Significance
Therapies for Leber hereditary optic neuropathy (LHON), in common with all disorders caused by mutated mtDNA, are inadequate, in part because of the absence of suitable animal models. To test a potential therapy, we introduced mutant human NADH ubiquinone oxidoreductase subunit 4 DNA directly into mitochondria of mouse zygotes to generate transgenic LHON mice. This mutation in mice caused the hallmark visual loss and optic neuropathy seen in LHON patients. We reversed the blindness by gene therapy with the wild-type allele. To our knowledge, this is the first description of a mouse model with the same genotype and phenotype as the human counterpart disorder. This innovative technology has implications not only for creating mouse models of mutant mtDNA but also for treating the mitochondrial dysfunction with gene therapy.
Keywords: gene therapy, adeno-associated virus, mitochondria, Leber hereditary optic neuropathy, blindness
Abstract
Considerable evidence supports mutations in mitochondrial genes as the cause of maternally inherited diseases affecting tissues that rely primarily on oxidative energy metabolism, usually the nervous system, the heart, and skeletal muscles. Mitochondrial diseases are diverse, and animal models currently are limited. Here we introduced a mutant human mitochondrial gene responsible for Leber hereditary optic neuropathy (LHON) into the mouse germ line using fluorescence imaging for tissue-specific enrichment in the target retinal ganglion cells. A mitochondria-targeted adeno-associated virus (MTS-AAV) containing the mutant human NADH ubiquinone oxidoreductase subunit 4 (ND4) gene followed by mitochondrial-encoded mCherry was microinjected into zygotes. Female founders with mCherry fluorescence on ophthalmoscopy were backcrossed with normal males for eight generations. Mutant human ND4 DNA was 20% of mouse ND4 and did not integrate into the host genome. Translated human ND4 protein assembled into host respiratory complexes, decreasing respiratory chain function and increasing oxidative stress. Swelling of the optic nerve head was followed by progressive demise of ganglion cells and their axons, the hallmarks of human LHON. Early visual loss that began at 3 mo and progressed to blindness 8 mo after birth was reversed by intraocular injection of MTS-AAV expressing wild-type human ND4. The technology of introducing human mitochondrial genes into the mouse germ line has never been described, to our knowledge, and has implications not only for creating animal models recapitulating the counterpart human disorder but more importantly for reversing the adverse effects of the mutant gene using gene therapy to deliver the wild-type allele.
Mitochondria are thought to have evolved from free-living aerobic bacteria. Most bacteria encode the majority of their genes in single circular chromosomes but maintain stable episomes such as plasmids independent of that chromosome. In mammals mitochondria have their own genomic DNA (mtDNA) encoding 37 genes (2 for ribosomal RNAs, 22 for tRNAs, and 13 for proteins) that are essential for cell function and viability. However, most mitochondrial proteins are encoded by nuclear genes and are imported into the organelle. The mammalian mitochondrial genome is transmitted by the egg from mother to child. These genes are buffered against the effect of mutations, because somatic cells typically contain around 1,000 copies of mtDNA. Although most cells are apparently uniform with respect to mtDNA composition, mammalian cells usually can tolerate a significant fraction of aberrant mtDNA (a condition called “heteroplasmy”) and retain respiratory chain function (1, 2). Considerable evidence supports mitochondrial mutations as the cause of maternally inherited diseases affecting tissues that rely primarily on oxidative energy metabolism, usually the central and peripheral nervous system, the heart, and skeletal muscles (1, 3–5). Patients frequently are heteroplasmic for the mutation, partially explaining why symptoms may be delayed until adolescence or even adulthood.
Most Leber hereditary optic neuropathy (LHON) cases are associated with mutations in one of three mitochondrial genes encoding subunits of NADH ubiquinone oxidoreductase (ND4) which is complex I of the mitochondrial respiratory chain (6–8). This enzyme contains 7 subunits encoded by mtDNA that are intimately associated with the inner mitochondrial membrane and 35 subunits that are encoded by nuclear DNA and imported into the organelle (9). The connection between LHON and mtDNA was established in 1988, when Wallace et al. reported a nucleotide transition from guanosine to adenosine at position 11778, which results in an arginine-to-histidine substitution in the ND4 subunit of complex I (6). Since then, several other mutations in genes for NADH dehydrogenase, cytochrome b, cytochrome oxidase, or ATP synthase subunits have been identified that also cause familial LHON (10). However, an animal model confirming the pathogenicity of the G11778A mutation has never been developed. Moreover, therapies for disorders caused by mutated mtDNA are inadequate, in large part because of the absence of animal models with mutated mtDNA and with the same genotype and phenotype as the human disorder that would help uncover the pathogenesis of disease and test potential avenues for treatment (11–13). Current procedures for the introduction of artificially mutagenized mtDNA into mitochondria have generated optic neuropathy in mice with a gene [ND6 G14600A (P25L)] that, when homoplasmic, is responsible for Leigh syndrome in humans (14).
Viruses have the ability to traverse the mitochondrial double membrane with subsequent access into the inner matrix (15), delivering DNA into the organelle (16). Taking advantage of this property, we redirected the adeno-associated virus (AAV) to target mitochondria efficiently, rather than its typical target, the nucleus, by adding the 23-aa cytochrome oxidase subunit 8 (COX8) presequence into the AAV2 capsid ORF (17, 18). Here we use this vector to deliver the mutant human G11778A ND4 DNA into zygotes to generate transgenic LHON mice.
