Significance
The nucleus is structured into chromatin and interchromatin compartments. Its sieve-like architecture permits individual proteins to rapidly diffuse while large macromolecular assemblies are corralled in the interchromatin. Herpesvirus capsids assemble in the nucleus and have to access the nuclear periphery for exit. It was hypothesized that nuclear herpesvirus capsids recruit filamentous actin and molecular motor protein to overcome nuclear entrapment. Here we use “ring-sheet” microscopy to track nuclear capsids with high spatiotemporal resolution. We report that nuclear herpesvirus capsids do not use directed motility. Instead, virus infection changes nuclear architecture, which allows capsids to reach the nuclear membranes by diffusion. Our findings illustrate a pathway for very large macromolecular assemblies to cross the nucleoplasm without directed motility.
Keywords: herpes, capsid, nucleus, ring sheet, light sheet
Abstract
The nuclear chromatin structure confines the movement of large macromolecular complexes to interchromatin corrals. Herpesvirus capsids of approximately 125 nm assemble in the nucleoplasm and must reach the nuclear membranes for egress. Previous studies concluded that nuclear herpesvirus capsid motility is active, directed, and based on nuclear filamentous actin, suggesting that large nuclear complexes need metabolic energy to escape nuclear entrapment. However, this hypothesis has recently been challenged. Commonly used microscopy techniques do not allow the imaging of rapid nuclear particle motility with sufficient spatiotemporal resolution. Here, we use a rotating, oblique light sheet, which we dubbed a ring-sheet, to image and track viral capsids with high temporal and spatial resolution. We do not find any evidence for directed transport. Instead, infection with different herpesviruses induced an enlargement of interchromatin domains and allowed particles to diffuse unrestricted over longer distances, thereby facilitating nuclear egress for a larger fraction of capsids.
The nucleus is structured into chromatin and interchromatin compartments, giving it a “sponge-like” appearance, with the chromatin representing the actual sponge material and the interchromatin regions representing the enclosed pores, tubes, tunnels, and cisterna. This conceptual model has important implications for how differently sized molecules could move inside the nucleus. Although small molecules like GFP, streptavidin, fluorescent dextrans, or mRNA can mostly freely diffuse throughout the nucleus (1–3), large macromolecular assemblies, such as 100-nm fluorescent beads and promyelocytic leukemia (PML) or Cajal bodies, are trapped in interchromatin spaces called corrals. These corrals slowly move as a result of chromatin dynamics, and particles can also escape over long time scales (4, 5). Herpesvirus capsids of 125-nm diameter are assembled in the nucleus and must move to the nuclear periphery to exit the nucleus by budding through the nuclear membranes (6). Similarly, large host cargo like ribonucleoprotein (RNP) particles may need to move through the nucleus before they exit the nucleus in the same way (7). Earlier, we and others provided data that suggested that herpesvirus capsids use an active, directed mechanism based on F-actin to transport through the nucleoplasm (8, 9) and/or that nuclear F-actin is involved in capsid assembly. However, we recently showed that herpesviruses do not induce nuclear F-actin in most cells, and, more importantly, that nuclear capsid motility is not dependent on nuclear F-actin (10). This led us to reinvestigate nuclear capsid motility by single particle tracking to address whether molecular motors power it.
During herpesvirus infection, the interchromatin domains enlarge and the nuclear volume increases as much as twofold (11). At the same time, viral replication compartments expand, move, and coalesce, but do not mix (12–16). By using a light-sheet modality we dubbed a ring-sheet, we were able to trace intranuclear herpes virus capsids with high spatial and temporal precision and low cytotoxicity. Our data show that nuclear herpesvirus capsids do not use a directed transport mechanism. Instead, we find by single particle tracking and analysis of capsid motility that capsids diffuse in enlarged nuclear spaces. We conclude that the previously described size increase of the interchromatin domain increases the likelihood of capsids to reach the nuclear envelope by diffusion.
Results
Ring-Sheet Microscopy Allows Recording of Rapid Nuclear Dynamics.
We found that conventional imaging modalities did not provide both the speed and sensitivity required to generate image data suitable for a high-fidelity recording of capsid motility. Recently, light-sheet microscopy has come into focus (17). Its basic principle, the separation of illumination and detection, enables selective imaging of parts of tissues or small organisms, but is also advantageous for smaller objects like single cells. Commonly used confocal microscopy illuminates the entire sample above and below the focal plane, and then filters out-of-focus emission. Instead, light-sheet microscopy illuminates only the focal plane, increasing contrast and reducing phototoxicity.
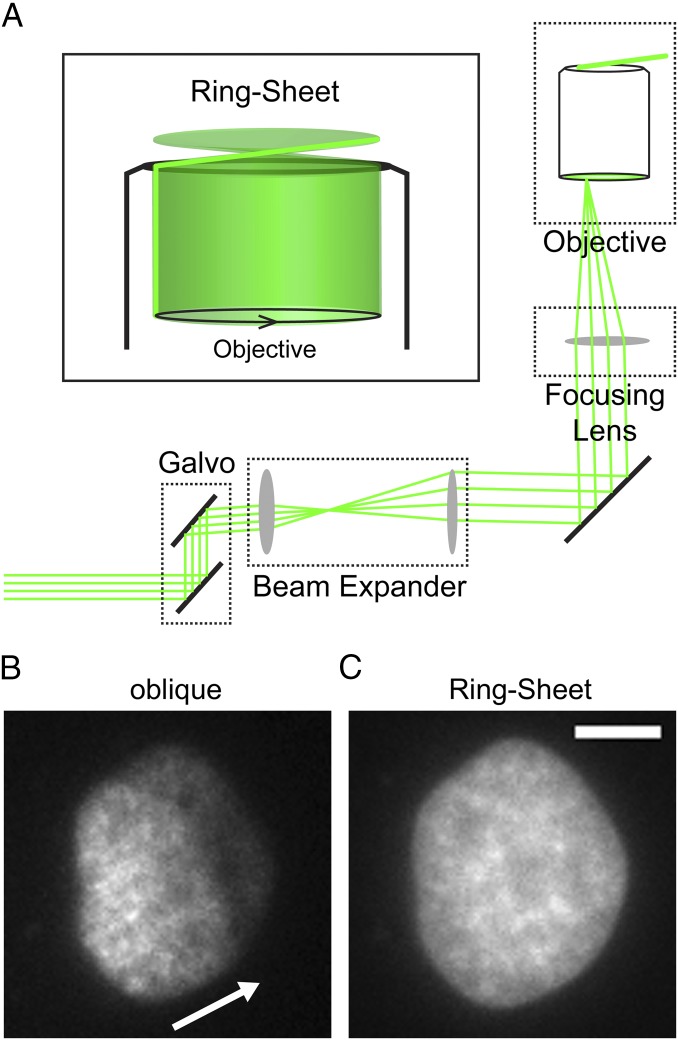
We developed a custom microscope that generates a rapidly rotating oblique light sheet by introducing a two-axis galvo system (Fig. 1A) into an otherwise standard objective-based total internal reflection fluorescence (TIRF) microscopy system (18). This allowed us to project a circle of light with variable diameter onto the back focal plane of the objective, resulting in a virtual ring-shaped sheet emanating from the objective (Fig. 1A). We therefore dubbed this modality a ring-sheet. Although conventional light-sheet techniques typically rely on two low-N.A. objectives, this modality uses only one high-N.A. objective to generate the illuminating light sheet and to detect the emission light. This has several advantages. First, the xy and especially the z axis resolutions are enhanced, as z-axis resolution (resolutionz) changes quadratically with increasing N.A. (Abbe resolutionz = 2λ/N.A.2). Second, in contrast to similar oblique techniques (19), ring-sheet microscopy does not suffer from fringing and shading artifacts because the illumination direction is rotated rapidly and averaged over the acquisition time. An example of how image quality is enhanced by averaging the illumination direction is shown in Fig. 1 B and C. In static oblique mode, the light sheet illuminated the sample from one direction (Fig. 1B, arrow). This induces shading artifacts as dense sample regions cast a shadow. This can be seen in Fig. 1B as decreasing intensity from the left to the right of the nucleus. In addition, interaction of imperfect optical surfaces with coherent laser light induces interference that overlays the sample with a pattern of dark and bright fringes. In contrast, the rapidly rotating ring-sheet illumination produces even excitation (Fig. 1C). In addition, ring-sheet microscopy increases the image contrast and reduces phototoxicity, which enabled us to image herpesvirus nuclear capsid motility with high spatial and temporal resolution for longer times.
Fig. 1.
