Significance
A high-resolution genetic lineage-tracing study in mice reveals that cKit identifies multipotent progenitors of cardiac neural crest (CNC) origin. Normally, the proportion of cardiomyocytes produced from this lineage is limited, not because of poor differentiation capacity as previously thought, but because of stage-specific changes in the activity of the bone morphogenetic protein pathway. Transient bone morphogenetic protein antagonism efficiently directs mouse iPSCs toward the CNC lineage and, consequently, the generation of cKit+ CNCs with full capacity to form cardiomyocytes and other CNC derivatives in vitro. These findings resolve a long-standing controversy regarding the role of cKit in the heart, and are expected to lead to the development of novel stem cell-based therapies for the prevention and treatment of cardiovascular disease.
Keywords: cardiac stem cells, cardiac neural crest, cardiomyogenesis, BMP signalling
Abstract
The degree to which cKit-expressing progenitors generate cardiomyocytes in the heart is controversial. Genetic fate-mapping studies suggest minimal contribution; however, whether or not minimal contribution reflects minimal cardiomyogenic capacity is unclear because the embryonic origin and role in cardiogenesis of these progenitors remain elusive. Using high-resolution genetic fate-mapping approaches with cKitCreERT2/+ and Wnt1::Flpe mouse lines, we show that cKit delineates cardiac neural crest progenitors (CNCkit). CNCkit possess full cardiomyogenic capacity and contribute to all CNC derivatives, including cardiac conduction system cells. Furthermore, by modeling cardiogenesis in cKitCreERT2-induced pluripotent stem cells, we show that, paradoxically, the cardiogenic fate of CNCkit is regulated by bone morphogenetic protein antagonism, a signaling pathway activated transiently during establishment of the cardiac crescent, and extinguished from the heart before CNC invasion. Together, these findings elucidate the origin of cKit+ cardiac progenitors and suggest that a nonpermissive cardiac milieu, rather than minimal cardiomyogenic capacity, controls the degree of CNCkit contribution to myocardium.
Heart development is a highly regulated process during which cell lineage diversification and growth programs are dynamically coordinated in temporal and spatial manners (1). These programs are activated sequentially, in parallel, or intersect to give rise to distinct heart domains. For example, the myocardial lineage originally develops from cardiac progenitors (CPs) of mesodermal origin (2–5), which form the first and second heart fields. However, later during morphogenesis, the cardiomyogenic program diverges and activates cardiomyocyte proliferation signals, along with CPs from the hemogenic endothelium, epicardial, cardiopulmonary, and cardiac neural crest (CNC) lineages, to produce new cardiomyocytes (1, 6–11). Gauging the relative contribution of each lineage for scaling their cardiomyogenic—and consequently therapeutic—capacity is a challenge. For example, many of the CP lineages are heterogeneous and incompletely characterized, and therefore cannot always be traced under a straightforward genetic fate-mapping experiment. Furthermore, it is unknown whether and how changes in the cardiac milieu (i.e., morphogens, tissue composition, and size) regulate the final proportions of heart muscle derived from each lineage.
cKit is a receptor tyrosine kinase that marks several cell lineages, including neural crest (NC), hematopoietic, and germ-line stem cells (12–15). Following the seminal description by Beltrami et al. (16) of clusters of cKit cells in the postnatal mammalian heart, several laboratories, including ours, suggested that cKit marks CPs (16–19), a finding that led to the clinical testing of these cells for heart repair (20). Recently, a straightforward genetic fate-mapping study showed that a relatively small proportion of murine myocardium is derived from cKit+ CPs, leading to the conclusion that the cardiomyogenic capacity of cKit+ CPs is functionally insignificant (21). However, the identity of cKit+ CPs and the mechanisms controlling their differentiation into cardiomyocytes remain controversial (22). Here, by using a high-resolution genetic lineage-tracing strategy, as well as induced pluripotent stem cell (iPSC)-based models of cardiogenesis, we demonstrate that cKit marks CNCs. Furthermore, we show that their relatively small contribution to myocardium during embryogenesis is not related to poor cardiomyogenic capacity, but rather to changes in the cardiac activity of the bone morphogenetic protein (BMP) pathway that prevent their differentiation into cardiomyocytes.
Results
Genetic Lineage-Tracing of cKit+ CPs.
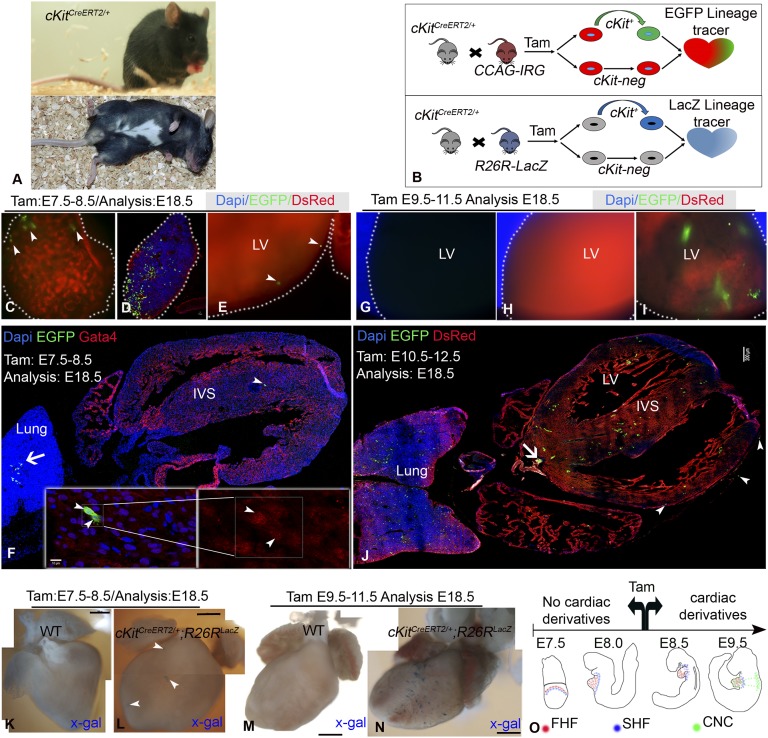
We used a well-characterized cKitCreERT2/+ mouse line to lineage-trace cKit+ CPs (23–25). cKitCreERT2/+ are healthy, fertile, and express the white spotting phenotype (12, 23, 24, 26) (Fig. 1A).
Fig. 1.
cKitCreERT2/+ lineage-tracing. (A) Phenotype of cKitCreERT2/+ mice. (B) Summary of the experimental design. (C–F) administration of TAM during E7.5–E8.5 (n = 10) marks testicular (C, arrowheads), pulmonary (D) and, rarely, immature cells in the myocardium (E and F, arrowheads). (G–J) Live tissue imaging of cKitCreERT2/+ (G), IRG (H), and cKitCreERT2;IRG (I) E18.5 littermates subjected to TAM during E9.5–E11.5 (n = 7). Widespread EGFP epifluorescence in ventricles and atria (I and J), lungs (J), OFT (J, arrow), epicardium (J, arrowheads). (K–N) Lineage-tracing in cKitCreERT2;R26RlacZ mice (n = 8). (O) Summary of cKit genetic fate-mapping. Panels F and J are confocal tile-scans. Panels K–N are photomerged image tiles. (Scale bars, 10 µm in D and F; 200 µm in J; 500px in K–N.) (Magnification, 100× in C, E, and G–I.)
We first investigated whether cKit marks mesodermal CPs (e.g., first- or second-heart field CPs; or primitive hemogenic lineage) (1), by administering pregnant mice carrying cKitCreERT2;IRG embryos with tamoxifen (TAM) from embryonic days (E)7.5 to E8.5 (Fig. 1B and Table S1). At E18.5, EGFP expression was detected in mesodermal cells (13, 14, 21, 26), including gonads, blood, and lungs (Fig. 1 C and D). At this stage of labeling (21), EGFP was rarely detected in the heart, and EGFP+ heart cells were noncardiomyocytes with rare colocalization with the cardiac transcription factor Gata4 (Fig. 1 E and F).
Table S1.
Exact sample sizes of genetic lineage-tracing experiments
| Litter ID no. | TAM started | TAM ended | Day of analysis | Strain | Litter size | Embryos with desired genotype |
| 1 | E7.5 | E8.5 | E18.5 | cKitCreERT2;IRG | 3 | 1 |
| 400 | E7.5 | E8.5 | E18.5 | cKitCreERT2;IRG | 7 | 3 |
| 283 | E7.5 | E8.5 | E18.5 | cKitCreERT2;IRG | 10 | 3 |
| 497 | E7.5 | E8.5 | E18.5 | cKitCreERT2;R26RLacZ | 10 | 3 |
| 237 | E9.5 | E11.5 | E18.5 | cKitCreERT2;IRG | 3 | 1 |
| 393 | E10.5 | E12.5 | E18.5 | cKitCreERT2;IRG | 6 | 2 |
| 453 | E10.5 | E12.5 | E18.5 | cKitCreERT2;IRG | 10 | 3 |
| 309 | E10.5 | E12.5 | E18.5 | cKitCreERT2;IRG | 10 | 1 |
| 364 | E9.5 | E11.5 | E18.5 | cKitCreERT2;R26RLacZ | 2 | 0 |
| 365 | E9.5 | E11.5 | E18.5 | cKitCreERT2;R26RLacZ | 3 | 2 |
| 366 | E9.5 | E11.5 | E18.5 | cKitCreERT2;R26RLacZ | 9 | 3 |
| 472 | E9.5 | E11.5 | E12.5 | cKitCreERT2;IRG;Isl1nLacZ | 8 | 1 |
| 18 | E9.5 | E11.5 | E12.5 | cKitCreERT2;IRG;Isl1nLacZ | 8 | 1 |
| 20 | E9.5 | E11.5 | E12.5 | cKitCreERT2;IRG;Isl1nLacZ | 9 | 1 |
| 23 | E9.5 | E11.5 | E12.5 | cKitCreERT2;IRG;Isl1nLacZ | 8 | 1 |
| 22 | E9.5 | E11.5 | E12.5 | cKitCreERT2;IRG;Isl1nLacZ | 7 | 2 |
| 774 | E9.5 | E11.5 | E16.5 | cKitCreERT2;IRG;Wnt1::Flpe;RC::Fela | 6 | 1 |
| 785 | E9.5 | E11.5 | E18.5 | cKitCreERT2;IRG;Wnt1::Flpe;RC::Fela | 8 | 1 |
| 787 | E8.5 | E9.5 | E10.5 | cKitCreERT2;IRG;Wnt1::Flpe;RC::Fela | 11 | 1 |
| 788 | E8.5 | E16.5 | E17.5 | cKitCreERT2;IRG;Wnt1::Flpe;RC::Frepe | 12 | 2 |
Summary of sample sizes of genetic lineage-tracing experiments. For the purpose of cKit genetic fate-mapping studies, a total of 150 mouse embryos from 20 different litters were analyzed. Thirty-three embryos carried the desired genotypes.
Next, to test whether cKit marks other cardiomyogenic lineages (e.g., proliferating cardiomyocytes; or CPs of the epicardial, CNC, and definitive hemogenic lineages) (1), we administered TAM to pregnant mice at selected time points during E9.5–E12.5 (Table S1). Cre-mediated recombination resulted in EGFP expression in embryonic melanoblasts, craniofacial cells (27), neural tube (NT), dorsal root ganglia (DRGs), blood, gastrointestinal cells, gonads, and pulmonary cells (Fig. 1 G–J). Unlike the fate-map of E7.5–E8.5 cKit-expressing cells, EGFP epifluorescence is detected within the cardiac outflow tract (OFT), epicardium, and myocardium (Fig. 1 G–J).