Mutant Human ND4 in Transgenic Mice
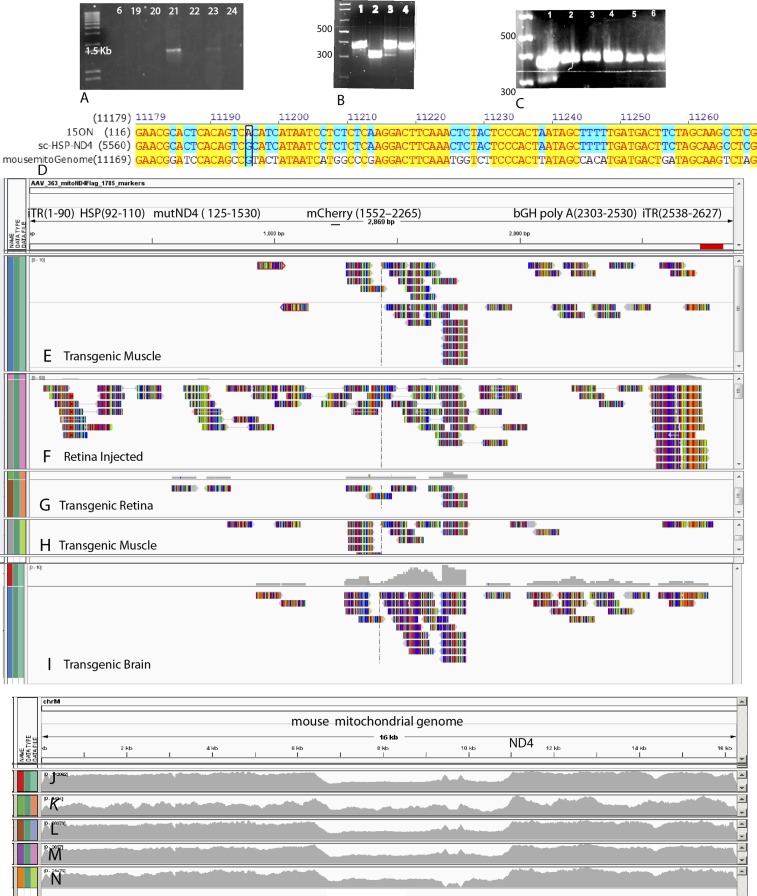
Chloramphenicol resistance has been used as a selectable marker for mitochondrial transformation, but chloramphenicol-resistant chimeric mice develop ocular abnormalities (19–26). Therefore, we inserted a mitochondrial-encoded mCherry gene downstream of the mutant human ND4 (mutND4) gene that was fused in frame with FLAG (mutND4FLAG). We cloned the entire cassette under the control of the mitochondrial heavy-strand promoter (HSP) into a self-complementary AAV (scAAV) backbone (sc-HSP-mutND4FLAG+mCherry) (Fig. 1A). A stop codon (AGA) was inserted between the two genes. After packaging with the mitochondria-targeted capsid (cox8GFP) (Fig. 1B) (17, 18), the recombinant AAV (rAAV) was injected into fertilized oocytes (Fig. 1 C and D). Transgenic mice (F0) were generated (Table 1) that contained varying levels of mCherry expression as detected by confocal laser-scanning ophthalmoscopy (CLSO) of live mice (Fig. 1 E and F). Three females with the highest levels of visible mCherry expression in the optic nerve head, ganglion cell, and retinal nerve fiber layers were backcrossed with normal C57BL/6 males for more than eight generations, resulting in 233 viable fertile mutND4 mice (Fig. 1 G and H). The phenotype of transgenic mice was grossly normal, except for a larger body size when compared with age-matched wild-type mice (Fig. 1I and Table 2). In 77% of the transgenic mice, CLSO showed red fluorescent particles in the retina and optic nerve head that increased as the mice aged and could be observed through multiple generations (Fig. 1 J–O). Although enabling serial in vivo imaging of red fluorescence (27), CLSO is far less sensitive than epifluorescence microscopy in visualizing mCherry.
Fig. 1.
Scheme of transgenic mice generation. (A) Mitochondrially encoded human mutant G11778A ND4 fused to FLAG (mutND4FLAG) and mitochondrially encoded mCherry were inserted into the scAAV backbone under the control of the human mitochondrial HSP. The stop codon AGA separated the two genes. (B–D)The construct was packaged in mitochondrially targeted cox8 MTS-scAAV (B) and was microinjected into fertilized eggs (C and D). (E) Offspring were screened for mCherry by CLSO (Inset). (F–H) Females with higher mCherry expression (F) were backcrossed with wild-type C57BL/6 males (G), generating mutant ND4 mice (H). (I)The transgenic mouse (arrow) was larger than a control mouse. (J–O) CLSO imaging of the optic nerve head (onh) and retinal nerve fiber layer (arrows) shows red fluorescent particles (arrows) in the retina of first-generation LHON mito-mice (F0) at 2 mo (2m) (J and K) and 11 mo (11m) (L) after birth and in their F1 progeny at 2m (M and N) and 7m (O) after birth. bGH-Poly(A), bovine growth hormone polyadenylation sequence; iTR, inverted terminal repeats.
Table 1.
Offspring of zygote injections
| Injection date (date of birth) | No. of embryos collected (no. of donor mice) | No. of embryos injected | No. of embryos lysed (% of injected embryos) | No. of embryos transferred | No. of mice born (% of embryos transferred) | No. of mutants (%) |
| 4/5/11 (4/24/11) | 96 (10) | 58 | 17 (29.3) | 42 | 16 (38.1) | 12 (77) |
| 4/7/11 (4/26/11) | 107 (10) | 89 | 41 (46.1) | 48 | 8 (16.7) | 6 (77) |
Table 2.
Weights of transgenic and control mice
| Age in months | Weight, g ± SD | |
| Transgenic | Control | |
| 1 | 16 ± 2.5 | 14.9 ± 2 |
| 3 | 26 ± 2.5 | 24.2 ± 1.6 |
| 6 | 35.3 ± 5.9 | 29.7 ± 7.4 |
| 9 | 46.8 ± 3.2 | 24.3 ± 1.6 |
| 13 | 52.5 ± 2.1 | 32 ± 2.2 |
To verify the presence of mutant human ND4, mitochondrial DNA was isolated from transgenic mouse tissues including the retina, optic nerve, brain, heart, liver, lung, spinal cord, and skeletal muscle (SI Appendix, Fig. S1). PCR with a forward primer nested in the mutant human ND4 and a reverse primer nested in the FLAG epitope generated a 1.4-kb band (Fig. 2A). PCR of optic nerve mitochondria with primers that amplify both human and mouse ND4 together were digested with the restriction enzyme ScaI that cut the human but not the mouse ND4 fragment and thus was diagnostic for the human ND4 (Fig. 2B, lane 3). In addition, laser capture microdissection of the retina showed the human ND4 Sca1 was digested in cells of the retinal ganglion cell (RGC) layer but not in cells of other retinal layers of transgenic mice or in any retinal layer of normal controls (Fig. 2C). Using quantitative PCR (qPCR), we found that mutant human ND4 DNA in the RGC layer was 20% of endogenous mouse ND4 but was much lower in the inner nuclear layer (0.08%), and no mutant human ND4 was detected in the outer nuclear layer of transgenic mice or in any retinal layer of control C57B/6 mice. Sequencing and alignment to wild-type human and mouse ND4 (Fig. 2D) further confirmed that the PCR product was the mutant human ND4.