Ring-sheet microscopy. (A) Schematic diagram of the optical path in a ring-sheet setup. A 2D galvo system steers a collimated laser beam of ∼1.25 cm diameter. A 2× beam expander expands the beam such that it fills a 2.5-cm-diameter, 200-mm focusing lens. The focusing lens focuses the beam at the back focal plane of the microscope objective (objective). As the galvo steers the beam in a ring, the objective generates a rotating sheet of light (Inset). (B and C) Comparison of static oblique and ring-sheet microscopy. (Scale bar: 5 µm.) A PtK2 cell was stained with 0.5 µg/mL Hoechst 33342 and imaged with (B) oblique microscopy, which uses a highly inclined directional beam (direction indicated by white arrow) or (C) ring-sheet microscopy. Compared with C, it can be clearly seen how the directional illumination induces shading artifacts and also suffers from interference fringes, which overlay a pattern of bright and dark regions onto the sample structure.
Nuclear Herpesvirus Capsids Do Not Use Directed Transport.
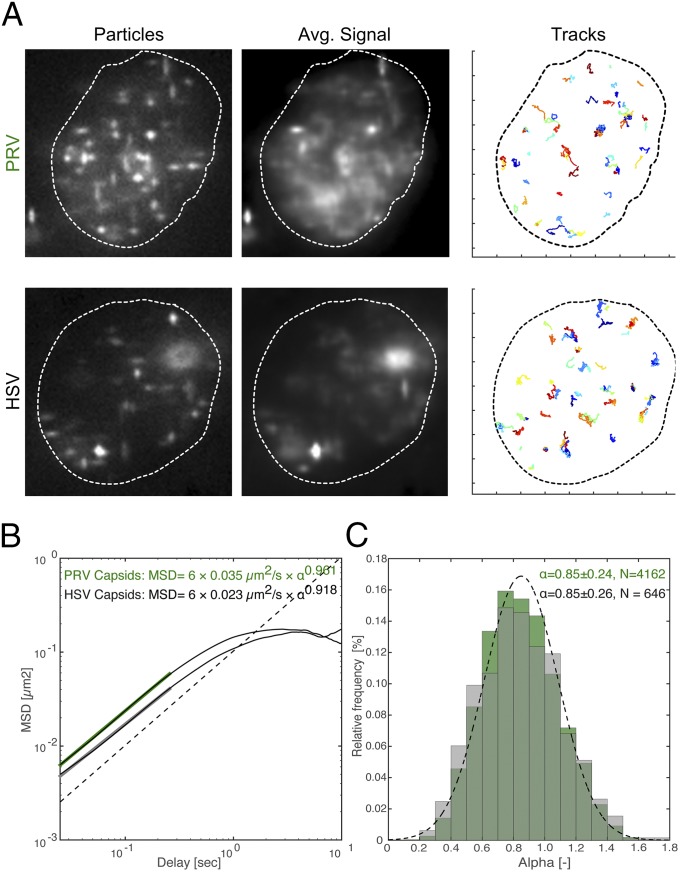
To quantify herpesvirus nuclear capsid dynamics, we infected PtK2 cells with pseudorabies virus (PRV; i.e., suid herpesvirus 1) or HSV-1 that expresses a fluorescent capsid protein (20–22). We recorded motility with ring-sheet microscopy as soon as the first capsids appeared in the nucleus [4 hours postinfection (hpi) or 5 hpi, respectively]. Imaging was done at a minimum of 36 frames per second by using a cylindrical lens to determine particle positions in 3D over time. Fig. 2A shows still images and average intensity projections over time of representative cells. The degree of smearing in the average intensity projection indicates particle motility (Movies S1 and S2). We were able to record 5,469 PRV tracks and 876 HSV-1 tracks (Table S1 shows a summary of the datasets used throughout this work). All tracks were gapless and 40 frames or longer, and localization accuracy was better than ±40 nm on all three axes (±SD of immobile particles).
Fig. 2.
The major mode of nuclear herpesvirus capsid motility is diffusion. PtK2 cells were infected with mEFGP-VP26–tagged PRV or EGFP-VP26–tagged HSV-1 at a multiplicity of infection (MOI) of 100. At 4 h or 5 h after infection, respectively, cells were stained with 0.5 µg/mL Hoechst 33342 and imaged with ring-sheet microscopy at a frame rate of 36 frames per second for 28 s (PRV and HSV). (A) Outlines indicate nuclear boundaries as visualized by Hoechst staining. The first column (particles) depicts a single representative frame showing capsid or bead positions. The second column (avg. signal) depicts the average projection over the whole imaging time, indicating particle motility. The third column (tracks) depicts all recorded tracks over 40 frames total length. Tick marks along the axis correspond to 2-µm increments. (B) The average MSD was calculated from 5,469 PRV (green) and 876 HSV-1 (black) tracks and plotted in log-log space. Its linear portion was fitted with an MSD of 6Dtα (D, diffusion coefficient; t, time) for a delay of approximately 270 ms. Slopes indicate the anomalous diffusion exponent α. The dotted line indicates a slope of 1. (C) The first 20% of each individual MSD curve was fitted with an MSD of 6Dtα to determine α. Only fits with an R2 > 0.8 were kept (4,162 of 5,469 tracks for PRV, 646 of 876 tracks for HSV-1), and the distributions of α values were plotted. The dotted line indicates a normal distribution with a mean of 0.85 and a SD of 0.24.
Table S1.
Datasets used for figures
| No. | Name | No. of tracks | Imaging conditions | Figures |
| 1 | PtK2/PRV-mEGFP-VP26 | 5,469 | RS, 27 ms/f 1,000 f | Figs. 2, 3, and 5 |
| 2 | PtK2/HSV-1-EGFP-VP26 | 876 | RS, 27 ms/f 1,000 f | Figs. 2 and 3 |
| 3 | PtK2/beads | 289 | O, 24 ms/f 1,000 f | Figs. 3 and 5 |
| 4 | PtK2/PRV/beads | 226 | O, 36 ms/f 1,000 f | Figs. 3 and 5 |
f, frame; O, static oblique illumination; RS, ring-sheet.
To determine whether nuclear herpesvirus capsids exhibited diffusion or directed transport, we first calculated the mean squared displacement (MSD) for all tracks. Plotting MSD curves in log-log space can be used as a diagnostic tool to distinguish diffusion from directed transport, because the MSD grows quadratically with time in the case of directed transport, but only linearly in the case of diffusion. Accordingly, the slope of a particle MSD undergoing pure directed transport is equal to 2, whereas a diffusing particle produces a slope equal to 1. Divergences from diffusion or a slope of 1 are denoted by an anomalous diffusion exponent, α (23).
To measure α, we averaged all 5,469 PRV and 876 HSV-1 MSD curves and fitted the short-time scale regime to determine the slope (up to a delay of ∼0.27 s; Fig. 2B). Alternatively, we also fitted the first 20% of individual curves (individual fits were excluded if R2 < 0.8, delay of ∼0.36 s; Fig. 2C). Both methods yielded very similar results, with mean values for α near 0.9 (α values between 0.9 and 1.1 are consistent with free diffusion). Thus, capsids appeared to move by nearly free diffusion over short time scales.
Fitting thousands of individual curves also gave us the opportunity to analyze the distribution of α values. A previous report (8) defined all fits with an α greater than 1.1 as directed. If a significant population of particles had moved by directed motility, we should observe a bimodal α distribution. Instead, the α values of PRV and HSV-1 have a normal distribution (Fig. 2C, dotted line), with a mean α value <1 (one-sample t test, P < 10−44), indicating that there was not a significant population of tracks that exhibit directed transport.
Herpesvirus Infection Enlarges Interchromatin Corrals.
Whereas the MSD curves for capsids were roughly linear at short time scales (delay < 1 s), they saturated over longer timescales, suggesting that particles had diffused within corrals. Previously, such confined diffusion of particles was observed in interchromatin spaces (1).
