s ratchet (49). Rates of gene loss from bird W chromosomes are comparable to Y degeneration in humans (figure 4i in ref. 1). More recent origins have been proposed for S. latifolia chromosomes (50), bringing the rate of degeneration in line with that in primates and birds. However, a more precise age estimate for S. latifolia sex chromosomes is needed before we can be confident that Y degeneration in Silene is as rapid as the fastest rates in animals.
Dosage Compensation.
We found that decreased Y expression correlates with a general increase in expression of X gametologs, consistent with dosage compensation (16). However, at the end of this continuum, where Y gametologs have no detectable expression, it is clear that not all X gametologs undergo a compensatory increase in expression. A bimodal distribution was apparent (Fig. 3D), with a subset of X-linked genes displaying dosage compensation, whereas the other genes showed no increase in transcription. This variable response to the loss of Y expression may explain both increased variance in X expression in males (16) and the lack of clear dosage compensation in some S. latifolia genes that have no actively expressed Y gametolog (18, 22). Some X-linked genes do not show a compensatory increase in expression, and this may be because reduced expression of these genes is of little detriment to males. It is still unclear whether dosage compensation is acting across large portions of the X. The six mapped genes that showed compensatory up-regulation of the X copy were all in the older stratum of the X chromosome (Fig. 4). This result runs counter to the observation that dosage compensation was independent of X:Y coding sequence divergence. However, the apparent clustering of up-regulated X gametologs might reflect the influence of factors unrelated to X:Y divergence, such as localized epigenetic alterations. Noncompensated genes were located in both old and young strata of the X chromosome (Fig. 4). Therefore, the possibility still remains for dosage compensation to be occurring on a gene-by-gene basis, as has been reported for other systems [eg, Schistosoma mansoni (51), Plodia interpunctella (52), and Ficedula albicollis (53); see also ref. 23].
Levels of dosage compensation were not correlated with silent X:Y-divergence, indicating that compensatory up-regulation of X-genes evolves rapidly or may be caused by preexisting mechanisms and does not have to evolve. Dosage compensation mechanisms are known to be present in species that have no sex chromosomes (24, 25). Compensatory up-regulation could be achieved via transcriptional buffering because of regulatory interactions in gene networks (26). Indeed, deletion of one gene copy in Drosophila does not necessarily lead to a 50% reduction in gene expression; rather, the average hemizygous gene expression is about two-thirds normal expression (54). If expression of a Y-linked gametolog was lost through degeneration of cis-regulatory regions, an increased availability of transcription factors may promote X-gametolog expression. Such transcriptional buffering mechanisms may explain why X up-regulation in S. latifolia was observed only for genes with reduced Y-expression, and not for genes truncated by stop codons.
Conclusions
Our analyses of white campion demonstrate that the previous finding of slower Y degeneration in plants compared with animals was an artifact of transcriptome-only analyses. The use of genomic sequencing in our study circumvented this bias and revealed a much greater extent of genetic degeneration in S. latifolia Y-linked genes than observed previously. We have demonstrated that Y degeneration is extensive in both older and younger evolutionary strata, indicating that degeneration proceeds rapidly once recombination is shut down, as in animals. Furthermore, our expression analyses confirmed the presence of incomplete gene-specific dosage compensation in S. latifolia, but it remains unclear whether X-up-regulation is a result of preexisting transcriptional buffering mechanisms or evolved in response to the loss of Y-expression. Our results resolve questions regarding the pace of genetic degeneration of plant Y chromosomes (18, 22, 33) and dosage compensation in S. latifolia (16, 18, 22). Further work will reveal whether genetic degeneration in other plants with heteromorphic sex chromosomes keeps pace with that in animals.
Materials and Methods
For detailed methods description, see SI Appendix Materials and Methods.
Genome Sequencing and Assembly.