Fig. 2.
Genetic analysis of transgenic mice. (A) Agarose gel electrophoresis of PCR-amplified human mutant ND4 in animal 21. (B) PCR products using primers amplifying both human and mouse ND4 were digested with Sca1 (lane 1 = control optic nerve from a normal C57BL6 mouse; lane 2 = vector DNA; lane 3 = 15ON, the optic nerve from transgenic mouse #15; lane 4 = undigested 15ON). Retinal microdissection PCR products using primers amplifying human and mouse ND4 show that only cells of the RGC layer of transgenic mice (lane 1), but not cells of the inner nuclear (lane 2) or outer nuclear layers (lane 3), were digested with Sca1. (C) Human ND4 was absent in the RGC layer (lane 4), inner nuclear layer (lane 5), and outer nuclear layer (lane 6) of normal controls. (D) Sequencing of amplified human ND4 and alignment to wild-type mouse and human ND4 inserted into our AAV vector demonstrating the G-to-A transition of mutant human ND4 in the optic nerve of animal 15 (15ON) and the conservation of the G nucleotide in mouse and human (boxed). (Nucleotides highlighted in yellow indicate homology of the sequenced PCR product, human ND4 and mouse ND4. Nucleotides highlighted in blue indicate homology between the sequenced PCR product and human ND4). (E–I) Alignment of the AAV vector next-generation sequencing reads of mitochondrial DNA extracted from transgenic mouse muscle (E), retina of a wild-type mouse directly injected with the MTS-AAV (F), transgenic mouse retina (G), another transgenic mouse muscle (H), and transgenic mouse brain (I). (J–N) Alignment of AAV vector sequences of mitochondrial DNA extracted from transgenic mouse muscle (J), retina directly injected with the MTS-AAV (K), transgenic retina (L), additional transgenic muscle (M), and transgenic brain (N) samples to the mouse mitochondrial genome show no evidence for homologous recombination of the mutant human ND4 or nonhomologous insertions of viral DNA into the mouse mitochondrial genome.
Mutant Human ND4 Does Not Integrate into Mouse Mitochondrial DNA
Because the AAV vector recently has been demonstrated to integrate viral DNA into the host mitochondrial genome (16), we next determined whether the mutant human ND4 integrated into the mouse mitochondrial genome. We performed next-generation sequencing of mtDNA extracted from the retina, brain, and muscles of 6-mo-old transgenic mice and the retinas of mice vitreally injected with the mitochondria-targeted scAAV-HSP mutant ND4 as a control. Next, we aligned the human sequences reading with the mouse mitochondrial genomes and also with the AAV genome, looking for reads that align one part to the virus and other to mouse or human mtDNA and determining the depth at each base level to look for variants. In each of the samples, we identified reads that mapped to the mitochondria-targeted AAV (MTS-AAV) containing mutant human ND4 (Fig. 2 E–I). Homologous recombination of the mutant human ND4 or nonhomologous insertions of rAAV DNA into the mouse mitochondrial genome (Fig. 2 J–N) were not detected by next-generation sequencing. A recent study also supports the lack of AAV integration into mitochondria (28). The absence of evidence that the viral genome integrated into the host mitochondrial DNA suggested that it remained episomal.
For the viral mutant human ND4 DNA to be transferred episomally in mitochondria, it likely would have to replicate. Therefore, we performed an in vitro DNA synthesis assay with mitochondria isolated from the livers of transgenic ND4 mice and wild-type controls, using α32P-dTTP and the other three unlabeled dNTPs. We found an ∼2-kb band that was absent in controls (Fig. 3A). Southern blotting using unlabeled dNTPs for the in vitro DNA synthesis reaction hybridized to a 32P-labeled oligonucleotide probe derived from the human ND4 gene (Fig. 3B). These findings suggest that the viral DNA is capable of replicating in mitochondria using endogenous cellular proteins (29).
Fig. 3.
Replication, integration, and in vivo effects of mutant ND4. (A) Autoradiogram of agarose gel of in vitro DNA synthesis assay of transgenic liver (lane 1) and control liver mitochondria (lane 2). (B) Autoradiogram of Southern blot hybridization to labeled probe detecting human ND4. (C–F) 2D BN-PAGE reacted against NDUFS4 (S4) and NDUFA9 (A9) subunits of complex I and FLAG (C), FLAG (D), ND4 (E), and mCherry (F). (G and H) Averaged PERG waveforms with the x axis in 100-ms increments (G) and scatterplot of PERG amplitudes (H) of transgenic mice. (I and J) Scatterplots of 3-mo-old transgenic mice with low PERG amplitudes at baseline show that amplitudes increased 1 (1M) and 3 (3M) mo after intravitreal injection of mitochondria-targeted scAAV containing the wild-type human ND4 (I) but remained low in eyes injected with sterile PBS (J). (K) Bar plot of PERG amplitudes recorded 1, 3, and 6 mo after injection of rAAV-COX8Cherry. (L and M) Scatterplots of serial SD-OCT of the RGC layer and IPL of the same group of mice at 5 (5M) to 7 (7M) mo (L) and at 8 (8M) to 11 (11M) mo (M). (N) OCT images of a transgenic mouse at age 3 (3m), 7 (7m), and 15 (15m) mo.