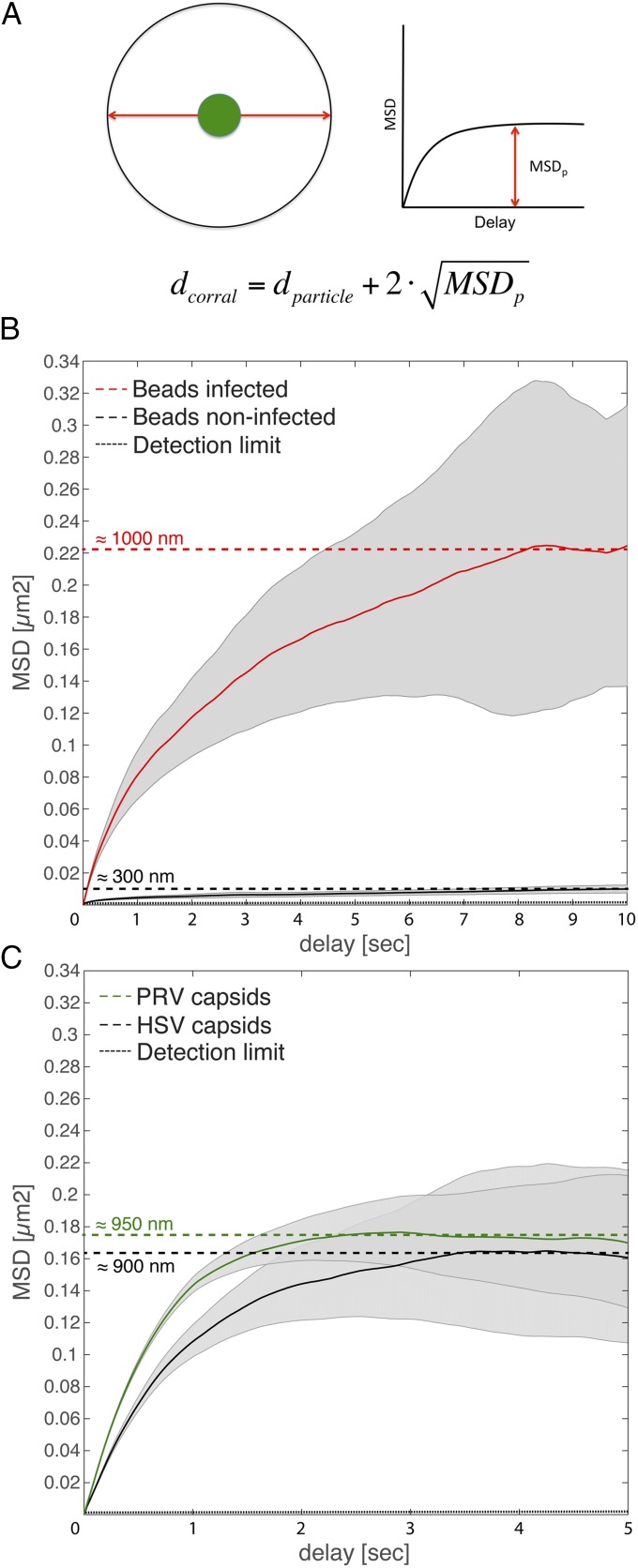
To test this hypothesis, we microinjected beads as inert nanoscale probes of the local nuclear architecture into infected or noninfected cells. After microinjection with PEG-passivated 100-nm beads, cells were allowed to recover overnight. The next day, cells were infected with PRV or imaged directly. We were able to record 289 bead tracks in noninfected cells and 226 bead tracks in infected cells. By plotting the average MSD curves for noninfected cells, the mean corral size can be calculated as depicted in Fig. 3A. Beads in noninfected cells diffused very little, and appeared to be confined to corrals of approximately 300 nm (Fig. 3B, black line), consistent with previous reports (5). This was also evident from the tracks themselves (Movie S3), as beads were able to move only very short distances. Surprisingly, the beads behaved very differently in the infected cells and moved throughout the nucleus (Movie S4). Plotting the MSD and calculating the mean corral size revealed that corrals in infected cells had been enlarged to ∼1,000 nm (Fig. 3B, red line). Reanalyzing the data of Fig. 2 to investigate longer time scales showed that HSV-1 and PRV capsids had diffused in corrals of approximately 900 nm (Fig. 3C). These results indicate that herpesvirus infection increases the corral size by at least threefold, and suggest that capsids move passively within these corrals similarly to inert beads.
Fig. 3.
Herpesvirus infection leads to increased nuclear corral size. PtK2 cells were infected with mEFGP-VP26–tagged PRV or EGFP-VP26–tagged HSV-1 at an MOI of 100 or microinjected with passivated, 100-nm latex beads and infected the next day. At 4 h or 5 h after infection, respectively, cells were stained with 0.5 µg/mL Hoechst 33342 and imaged with ring-sheet microscopy at a frame rate of 36 frames per second for 28 s (PRV and HSV) or imaged with static oblique microscopy at 42 frames per second for 24 s (noninfected beads) or at 28 frames per second for 36 s. (A) Estimation of corral sizes. A particle (green) is trapped in an idealized spherical corral. Its maximum mean displacement is the radius of the sphere minus the radius of the particle (red arrow). Its MSD curve will therefore saturate. The mean corral diameter can be estimated by doubling the square root of MSDp and adding the particle diameter. (B) Average MSD curves as well as their SE (gray) of bead tracks from noninfected cells (black line) and from PRV-infected cells (red line) were plotted in linear space. Their saturation points were determined (dotted lines), and approximate corral sizes are indicated. (C) The same MSD dataset as used in Fig. 1 (green, PRV; black, HSV-1) was plotted in linear space, and their saturation point estimated. The dashed line indicates the detection limit of 40 nm2.
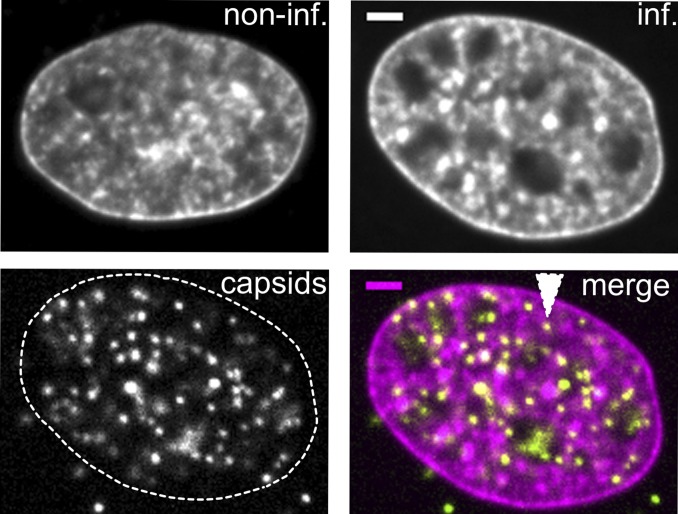
The use of Hoechst to stain chromatin in noninfected and infected cells also suggested an increase in the apparent corral size (Fig. 4). Infected nuclei exhibited large areas depleted of DNA and chromatin that often harbored capsids. However, the capsids were not exclusively located in these large spaces but also in much smaller chromatin-depleted regions throughout the nucleoplasm (e.g., Fig. 4, white triangle). These observations of enlarged interchromatin regions and a more porous chromatin structure are consistent with a previous report describing a marginalization of host chromatin and an expansion of interchromatin compartments during HSV-1 infection (11).
Fig. 4.
Nuclear capsids localize to enlarged nuclear corrals. PtK2 cells were left noninfected (noninf) or infected with mEGFP-VP26 tagged PRV at an MOI of 100 (inf). At 4 h postinfection, cells were fixed and stained with 0.5 µg/mL Hoechst 33342 Abs imaged with spinning-disk microscopy. After infection, chromatin structure becomes more porous and capsids can be found in large and small (white arrow) corrals. (Scale bars: 2 µm.)
Capsid “Hopping” Between Corrals Is Nondirected.
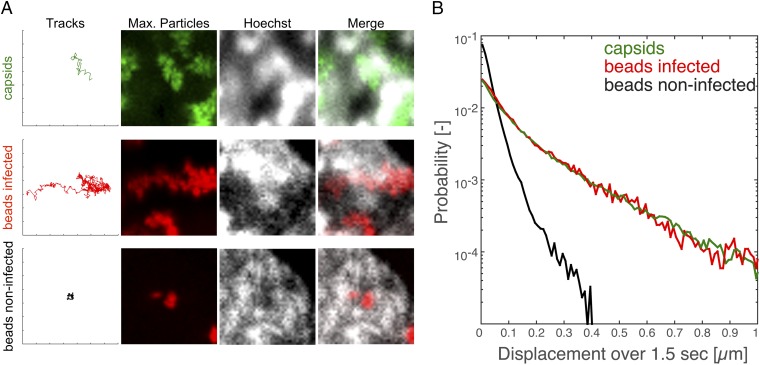
Our results indicate that herpesvirus capsid motility is based on diffusion, and not on directed transport. However, it is possible that a small fraction of directed motility might have been averaged out in the aforementioned analyses. Indeed, we observed a small fraction of tracks that seemed to be more directed, and even moved through chromatin-enriched regions (Fig. 5A and Movies S5–S7). We dubbed this behavior “hopping” because it often looked as if particles could “hop” between corrals. This capsid motion might have been caused by specific recruitment of motor complexes to viral capsids, to biopolymerization, or to other directed transport mechanisms. Alternatively, the hopping motion could have been caused by structural rearrangements and random diffusive motion within the nucleus. To distinguish between these possibilities, we compared the tracks of capsids to tracks from microinjected beads in infected cells; PEG-passivated beads are unable to bind to molecular motors and should passively report on local fluctuating motion. Consistent with purely random capsid motion, bead behavior in infected cells was indistinguishable from capsids. Beads in infected nuclei exhibited the same hopping behavior as capsids characterized by apparent directed motion over several hundred nanometers (Fig. 5A). To quantify this behavior, we calculated the probability of beads or capsids (i.e., we reanalyzed the data shown in Fig. 3) to move a particular distance over a specific period. If capsids use an active transport mechanism, they should have a higher probability to move larger distances. However, we did not find any difference between capsids and beads in infected cells. Because the beads we used were passivated and previously reported to be biologically inert (5, 24), and our observed bead dynamics in noninfected cells matched previous reports (5), we conclude that the capsid hopping behavior was not caused by an active, directed transport mechanism, but most likely by random diffusive motion.