Genomic sequencing was conducted for one female and one male from a highly inbred line (41), using paired-end, Illumina HiSeq and MiSeq (SI Appendix, Table S1). Only the female sequence data were used for the de novo assembly of the female draft genome. To identify male-specific regions, we used cortex_var (55) to compare male and female genome sequences.
Genetic Map.
Mapping was performed in an F2 cross between S. latifolia male (mSa984) and female (fSa985) individuals. RNA was extracted from actively growing shoots with flower buds for grandparental (F0) and parental (F1) males and females, as well as 52 F2 progeny (20 males and 32 females). For each individual, a separate RNAseq library was prepared and sequenced (SI Appendix, Table S1). After genotyping with samtools v1.1, SNPs that either were homozygous and different between F0s or were heterozygous in either F1 parent were selected and used to identify sex-linked contigs (SI Appendix, Fig. S1), determine linkage groups (R/qtl), and perform genetic mapping (Joinmap v4.1). To construct a hybrid physical/genetic map, genetically mapped genes were located on the genomic contigs with blat.
Gene Expression and Sequence Divergence Analyses.
Gene expression was measured from RNAseq data for the individuals from the genetic cross (including parents, F1 and F2 progeny) and three S. vulgaris individuals, using RSEM (56). Sequence divergence was calculated from pairwise alignments of autosomal genes (S. latifolia + S. vulgaris) and tripartite alignments of genes that are sex-linked in S. latifolia (X + Y + S. vulgaris). Gaussian mixed model analysis was performed using the mclust package in R (57).
Supplementary Material
Acknowledgments
We are grateful to Yusuke Kazama for providing S. latifolia seeds, Zamin Iqbal and Bruno Nevado for their advice on computational analyses, and Wellcome Trust Centre for Human Genetics (Oxford, United Kingdom) staff for Illumina sequencing. This work was funded by Biotechnology and Biological Sciences Research Council, United Kingdom Grant BB/K016539/1 (to D.A.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database, (accession nos. PRJNA289891, PRJNA289919, PRJNA290193).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508454112/-/DCSupplemental.
References
- 1.Zhou Q, et al. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science. 2014;346(6215):1246338. doi: 10.1126/science.1246338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veyrunes F, et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008;18(6):965–973. doi: 10.1101/gr.7101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renner SS, Ricklefs RE. Dioecy and its correlates in the flowering plants. Am J Bot. 1995;82(5):596–606. [Google Scholar]
- 4.Charlesworth D. Plant sex chromosome evolution. J Exp Bot. 2013;64(2):405–420. doi: 10.1093/jxb/ers322. [DOI] [PubMed] [Google Scholar]
- 5.Westergaard M. The mechanism of sex determination in dioecious flowering plants. Adv Genet. 1958;9:217–281. doi: 10.1016/s0065-2660(08)60163-7. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z, et al. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature. 2004;427(6972):348–352. doi: 10.1038/nature02228. [DOI] [PubMed] [Google Scholar]
- 7.Akagi T, Henry IM, Tao R, Comai L. Plant genetics. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science. 2014;346(6209):646–650. doi: 10.1126/science.1257225. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, et al. Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc Natl Acad Sci USA. 2012;109(34):13710–13715. doi: 10.1073/pnas.1207833109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navajas-Pérez R, et al. The evolution of reproductive systems and sex-determining mechanisms within rumex (polygonaceae) inferred from nuclear and chloroplastidial sequence data. Mol Biol Evol. 2005;22(9):1929–1939. doi: 10.1093/molbev/msi186. [DOI] [PubMed] [Google Scholar]