Expressed Human mutND4 Is Assembled into Complex I
For the AAV-transferred mutant human ND4 to have mitochondrial toxicity, it would have to be expressed and integrate into the murine complex I. To test the assembly of the expressed mutant human ND4FLAG into respiratory complex I [as previously demonstrated for wild-type ND4 that was injected directly into the eye (17)], mitochondria from transgenic mouse liver were isolated for 2D blue native PAGE (2D BN-PAGE). We found that signals with antibodies against subunits of complex I, including NDUFS4 and NDUFA9, migrated in the same vertical plane as signals seen with the FLAG antibody (Fig. 3C). FLAG immunoblotting generated two bands (Fig. 3D), one with the molecular weight of the mutant human ND4 and another higher-molecular-weight band consistent with a fusion protein containing both mutant ND4FLAG and mCherry. These two bands migrated at the same horizontal position, respectively, as ND4 (Fig. 3E) and mCherry (Fig. 3F). The smaller band in the ND4 immunoblot (Fig. 3E) may be endogenous mouse ND4. The mutant ND4FLAG–mCherry fusion may have been generated by the lack of the correct secondary structure required by mitochondrial RNase P and nucleases for precise endonucleolytic excision of mitochondrial mRNAs by processing tRNAs from the primary transcript (30–33). Another possible explanation for the fusion protein is that the stop codon (AGA) we inserted between the ND4FLAG and mCherry genes without a tRNA may have allowed some read through. Taken together, our data show that mutant human ND4 assembles into the murine complex I.
Human mutND4 and mCherry Tissue Expression
Longitudinal sections of the retina reacted with antibodies against FLAG, mCherry, the mitochondrial marker VDAC/porin, and the RGC marker Thy1.2 demonstrated expression of ND4FLAG and mCherry in RGCs (SI Appendix, Fig. S2). In addition, mCherry expression was detected in the optic nerve, brain, heart, liver, and skeletal muscle of transgenic mice (SI Appendix, Fig. S3).
Expressed mutND4 Causes Visual Loss That Is Reversed by Gene Therapy
Next, we assessed the effects of mutND4 on visual function. Pattern electroretinograms (PERGs), a sensitive measure of RGC dysfunction, showed progressive declines in amplitude after 3 mo, dropping to noise levels by 8–11 mo after birth (Fig. 3 G and H). To intervene after the onset of visual loss, we injected both eyes of mice with low PERG amplitudes with mitochondria-targeted scAAV containing the wild-type human ND4. Littermates with similar low PERG amplitudes were injected with sterile PBS. One month and 3 mo later, mice treated by wild-type ND4 had significantly improved amplitudes (Fig. 3I) compared with their baseline and compared with littermates injected with PBS, in which PERG amplitudes remained low (Fig. 3J). Control wild-type mice injected with scAAV-mCherry maintained normal visual function at 1, 3, and 6 mo (Fig. 3K), indicating that intravitreal injection and reporter gene expression had no adverse effects on vision. Thus, the visual loss induced by the mutant ND4 was reversed by gene therapy with the wild-type allele.
MutND4 Causes Retinal Degeneration
The retinas of transgenic mutant ND4 mice degenerated with age. By 5–7 mo after birth, spectral domain optical coherence tomography (SD-OCT) measurements in live mice showed a statistically significant decrease in the thickness of the RGC layer and the inner plexiform layer (IPL) (P = 0.0089) (Fig. 3L). The decrease became less pronounced but was still statistically significant from 8 to 11 mo of age (P = 0.026) (Fig. 3M). Representative SD-OCT images of the same animal illustrate the progressive thinning of the RGC layer and IPL in transgenic mutND4 mice with advancing age (Fig. 3N). No changes were detected in wild-type mice from 2 to 12 mo of age (34).
MutND4 Causes Loss of Axons and RGCs
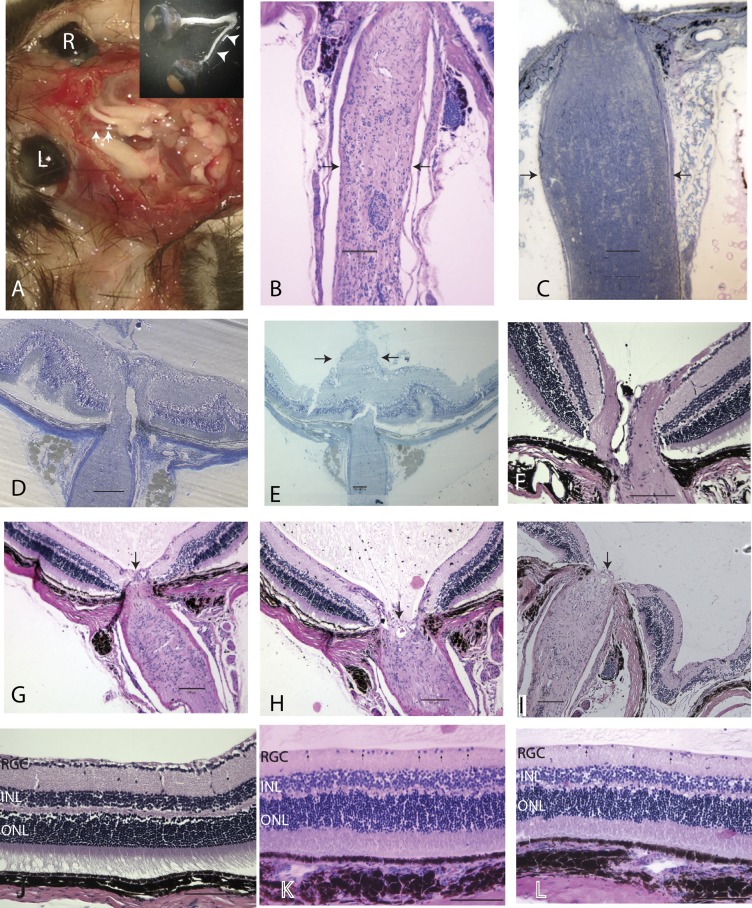
Postmortem, we found severe optic atrophy in transgenic mutND4 mice examined 2 y after birth (Fig. 4A). Longitudinal sections of these optic nerves showed a marked reduction in the diameter of the optic nerve (Fig. 4B) compared with age-matched controls (Fig. 4C). Optic nerve head swelling began as early as 1 mo after birth, and swelling increased gradually (Fig. 4D), mushrooming into the vitreous cavity by 3 mo (Fig. 4E). (For comparison, a normal optic nerve head is shown in Fig. 4F.) Swelling was followed by a progressive loss of the optic nerve head tissues by 22–26 mo of age (Fig. 4 G–I). (A normal retina is shown in Fig. 4J.) In addition, progressive RGC loss and IPL thinning were also evident as mice aged from 3 to 22 mo (Fig. 4K) and 26 mo (Fig. 4L).