Fig. 5.
Displacement probabilities for capsids and beads are similar over longer time scales. PtK2 cells were infected with mEFGP-VP26–tagged PRV at an MOI of 100, microinjected with passivated 100-nm latex beads, or microinjected and infected. At 4 h after infection, cells were stained with 0.5 µg/mL Hoechst 33342 and imaged with ring-sheet microscopy at a frame rate of 36 frames per second for 28 s (PRV) or imaged with static oblique microscopy at 42 frames per second for 24 s (noninfected beads) or at 28 frames per second for 36 s (infected beads). (A) Tracks showing events of seemingly directed motility through chromatin-enriched regions for both capsids (first row) and microinjected beads in infected cells (second row). In contrast, microinjected beads in noninfected cells (third row) move much less. Tick marks along the axis correspond to 1-µm increments. (B) The displacements of capsids and beads in infected and noninfected cells over a delay of 1.5 s was calculated and plotted in semilog space depicting the probability for a particle to move a certain distance over 1.5 s (5,469 tracks for PRV, 289 tracks for noninfected beads, 226 tracks for infected beads).
Uniform Distribution of Capsid Assembly and Flattened Nuclear Geometry Enables Efficient Capsid Flux to the Nuclear Membrane.
Intranuclear capsids have to reach the inner nuclear membrane for egress. Our data indicate that nuclear capsid dynamics are purely stochastic processes that do not involve directed transport. This conclusion is surprising because directed motility is necessary for long-distance viral movement through the cytoplasm (25). After nuclear egress, capsids must travel long distances through the viscous cytoplasm to reach the plasma membrane, necessitating active transport processes by motor proteins along filament tracks. In contrast, capsids assembling in the nucleus do not have a single point of origin. Rather, capsid subunits should be small enough to freely diffuse throughout the nucleus (1), and capsid assembly is evenly distributed throughout the nuclear volume. Thus, capsids that assemble near the nuclear membrane do not need to traverse the entire nuclear volume and could reach the nuclear membrane without the need for active transport mechanisms.
To investigate this concept, we evaluated temporal SD intensity projections of PRV capsid signals and found that capsid motility was distributed over most of the nucleus at 4 hpi (Fig. 6A and Movie S8). Of course, only particles in corrals near the nuclear membrane would be able to access the nuclear membrane directly for nuclear egress. We therefore determined what volume fraction of the nucleus was within 0.9 μm (i.e., the previously determined mean corral diameter) from the nuclear membrane. We measured the dimensions of 39 infected PtK2 nuclei at 4 hpi and found that they can be approximated as an ellipsoid of 16.5 ± 2.3 µm length by 10.5 ± 1.9 µm width by 3.3 ± 0.5 µm height (mean ± SD). Because the nuclei are relatively flat, a 0.9-µm region around the nuclear membranes accounts for approximately 66% (range, 59–76%) of the nuclear volume. (Similarly, the region within 0.9 µm of the nuclear membranes accounts for 50% of the nuclear volume, for a spherical nucleus of the same volume given earlier.) Accordingly, the majority of the capsids might have instant access to the nuclear membrane through diffusion within 0.9-µm corrals. In comparison, if virus infection would not change nuclear architecture, capsids could access a volume fraction of only 31% (range, 27–37%) based on the dimensions of noninfected cells [16.1 ± 2.1 µm length by 10.3 ± 2.1 µm width by 2.5 ± 0.4 µm height (mean ± SD from 69 cells)] and a corral size of approximately 0.3 µm. The infection-induced large-scale changes in nuclear architecture therefore result in a more than twofold gain in membrane accessible volume fraction.
Fig. 6.
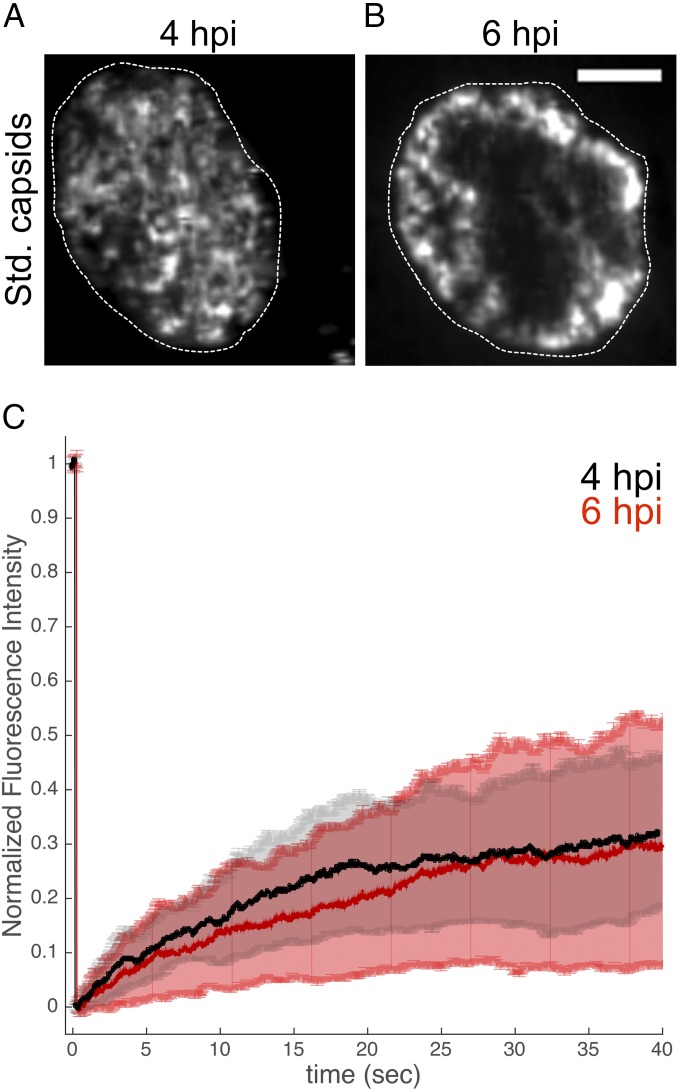
Virus-induced reorganization of the nuclear structure leads to a concentration of capsids near nuclear membranes. (A and B) PtK2 cells were infected with mEFGP-VP26–tagged PRV at an MOI of 100, stained with 0.5 µg/mL Hoechst 33342, and imaged with ring-sheet microscopy at a frame rate of 36 frames per second for 28 s at 4 hpi (B) or 6 hpi (C). Regions of capsid motility are represented as the SD of signals at each pixel over the entire acquisition time (Std. capsids). Outlines depict nuclear dimensions as determined by Hoechst staining. (Scale bar: 5 µm.) (C) An ∼2 µm region was bleached into cells at 4 hpi (black) or 6 hpi (red), and FRAP was measured.
Corrals Can Be Rapidly Replenished with Capsids.
Capsids are able to cross from corral to corral by hopping (Fig. 5). However, the impact of this phenomenon on the ability of capsids to replenish the membrane-proximal region from the interior nuclear volume is hard to estimate because it depends on the local concentration of capsids. To measure this flux, we performed fluorescence recovery after photobleaching (FRAP) experiments, in which we bleached nuclear regions∼2 µm in diameter (corresponding to a maximum of four tightly packed corrals) and then determined the rate of recovery of fluorescent capsid protein. We measured a half time of 16 s for the mobile fraction (36% of fluorescence) to recover (Fig. 6C). However, FRAP analysis might be less reliable in this case because the recovery of soluble capsid protein and assembled capsids are measured. We therefore also manually determined the time for a capsid to arrive in the middle of the bleached region. Among 27 cells, capsids moved through the center of the bleached area in 21 cells after 12 ± 2 s (mean ± SD; Movie S9). This indicates that it takes only approximately 10 s to replenish membrane-associated corrals from the nuclear interior.
Motile PRV Capsids Concentrate Near the Nuclear Envelope at Later Times During Infection.
As herpesvirus infection progresses, nuclear chromatin is dynamically marginalized and modified with progressing infection (11, 26, 27). We therefore investigated whether PRV capsid motility changed between 4 hpi and 6 hpi. However, the capsid density increased so strongly during this period that tracking of individual capsids became impossible. We chose to depict capsid motility by the temporal SD of signal fluctuations. Here, only large fluctuations of intensity caused by rapidly moving capsids will produce a signal, whereas the background of soluble VP26 evenly distributed in the nuclear volume is deemphasized. This revealed that capsid motility was often restricted to a “shell” surrounding a capsid-depleted nuclear interior (Fig. 6B and Movie S10). We measured the motility in such capsid-filled shells by FRAP and found almost no difference at 6 hpi (36% mobile fraction, 16 s t1/2) compared with 4 hpi (Fig. 6C). Also, manual counting of the time until the first particle had moved through the center of the bleached area gave similar results (15 of 25 cells showed particles moving through the middle after 14 ± 3 s, mean ± SD; Movie S11). The enrichment of motile capsids in the margins might bring an even greater fraction of virus capsids into close proximity of nuclear membranes.