To rule out a limited transgene expression, we also performed fate-mapping using the R26 promoter-driven R26RlacZ allele. The results were similar using this reporter compared with EGFP (Fig. 1 K–N).
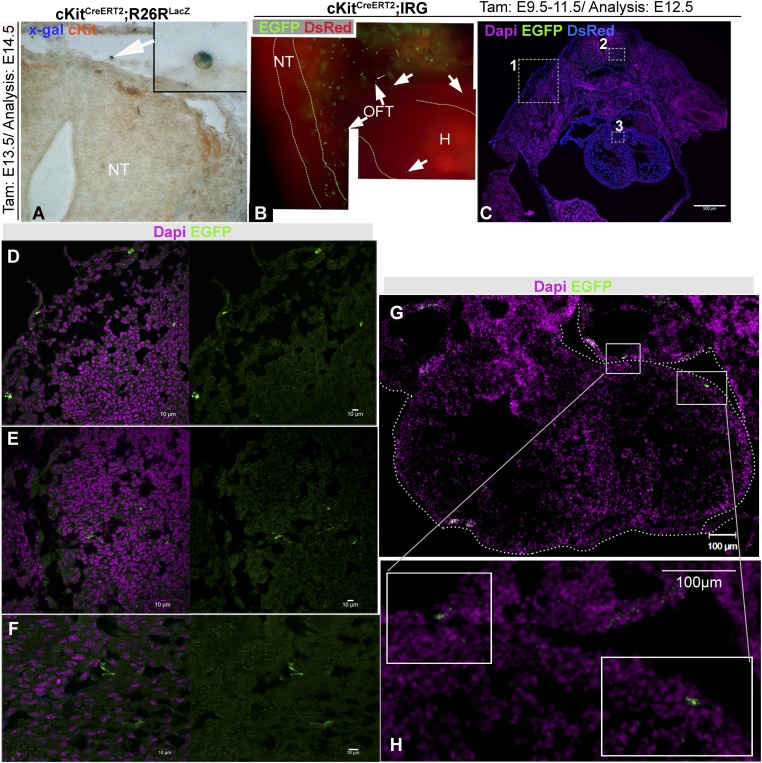
To identify the original population of CPs, we administered TAM at selected time points between E9.5 and E13.5, and collected embryos 24 h after the last injection (Fig. S1A). Using live epifluorescence and immunofluorescence (IF) imaging, we detected Cre-recombined cells in the NT, skin, lungs, gut, conotruncus, OFT, and epicardium, but not within the myocardium (Fig. S1). These findings suggest that the cKitCreERT2/+-labeled heart cells during the period of TAM-induced recombination do not arise within the myocardium [i.e., differentiated cardiomyocytes (28) or transdifferentiating cardiac fibroblasts (29) and hemogenic progenitors (6, 7)], and derived from an extracardiac CP lineage.
Fig. S1.
cKit is not expressed in the embryonic myocardium at the time of TAM administration. (A) Colocalization of X-gal with anti-cKit immunohistochemistry (arrow) in an E13.5 NT of a cKitCreERT2/R26RlacZ embryo. (Magnification, 200×.) Inset depicts a higher-magnification image of the indicated cell. (B) Live embryo imaging of EGFP and DsRed epifluorescence in an E12.5 cKitCreERT2;IRG embryo. Two EGFP+ cells are detected in proximity to the OFT and two more in the epicardium (arrows). EGFP expression is absent in the myocardium. In contrast, strong expression of EGFP is seen in the NT and the skin. (C–F) A transverse section from an E12.5 cKitCreERT2;IRG mouse embryo in which immunohistochemistry against EGFP has been performed. EGFP cells are detected in the skin (Inset 1 and D in higher magnification), neural tube (Inset 2 and E in higher magnification), and the conotruncus (Inset 3 and F in higher magnification). No EGFP signal is detected in the myocardium. (G and H) A transverse section of an E12.5 cKitCreERT2;IRG heart illustrating expression of EGFP in the epicardium and left atrium. No signal is detected in the myocardium. Panel B is a photomerged image tile. (Magnification, 100×/tile.) Panel C is a confocal tile-scan. Ht, Heart. (Scale bars, 10 µm in D–F, and 100 µm in G–H.)
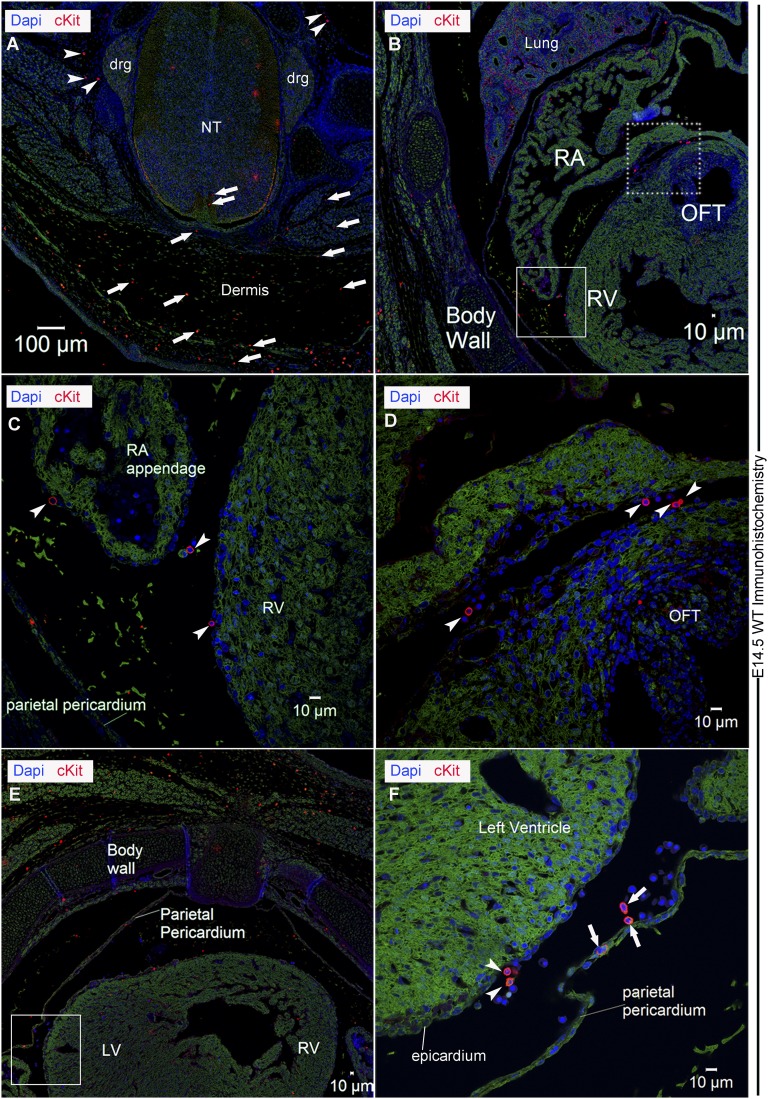
Finally, similar to cKitCreERT2/+ reporters, cKit IF in cKit+/+ embryos marked cells within the NT, skin, lung, OFT, and epicardium, but not differentiated cardiomyocytes (Fig. S2).
Fig. S2.
Expression of cKit in the cKit+/+ developing mouse heart. Representative confocal immunofluorescence of cKit antibody localization in E14.5 wild-type mouse embryos. (A) Consistent with cKitCreERT2/+ genetic fate map, CNCkit (red fluorescence) are detected in the NT, DRG, as well as in the dermis. CNCkit are detected in proximity of both the dorsolateral (arrows) and ventral (arrowheads) sites of the NT. (B) cKit expression in the embryonic lungs, RA, OFT, and epicardium (boxes). (C) Magnification of the solid box in B, highlights CNCkit (arrowheads) in the RA appendage, pericardial and epicardial walls. (D) Magnification of the dashed box in B, highlights CNCkit (arrowheads) accessing the RA, epicardium, and OFT. (E) CNCkit are dispersed in dermis, parietal pericardium, epicardium, and cardiac ventricular walls. (F) Magnification of the boxed area in E, illustrates CNCkit accessing the LV through the pericardial (arrows) and epicardial (arrowheads) walls. (Scale bars, 100 µm in A and 10 µm in B–F.) RA, right atrium; RV, right ventricle.
Collectively, our findings suggest that cKit marks a CP lineage that emerges at ∼E9.5 and contributes to the development of the mouse heart.
Intersectional Genetic Fate-Mapping of cKit and Wnt1 Protooncogenes.
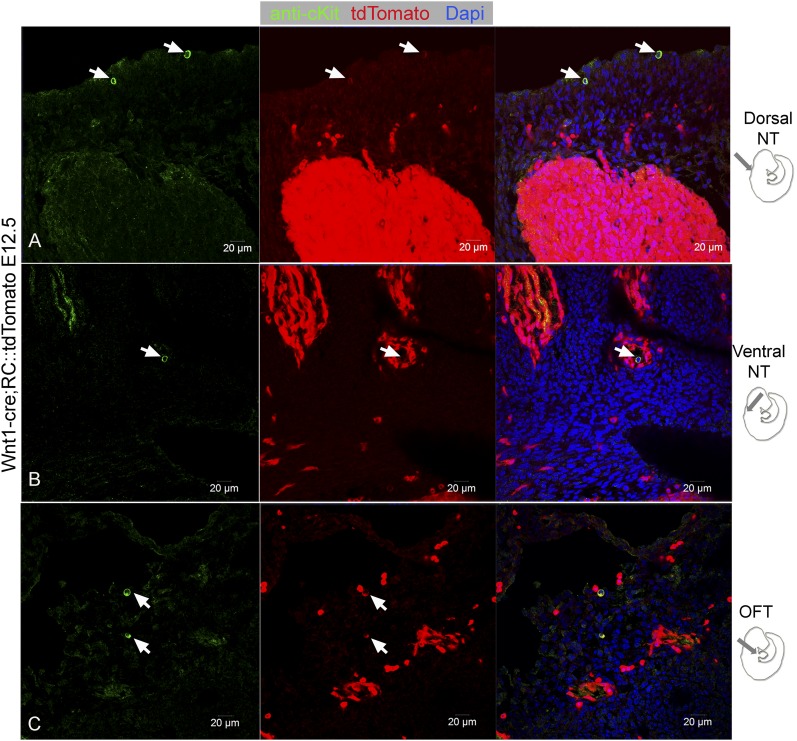
Because our findings are consistent with a CNC origin of cKit+ CPs, we used a well-established CNC-specific mouse, the Wnt1-Cre;RC::tdTomato (30–32), to examine the expression of cKit in CNC CPs (8, 10, 11, 31–33). IF against cKit illustrated its colocalization with tdTomato in various NC-derived tissues of E12.5 Wnt1-Cre;RC::tdTomato embryos, including the NT and the heart, (Fig. S3). However, compared with the cKit−/tdTomato+ cells, cKit+/tdTomato+ population exhibited a weak expression of tdTomato (Fig. S3).
Fig. S3.
cKit immunohistochemistry labels weakly expressing tdTomato+ cells in Wnt1-Cre;RC::tdTomato mouse embryos. Confocal immunofluorescence analysis following anti-cKit immunohistochemistry in E12.5 Wnt1-Cre;RC::tdTomato embryos illustrates colocalization of cKit in a population of cells with weak tdTomato epifluorescence, located dorsally (A, arrows) and ventrally from the NT (B, arrow), as well as within the outflow tract (C, arrows). A total of n = 3 Wnt1-Cre;RC::tdTomato embryos were analyzed. (Scale bars, 20 µm.)