Fig. 4.
Ocular histopathology. (A) Gross dissection specimen of a 24-mo-old transgenic mouse with marked thinning of the entire left optic nerve (arrowheads) from the globe to the optic chiasm. (B and C) Longitudinal sections of atrophic optic nerve (arrows) (B) and the optic nerve (arrows) of an age-matched normal mouse (C). (D and E) Optic nerve head swelling 1 mo (D) and 3 mo (E) after birth. (F) A normal optic nerve head. (G–I) Loss of the optic nerve head tissues (arrow) at age 22 mo (G and H) and 26 mo (I). (J–L) Longitudinal retinal sections of a normal mouse (J) and transgenic mice at 22 mo (K) and 26 mo (L) of age. Arrows point to cells in the ganglion cell layer. (Scale bars, 100 µm.) INL, inner nuclear layer; ONL, outer nuclear layer.
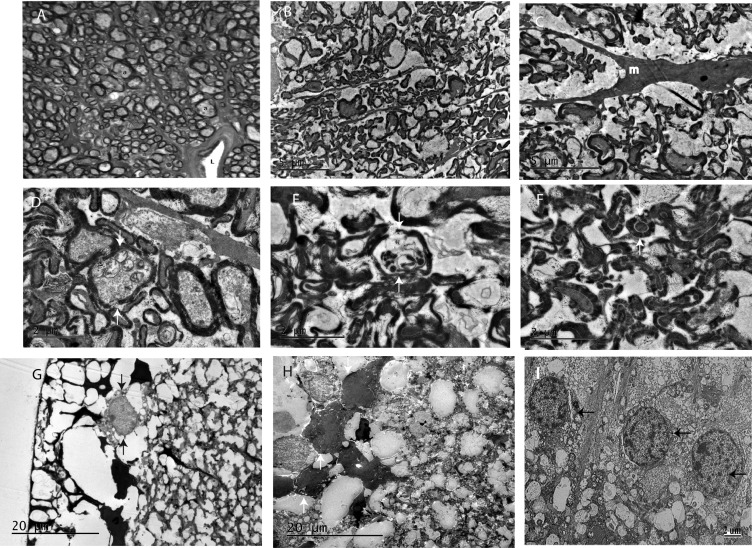
Ultrastructural analysis confirmed that the optic nerve atrophy seen on light micrographs was caused by the loss of axons. Age-matched wild-type mice injected with AAV-GFP have a normal complement of myelinated optic nerve axons (Fig. 5A), but transgenic mice with severely atrophic nerves had far fewer axons (Fig. 5 B and C). Swollen axons contained mitochondria with loss of cristae (Fig. 5D), and axons were undergoing Wallerian degeneration (Fig. 5E). Interestingly, we found that thick sheaths of myelin, a characteristic of large axons, surrounded small-caliber axons (Fig. 5F). The retinas of transgenic mutND4 mice revealed that RGCs were lost (Fig. 5G). Many cells in the RGC layer had condensation of the cytoplasm and nuclear chromatin consistent with apoptosis (Fig. 5H). (For comparison, RGCs of a normal control are shown in Fig. 5I.) These severe pathological features were absent in the brain (SI Appendix, Fig. S4A), heart (SI Appendix, Fig. S4B), and skeletal muscle (SI Appendix, Fig. S4C) of transgenic mutant ND4 mice.
Fig. 5.
Optic nerve and retinal ultrastructure. (A) Transmission electron micrograph of an optic nerve from an age-matched wild-type mouse injected with AAV-GFP showing normal myelinated optic nerve axons (a). L, lumen of a blood vessel. (B–E) Transgenic optic nerves with few axons (m, microglial) (B and C), swollen axons (arrows) containing mitochondria with loss of cristae (arrows) (D), and axons undergoing Wallerian degeneration (arrows) (E). (F) Axons of small caliber surrounded by thick sheaths of myelin (arrows). (G) There were few RGCs in retinas of transgenic mutND4 mice. (H) Cells in the RGC layer with condensation of the cytoplasm and nuclear chromatin (arrows) in transgenic mutant ND4 mice. (I) Retinal ganglion cells (arrows) of a normal mouse retina.
Oxidative Stress in Mutant ND4 Mice
Next, we tested for mitochondrial oxidative stress using the cell permeate reagent 2′,7′-dichlorofluorescein diacetate (DCFDA) that exhibits green fluorescence in the presence of reactive oxygen species. One microliter of DCFDA was injected into the vitreous, and the eye was examined 3 h later. In transgenic retinas, many green fluorescent particles were seen in the RGC, ILP, and inner nuclear layer (SI Appendix, Fig. S5 A and B) in comparison with age-matched normal controls (SI Appendix, Fig. S5C). Expression of mCherry (without antibody detection) in transgenic mice (SI Appendix, Fig. S5 D and E), absent in controls (SI Appendix, Fig. S5F), was seen in DCFDA+ cells (SI Appendix, Fig. S5 G and H). Quantitative analysis revealed that 40% of cells in the RGC layer of mutant ND4 mice were positive for reactive oxygen species particles, whereas in controls this value was only 8% (P < 0.0001) (SI Appendix, Fig. S5I). Thus, the presence of the mutant ND4 increased oxidative stress in transgenic mice, suggesting oxidative injury as a pathway for neurodegeneration (35, 36).
Respiratory Chain Function in Mutant ND4 Mice
Because the optic nerve is highly dependent on oxidative phosphorylation and is adversely impacted by oxidative stress, we measured complex I activity and the rate of ATP synthesis by chemiluminescence with a modified luciferin-luciferase assay in freshly extracted digitonin-permeabilized optic nerve homogenates using the complex I substrates malate and pyruvate. We found that complex I activity was reduced in the optic nerves of transgenic mice relative to wild-type controls (P = 0.0114) (SI Appendix, Fig. S5J). We also found that the rate of complex I-dependent ATP synthesis was reduced significantly in the optic nerves (P = 0.006) of transgenic mice (SI Appendix, Fig. S5K). Thus, substantial energy failure was detected in transgenic mutant ND4, mice similar to the levels of G11778A LHON cells (37).