PRV Viral Replication Compartment Maturation Correlates with Capsid Marginalization.
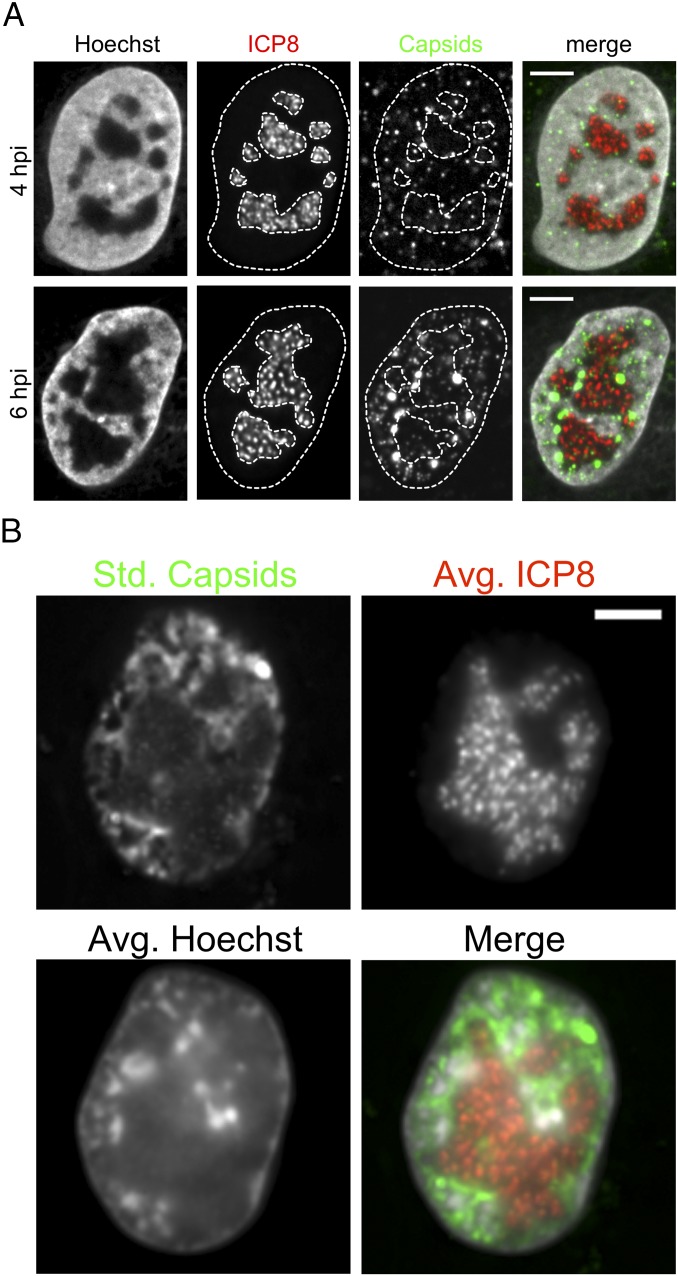
The viral DNA replication compartment might displace host chromatin and virus capsids at 6 hpi (28). To visualize the viral replication compartment, we generated a PRV mutant that expresses a fluorescently tagged ICP8, the viral single-stranded viral DNA-binding protein. Viral replication compartments expand and coalesce with progressing infection, whereas the host chromatin is marginalized (11, 15, 16). Indeed, ICP8, which indicates the replication compartments, was localized in the center of the nuclei, and the PRV capsids were reduced in these regions (Fig. 7A). Live-cell microscopy revealed that capsids moved mostly within the 1–2-µm-thick marginalized chromatin that surrounded the replication compartments (Fig. 7B and Movie S12).
Fig. 7.
Replication compartment expansion marginalizes capsids to the nuclear periphery. PtK2 cells were infected with a PRV mutant expressing fluorescent capsids (mRFP-VP26) and ICP8-mEGFP. (A) Cells at 4 and 6 hpi were fixed and stained with 0.5 µg/mL Hoechst 33342 and imaged with spinning-disk microscopy. Nucleus and replication compartment outlines are depicted as dotted lines. (B) Cells at 6 hpi were stained with Hoechst and imaged live with ring-sheet microscopy at a frame rate of 12 frames per second (28 ms per channel) for 67 s. Capsid motility is depicted as the SD of capsid signals over time (std. capsids), and the viral replication compartment as indicated by ICP8 is shown as an average projection over time (avg. ICP8). Nuclear chromatin is also shown as an average projection over time (avg. Hoechst). The merge image shows capsid signals in green, ICP8 in red, and chromatin in gray. (Scale bars: 5 µm.)
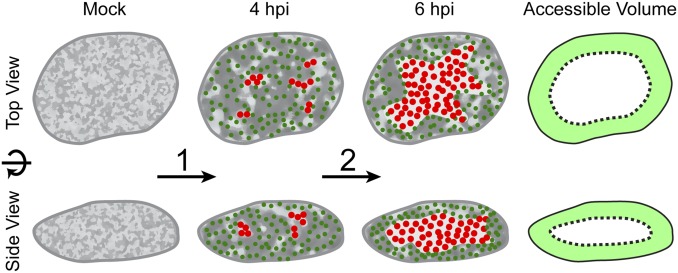
In summary, our data suggest a model in which herpesvirus capsids first can assemble throughout the entire nuclear volume, while concomitantly the host chromatin becomes more porous and thus facilitates diffusion of capsids (Fig. 8, mock to 4 hpi). At this stage, as much as two thirds of the capsid volume can access the nuclear membranes directly by diffusion in 0.9-µm corrals (Fig. 8, accessible volume). The remaining third can access the nuclear envelope by hopping. Later, an expansion of the viral replication compartments marginalizes and concentrates motile capsids into a shell-like structure adjacent to the nuclear envelope, strongly minimizing the space that has to be bridged by diffusion (Fig. 8, 4 hpi to 6 hpi).
Fig. 8.
Viral remodeling of the nuclear architecture allows efficient flux of nuclear capsids in the absence of a directed transport system. Schematic depiction of the spatial changes during PRV infection that allow capsids to diffuse efficiently to site of nuclear egress. First (1), the interchromatin compartments enlarge, which leads to an increase in corral size (light gray regions). Capsids form and populate the enlarged corrals (green dots; 4 hpi). Over time (2), replication compartments form (red dots) enlarge and coalesce. At 6 hpi, chromatin is marginalized and most of the chromatin-depleted region in the center of the nucleus is occupied by the replication compartment. Capsids now mainly move in the peripheral corrals near the nuclear membranes, bringing them nearer to sites of egress. The accessible volume fraction is depicted as light green area, showing that a large fraction of the nuclear volume has access to nuclear membranes in 3D.
Discussion
The past decade has brought huge advances in live cell microscopy to enable dynamic multichannel imaging of several nuclear structures (29). Still, imaging the nuclear egress of virus particles remains a challenge because of large amounts of soluble capsid proteins that reduce the contrast to high particle densities, which demands fast acquisition, and to phototoxicity and bleaching, which must be counterbalanced by very sensitive cameras. Recently, light-sheet microscopy has provided major improvements with regard to contrast and reduced phototoxicity (30). Nevertheless, there are downsides to the classical two-objective light-sheet architecture, as geometrical constraints typically do not permit the use of high-N.A. objectives. Therefore, we have modified the classical objective-based TIRF architecture (18) such that a rotating oblique light sheet is generated. This setup has several advantages. First, it requires only one objective, which makes it compatible with many existing microscope architectures. Second, an objective with a high N.A. increases resolution especially in the z dimension and allows better particle identification at high particle concentrations. Third, the rotating illumination eliminates shading and laser interference artifacts that cannot be avoided in oblique microscopy. Most importantly, sample contrast is enhanced while phototoxicity is reduced. Together, these features enabled the presented study. There are, however, caveats: light-sheet thickness correlates with illumination area (19) in oblique geometries, and more work is needed to characterize the generated oblique light sheet and its effects.
The nucleus relies on the inward and outward flux of molecules to fulfill its various biological duties. Its sponge-like interior, however, constrains the movement of larger complexes (1). Intermediate-sized complexes, like RNA molecules (31, 32) and ribosome subunits (33), are able to diffuse within and frequently hop between subnuclear corrals. However, diffusion of very large particles like PML bodies, Cajal bodies (4), or microinjected 100-nm latex beads (5) is strongly confined to corrals with a diameter of approximately 300 nm (5). Larger translocations are only possible if the corral itself moves as a result of large-scale rearrangements of the nucleus. Some of these nuclear bodies might be able to dynamically assemble and disperse (34), such that long-range motility of the whole complex is not needed. In contrast, herpesvirus capsids and possibly cellular RNP particles (7) assemble in the nucleoplasm and need to reach the nuclear envelope for nuclear egress.