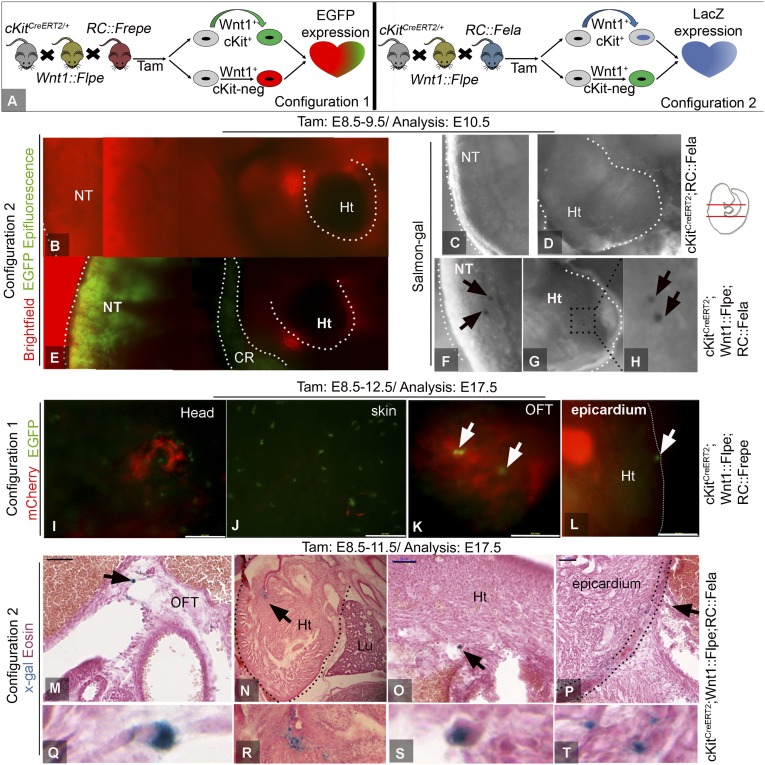
We therefore performed NC lineage-restricted genetic fate-mapping of cKit. We generated a novel mouse carrying two recombinase systems (Cre-loxP and Flp-FRT), which enables intersectional genetic fate-mapping of cKitCreERT2/+ in the Wnt1-expressing CNC lineage and its derivatives (34). Two previously established dual-recombinase responsive indicator alleles, the RC::Fela (35) and RC::Frepe (36), and a novel Wnt1-Flpe recombinase driver line, the Wnt1::Flpe4351, were used (Fig. 2A).
Fig. 2.
Intersectional genetic fate-mapping of cKit and Wnt1. (A) schematic of the two different approaches of the study. (B–D) Live epifluorescence imaging (B), followed by salmon-gal histochemical detection of nLacZ (C and D), in an E10.5 cKitCreERT2;RC::Fela embryo exposed to TAM during E8.5–E9.5. The flp- and intersectional indicators are not expressed in the absence of Flpe and Flpe/Cre-mediated recombination, respectively. (E–H) A Wnt1::Flpe;cKitCreERT2;RC::Fela littermate exhibits widespread GFP epifluorescence (E) and a few salmon-gal+ cells in the NT and heart. (I–L) Live embryo imaging of mCherry and GFP epifluorescence in a E17.5 Wnt1::Flpe;cKitCreERT2;RC::Frepe embryo. EGFP+ CNCkit in the craniofacial region (I), skin (J), OFT (K), and the epicardial wall of the heart (L). (M and T) X-gal+ CNCkit derivatives in the OFT (M and G), heart (N, O, R, and S), and epicardium (P and T) of E17.5 Wnt1::Flpe;cKitCreERT2;RC::Fela embryos. Arrows in M–P are depicted in higher magnification in panels Q–T, respectively. Panels B, E are photomerged image tiles. OFT, outflow tract; Ht, heart; Lu, lung. (Scale bars, 50 µm in M–P and 300px in I–L.) (Magnification, 100× in B–H and Q–T.)
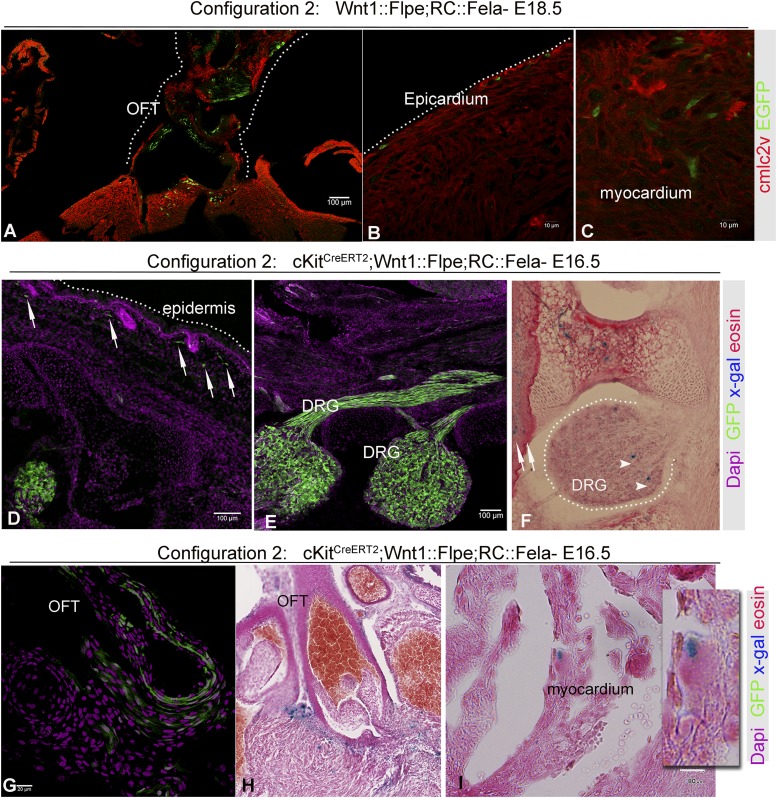
Timed-pregnant cKitCreERT2;Wnt1::Flpe;RC::Fela or cKitCreERT2;Wnt1::Flpe;RC::Frepe mice were administered TAM during E8.5–E11.5 and embryo analysis was performed at selected time points between E10.5 and E18.5 (Table S1). Wnt1::Flpe-mediated recombination resulted in extensive labeling with the flp indicators in the craniofacial region, melanoblasts, gut, DRG, OFT, as well as in cells within the epicardium and myocardium (Fig. 2 and Fig. S4). When both Wnt1::Flpe and cKitCreERT2/+ were expressed, Flpe indicators persisted in all NC-derived tissues, including the DRG, skin, heart, and OFT (Fig. 2 and Fig. S4 A–E). However, expression of intersectional indicators was also detected in the craniofacial region, melanocytes, DRG, OFT, as well as in cells within the epicardium and myocardium (Fig. 2 and Fig. S4). Importantly, expression of intersectional indicators in the lung was not documented (Fig. 2N), consistent with the hypothesis that the heart and lung cKit+ progenitors are of different origins. These studies illustrate a lineal relationship between the cKit+ CPs and Wnt1+ CNCs (CNCkit).
Fig. S4.
Intersectional genetic fate map of cKit and Wnt1. A-C, Wnt1::Flpe4351 reliably marks the cardiac neural crest as indicated by the expression of EGFP in the OFT (A), epicardium (B), and myocardium (C) of an E18.5 Wnt1::Flpe;Rc::Fela heart. (D–I) When both Wnt1::Flpe and cKitCreERT2/+ become activated in cKitCreERT2;Wnt1::Flpe;Rc::Fela embryos, expression of EGFP is still widely expressed in the NC derivatives, including melanoblasts (D, arrows), DRGs (E), and OFT (G). In addition, a population of nLacZ+ cells are also present within the skin (F, arrows), DRGs (F, arrowheads), OFT (H) and, rarely, within the compact myocardium (I). [Scale bars, 100 µm (A, D, and E), 10 µm (B and C), 20 µm (G) and 80 µm (I).] (Magnification, 100× in H.)
CNCkit Derivatives in the Heart.
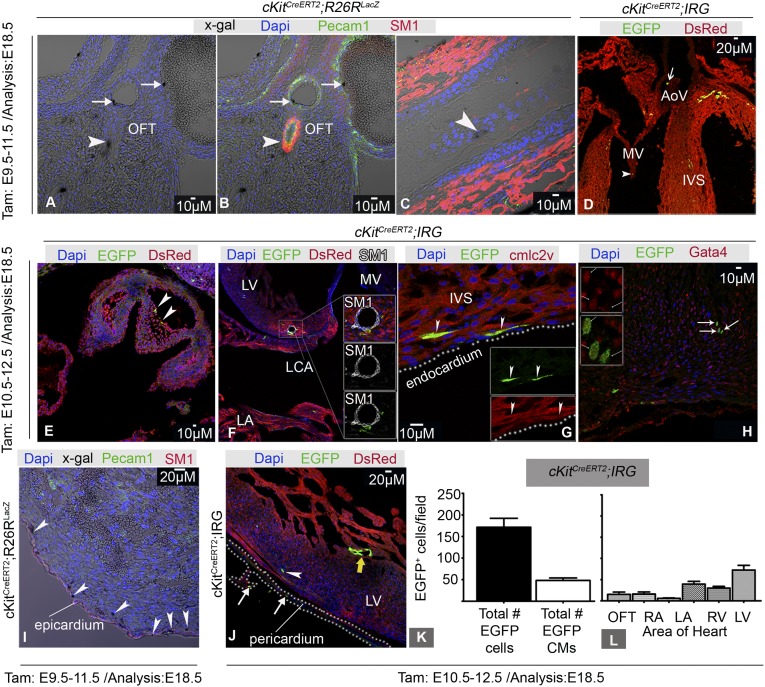
Expression of Cre-reporters was detected in all expected cardiac NC derivatives (37), including the OFT (Figs. 1J and 3 A and B, and Figs. S2B and S5), the tunica media of the aortic arch (Fig. 3C), cardiac and aortic valves (Fig. 3 D and E), atria (Fig. 1K), inflow tract, satellite glial progenitors, and sensory cells (Fig. S5 A and B and Movie S1). Consistent with their CNC origin (38), CNCkit contributed to endothelium and smooth muscle layers of the OFT (Fig. 3 A and B), although coronary vascular cell differentiation was not observed (Fig. 3F).
Fig. 3.
Heart derivatives of CNCkit. (A and B) Colocalization of X-gal with SM1 (arrowhead) and Pecam1 (arrows) in the OFT. (C) X-gal+ cells (arrowhead) within the aortic tunica media. (D) EGFP+ derivatives within the mitral valve (arrowhead) and aortic valve (arrow). (E) EGFP+ derivatives in the aortic valve (arrowheads). (F) CNCkit are associated with, but do not contribute to, coronary vasculature. (G) EGFP+/cmlc2v+ ventricular cardiac myocytes. (H) EGFP+ cardiac derivatives coexpress Gata4+. (I and J) EGFP+ pericardium (arrows), epicardium (arrowheads), and endocardium (J, yellow arrow). (K) Cardiomyocytic versus noncardiomyocytic EGFP+ derivatives in the heart. (L) Distribution of EGFP+ cells in the heart [n = 3 embryos; 11 sections (K and L)]. AoV, aortic valve; CM, cardiomyocytes; cmlc2v, cardiac muscle light chain 2v; LA, left atrium; LV, left ventricle; LCA, left coronary artery; MV, mitral valve; SM1, smooth muscle myosin heavy chain. Values represent means ± SEM.