Brain
Because lesions of the white matter sometimes can be seen in LHON patients, we examined the brains of transgenic mice and controls using MRI. We found no lesions in transgenic mice, and their scans looked similar to those of controls (SI Appendix, Fig. S6). Mice did not show ventriculomegaly. Last, we measured intracranial pressure of transgenic mice and controls to exclude papilledema as the cause of optic nerve head swelling and optic neuropathy, and we found no differences between the two groups (Table 3).
Table 3.
Intracranial pressure measurements
| Control intracranial pressure, mm Hg | Transgenic ND4 intracranial pressure |
| 7.75 | 17.5 |
| 17.5 | 16.5 |
| 16.5 | 16.5 |
| 11.5 | 4.5 |
| Average ± SD 13.3 ± 4.5 | Average ± SD 13.8 ± 6.2 |
Discussion
We show here that the consequences of germline transfer of mutant human G11778A ND4 are visual loss and optic neuropathy in mice, as seen in man. That these adverse effects were reversed by gene therapy with the wild-type allele further confirms the pathogenicity of this mutation, whose association with LHON was discovered almost three decades ago, and suggest a pathway toward treatment that also could be applied to other mitochondrial diseases. Previous animal models of G11778A LHON used consecutive intraocular injections of an AAV containing the disease-inducing mutant ND4 with the rescue wild-type allele (38, 39). The mechanism of progressive visual failure induced by mutant ND4 may be mediated by accumulating mitochondrial oxidative stress resulting in energy failure that primes neurodegeneration (40). In a different disease, Leber congenital amaurosis, gene therapy improved visual function but did not prevent progressive neurodegeneration of the retina (41). A combinatorial approach with antioxidants, antiapoptotic genes, or other agents ultimately may be needed to target the neurodegeneration of LHON sustainably (35, 36).
Unlike most mitochondrial diseases, which are heteroplasmic (i.e., having both mutated and normal mitochondrial DNA), most cases of human LHON are homoplasmic, with only ∼15% being heteroplasmic (42–44). Still, the LHON phenotype has been reported with as little as 33% mutated mitochondrial DNA (45). Therefore, in these people and in rodent models (38, 39), heteroplasmy is sufficient to induce the phenotype of visual loss and optic disk edema followed by progressive demise of retinal ganglion cells and their axons in the optic nerve. We found mutant human ND4 DNA levels were one fifth of the levels of endogenous mouse ND4; this dominant-negative effect of the mutant human ND4 expressed in the mouse is unlike most human diseases, in which the threshold of mutant to wild-type mitochondrial DNA usually must exceed 70–90% to be symptomatic. However, at least one human disorder has been shown to be caused with as little as 4–8% mutant mitochondrial DNA (46).
We were unable to find any recombination of the mutant human ND4 within the mouse mitochondrial genome, suggesting that the transferred human gene remained episomal. Although we detected some evidence for the replication of viral genomes, as would be expected with tissue expression after picoliter AAV injection into zygotes, the absence of integration into the mouse mitochondrial genome suggests that this replication did not occur by the “lagging-strand” mechanism characteristic of mitochondrial DNA synthesis but perhaps occurred by replication of viral DNA with an endogenous REP-like protein during the development of the embryo. Most of the 13 proteins encoded by the mitochondrial genome have some codons that are different from the nuclear genetic code; thus human ND4 can be translated in the mitochondria only where TGA encodes tryptophan but is a stop codon in the nucleus, where only a 13 N-terminal amino acid peptide would be generated. Taken together, these findings indicate that mutant human ND4 remained episomal within mitochondria where the human ND4 was transcribed and translated.
Clearly, relevant animal models of disease are a requisite in the quest to develop a clinically effective therapy for mitochondrial diseases. Using electrofusion technology, Lin et al. (14) fused enucleated cytoplasts of cybrids to mouse embryonic stem cells and generated transgenic mice homoplasmic for ND6 G13997A resulting in a P25L substitution. As in LHON these transgenic mice showed loss of optic nerve axons, but unlike LHON they also had a reduction in photoreceptor function as determined by electroretinography but, surprisingly, had normal visual acuity. In addition, it was reported that, rather than causing LHON in humans, the ND6P25L mutation causes Leigh syndrome (14). Our procedure is able to introduce mutant human DNA into the mitochondria of mice over multiple generations. Using this methodology, we also generated a mouse line with the human T8993G ATP6 responsible for human Leigh’s syndrome and neurogenic ataxia and retinitis pigmentosa. This mutation resulted not only in visual loss but also in paralysis and eventually death of 78% of mutant ATP6 mice (47). Lethality was not seen in mutant ND4 mice. In summary, the application of our methodology to other pathogenic mtDNA mutations may generate various transgenic mitochondrial mice for precise study of the pathogenesis of these devastating mitochondrial diseases and the testing of new therapies for combatting them.
Materials and Methods
Plasmids.
scHSP-mutND4FLAG+mCherry was constructed as previously described (17, 18). In brief, human mutND4FLAG and mitochondrial-encoded mCherry (mCherry) were generated by site-directed mutagenesis (Quikchange II XL site-directed mutagenesis kit; Stratagene) with the substitution of A for G at 11778 in ND4 and A for C at 559 in mCherry. Both then were cloned into scAAV backbones under the control of the mitochondrial HSP, where ND4FLAG is followed by mCherry with a stop codon between two genes. Primers used for site-directed mutagenesis were mutND4: cga acg cac tca cag tcA cat cat aat cct ctc tc (sense) and ga gag agg att atg atg Tga ctg tga gtg cgt tcg (antisense); mCherry: gcg gat caa gca gCg gct gaa gct gaa gg (sense) and cc ttc agc ttc agc cGc tgc ttg atc cgc (antisense) (the mutated nucleotide is indicated by a capital letter).
Animal Experiments.