Strikingly, fluorescently labeled herpesvirus capsids are surprisingly motile in the nuclei compared with microinjected beads of similar size in uninfected nuclei. Previous reports have suggested that herpesvirus capsids use an F-actin–dependent, directed, and active transport mechanism to propel through the nuclear interior (8, 9). However, further study of this hypothesis has been missing for more than a decade. We recently reinvestigated this idea, and did not find any evidence that herpesviruses induce nuclear F-actin (10).
First, we find that capsids diffuse over short time scales (<1 s), but are corralled over longer time scales. A previous report (8) was not able to analyze very short time scales because of technical limitations of their microscopy approach, but found α values indicating directed motility by fitting time scales ranging from 0.5 to 2.5 s. Our data do not support this, as the MSD curves we measured plateau at such longer time scales (Fig. 1B). Our data show clear evidence for corralling at longer time scales, consistent with previous reports on other large nuclear complexes (4) with the marked difference that herpesvirus infection increases corral size threefold.
These results suggest a model in which herpesvirus infection increases the porosity of the nucleus. Diffusion in these enlarged corrals allows the majority of capsids to have direct access to the nuclear envelope. Although the motility of capsids was indistinguishable from that of the inert tracer particles, it is possible that the motion of these and other nuclear components could reflect nonthermal kicks as a result of active cellular or viral processes known as “active diffusion” (35); active diffusion could allow capsids to hop between corrals, resulting in the observed replenishment of capsids from the interior nuclear volume over tens of seconds.
In addition, the expansion of the viral DNA replication compartments over time excludes herpesvirus capsids from the interior nucleoplasm, enriching them in corrals in close proximity to the nuclear envelopes. Such a marginalization of host chromatin during infection is a very compelling phenotype that might not be unique to the herpesviruses. Other DNA viruses replicating in the nucleus, like parvoviruses and baculoviruses, show similar effects (36, 37). It has been proposed (36) that depletion attraction plays a role, as virus infection might introduce molecules that act as crowders (38). Phase separation between viral structures and host chromatin could also play a role, as a number of physiological nuclear bodies appear to represent condensed liquid phases of nucleoplasm (39). However, the compartmentalization of capsids, viral DNA, and cellular chromatin might be more complex: We find that motile PRV capsids are deenriched from replication compartments, which has also been reported for murine cytomegalovirus capsids (40) and parvovirus capsids (36). However, HSV-1 capsids seem to be instead enriched in late replication compartments (41–43). It is possible that the balance between host chromatin and replication compartment density plays a role. PRV seems to induce chromatin marginalization, but at the same time does not reduce capsid motility significantly, as our FRAP data show. This is only possible if corral size does not decrease, meaning that the marginalized chromatin maintains its porosity. Increasing the replication compartment size and density might then lead to an exclusion of capsids from the replication compartments and their enrichment near the nuclear envelope. HSV-1, on the contrary, might compact the marginalized chromatin, while having a less dense replication compartment, displacing HSV-1 capsids from the chromatin into the replication compartment. In future studies, it will be important to investigate by what molecular mechanism the nuclear architecture is modulated, and what affect this has on the nuclear egress of HSV-1 capsids.
Future technological developments might enable an even more detailed analysis. Particle detection algorithms that can deal with very high signal densities are an active field of research to allow better and faster superresolution localization microscopy (44). These approaches might also allow the tracking of single capsids at very high concentrations later in infection. Moreover, we do not know if our model applies to other herpesvirus subfamilies, like the β-herpesviruses (e.g., human cytomegalovirus), which have much longer life cycles and therefore might be able to reorganize the host environment even more profoundly. It is possible that cytomegaloviruses use extensive nuclear membrane infoldings (45) to increase the surface-accessible volume fraction and compensate for increased nuclear height. Capsid-tagged cytomegaloviruses are available now (46), and future studies are aimed at understanding the differences between these subfamilies.
Our model has one more important implication: If motility is stochastic and no mechanism actively delivers capsids to the nuclear membrane, there might be molecular mechanisms to retain capsids that have arrived at the nuclear membranes. Recent reports (40, 47) have suggested that nuclear egress is a two-part process: First, UL31, the soluble partner of the nuclear egress complex (PRV, Uniprot Q7TBN7), binds capsids in the nucleoplasm. Afterward, UL31 acts an adapter that mediates binding to UL34, the membrane-bound partner of the nuclear egress complex (PRV, Uniprot G3G8X8). This UL31/UL34 interaction may provides the high affinity/avidity interactions needed to retain diffusing capsids upon stochastic collision with the nuclear membrane.
In conclusion, our findings suggest that remodeling of the nuclear structure is needed to allow very large nuclear particles to cross the nuclear space in the absence of directed transport. It is worth investigating if a similar physiological mechanism exists for movement of large nuclear complexes, such as large RNP particles (7).
Materials and Methods
Cells and Viruses.
PtK2 cells were acquired from American Type Culture Collection (ATCC CCL-56), cultured in ATCC EMEM (ATCC 30–2003) supplemented with 10% FBS, and kept at standard cell culture conditions (5% CO2, 37 °C). Cells were split twice per week at a 1:4 ratio and not cultured for more than 20 passages.
PRV943c is a PRV443 derivative (22) constructed by first replacing the GFP gene with an mRFP gene by homologous recombination that resulted in PRV180 (48). The mRFP gene was then replaced with monomeric EGFP (mEGFP, A206K) gene by homologs recombination as described previously (48), resulting in PRV943c. This virus produces fewer assemblon-like structures, which recently were shown to be the result of dimerizing fluorescent proteins (43).
PRV926, coding for a C-terminal fusion of UL29 (ICP8) to mEGFP and mRFP-VP26, was constructed by homologous recombination of a synthetic construct consisting of the C-terminal region of UL29 (bp 21607–22108 of GenBank ID code JF797219.1), a linker region (acttctataggcatatcctcttccgggtccttgtccttgtccttctccgaagggcga), an XbaI site, the mEGFP coding region without the start codon, a BglII site, and a region downstream of the UL29 stop codon (bp 21106–21603 of GenBank ID code JF797219.1) with PRV Becker (GenBank ID code JF797219.1), resulting in PRV 927. PRV 927 was then crossed with PRV180 coding for mRFP-VP26 by homologous recombination in coinfected cells, and selection of mEGFP and mRFP positive plaques, resulting in PRV926.
HSV1(17+)Lox-eGFPVP26 was generated according to Nagel et al. (21) on an improved BAC backbone HSV1(17+)Lox (49).
Passivation of Beads.
One-hundred-nanometer yellow-green (F8803) and red fluorescent (F8801) carboxylated latex beads (Life Technologies) were passivated with PEG according to Feric et al. (24).
Microinjection.
Cells were microinjected with a custom-built system consisting of a 5242 Microinjector (Eppendorf) connected to pressurized nitrogen tank, an MP225 motorized stage (Sutter), and an Eppendorf needle holder. Eppendorf Femtotips were loaded from the back with 5 µL bead suspension using Eppendorf loading tips. Femtotips were checked for clean loading and air bubbles with a stereomicroscope. Before microinjection, the medium was changed to prewarmed cell culture medium plus 20 mM Hepes pH buffer. Cells were then transferred to a prewarmed cell culture insert. Nuclei were visualized with phase-contrast microscopy and injected with continuous holding pressure of 60–120 mP by manually driving the tip into the nuclei. Afterward, the medium was changed to full cell culture medium and cells were allowed to recover overnight in all cases before infection or imaging.
Static Oblique Microscopy.
Microinjected cells were imaged with static oblique microscopy by using a commercial TIRF/STORM system (Nikon) equipped with an Agilent laser source (Agilent) producing approximately 65 mW of 488-nm and 561-nm light at the fiber exit, a TRF89902 Quad band filter set (Chroma), an Nikon APO TIRF 100× (N.A. 1.49) objective, and an iXon Ultra EM-CCD camera (Andor), run through Nikon NIS AR software in triggered acquisition mode. Lasers at 488 and 561 nm were used at no more than 10% of their maximal output with a frame rate of 28–80 per second. Oblique illumination was achieved by first focusing on virus particles placed on the cover glass surface and then increasing the laser angle until capsid signal intensity sharply decreased. Then, the angle was slowly reduced again until particles just at the coverslip surface were illuminated. This angle was noted as the TIRF angle. For each cell, the laser angle was set back to the TIRF angle and then reduced until the nucleus was just illuminated (i.e., oblique illumination). Focus and laser angle were adjusted from cell to cell to optimize contrast and reduce fringing artifacts.
Spinning-Disk Confocal Microscopy.
Measurements of cell dimensions on live cells and imaging of fixed cells [fixed for 10 min with 4% (wt/vol) paraformaldehyde in Dulbecco's phosphate-buffered saline (DPBS)] were performed on a spinning disk confocal microscope based on a Nikon Ti inverted stand coupled to a Yokogawa CSU 21 spinning disk (Yokogawa) and connected through an Andor TuCam adapter to a Hamamatsu Flash 4.0 (C11440) sCMOS (Hamamatsu). The objective used was a Nikon Plan Apo 60×, N.A. 1.4, DIC N2 objective, resulting in magnification of 108 nm per pixel. An Agilent laser source supplied 405-, 488-, and 561-nm laser light, and the following band-pass emission filters were used: 435–485 nm (Hoechst), 515–555 nm (GFP), and 590–650 nm (mRFP).