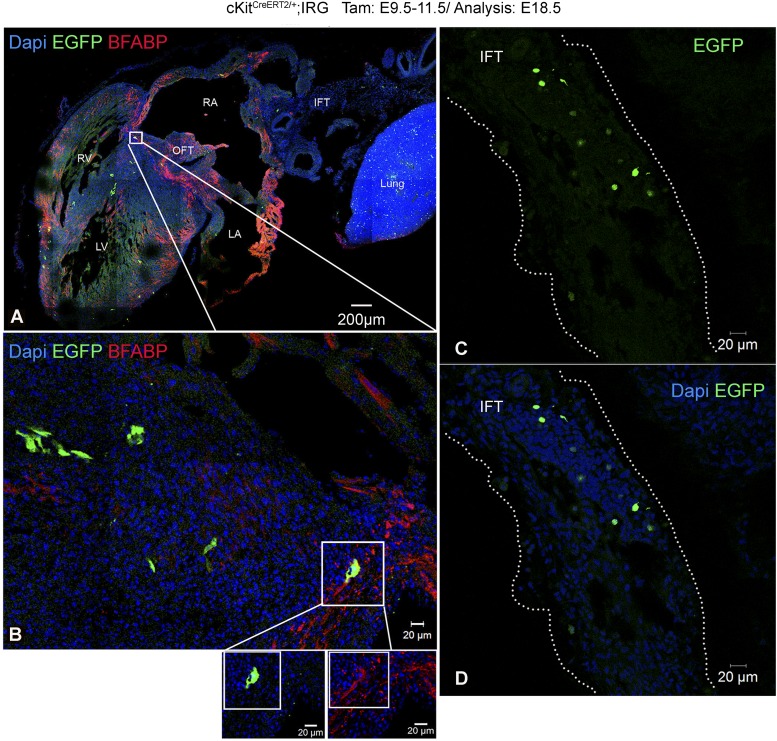
Fig. S5.
CNCkit derivatives in the heart and their identity. (A) Tilescan image illustrating extensive contribution of EGFP+ CNCkit derivatives in the lung, IFT, atria, ventricles and OFT. Notably, EGFP+ cells are consistently detected to be closely associated with BFABP+ satellite glial progenitors. Occasionally, EGFP and BFABP colocalize, illustrating that CNCkit contribute to glia and glial progenitor lineages (B, boxed area). (C and D) Higher magnification of the IFT from A, illustrating extensive contribution from CNCkit. BFABP, brain fatty acid-binding protein; IFT, inflow tract; LA, left atrium; LV, left ventricle; RA, right atrium; RV, right ventricle.
In agreement with previous reports in zebrafish (8, 11) and mice (10, 21, 31–33), our analysis with the Wnt1-Cre (Fig. S6 and Movie S2) and cKitCreERT2/+ alleles suggests that CNCs contribute to the myocardial lineage. Particularly, we documented contribution of CNCkit to atrial and ventricular cardiomyocytes (29.9% ± 3.1% of total EGFP+ derivatives) (Fig. 3 K and L), and pericardial, endocardial, and epicardial cells (Figs. 1 I, J, and N, and 3 G–K, and Movie S3). The majority of CNCkit-derived cardiomyocytes was localized in the interventricular septum (Figs. 1J and 3 K and L), which, unlike the left and right ventricular myocardium, is partly derived from mesoderm posterior 1 homolog nonexpressing (Mesp1−) CPs of undefined origin (9).
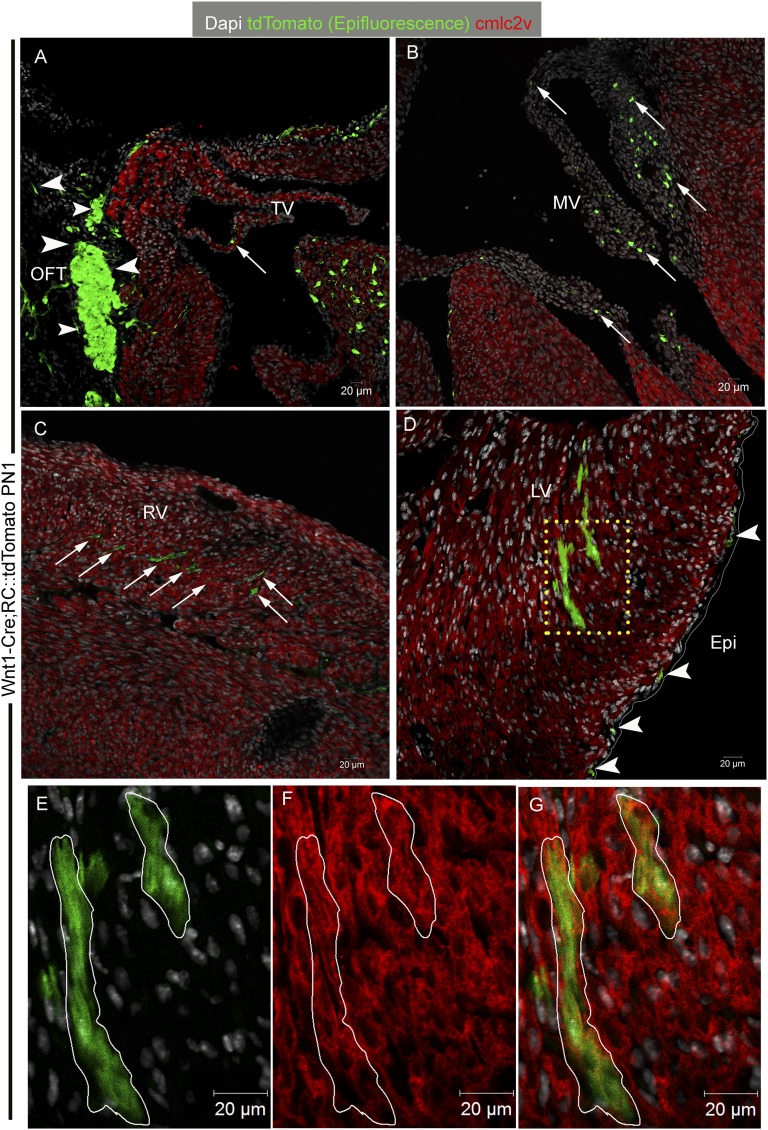
Fig. S6.
Wnt1-expressing CNCs contribute cardiomyocytes in the murine heart. Similar to the CNCkit derivatives, confocal microscopy of tdTomato (epifluorescence; pseudocolored green) in postnatal day 1 (PN1) Wnt1-Cre;RC::tdTomato mouse hearts illustrates that the Wnt1-expressing CNCs contribute extensively in the OFT (A, arrowheads), tricuspid valve (A, arrow), and mitral valve (B, arrows). In addition, tdTomato+ cells are detected within the ventricular myocardium (C, arrows, and D; yellow Inset) and epicardium (D, arrowheads). Immunostaining against the cardiomyocyte-specific marker cmlc2v illustrates that, similar to the CNCkit, the Wnt1-expressing CNCs contribute both noncardiomyocytes (C, arrows; tdTomato+/cmlc2v−) and fully differentiated cardiomyocytes (D, Inset; tdTomato+/cmlc2v+) in the mouse ventricle. (E–G) Higher magnification of the tdTomato+/cmlc2v+ clone of ventricular cardiomyocytes in the yellow Inset in D. (Scale bars, 20 µm.)
CNCkit Identity.
To better characterize the identity of CNCKit, we studied the expression of micropthalmia-associated transcription factor (Mitf), a direct target and transactivator of cKit signaling, expressed not only in cranial NC derivatives and mast cells but also in cardiomyocytes (39). IF analysis demonstrated that Mitf is also expressed in CNCkit and their cardiomyocytic derivatives (Fig. S7 A–C). However, EGFP+ cells in the heart did not express the melanocyte-specific markers tyrosinase or trp1, suggesting that Mitf+ CNCkit derivatives in the heart are not melanocytes (Fig. S7C).
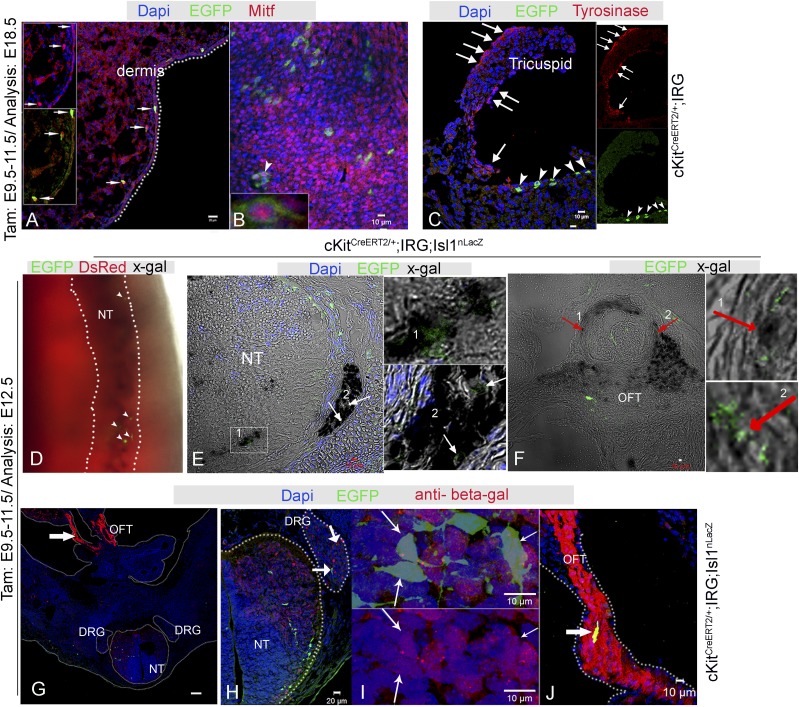
Fig. S7.
Mitf and Isl1 expression in CNCkit. (A) EGFP+ melanoblasts in the skin (arrows) coexpress Mitf. (B) EGFP+ derivatives in the heart (arrowhead, Inset) coexpress Mitf. (C) Tyrosinase+ melanocytes in the tricuspid valve (arrows) do not colocalize with EGFP (arrowheads). (D) Colocalization of X-gal and EGFP epifluorescence in cKitCreERT2;IRG;Isl1nLacZ embryos. (Magnification, 100×.) (E) X-gal and EGFP colocalization in the NT (Inset, 1) and DRGs (arrows, 2). (F) X-gal and EGFP colocalization (1, 2, arrows) in the OFT. (G) Colocalization of EGFP and β-gal in the NT, DRGs, and OFT (arrow). (H) EGFP and β-gal colocalization in NT and DRGs. (I) EGFP+/β-gal+ neurons (arrows) in the NT. (J) Higher magnification of the OFT in (G, arrow). [Scale bars, 10 µm (B, C, E, F, I, J), 100 μm (G), and 20 µm (A and H).]
Next, we investigated the expression of Isl1, a homeobox transcription factor that specifies the majority of the mammalian CP lineages, including CNCs (36). Accordingly, cKitCreERT2;IRG mice were crossed with mice carrying an Isl1 nuclear lacZ (Isl1nLacZ) allele (40). When pregnant mice were administered TAM from E9.5–E11.5, (Table S1), colocalization of EGFP and nLacZ was documented in cells of the NT, DRGs, and the OFT (Fig. S7 D–J) in E12.5 embryos.
Transient BMP Antagonism Induces Cardiomyogenesis in CNCkit.
To determine the full cardiomyogenic capacity of cKit+ CPCs (Fig. 1), we established iPSCs from cKitCreERT2;IRG mice (iPSCkit). We induced cardiomyogenesis in iPSCs with either ascorbic acid (AA), which drives cardiogenesis partly via an intermediate differentiation stage into cKit+/Nkx2.5+ CPs (15, 41), or BMP antagonism, a signaling pathway that regulates the development of both mesodermal and NC lineages (Fig. 4A) (2, 5, 31, 42–46).