Injections of mitochondria-targeted viral vectors into fertilized oocytes were performed using a standard microinjection protocol. Briefly, hybrid (C57BL/6J × DBA/2J) F1 mice, referred to herein as “B6D2F1 mice,” were obtained from The Jackson Laboratory for production of fertilized oocytes. Superovulation was stimulated by i.p. injection of B6D2F1 females with 5 IU pregnant mare serum gonadotropin (National Hormone and Peptide Program, National Institute of Diabetes and Digestive and Kidney Diseases) followed 46–48 h later by injection of 5 IU human chorionic gonadotropin (HCG) (Sigma-Aldrich). Following HCG injection, females were mated with B6D2F1 stud males, and oocytes were harvested the following day. Approximately 1–2 pL of the MTS-AAV-HSP-mutantND4Flag+mCherry virus [1.06 × 1012 vector genomes (vg)/mL] was microinjected into fertilized oocytes using a continuous flow injection mode. Surviving eggs were implanted into the ampulla of pseudopregnant Swiss Webster (Taconic) recipient females. The resulting offspring were analyzed for the presence of the transgene after weaning. The viral injections resulted in the offspring listed in Table 1. A summary of the number of animals used for each experiment is provided in Table 4. Mouse weights are given in Table 2. All animal procedures were conducted facilities accredited by the Association for Assessment and Accreditation of Laboratory Animal Care and in accordance with the University of Miami Institutional Animal Care and Use Committee policies (48).
Table 4.
Animal utilization
| Experiment | No. of animals |
| PERG | 223 |
| OCT | 223 |
| CLSO | 223 |
| PCR | 55 |
| BN-PAGE | 10 |
| Immunohistochemistry | 50 |
| Transmission electron microscopy | 10 |
| Next-generation sequencing | 4 |
| Respiration | 37 |
| Rescue of visual loss | 23 |
| In vitro DNA synthesis | 10 |
| MRI | 6 |
| Intracranial pressure measurements | 8 |
PERG, OCT, and CLSO Imaging.
Mitochondrial mCherry fluorescence was visualized using a Heidelberg laser-scanning ophthalmoscope outfitted with special filters for red or green fluorescence. PERGs were performed according to previous reports (49). Averaged PERGs were automatically analyzed to evaluate the major positive and negative waves with Sigma Plot (Systat Software Inc.). Retinal images were visualized with in vivo spectral-domain (SD) OCT (Bioptigen, Inc.) and then were analyzed with semiautomated custom software written using MATLAB software (MathWorks, Inc.).
Intracranial Pressure Measurements.
Mice were anesthetized with a ketamine/xylazine mixture. A linear scalp incision was made in the skin. A 0.6-mm bur was used to drill a hole at the bregma. An OpSens fiberoptic pressure sensor (L222-0697-01) that was connected to an OpSens monitor was inserted through the burr hole into the brain. Diastolic and systolic measurements were taken and averaged.
MRI.
Mice were anesthetized with an Engler Veterinary anesthesia delivery system ADS 1000, flow rate of 2 liters per min and O2 at 2%. MRI was performed with a Siemens Medical Systems 3.0-Tesla Trio Model using a Syngo MR B19 DHHS 16-channel phase-array coil. The pulse sequence used was T2 fast spin echo 3-D, echo train length 21, repetition time 2,000, echo time 84, average 12, slice thickness 0.4 mm, field of view 46, voxel size 0.4 × 0.4 × 0.4 mm.
PCR.
Mitochondria were isolated from the retina, optic nerve, brain, spinal cord, liver, lung, skeletal muscle, and heart. DNA was extracted with the DNeasy blood and tissue kit (Qiagen). PCR and qPCR were performed with primers as previously described (17). In addition PCR was performed with primers forward 5′-TATGACT(A+C)CC(A+T)AAAGCCCATGT-3′ and reverse 5′-AGTTTGCTAGGCAGAATAG-3′ that amplified both human and mouse ND4 together. We then cut the ND4 fragment with ScaI, which is diagnostic for the human ND4.
Reversal of Visual Loss.
Mice (3 mo of age) with low PERG amplitudes (n = 23) were sedated by inhalation with 1.5–2% isoflurane for the intraocular injections. A local anesthetic (proparacaine hydrochloride) was applied topically to the cornea, and pupils were dilated by applying a drop of tropicamide. A 32-gauge needle attached to a Hamilton syringe was inserted through the pars plana, and 1 μL of mitochondria-targeted scAAV2 containing the wild-type human ND4 (scAAV2-HSP-ND4) packaged with triple-mutant VP3 (titer = 4.4 × 1011 vg/mL) was injected in both eyes of 14 mice with visual loss indicated by low PERG amplitudes. Nine mice with similar low PERG amplitudes received injections of sterile PBS into both eyes. PERGs were performed at baseline and then again at 1 and 3 mo after intravitreal injections.
Immunohistochemistry, Histology, and Light and Electron Microscopy.
Mice were anesthetized with isoflurane/oxygen and perfused transcardially with 1× PBS followed by 4% paraformaldehyde in PBS (for light microscopy) or 4% paraformaldehyde/2.5% glutaraldehyde in PBS (for electron microscopy). Tissues were collected and were cryoprotected in graded concentrations of sucrose and sectioned at 10 µm on a Leica CM1850 cryostat (for light microscopy) or were embedded in EPON resin (Ted Pella) after postfixing in osmium tetroxide (for electron microscopy). Longitudinal retinal sections were used for staining. The following antibodies were used: Cy3- or FITC-conjugated FLAG (1:100; Sigma), Porin (1:100; Abcam), Thy1.2 (1:200; Abcam), mCherry (1:500; Abcam), and secondary antibodies that included anti-rat 488, anti-mouse 488, anti-rabbit 647, and anti-mouse cy3 (1:600; Invitrogen). Images were taken with a Leica TCS SP5 confocal microscope. Ultrathin sections (90 nm) were placed on nickel grids. Examinations were made with a Hitachi H-7600 or H-7650 transmission electron microscope operating at 80 kV.
Laser Microdissection.
The retinas of transgenic or normal C57BL/6 mice were fixed overnight at 4 °C in 4% paraformaldehyde/PBS and equilibrated in 30% sucrose/PBS overnight at 4 °C. After embedding into OCT and frozen solid in liquid nitrogen, retinas were cut into 8-μm sections and placed on director slides (Expression Pathology). Laser capture microdissection was performed using a Leica LMD6500. RGCs and cells from the inner and outer nuclear layers were excised and collected for DNA extraction.