Ring-Sheet Microscopy.
Ring-sheet microscopy was performed on a custom-built system. Its basic layout resembles a classic objective-based TIRF setup (18) but uses a galvo system for laser beam steering. We incorporated four laser lines: Two custom-built 405-nm and 638-nm diode lasers using 65-mW SLD3234VF (Sony), 120-mW HL63603TG (Oclaro) laser diodes, and 50-mW 473-nm and 100-mW 532-nm diode pumped solid state (DPSS) lasers (Dragonlasers) combined by dichroic mirrors into one laser path. The DPSS lasers were shuttered with LST200SLP laser shutters (nmLaser), whereas the diode lasers were directly modulated by using TLD001 drivers (Thorlabs) through a 12-bit, NI-PCI-6713 analog output data acquisition (DAQ) board coupled to a NI BNC2110 breakout board (National Instruments). All laser beams were individually expanded and spatially filtered by using Keplerian beam expanders with 15-µm pinholes (Thorlabs) inserted at the focal point, resulting in a clean 10-mm-diameter Gaussian beam. This beam was then guided onto a large beam diameter dual-axis scanning galvanometer system (GVS212; Thorlabs) and afterward sent through another 2× Keplerian beam expander before being focused by a 200-mm achromatic lens onto the back focal plane of a Nikon 100× 1.45 Plan Apochromat (CFI Plan Apochromatλ DM, 100×, oil). A quad-band filter set consisting of an Di01-R405/488/532/635–25 × 36 dichroic mirror and a FF01-446/510/581/703–25 emission filter (Semrock) was used for fluorescence detection. The objective was mounted on an IX50 microscope stand (Olympus) attached to an Andor iXON 897 EM-CCD camera, resulting in magnification of 178 nm per pixel. Triggered acquisition was performed through µManager (50) coupled to an Arduino Uno (www.arduino.cc) acting as a signal generator. The galvo system was controlled through a custom Matlab (Mathworks) script generating two 180° shifted sine functions. The amplitude (voltage) of these curves determined the diameter of the ring projected onto the back focal plane of the objective. Signals were put out through a 12-bit analog National Instruments DAQ board (NI-PCI-6713) coupled to a breakout board (NI BNC2110). The circle at the back focal plane was projected at 1,000 rounds per second, resulting in more than 30 projections per acquired frame.
Live-Cell Imaging.
For live cell imaging, cells were seeded in glass-bottomed dishes (MatTek) that were coated with Fibronectin F1141-G (Sigma-Aldrich) diluted 1:100 in DPBS, incubated for 1 h at 37 °C and then washed twice with DPBS and air-dried. Physiological conditions during imaging were maintained by using a live cell chamber and an objective heater such that the temperature at the center of the cell culture dish was kept at 37 °C. The pH was kept constant by using humidified 5% CO2. All cells were stained with 0.5 µg/mL Hoechst 33342 (Life Technologies) immediately before imaging.
Virus Infection.
Cells were infected with 100 plaque-forming units per cell for 1 h, and medium was exchanged afterward. PRV-infected cells were incubated for a total of 4 or 6 h as indicated before imaging. HSV-infected cells were incubated for 5 h before imaging.
Image Manipulation.
All data presented in this work was Kalman-filtered (settings: acquisition noise, 0.05; bias, 0.80) in Fiji (51) to suppress spurious noise.
Particle Tracking.
Particle tracking was performed first by determining the positions of fluorescent particles through QuickPALM (52) in Fiji (51) with following settings: minimum SNR, 5; maximum FHWM, 5 pixels for capsids and 3 pixels for beads; image plane pixel size, 178 nm; minimum symmetry, 0%; local threshold, 20% maximum intensity; maximum iterations per frame, 100,000; and threads, 50. Particle positions and astigmatism information were then fed into a custom-written Matlab script which calls Simpletracker (Jean-Yves Tinevez, www.mathworks.com/matlabcentral/fileexchange/34040-simple-tracker) and MSD Analyzer (53) to first generate tracks from particle positions and then calculate the MSD, as well as the diffusion coefficient and α, the anomalous diffusion coefficient. The maximum allowed linking distance between two points was 0.5 µm, and particles had to be present in each consecutive frame (i.e., no gap closing). The minimum allowed track length was 40 frames (scripts can be found as Datasets S1 and S2).
z-Calibration.
The z position of virus particles was determined by introducing a 500-mm cylindrical lens (LJ1144RM-A; Thorlabs) approximately 3.5 cm in front of the camera entrance port. To translate the resulting astigmatism into z information, a z-calibration curve was generated. To reduce the localization error typically introduced by refractive index mismatch between glass and cells, we used fixed and infected cells instead of beads on glass and moved their nuclei through the focus with a closed-loop, 12V40SG piezo-driven objective holder (Piezo Jena) controlled in Matlab through a 12-bit DAQ board (NI-PCI-6713) coupled to a breakout board (NI BNC2110). Particle positions and their astigmatism were then determined through Quickpalm, and particle identities were determined through a modified tracking script. A calibration curve ranging over approximately 1 µm was then constructed from averaged astigmatism vs. z-position curves (scripts can be found as Datasets S3–S5).
Determination of Displacement Probabilities.
Displacement probabilities were calculated according to a previous study (54).
FRAP.
FRAP experiments were performed on the ring-sheet microscope by illuminating a ∼2-µm-diameter region with 405 nm laser light for 5 s. Cells were imaged afterward in ring-sheet mode. A Matlab script was used to control laser illumination and ring illumination through a 12-bit DAQ board (NI-PCI-6713) coupled to a breakout board (NI BNC2110). Data were analyzed by using EasyFRAP (55). Data were full-scale normalized and single-term fitted.
Supplementary Material
Acknowledgments
We thank Zsolt Ruzsics (University of Freiburg) and Ulrich Koszinowski (Ludwig-Maximilians-Universität Munich) for support at the early stages of this project; Rudolf Bauerfeind (Institute of Cell Biology, Hannover Medical School) for very helpful discussions; Halina Staniszewska Goraczniak for excellent technical support; Gary S. Laevsky (Princeton Molecular Biology Imaging Facility) for imaging support and helpful discussions; Evangelos G. Gatzogiannis (Princeton Lewis Sigler Imaging Facility) for invaluable advice; and Jean-Yves Tinevez (Institut Pasteur, Paris) for providing Simple Tracker and MSD analyzer through the Matlab File Exchange. This work was supported by National Institutes of Health (NIH) Grants NS033506 and NS060699 (to L.W.E.); NSF CAREER 1253035 and NIH Grant 1DP2GM105437-01 (to C.P.B. and M.F.); the Niedersachsen-Research Network on Neuroinfectiology of the Ministry of Science and Culture of Lower Saxony, Germany (B.S.); German Research Foundation Fellowship BO 4158/1-1 (to J.B.B.); and American Cancer Society Fellowship PF-13-050-01-MPC (to I.B.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513876112/-/DCSupplemental.