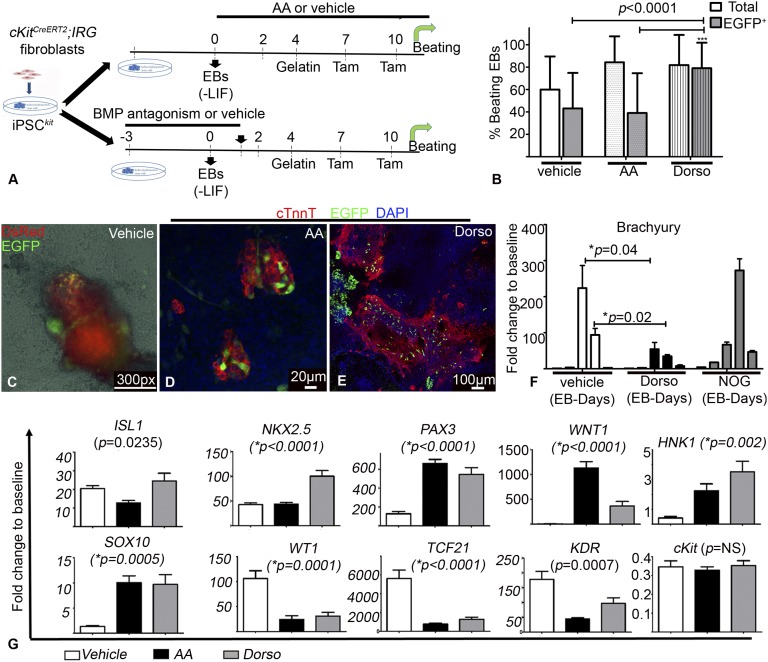
Fig. 4.
Transient BMP antagonism in iPSCkit induces CNCkit and suppresses the epicardium. (A) Schematic of the experimental approach. (B) Quantification of the percentage of beating EBs, and the percentage of beating EBs that are EGFP+ following Cre-recombination. (C) Live fluorescent imaging of a vehicle-treated spontaneously beating EB, coexpressing EGFP and DsRed. (D and E) Confocal microscopy of EBs following treatment with AA (D) or Dorso (E), illustrates that the EGFP+ cKitCreERT2/+ derivatives within the EBs are cTnnT+ cardiomyocytes. (F) Gene-expression analysis of Brachyury during the time-course of iPSCkit differentiation into cardiomyocytes following treatment with vehicle, Dorso, or NOG. (G) Comparison of the expression profiles of cardiac mesoderm- and CNC-related genes in day 11 EBs, in response to treatment with vehicle, AA, or Dorso. Compared with controls, AA and Dorso enhance cardiomyogenesis via a significant induction in CNC-related genes while suppressing proepicardial and endothelial progenitor genes. In addition, Dorso significantly enhances the expression of ISL1 and NKX2.5. cTnnt, cardiac troponin T; values represent means ± SEM.
Both AA or BMP antagonists enhanced cardiac differentiation into spontaneously contracting embryoid bodies (EBs) compared with controls (Fig. 4B) (P = 0.0073). When EBs were treated with 4-OH TAM during differentiation (Fig. 4A), we detected that 79.05% ± 4.9% of the beating EBs generated via transient BMP antagonism were EGFP+, compared with 39.03% ± 7.7% and 43.14% ± 6.9% EGFP+ beating EBs following treatment with AA or vehicle, respectively (Fig. 4 B–E and Movie S4) (P < 0.0001), suggesting that endogenous BMP signaling may limit cardiomyogenesis from cKit+ CPs.
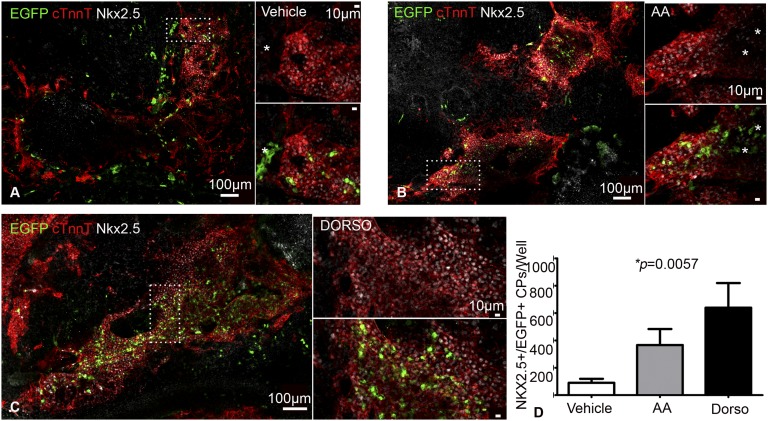
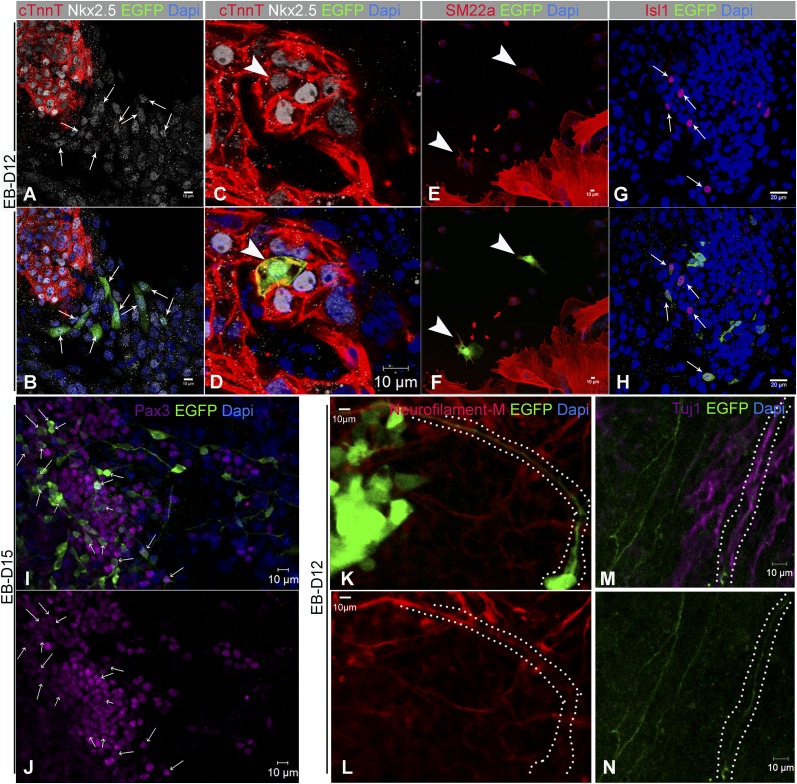
Compared with control, AA, and Noggin (NOG)-treated iPSCkit, Dorsomorphin (Dorso) enhanced cardiomyogenesis by significantly repressing Brachyury transcription (Fig. 4F and Fig. S8), while up-regulating ISL1, NKX2-5 (Fig. 4F) (43). In addition, we detected a significant up-regulation in CNC-related genes, including PAX3 and WNT1 (Fig. 4G and Fig. S8 E–H), whereas the expression of the proepicardial genes WT1, TCF21, and TBX18 (2), as well as the coronary endothelium marker KDR (Fig. 4G and Fig. S8), were significantly down-regulated compared with controls. Finally, expression of cKit increased significantly over time (Fig. S8D), although the level of expression was similar between the different treatment groups (Fig. 4G). As previously shown (15, 41), IF analysis confirmed that myocardial specification of cKit+ CPs commenced via coexpression of NKX2.5 (Fig. 5 and Fig. S9 A–D). Remarkably, BMP antagonism enhanced the development of EGFP+/NKX2.5+ progenitors by ∼sevenfold (Fig. 5, Fig. S9 A–D, and Movie S5) (P = 0.0057). Moreover, iPSCkit-derived CNCkit gave rise to all CNC derivatives, including EGFP+ smooth muscle cells (Fig. S9 E and F), Isl1+ (Fig. S9 G and H), and Pax3+ CPs (Fig. S9 I and J), while innervating the beating EBs with neurofilament-M+ and Tuj1+ neurons (Fig. S9 K–N).
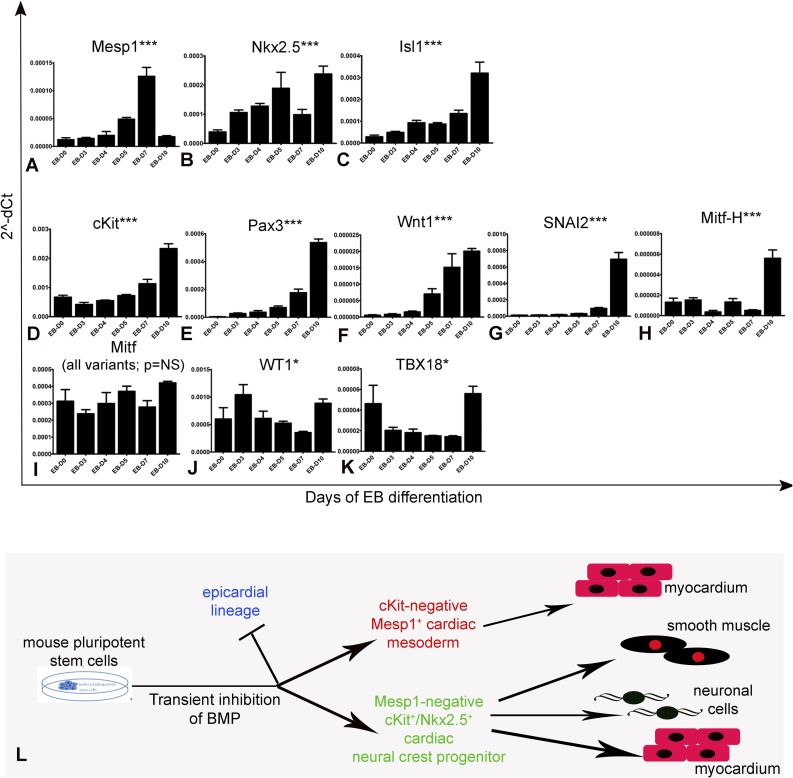
Fig. S8.
Transient BMP antagonism induces CNCkit and suppresses epicardium. (A–J) NOG promotes the generation of cardiac mesoderm from iPSCkit as indicated by the transient induction of Mesp1 (A) and up-regulation in the expression of Nkx2.5 (B) and Isl1 (C) at ∼EB- day 7. Subsequently, establishment of cardiac mesoderm is followed by the induction of CNC at ∼EB day 10, as indicated by the dramatic up-regulation in the expression of cKit (D), Pax3 (E), Wnt1 (F), SNAI2 (G), and Mitf-H (H). Notably, the noncardiac variants of Mitf (variants 1 and 2) remain unchanged. (J–K) Expression of the proepicardial lineage markers WT1 and TBX18 is significantly repressed before reaching baseline values by EB day 10. (L) Summary of the iPSC-based lineage tracing experiments. Values represent means ± SEM. ***P < 0.0001, *P < 0.05. n = 3 per group.
Fig. 5.
Derivation of CNCkit from mouse iPSCs. (A–C) Representative confocal immunofluorescence images illustrating EGFP+/NKX2.5+ derivatives within cTnnT+ EBs of vehicle-treated (A), AA-treated (B), or Dorso-treated (C) iPSCkit. Note that several of the EGFP+ cells in the vehicle- and AA-treated groups are NKX2.5− (asterisks). (Insets) Higher magnification. (D) Quantitation of EGFP+/NKX2.5+ cells between groups (n = 9 per group). Values represent means ± SEM.
Fig. S9.