- 10.Bergero R, Forrest A, Kamau E, Charlesworth D. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: Evidence from new sex-linked genes. Genetics. 2007;175(4):1945–1954. doi: 10.1534/genetics.106.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachtrog D. Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 2013;14(2):113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 2009;24(2):94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286(5441):964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 14.Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. Comparative sex chromosome genomics in snakes: Differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 2013;11(8):e1001643. doi: 10.1371/journal.pbio.1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355(1403):1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muyle A, et al. Rapid de novo evolution of X chromosome dosage compensation in Silene latifolia, a plant with young sex chromosomes. PLoS Biol. 2012;10(4):e1001308. doi: 10.1371/journal.pbio.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergero R, Charlesworth D. Preservation of the Y transcriptome in a 10-million-year-old plant sex chromosome system. Curr Biol. 2011;21(17):1470–1474. doi: 10.1016/j.cub.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 18.Chibalina MV, Filatov DA. Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr Biol. 2011;21(17):1475–1479. doi: 10.1016/j.cub.2011.07.045. [DOI] [PubMed] [Google Scholar]
- 19.Hough J, Hollister JD, Wang W, Barrett SC, Wright SI. Genetic degeneration of old and young Y chromosomes in the flowering plant Rumex hastatulus. Proc Natl Acad Sci USA. 2014;111(21):7713–7718. doi: 10.1073/pnas.1319227111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bull JJ. Evolution of Sex Determining Mechanisms. Benjamin/Cummings Publ.; San Francisco, CA: 1983. [Google Scholar]
- 21.Charlesworth D. Plant sex chromosomes. Genome Dyn. 2008;4:83–94. doi: 10.1159/000126008. [DOI] [PubMed] [Google Scholar]
- 22.Bergero R, Qiu S, Charlesworth D. Gene loss from a plant sex chromosome system. Curr Biol. 2015;25(9):1234–1240. doi: 10.1016/j.cub.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Mank JE, Hosken DJ, Wedell N. Some inconvenient truths about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution. 2011;65(8):2133–2144. doi: 10.1111/j.1558-5646.2011.01316.x. [DOI] [PubMed] [Google Scholar]
- 24.Birchler JA, Newton KJ. Modulation of protein levels in chromosomal dosage series of maize: The biochemical basis of aneuploid syndromes. Genetics. 1981;99(2):247–266. doi: 10.1093/genetics/99.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: Genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24(8):390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Mank JE. Sex chromosome dosage compensation: Definitely not for everyone. Trends Genet. 2013;29(12):677–683. doi: 10.1016/j.tig.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Vicoso B, Bachtrog D. Progress and prospects toward our understanding of the evolution of dosage compensation. Chromosome Res. 2009;17(5):585–602. doi: 10.1007/s10577-009-9053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vicoso B, Bachtrog D. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 2015;13(4):e1002078. doi: 10.1371/journal.pbio.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julien P, et al. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 2012;10(5):e1001328. doi: 10.1371/journal.pbio.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GA. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci USA. 2012;109(14):5346–5351. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gschwend AR, et al. Rapid divergence and expansion of the X chromosome in papaya. Proc Natl Acad Sci USA. 2012;109(34):13716–13721. doi: 10.1073/pnas.1121096109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bachtrog D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics. 2008;179(3):1513–1525. doi: 10.1534/genetics.107.084012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charlesworth D. Sex chromosome origins and evolution. In: Pagel MPA, editor. Evolutionary Genomics and Proteomics. Sinauer Associates; Sunderland: 2008. pp. 207–240. [Google Scholar]
- 34.Armstrong SJ, Filatov DA. A cytogenetic view of sex chromosome evolution in plants. Cytogenet Genome Res. 2008;120(3-4):241–246. doi: 10.1159/000121073. [DOI] [PubMed] [Google Scholar]
- 35.Bernasconi G, et al. Silene as a model system in ecology and evolution. Heredity (Edinb) 2009;103(1):5–14. doi: 10.1038/hdy.2009.34. [DOI] [PubMed] [Google Scholar]