References
- 1.Görisch SM, Lichter P, Rippe K. Mobility of multi-subunit complexes in the nucleus: Accessibility and dynamics of chromatin subcompartments. Histochem Cell Biol. 2005;123(3):217–228. doi: 10.1007/s00418-005-0752-y. [DOI] [PubMed] [Google Scholar]
- 2.Grünwald D, et al. Probing intranuclear environments at the single-molecule level. Biophys J. 2008;94(7):2847–2858. doi: 10.1529/biophysj.107.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shav-Tal Y, et al. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304(5678):1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Görisch SM, et al. Nuclear body movement is determined by chromatin accessibility and dynamics. Proc Natl Acad Sci USA. 2004;101(36):13221–13226. doi: 10.1073/pnas.0402958101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tseng Y, Lee JSH, Kole TP, Jiang I, Wirtz D. Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking. J Cell Sci. 2004;117(pt 10):2159–2167. doi: 10.1242/jcs.01073. [DOI] [PubMed] [Google Scholar]
- 6.Mettenleiter TC, Müller F, Granzow H, Klupp BG. The way out: What we know and do not know about herpesvirus nuclear egress. Cell Microbiol. 2013;15(2):170–178. doi: 10.1111/cmi.12044. [DOI] [PubMed] [Google Scholar]
- 7.Speese SD, et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell. 2012;149(4):832–846. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forest T, Barnard S, Baines JD. Active intranuclear movement of herpesvirus capsids. Nat Cell Biol. 2005;7(4):429–431. doi: 10.1038/ncb1243. [DOI] [PubMed] [Google Scholar]
- 9.Feierbach B, Piccinotti S, Bisher M, Denk W, Enquist LW. Alpha-herpesvirus infection induces the formation of nuclear actin filaments. PLoS Pathog. 2006;2(8):e85. doi: 10.1371/journal.ppat.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosse JB, et al. Nuclear herpesvirus capsid motility is not dependent on f-actin. mBio. 2014;5(5):e01909–14. doi: 10.1128/mBio.01909-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monier K, Armas JC, Etteldorf S, Ghazal P, Sullivan KF. Annexation of the interchromosomal space during viral infection. Nat Cell Biol. 2000;2(9):661–665. doi: 10.1038/35023615. [DOI] [PubMed] [Google Scholar]
- 12.Quinlan MP, Chen LB, Knipe DM. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36(4):857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- 13.Rixon FJ, Atkinson MA, Hay J. Intranuclear distribution of herpes simplex virus type 2 DNA synthesis: Examination by light and electron microscopy. J Gen Virol. 1983;64(Pt 9):2087–2092. doi: 10.1099/0022-1317-64-9-2087. [DOI] [PubMed] [Google Scholar]
- 14.Randall RE, Dinwoodie N. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: Evidence that ICP 4 is associated with progeny virus DNA. J Gen Virol. 1986;67(pt 10):2163–2177. doi: 10.1099/0022-1317-67-10-2163. [DOI] [PubMed] [Google Scholar]
- 15.Chang L, et al. Herpesviral replication compartments move and coalesce at nuclear speckles to enhance export of viral late mRNA. Proc Natl Acad Sci USA. 2011;108(21):E136–E144. doi: 10.1073/pnas.1103411108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobiler O, Brodersen P, Taylor MP, Ludmir EB, Enquist LW. Herpesvirus replication compartments originate with single incoming viral genomes. MBio. 2011;2(6):e00278-11. doi: 10.1128/mBio.00278-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stelzer EHK. Light-sheet fluorescence microscopy for quantitative biology. Nat Methods. 2015;12(1):23–26. doi: 10.1038/nmeth.3219. [DOI] [PubMed] [Google Scholar]
- 18.Shroff H, White H, Betzig E. Photoactivated Localization Microscopy (PALM) of Adhesion Complexes. Wiley; Hoboken, NJ: 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5(2):159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 20.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72(9):7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagel C-H, et al. Nuclear egress and envelopment of herpes simplex virus capsids analyzed with dual-color fluorescence HSV1(17+) J Virol. 2008;82(6):3109–3124. doi: 10.1128/JVI.02124-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith GA, Gross SP, Enquist LW. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc Natl Acad Sci USA. 2001;98(6):3466–3470. doi: 10.1073/pnas.061029798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saxton MJ. Anomalous diffusion due to obstacles: A Monte Carlo study. Biophys J. 1994;66(2 pt 1):394–401. doi: 10.1016/s0006-3495(94)80789-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feric M, Brangwynne CP. A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat Cell Biol. 2013;15(10):1253–1259. doi: 10.1038/ncb2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sodeik B. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 2000;8(10):465–472. doi: 10.1016/s0966-842x(00)01824-2. [DOI] [PubMed] [Google Scholar]
- 26.Crouse HV, Coriell LL, Blank H, Scott TFM. Cytochemical studies on the intranuclear inclusion of herpes simplex. J Immunol. 1950;65(1):119–128. [PubMed] [Google Scholar]
- 27.Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol. 2008;6(3):211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 28.de Bruyn Kops A, Knipe DM. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55(5):857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 29.Grünwald D, Singer RH, Rout M. Nuclear export dynamics of RNA-protein complexes. Nature. 2011;475(7356):333–341. doi: 10.1038/nature10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siebrasse JP, Kaminski T, Kubitscheck U. Nuclear export of single native mRNA molecules observed by light sheet fluorescence microscopy. Proc Natl Acad Sci USA. 2012;109(24):9426–9431. doi: 10.1073/pnas.1201781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Politz JC, Tuft RA, Pederson T, Singer RH. Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr Biol. 1999;9(6):285–291. doi: 10.1016/s0960-9822(99)80136-5. [DOI] [PubMed] [Google Scholar]
- 32.Smith CS, et al. Nuclear accessibility of β-actin mRNA is measured by 3D single-molecule real-time tracking. J Cell Biol. 2015;209(4):609–619. doi: 10.1083/jcb.201411032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Politz JCR, Tuft RA, Pederson T. Diffusion-based transport of nascent ribosomes in the nucleus. Mol Biol Cell. 2003;14(12):4805–4812. doi: 10.1091/mbc.E03-06-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol. 2012;24(3):415–422. doi: 10.1016/j.ceb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brangwynne CP, Koenderink GH, MacKintosh FC, Weitz DA. Cytoplasmic diffusion: molecular motors mix it up. J Cell Biol. 2008;183(4):583–587. doi: 10.1083/jcb.200806149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ihalainen TO, et al. Parvovirus induced alterations in nuclear architecture and dynamics. PLoS One. 2009;4(6):e5948. doi: 10.1371/journal.pone.0005948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laakkonen JP, et al. Baculovirus-mediated immediate-early gene expression and nuclear reorganization in human cells. Cell Microbiol. 2008;10(3):667–681. doi: 10.1111/j.1462-5822.2007.01074.x. [DOI] [PubMed] [Google Scholar]
- 38.Hancock R. The crowded nucleus. Int Rev Cell Mol Biol. 2014;307:15–26. doi: 10.1016/B978-0-12-800046-5.00002-3. [DOI] [PubMed] [Google Scholar]
- 39.Zhu L, Brangwynne CP. Nuclear bodies: The emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol. 2015;34:23–30. doi: 10.1016/j.ceb.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pogoda M, et al. Characterization of conserved region 2-deficient mutants of the cytomegalovirus egress protein pM53. J Virol. 2012;86(23):12512–12524. doi: 10.1128/JVI.00471-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Bruyn Kops A, Knipe DM. Preexisting nuclear architecture defines the intranuclear location of herpesvirus DNA replication structures. J Virol. 1994;68(6):3512–3526. doi: 10.1128/jvi.68.6.3512-3526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Bruyn Kops A, Uprichard SL, Chen M, Knipe DM. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology. 1998;252(1):162–178. doi: 10.1006/viro.1998.9450. [DOI] [PubMed] [Google Scholar]
- 43.Nagel C-H, Döhner K, Binz A, Bauerfeind R, Sodeik B. Improper tagging of the non-essential small capsid protein VP26 impairs nuclear capsid egress of herpes simplex virus. PLoS One. 2012;7(8):e44177. doi: 10.1371/journal.pone.0044177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sage D, et al. Quantitative evaluation of software packages for single-molecule localization microscopy. Nat Methods. 2015;12(8):717–724. doi: 10.1038/nmeth.3442. [DOI] [PubMed] [Google Scholar]
- 45.Buser C, Walther P, Mertens T, Michel D. Cytomegalovirus primary envelopment occurs at large infoldings of the inner nuclear membrane. J Virol. 2007;81(6):3042–3048. doi: 10.1128/JVI.01564-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bosse JB, et al. A beta-herpesvirus with fluorescent capsids to study transport in living cells. PLoS One. 2012;7(7):e40585. doi: 10.1371/journal.pone.0040585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Funk C, et al. The Herpes simplex virus protein pUL31 escorts nucleocapsids to sites of nuclear egress, a process coordinated by its N-terminal domain. PLoS Pathog. 2015;11(6):e1004957. doi: 10.1371/journal.ppat.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.del Rio T, Ch’ng TH, Flood EA, Gross SP, Enquist LW. Heterogeneity of a fluorescent tegument component in single pseudorabies virus virions and enveloped axonal assemblies. J Virol. 2005;79(7):3903–3919. doi: 10.1128/JVI.79.7.3903-3919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagel C-H, Pohlmann A, Sodeik B. Construction and characterization of bacterial artificial chromosomes (BACs) containing herpes simplex virus full-length genomes. Methods Mol Biol. 2014;1144(chap 4):43–62. doi: 10.1007/978-1-4939-0428-0_4. [DOI] [PubMed] [Google Scholar]
- 50.Edelstein AD, et al. Advanced methods of microscope control using μManager software. J Biol Methods. 2014;1(2):e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henriques R, et al. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat Methods. 2010;7(5):339–340. doi: 10.1038/nmeth0510-339. [DOI] [PubMed] [Google Scholar]
- 53.Tarantino N, et al. TNF and IL-1 exhibit distinct ubiquitin requirements for inducing NEMO-IKK supramolecular structures. J Cell Biol. 2014;204(2):231–245. doi: 10.1083/jcb.201307172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toyota T, Head DA, Schmidt CF, Mizuno D. Non-Gaussian athermal fluctuations in active gels. Soft Matter. 2011;7(7):3234–3239. [Google Scholar]
- 55.Rapsomaniki MA, et al. easyFRAP: An interactive, easy-to-use tool for qualitative and quantitative analysis of FRAP data. Bioinformatics. 2012;28(13):1800–1801. doi: 10.1093/bioinformatics/bts241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.