Derivation of CNCkit from mouse iPSCs following NOG-mediated BMP antagonism. (A and B) A cluster of iPSCkit-derived EGFP+/Nkx2.5+, undifferentiated (cTnnT−) CNCkit (arrows) next to a cluster of non-CNCkit-derived (EGFP−) cTnnT+ differentiated cardiomyocytes. (C and D) An iPSCkit-derived CNCkit fully differentiated into cTnnT+/Nkx2.5+ cardiomyocyte (arrowhead). (E and F) Differentiation of iPSCkit-derived CNCkit into SM22a+ smooth muscle cells. (G and H) Colocalization of iPSCkit-derived CNCkit with Isl1. (I and J) iPSCkit-derived CNCkit generate Pax3+ progenitors. (K and L) EGFP+/neurofilament M+ motorneurons within a spontaneously beating EB. (M and N) An EGFP+/Tuj1+ neuron within the beating EBs. n = 3 per group. (Scale bars, 10 µm in A–F and I–N; 20 µm in G and H.)
Discussion
The major findings are that cKit marks CPs of CNC origin, which enter the embryonic mouse heart at ∼E9.5 and contribute a relatively small proportion of myocardium and other derivatives of the CNC, but not coronary vascular cells. In addition, we show that CNCkit CPs with cardiomyocyte differentiation capacity can be derived in vitro from mouse iPSCs following transient antagonism of the BMP pathway, which drives the stage-specific differentiation of iPSCs toward the cardiac mesodermal and CNC lineages.
Our findings confirm previous developmental studies in mice showing that, during gastrulation, cKit is expressed in extraembryonic mesoderm and embryonic ectoderm, but not mesodermal CPs (4, 13, 14, 42). Furthermore, the findings are in agreement with previous reports supporting the existence of cKit+ CPs, which do not contribute to coronary endothelium (15), as well as with recent endothelial lineage fate-mapping analyses suggesting that the coronary endothelium is unlikely to originate from cKit+ cells (6, 29).
Notably, although much controversy exists over the contribution of CNCs to the myocardium (8, 10, 11, 30–33), our studies with the cKitCreERT2/+, Wnt1-Cre, and Wnt1::Flpe alleles strongly support the hypothesis that the mammalian CNC holds full cardiomyogenic capacity, which our findings now suggest is undermined by developmental changes in the activity of BMP and Wnt pathways preceding their invasion in the heart (5, 31, 45). It is also noteworthy that a pool of multipotent postmigratory NC progenitors (47, 48), some of which express cKit (49), has been recorded in other tissues; hence, it would be interesting to examine their relationship to CNCkit.
Our study differs from a recent cardiac genetic fate-map of cKit, using different cKit alleles (21). First, in contrast to findings presented here and elsewhere (6, 15, 29), van Berlo et al. (21) reported that cKit CPs contribute extensively to coronary endothelium. However, it is noteworthy that mutations in the mouse W/cKit locus have not been associated with tangible cardiovascular defects (12), as would be likely if cKit+ CPs comprised a major source of coronary vascular cells. Second, van Berlo et al. (21) concluded that the minimal cardiomyocyte contribution of cKit CPs reflects minimal differentiation capacity. However, although our study agrees that the in vivo cardiomyocyte contribution of cKit+ CPs is lower than expected from previous reports (50, 51), we show that this is not a result of minimal differentiation capacity, but rather, because of their developmental origin in the CNC, which comprises a minor contributor of cardiomyocytes to the mammalian heart. Importantly, using iPSC modeling we demonstrate that differentiation of CNCkit to cardiomyocytes requires the BMP signaling pathway, which also directs differentiation of mesodermal CPs to the myocardium. This finding suggests that, although CNCkit hold full cardiomyocyte differentiation capacity, their in vivo contribution is repressed by spatiotemporal changes in BMP activity, which render the cardiac milieu nonconducive for cardiomyocyte differentiation during CNC invasion to the heart (45).
Our findings have several important implications. First, they resolve the current controversy over the existence and cardiomyogenic capacity of cKit+ CPs (22). We show that cKit+ CPs invested within the developing heart are fully capable of producing new cardiomyocytes, both in vivo and in vitro. Therefore, coupled with the findings from many laboratories that cKit+ CPs are present in the postnatal heart, they represent an important therapeutic target for heart regeneration (17–19, 52, 53). For example, the activity of BMP in the damaged myocardium could be modulated pharmacologically, or via transplantation of cells capable of regulating BMP activity, to support production of myocardium from endogenous or exogenously supplied cKit+ CPs (18, 52).
Second, the findings advance our understanding of the cellular and molecular mechanisms underlying mammalian cardiomyogenesis, by illustrating a previously unknown relationship between the spatiotemporal modulation of the BMP pathway and the generation of myocardium from mesodermal and CNC CPs. Third, the generation of CNCs from iPSCs provides a unique opportunity to study and understand the biology and function of CNCs, as well as to test their regenerative capacity in novel cell-based therapeutic strategies. Finally, considering the technical limitations often associated with conditional gene-targeting approaches, our findings do not exclude the possibility that, in addition to CNCkit, the adult heart contains other cKit+ cells with full cardiovascular differentiation capacity, as those reported by others (21, 28, 50, 51), which may have remained undetectable with our reagents.
In conclusion, our findings support the hypothesis that the mammalian heart is invested with a cKit+ CP lineage, with full capacity to generate cardiomyocytes in vivo and in vitro, and therefore provide an important therapeutic target for the prevention and treatment of heart disease. Modulation of the activity of the BMP pathway in the heart may enhance the therapeutic regeneration of damaged myocardium from CNCkit.
Materials and Methods
All animals were maintained in an Association for Assessment and Accreditation of Laboratory Animal Care-approved animal facility at the University of Miami, Miller School of Medicine, and procedures were performed using Institutional Animal Care and Use Committee-approved protocols according to NIH standards. cKitCreERT2/+ mice were developed as previously described (24). The Wnt1-Cre, RC::tdTomato, IRG, and R26RLacZ mouse lines were purchased from Jackson Laboratories. The Isl1nLacZ mice have been described elsewhere (40). Wnt1::Flpe, RC::Fela and RC::Frepe mice were developed as previously described (35, 36). iPSCkit were generated from adult cKitCreERT2/IRG tail-tip fibroblasts. Genotyping, TAM injections, gene-expression analysis, lineage-tracing, and histological analysis of mouse embryos was performed as previously described (24). See SI Materials and Methods for more detailed discussion.
SI Materials and Methods
Mice.
cKitCreERT2/+ mice were developed at the Medizinische Klinik und Poliklinik der Technischen Universität München, in Germany (24). The endogenous cKit locus was targeted by homologous recombination in embryonic stem (ES) cells as previously described for the Rosa26 locus (24). In brief, the targeting vector with the CreERT2 cassette, an internal ribosome entry site and an frt-flanked neomycin resistance cassette was electroporated into 129S6 ES cells. Homologous recombination and single-copy insertion was verified by Southern blot analysis and correctly targeted ES cells were injected into C57BL/6J blastocysts.
Wnt1::Flpe4351 were generated as previously described (36, 54). Briefly, sequences encoding a synthetic intron fused to Flpe-encoding sequence were inserted into the EcoRV site of WEXP2 harboring 11 kb of the Wnt1 gene. The SalI fragment was purified and injected into B6SJLF2 fertilized eggs using standard methods to generate transgenic mice, which were subsequently backcrossed to achieve a largely C57BL6 genetic background. All experiments involving these mice were approved by the University of Miami Institutional Animal Care and Use Committee.
The previously established Wnt1-Cre; the Cre reporter mouse lines RC::tdTomato (or Ai14); IRG; and R26RLacZ; were from Jackson Laboratories. The RC::Frepe and RC::Fela mice have been previously described (34–36). Expression of the reporter genes is driven by the CAG promoter in the IRG mouse; the R26-CAG allele (RC) in the tdTomato, RC::Frepe and RC::Fela mice; and the R26 promoter in the R26RLacZ mice. The Isl1nLacZ mice were kindly provided by Sylvia Evans, at the University of California, San Diego, CA and have been previously described (40). The Isl1nLacZ knock-in vector contains a LoxP-flanked nLacZ reporter gene followed by humanized Renilla GFP. However, cells carrying this transgene do not express GFP, either before or after Cre-mediated excision of the floxed nLacZ reporter (40). The R26RLacZ and RC::TdTomato were bred to homozygosity. All other strains used in the study are heterozygous.
For lineage-tracing experiments, cKitCreERT2/+ mice were crossed to IRG or R26RLacZ Cre-reporters and embryos were studied at selected time points following TAM-induced recombination. Wnt1-Cre mice were crossed to RC::tdTomato and embryos carrying the Wnt1-Cre;RC::tdTomato genotype were studied at E12.5. For the generation of embryos carrying a cKitCreERT2;IRG;Isl1nLacZ genotype, male cKitCreERT2;IRG mice were crossed to female Isl1nLacZ. For intersectional genetic fate-mapping studies, different crosses between male and female cKitCreERT2/+, RC::Frepe, RC::Fela, and Wnt1::Flpe were used to obtain embryos with the desired cKitCreERT2;Wnt1::Flpe;RC::Fela or cKitCreERT2;Wnt1::Flpe;RC::Frepe genotypes.
All animals were maintained in an Association for Assessment and Accreditation of Laboratory Animal Care-approved animal facility at the University of Miami, Miller School of Medicine, and procedures were performed using Institutional Animal Care and Use Committee-approved protocols according to NIH standards.
Genotyping.
Genomic DNA was isolated from mouse tails, as previously described (27). Genotyping was performed by PCR (Fermentas) according to the manufacturer’s instructions. The following primers were used: For cKitCreERT2/+ CCTCCACCATAAGCCGAATA, CCTTCGAGGTGGTAGGCATG and CCCCATTGTATGGGATCTGATC; for Isl1nLacZ: ACTATTTGCCACCTAGCCACAGCA and GACAGTATCGGCCTCAGGAA; For Wnt1::Flpe: AGCAACCACAGTCGTCAGAACC and TGGCTCATGGTGGAAGCTTGG; For RC::Frepe and RC::Fela: GCTGTTGTAGTTGTACTCCAGC and TACGGCAAGCTGACCCTGAAGTTC. The Wnt1-Cre, RC::tdTomato, IRG, and R26RlacZ genotyping protocols are available from Jackson Laboratories. X-gal and live epifluorescence were also used to validate PCR data. In addition, the white-spotting phenotype of cKitCreERT2/+ was considered an additional marker for genotyping.
Tamoxifen Injection.
For genetic fate-mapping, CreERT2 was activated by intraperitoneal injections of 100 µL of TAM (Sigma), dissolved in peanut oil (Sigma) at a concentration of 20 mg/mL at desired time points, as previously described (40).
Briefly, for assessing the role of cKit in first- or second-heart field progenitors, mice carrying cKitCreERT2;IRG or cKitCreERT2;R26RLacZ embryos received a daily injection of TAM for 2 consecutive days, during E7.5–E8.5. Embryos were harvested at E18.5. For assessing the expression of cKit in NC cells, mice carrying cKitCreERT2;IRG or cKitCreERT2;R26RLacZ embryos received a daily injection of TAM for 3 consecutive days, either during E9.5–E11.5 or during E10.5–E12.5. Embryos were harvested at E12.5 or E18.5. To trace the coexpression of Isl1-driven nLacZ and cKitCreERT2;IRG-driven EGFP in cKitCreERT2;IRG;Isl1nLacZ embryos, microscopic analysis was performed within 24 h after the last injection of TAM. This strategy allowed us to detect cells in which Cre-mediated recombination was induced for a period that was sufficient to report expression of EGFP before expression of nLacZ disappear. The same approach was used for intersectional genetic fate-mapping in mice carrying cKitCreERT2;Wnt1::Flpe;RC::Fela or cKitCreERT2;Wnt1::Flpe;RC::Frepe embryos.
Mouse Embryo Dissections.