- 36.Desfeux C, Maurice S, Henry JP, Lejeune B, Gouyon PH. Evolution of reproductive systems in the genus Silene. Proc Biol Sci. 1996;263(1369):409–414. doi: 10.1098/rspb.1996.0062. [DOI] [PubMed] [Google Scholar]
- 37.Filatov DA. Evolutionary history of Silene latifolia sex chromosomes revealed by genetic mapping of four genes. Genetics. 2005;170(2):975–979. doi: 10.1534/genetics.104.037069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergero R, Qiu S, Forrest A, Borthwick H, Charlesworth D. Expansion of the pseudo-autosomal region and ongoing recombination suppression in the Silene latifolia sex chromosomes. Genetics. 2013;194(3):673–686. doi: 10.1534/genetics.113.150755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Filatov DA, Monéger F, Negrutiu I, Charlesworth D. Low variability in a Y-linked plant gene and its implications for Y-chromosome evolution. Nature. 2000;404(6776):388–390. doi: 10.1038/35006057. [DOI] [PubMed] [Google Scholar]
- 40.Laporte V, Filatov DA, Kamau E, Charlesworth D. Indirect evidence from DNA sequence diversity for genetic degeneration of the Y-chromosome in dioecious species of the plant Silene: The SlY4/SlX4 and DD44-X/DD44-Y gene pairs. J Evol Biol. 2005;18(2):337–347. doi: 10.1111/j.1420-9101.2004.00833.x. [DOI] [PubMed] [Google Scholar]
- 41.Kazama Y, et al. Organization of the KpnI family of chromosomal distal-end satellite DNAs in Silene latifolia. J Plant Res. 2003;116(4):317–326. doi: 10.1007/s10265-003-0106-6. [DOI] [PubMed] [Google Scholar]
- 42.Bennett MD, Leitch IJ. 2012 Plant DNA C-values database. Available at www.kew.org/cvalues/. Accessed August 5, 2015.
- 43.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 44.Bellott DW, et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508(7497):494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bachtrog D, Hom E, Wong KM, Maside X, de Jong P. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 2008;9(2):R30. doi: 10.1186/gb-2008-9-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamato KT, et al. Gene organization of the liverwort Y chromosome reveals distinct sex chromosome evolution in a haploid system. Proc Natl Acad Sci USA. 2007;104(15):6472–6477. doi: 10.1073/pnas.0609054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDaniel SF, Neubig KM, Payton AC, Quatrano RS, Cove DJ. Recent gene-capture on the UV sex chromosomes of the moss Ceratodon purpureus. Evolution. 2013;67(10):2811–2822. doi: 10.1111/evo.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Q, Bachtrog D. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science. 2012;337(6092):341–345. doi: 10.1126/science.1225385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gordo I, Charlesworth B. On the speed of Muller’s ratchet. Genetics. 2000;156(4):2137–2140. doi: 10.1093/genetics/156.4.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rautenberg A, Hathaway L, Oxelman B, Prentice HC. Geographic and phylogenetic patterns in Silene section Melandrium (Caryophyllaceae) as inferred from chloroplast and nuclear DNA sequences. Mol Phylogenet Evol. 2010;57(3):978–991. doi: 10.1016/j.ympev.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Vicoso B, Bachtrog D. Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol Evol. 2011;3:230–235. doi: 10.1093/gbe/evr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrison PW, Mank JE, Wedell N. Incomplete sex chromosome dosage compensation in the Indian meal moth, Plodia interpunctella, based on de novo transcriptome assembly. Genome Biol Evol. 2012;4(11):1118–1126. doi: 10.1093/gbe/evs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uebbing S, Künstner A, Mäkinen H, Ellegren H. Transcriptome sequencing reveals the character of incomplete dosage compensation across multiple tissues in flycatchers. Genome Biol Evol. 2013;5(8):1555–1566. doi: 10.1093/gbe/evt114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malone JH, et al. Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol. 2012;13(4):r28. doi: 10.1186/gb-2012-13-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat Genet. 2012;44(2):226–232. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fraley C, Raftery AE, Murphy TB, Scrucca L. 2012. mclust Version 4 for R. in Technical Report (Department of Statistics, University of Washington, Seattle, WA), pp 1–57.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.