Females with a vaginal plug were considered at E0.5, as previously described (40). Embryos at different time points were harvested in ice-cold HBSS (Gibco). For E12.5 and E18.5 embryos carrying the cKitCreERT2;IRG Wnt1-Cre;RC::tdTomato, cKitCreERT2;Wnt1::Flpe;RC::Fela or cKitCreERT2;Wnt1::Flpe;RC::Frepe alleles, live-tissue imaging was performed immediately after dissection under a fluorescence microscope (Olympus) and expression of EGFP, DsRed, tdTomato, or mCherry epifluorescence were photodocumented. Samples were then fixed for 1–1.5 h in 4% (vol/vol) PFA (EMS) at room temperature followed by overnight incubation in 30% (wt/vol) sucrose (Calbiochem) at 4 °C. For embryos carrying the RC::Fela allele, tissues were fixed for 20 min in 0.2% glutaraldehyde/0.4% PFA. The next day, samples were embedded in OCT (EMS) and flash-frozen in liquid nitrogen. Cryosectioning was performed as previously described (40).
For cKit immunohistochemical analyses, E12.5–E14.5 wild-type mouse embryos were harvested and fixed overnight in 10% (vol/vol) buffered formalin. The next day, they were embedded in paraffin and processed for immunohistochemistry, as previously described (18).
Generation of iPSCs.
Mouse iPSCkit were generated from adult tail-tip fibroblasts obtained from a 3.5-mo-old female cKitCreERT2;IRG mouse, using a commercially available kit (STEMCCA, Millipore). Briefly, tail-tip fibroblasts at passage one were plated in gelatin-coated 12-well plates at a density of 2 × 104 cells per well (day 0) with DMEM (Gibco), 2 mM l-glutamine, 10% (vol/vol) FBS (Gibco), and 1% penicillin-streptomycin (Gibco). The next day (day 1), fibroblasts were transduced with STEMCAA at a multiplicity of infection of 100 on day 1, followed by a repeated infection at an multiplicity of infection of 75 on day 2. Approximately 48 h later, cells were collected by trypsinization and replated on fresh plates coated with irradiated mouse embryonic fibroblasts (MEFs; Millipore) and fed with mouse ES medium [DMEM, 2 mM l-glutamine, 15% (vol/vol) FBS (Gibco), 0.1 mM nonessential amino acids (Gibco), 0.1 mM 2-mercaptoethanol (Gibco), and 1,000 units/mL LIF (Millipore)] supplemented with a cocktail of small molecules to enhance reprogramming (Mouse iPS reprogramming Boost Supplement, Millipore). iPS colonies began to emerge on day 7. Each colony was manually picked and individually transferred to fresh plates coated with MEFs. A total of 32 iPSCkit clones were generated, 25 of which stably expressed the IRG cassette as indicated by DsRed fluorescence. Clones 8, 21, and 31 were selected to be further expanded for the purpose of the study. All iPSC lines where grown on MEFs and adapted gradually to a modified ES+2i medium [DMEM, l-glutamine, 15% (vol/vol) knockout serum replacement (Gibco), 0.1 mM nonessential amino acids, 0.1 mM 2-mercaptoethanol, 1,000 units/mL LIF, 1µM PD0325901 (Tocris) and 3 µM CHIR99021 (Tocris)].
The expression of pluripotency markers from iPSCkit was evaluated using a commercially available iPSC immunohistochemical characterization kit (Abcam) and an alkaline phosphatase live stain (Invitrogen).
Differentiation of iPSCs.
Two different methods were used to drive differentiation of iPSCs into cardiomyocytes. In the first method, iPSCkit were trypsinized into single cells and plated for 30–45 min with ES+2i medium on 100-mm2 gelatin-coated dishes to deplete MEFs. Feeder-depleted MEFs were then resuspended at a concentration of ∼4–8 × 104 cells/mL and allowed to grow for 3 d in MEF-conditioned differentiation medium [DM; IMDM (GIBCO), l-glutamine, 20% (vol/vol) FBS (Gibco), 0.1 mM nonessential amino acids, 0.1 mM 2-mercaptoethanol], supplemented with 2,000 units/mL LIF (day −3 to day 0). On day 0, iPSCkit were trypsinized and resuspended at a concentration of 2.5 × 104 cells/mL to generate EBs via the hanging-drop method, as previously described (15). EBs were then grown in hanging drops for 2 d in DM supplemented with 50 µg/mL AA or PBS, without LIF. On day 2, EBs were transferred into Petri dishes with DM+ AA or PBS and were grown in suspension for 2 more days. On day 4, EBs were transferred on gelatin-coated dishes with DM+ AA or PBS. Cells were fed every other day with DM+ AA or PBS. The first spontaneously beating cells emerged between late-afternoon of day 10 to day 12.
For the second method, feeder-depleted iPSCs were grown in MEF-conditioned DM for 3 d (day −3 to day 0), supplemented with 2,000 units/mL LIF, and 150 ng/mL recombinant mouse NOG (Noggin-FC; R&D) or 2 µM Dorso (Dorsomorphin; Tocris) or PBS, as previously described (43, 45). On day 0, EBs were generated as previously described in DM+ NOG or Dorso or PBS, without LIF. On day 1, treatment with NOG, Dorso, or PBS was discontinued and EBs were transferred into new Petri dishes or 12-well low-cluster plates (Corning) with fresh DM medium. EBs were grown in suspension for 2 more days before they were transferred into gelatin-coated plates.
To induce Cre-mediated recombination, (Z)-4-hydroxytamoxifen (Abcam) was supplemented to the cells at selected time points at a final concentration of 1 µM.
X-Gal and Salmon-Gal Histochemistry.
X-gal histochemistry was performed using a β-galactosidase staining kit in fixed intact mouse embryonic hearts or embryos carrying the cKitCreERT2;R26RLacZ or Isl1nLacZ alleles, as well as their respective littermates, according to the manufacturers’ instructions (Molecular Probes). Briefly, samples were incubated overnight in X-gal solution at 37 °C. The next day, X-gal was washed away with PBS, samples were photodocumented under a light microscope (Nikon), and incubated overnight in 30% (wt/vol) sucrose at 4 °C. Fixed tissues were then embedded in OCT and processed for cryosectioning. In some instances of cKitCreERT2;IRG;Isl1nLacZ and cKitCreERT2;Wnt1::Flpe;RC::Fela embryos, X-gal was performed after cryosectioning, following EGFP immunostaining. Staining with salmon-gal was performed as previously described (55). Briefly, samples were fixed for ∼20 min at room temperature in 0.2% glutaraldehyde/0.4% PFA, 5 mM EGTA and 2 mM MgCl2 in 0.1 M phosphate buffer (pH 7.3), followed by 3× 20-min washes with wash buffer [0.1% sodium deoxycholate, 0.2% IGEPAL, 2 mM MgCl2 and 0.1 M phosphate buffer (pH 7.3)]. Embryos were then incubated for 3–48 h in staining solution consisted of wash buffer supplemented with 1 mg/mL salmon-gal (Indofine) and 0.4 mM of Tetranitro blue tetrazolium salts dissolved in absolute ethanol (TCI).
Immunofluorescence Confocal Microscopy.
For cryosections, 10-μm-thick samples were postfixed for 10 min with 4% (vol/vol) PFA. For paraffin-embedded tissues, 4- to 5-μm-thick tissue sections were deparaffinized and rehydrated, as previously described (18). Antigen unmasking was performed by microwaving the slides for 2× 10 min in citrate buffer Solution, pH = 6 (Dako). Sections were then blocked for 1 h at room temperature with 10% (vol/vol) normal donkey serum (Chemicon International), followed by overnight incubation at 4 °C with the primary antibody.
The following antibodies were used: cKit (DAKO, eBiosciences, and R&D), EGFP (Abcam, Aves, Molecular Probes), β-galactosidase (Molecular Probes), MITF, PECAM1, α-smooth muscle actin (Sigma), antismooth muscle myosin heavy chain (SM1; Kamiya Biomedical), SM22a, cardiac troponin-I, cardiac troponin-T, Calponin, Tyrosinase, Oct4, Sox2, Nanog, SSEA1 (Abcam), cardiac myosin light chain-2v (cmlc2v), neurofilament-M, BFABP, Wnt1 (Novus Biologicals), Nkx2.5 (R&D and Santa Cruz Biotechnologies), GATA-4 (Santa Cruz Biotechnologies), Pax3, Isl-1 (40.2D6, Developmental Studies Hybridoma Bank), Factor VIII-related antigen (Biocare Medical), Tuj1 anti-Cre (Covance). Subsequently, the antibodies were visualized by incubating the sections for 1 h at 37 °C with FITC, Cy3 and Cy5- conjugated F(ab′)2 fragments of affinity-purified secondary antibodies (Jackson Immunoresearch) or Alexa 488 and Alexa 546 dyes (Molecular Probes). For MITF and SM1, tyramide signal amplification was used according to the manufacturer’s instructions (Perkin-Elmer). Slides were counterstained with DAPI, mounted with ProLong Antifade Gold reagent (Molecular Probes), and stored at 4 °C until further examination. Microscopic evaluations and image acquisitions were performed with a Zeiss LSM-710 Confocal Microscope (Carl Zeiss MicroImaging). The Zeiss ZEN software (v2009, Carl Zeiss Imaging Solutions) was used.
Gene-Expression Analysis.
Total RNA was extracted from iPSCkit at selected time points before and during the course of their differentiation into cardiomyocytes, using the RNeasy mini plus kit, according to the manufacturers’ instructions (Qiagen). cDNA synthesis was performed using the high-capacity cDNA reverse-transcription kit, according to the manufacturer’s instructions (Applied Biosystems). Quantitative PCR was performed using Taqman Universal Master mix in a iQ5 real-time PCR detection system (Bio Rad). All samples were run in triplicates and normalized to a GAPDH endogenous control. Relative fold-change was calculated using the 2−ΔΔCt method. The IDs for the Taqman Gene-expression assays are the following: GAPDH, Mm99999915_g1; BRACHYURY, Mm01318252_m1; MESP1, Mm00801883_g1; NKX2.5, Mm01309813_s1; ISL1, Mm00517585_m1; HNK1, Mm00661498_m1; SNAI2, Mm00441531_m1; CKIT, Mm00445212_m1; WNT1, Mm01300555_g1; PAX3, Mm00435491_m1; MITF-H (Variant 3), Mm01182481_m1; MITF (Variants. 1,2,3), Mm00434954_m1, SOX10, Mm01300162_m1; TBX18, Mm00470177_m1; WT1, Mm01337048_m1; TCF21, Mm00448961_m1; KDR, Mm01222421_m1.
Statistical Analysis.
For genetic lineage-tracing experiments, we estimate that to detect a minimum difference of 30 cardiomyocytic derivatives per cryosection between groups, with an expected SD of ±6 cardiomyocytes with a power of 90% and a 0.05 α-level, at least two embryos per group are necessary. Here, we used 10 embryos to perform genetic lineage-tracing of cKit during E7.5–E8.5; 12 embryos during E9.5–E12.5; 6 embryos of the ckitCreERT2;IRG:Isl1nLacZ genotype; and 5 embryos for intersectional genetic fate-mapping. Randomization and blinding were not applicable for this animal study. Statistical analyses were performed using GraphPad Prism v5.00 for Windows. A one-way ANOVA followed by Tukey’s post hoc tests were used for comparing changes in gene expression and beating EBs. Differences in the generation of NKX2.5+/EGFP+ progenitors between groups were compared using a Kruskall–Wallis test, followed by a Dunn’s post hoc analysis. All data met the assumptions of the tests. P < 0.05 was considered statistically significant. All values are reported as mean ± SEM.
Supplementary Material
Acknowledgments
We thank Dr. Sylvia Evans for providing the Isl1nLacZ mice. This study was funded by National Institutes of Health Grants R01 HL107110, R01 HL094849, R01 HL110737, R01 HL084275, and 5UM HL113460 (to J.M.H.); grants from the Starr foundation and the Soffer Family Foundation (to J.M.H.); and Deutsche Forschungsgemienschaft Grant SA 1374/1-3 (to D.S.).
Footnotes
Conflict of interest statement: K.E.H. and J.M.H. report having a patent for cardiac cell-based therapy. K.E.H. and J.M.H. own equity in Vestion Inc. and are members of the scientific advisory board and consultants of Vestion, Inc. J.M.H. is a board member of Vestion Inc. Vestion Inc. did not participate in funding this work. The other authors report no conflicts.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517201112/-/DCSupplemental.
References
- 1.Rana MS, Christoffels VM, Moorman AF. A molecular and genetic outline of cardiac morphogenesis. Acta Physiol (Oxf) 2013;207(4):588–615. doi: 10.1111/apha.12061. [DOI] [PubMed] [Google Scholar]
- 2.Witty AD, et al. Generation of the epicardial lineage from human pluripotent stem cells. Nat Biotechnol. 2014;32(10):1026–1035. doi: 10.1038/nbt.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: Human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10(1):16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453(7194):524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 5.Jain R, et al. HEART DEVELOPMENT. Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts. Science. 2015;348(6242):aaa6071. doi: 10.1126/science.aaa6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fioret BA, Heimfeld JD, Paik DT, Hatzopoulos AK. Endothelial cells contribute to generation of adult ventricular myocytes during cardiac homeostasis. Cell Reports. 2014;8(1):229–241. doi: 10.1016/j.celrep.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Handel B, et al. Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium. Cell. 2012;150(3):590–605. doi: 10.1016/j.cell.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li YX, et al. Cardiac neural crest in zebrafish embryos contributes to myocardial cell lineage and early heart function. Dev Dyn. 2003;226(3):540–550. doi: 10.1002/dvdy.10264. [DOI] [PubMed] [Google Scholar]
- 9.Kitajima S, Miyagawa-Tomita S, Inoue T, Kanno J, Saga Y. Mesp1-nonexpressing cells contribute to the ventricular cardiac conduction system. Dev Dyn. 2006;235(2):395–402. doi: 10.1002/dvdy.20640. [DOI] [PubMed] [Google Scholar]
- 10.Tomita Y, et al. Cardiac neural crest cells contribute to the dormant multipotent stem cell in the mammalian heart. J Cell Biol. 2005;170(7):1135–1146. doi: 10.1083/jcb.200504061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato M, Yost HJ. Cardiac neural crest contributes to cardiomyogenesis in zebrafish. Dev Biol. 2003;257(1):127–139. doi: 10.1016/s0012-1606(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 12.Reith AD, et al. W mutant mice with mild or severe developmental defects contain distinct point mutations in the kinase domain of the c-kit receptor. Genes Dev. 1990;4(3):390–400. doi: 10.1101/gad.4.3.390. [DOI] [PubMed] [Google Scholar]
- 13.Orr-Urtreger A, et al. Developmental expression of c-kit, a proto-oncogene encoded by the W locus. Development. 1990;109(4):911–923. doi: 10.1242/dev.109.4.911. [DOI] [PubMed] [Google Scholar]
- 14.Kataoka H, et al. Expressions of PDGF receptor alpha, c-Kit and Flk1 genes clustering in mouse chromosome 5 define distinct subsets of nascent mesodermal cells. Dev Growth Differ. 1997;39(6):729–740. doi: 10.1046/j.1440-169x.1997.t01-5-00009.x. [DOI] [PubMed] [Google Scholar]
- 15.Wu SM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127(6):1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 16.Beltrami AP, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114(6):763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 17.Kanashiro-Takeuchi RM, et al. Activation of growth hormone releasing hormone (GHRH) receptor stimulates cardiac reverse remodeling after myocardial infarction (MI) Proc Natl Acad Sci USA. 2012;109(2):559–563. doi: 10.1073/pnas.1119203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatzistergos KE, et al. Bone marrow mesenchymal stem cells stimulate cardiac stem cell proliferation and differentiation. Circ Res. 2010;107(7):913–922. doi: 10.1161/CIRCRESAHA.110.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Florea V, et al. Agonists of growth hormone-releasing hormone stimulate self-renewal of cardiac stem cells and promote their survival. Proc Natl Acad Sci USA. 2014;111(48):17260–17265. doi: 10.1073/pnas.1420375111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chugh AR, et al. Administration of cardiac stem cells in patients with ischemic cardiomyopathy: The SCIPIO trial: Surgical aspects and interim analysis of myocardial function and viability by magnetic resonance. Circulation. 2012;126(11) Suppl 1:S54–S64. doi: 10.1161/CIRCULATIONAHA.112.092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Berlo JH, et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509(7500):337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mummery CL, Lee RT. Is heart regeneration on the right track? Nat Med. 2013;19(4):412–413. doi: 10.1038/nm.3158. [DOI] [PubMed] [Google Scholar]
- 23.Heger K, et al. CreER(T2) expression from within the c-Kit gene locus allows efficient inducible gene targeting in and ablation of mast cells. Eur J Immunol. 2014;44(1):296–306. doi: 10.1002/eji.201343731. [DOI] [PubMed] [Google Scholar]
- 24.Klein S, et al. Interstitial cells of Cajal integrate excitatory and inhibitory neurotransmission with intestinal slow-wave activity. Nat Commun. 2013;4:1630. doi: 10.1038/ncomms2626. [DOI] [PubMed] [Google Scholar]
- 25.Schönhuber N, et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat Med. 2014;20(11):1340–1347. doi: 10.1038/nm.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson YM, Richards KL, Ford-Perriss ML, Panthier JJ, Murphy M. Neural crest cell lineage segregation in the mouse neural tube. Development. 2004;131(24):6153–6162. doi: 10.1242/dev.01533. [DOI] [PubMed] [Google Scholar]
- 27.Goldstein BJ, et al. Adult c-Kit(+) progenitor cells are necessary for maintenance and regeneration of olfactory neurons. J Comp Neurol. 2014;523(1):15–31. doi: 10.1002/cne.23653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tallini YN, et al. c-kit expression identifies cardiovascular precursors in the neonatal heart. Proc Natl Acad Sci USA. 2009;106(6):1808–1813. doi: 10.1073/pnas.0808920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ubil E, et al. Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature. 2014;514(7524):585–590. doi: 10.1038/nature13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127(8):1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 31.Stottmann RW, Choi M, Mishina Y, Meyers EN, Klingensmith J. BMP receptor IA is required in mammalian neural crest cells for development of the cardiac outflow tract and ventricular myocardium. Development. 2004;131(9):2205–2218. doi: 10.1242/dev.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown CB, et al. PlexinA2 and semaphorin signaling during cardiac neural crest development. Development. 2001;128(16):3071–3080. doi: 10.1242/dev.128.16.3071. [DOI] [PubMed] [Google Scholar]
- 33.Tamura Y, et al. Neural crest-derived stem cells migrate and differentiate into cardiomyocytes after myocardial infarction. Arterioscler Thromb Vasc Biol. 2011;31(3):582–589. doi: 10.1161/ATVBAHA.110.214726. [DOI] [PubMed] [Google Scholar]
- 34.Dymecki SM, Ray RS, Kim JC. Mapping cell fate and function using recombinase-based intersectional strategies. Methods Enzymol. 2010;477:183–213. doi: 10.1016/S0076-6879(10)77011-7. [DOI] [PubMed] [Google Scholar]
- 35.Jensen P, et al. Redefining the serotonergic system by genetic lineage. Nat Neurosci. 2008;11(4):417–419. doi: 10.1038/nn2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engleka KA, et al. Islet1 derivatives in the heart are of both neural crest and second heart field origin. Circ Res. 2012;110(7):922–926. doi: 10.1161/CIRCRESAHA.112.266510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutson MR, Kirby ML. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin Cell Dev Biol. 2007;18(1):101–110. doi: 10.1016/j.semcdb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waldo KL, Kumiski DH, Kirby ML. Association of the cardiac neural crest with development of the coronary arteries in the chick embryo. Anat Rec. 1994;239(3):315–331. doi: 10.1002/ar.1092390310. [DOI] [PubMed] [Google Scholar]
- 39.Tshori S, et al. Transcription factor MITF regulates cardiac growth and hypertrophy. J Clin Invest. 2006;116(10):2673–2681. doi: 10.1172/JCI27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y, et al. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304(1):286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christoforou N, et al. Mouse ES cell-derived cardiac precursor cells are multipotent and facilitate identification of novel cardiac genes. J Clin Invest. 2008;118(3):894–903. doi: 10.1172/JCI33942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8(2):228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 43.Hao J, et al. Dorsomorphin, a selective small molecule inhibitor of BMP signaling, promotes cardiomyogenesis in embryonic stem cells. PLoS One. 2008;3(8):e2904. doi: 10.1371/journal.pone.0002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi M, Stottmann RW, Yang YP, Meyers EN, Klingensmith J. The bone morphogenetic protein antagonist Noggin regulates mammalian cardiac morphogenesis. Circ Res. 2007;100(2):220–228. doi: 10.1161/01.RES.0000257780.60484.6a. [DOI] [PubMed] [Google Scholar]
- 45.Yuasa S, et al. Transient inhibition of BMP signaling by Noggin induces cardiomyocyte differentiation of mouse embryonic stem cells. Nat Biotechnol. 2005;23(5):607–611. doi: 10.1038/nbt1093. [DOI] [PubMed] [Google Scholar]
- 46.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27(3):275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baggiolini A, et al. Premigratory and migratory neural crest cells are multipotent in vivo. Cell Stem Cell. 2015;16(3):314–322. doi: 10.1016/j.stem.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 48.Buitrago-Delgado E, Nordin K, Rao A, Geary L, LaBonne C. NEURODEVELOPMENT. Shared regulatory programs suggest retention of blastula-stage potential in neural crest cells. Science. 2015;348(6241):1332–1335. doi: 10.1126/science.aaa3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Motohashi T, Kitagawa D, Watanabe N, Wakaoka T, Kunisada T. Neural crest-derived cells sustain their multipotency even after entry into their target tissues. Dev Dyn. 2014;243(3):368–380. doi: 10.1002/dvdy.24072. [DOI] [PubMed] [Google Scholar]
- 50.Ellison GM, et al. Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell. 2013;154(4):827–842. doi: 10.1016/j.cell.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 51.Ferreira-Martins J, et al. Cardiomyogenesis in the developing heart is regulated by c-kit-positive cardiac stem cells. Circ Res. 2012;110(5):701–715. doi: 10.1161/CIRCRESAHA.111.259507. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Williams AR, et al. Enhanced effect of combining human cardiac stem cells and bone marrow mesenchymal stem cells to reduce infarct size and to restore cardiac function after myocardial infarction. Circulation. 2013;127(2):213–223. doi: 10.1161/CIRCULATIONAHA.112.131110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karantalis V, et al. Synergistic effects of combined cell therapy for chronic ischemic cardiomyopathy. J Am Coll Cardiol. 2015 doi: 10.1016/j.jacc.2015.08.879. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Echelard Y, Vassileva G, McMahon AP. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development. 1994;120(8):2213–2224. doi: 10.1242/dev.120.8.2213. [DOI] [PubMed] [Google Scholar]
- 55.Sundararajan S, Wakamiya M, Behringer RR, Rivera-Pérez JA. A fast and sensitive alternative for β-galactosidase detection in mouse embryos. Development. 2012;139(23):4484–4490. doi: 10.1242/dev.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.