BN/PAGE and Immunoblotting.
BN/PAGE was performed using the Invitrogen NativePAGE Gel system with minor modifications. In brief, mitochondria from the liver were enriched and used for BN/PAGE gel as described in a previous report (17). Electrophoresis was performed according to the manufacturer’s specifications (Invitrogen). BN-PAGE strips were equilibrated and applied to the 2D SDS gel as described by Invitrogen. Samples were separated in the second dimension and transferred to PVDF membranes (Immobilon; Millipore) using the semitransfer system (Bio-Rad). The following antibodies were used: anti-FLAG (1:1,000; Sigma), anti-ND4 (1:200; Santa Cruz), anti-mCherry (1:1,000; Abcam), and mouse monoclonal antibodies from MitoSciences, including anti-NDUFS4 and anti-NDUFA9 at a concentration of 1:1,000. Secondary probing with anti-mouse or anti-rabbit HRP-conjugated antibodies (1:5,000; Sigma) was performed for 1 h, followed by detection using ECL reagents (Amersham) and a FUJI Film Imaging system.
Respiratory Chain Function.
Mice were euthanized, and the optic nerves were collected and freeze thawed. Assays of complex I activity were performed on the optic nerve in triplicate by the reduction of cytochrome c with NADH. For assays of ATP synthesis rate, mice were euthanized, and the optic nerves were collected and placed immediately in DMEM with 10% FBS on ice. The rate of ATP synthesis was measured by chemoluminescence with a modified luciferin-luciferase assay in homogenized optic nerves digitonin-permeabilized with the complex I substrates malate and pyruvate in real time with an Optocom I luminometer (MGM Instruments). In brief, the optic nerves (pooled from three mice) were put in 300 μL buffer A (150 mM KCl, 25 mM Tris⋅HCl, 2 mM EDTA, 0.1% BSA, 10 mM K3PO4, 0.1 mM MgCl2, pH 7.4) and were homogenized for 10 s with a handheld Omni TH tissue homogenizer set at 19,500 rpm. A 100-μL aliquot of the cell suspension was used for the assay. The protein concentration of each suspension was quantitated using the protein assay kit (Bio-Rad).
Oxidative Stress.
One microliter of DCFDA (2 mM) was injected into the vitreous cavity of transgenic mice and normal controls. Three hours later mice were euthanized and perfused with PBS. Eyes were enucleated and immersion fixed in 4% formaldehyde for 15 min, cryosectioned, and examined by confocal microscopy. The number of DCFDA+ cells in the ganglion cell layer was counted.
In Vitro DNA Synthesis.
In vitro DNA synthesis using dTTP α32P and the three other unlabeled dNTPs was performed using a kit (Invitrogen) on freshly isolated liver mitochondria from transgenic mutant ND4 and wild-type mice. Reactions were carried out for 3–12 h at 37 °C, run on a 1% agarose gel, and visualized by autoradiography. Southern blot analysis of unlabeled dNTPs of the DNA synthesis reaction was performed. Signal was detected using a radiolabeled probe made according to the manufacturer’s specifications (Promega end-labeling kit) for the addition of [α-32P]dATP to the oligonucleotide 5′TAGTCATATTAAGTTGTTGG-3′.
Statistical Analysis.
Statistical analysis was performed using the GraphPad Prism Statistical program. The one-way ANOVA and t test were used, taking P < 0.05 as statistically significant.
Supplementary Material
Acknowledgments
We thank Mabel Wilson for editing the manuscript and Donna Bortner and Debra Banner (TransViragen, Inc.) for production of the transgenic mice. This work was supported by NIH Grants R01 EY017141, EY012355, and P30-EY014801 and by an unrestricted grant to Bascom Palmer Eye Institute from Research to Prevent Blindness, Inc.
Footnotes
Conflict of interest statement: W.W.H. and the University of Florida have a financial interest in AGTC, a company that may commercialize the use of adeno-associated virus as a therapeutic.
This article is a PNAS Direct Submission. J.G.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506129112/-/DCSupplemental.
References
- 1.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283(5407):1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 2.Hauswirth WW, Laipis PJ. Mitochondrial DNA polymorphism in a maternal lineage of Holstein cows. Proc Natl Acad Sci USA. 1982;79(15):4686–4690. doi: 10.1073/pnas.79.15.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moraes CT, et al. Molecular analysis of the muscle pathology associated with mitochondrial DNA deletions. Nat Genet. 1992;1(5):359–367. doi: 10.1038/ng0892-359. [DOI] [PubMed] [Google Scholar]
- 4.Johns DR. Seminars in medicine of the Beth Israel Hospital, Boston. Mitochondrial DNA and disease. N Engl J Med. 1995;333(10):638–644. doi: 10.1056/NEJM199509073331007. [DOI] [PubMed] [Google Scholar]
- 5.Shoffner JM, Wallace DC. Heart disease and mitochondrial DNA mutations. Heart Dis Stroke. 1992;1(4):235–241. [PubMed] [Google Scholar]
- 6.Wallace DC, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242(4884):1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 7.Howell N. Leber hereditary optic neuropathy: Mitochondrial mutations and degeneration of the optic nerve. Vision Res. 1997;37(24):3495–3507. doi: 10.1016/S0042-6989(96)00167-8. [DOI] [PubMed] [Google Scholar]
- 8.Kogelnik AM, Lott MT, Brown MD, Navathe SB, Wallace DC. MITOMAP: A human mitochondrial genome database. Nucleic Acids Res. 1996;24(1):177–179. doi: 10.1093/nar/24.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sazanov LA, Peak-Chew SY, Fearnley IM, Walker JE. Resolution of the membrane domain of bovine complex I into subcomplexes: Implications for the structural organization of the enzyme. Biochemistry. 2000;39(24):7229–7235. doi: 10.1021/bi000335t. [DOI] [PubMed] [Google Scholar]
- 10.Howell N. Leber hereditary optic neuropathy: Respiratory chain dysfunction and degeneration of the optic nerve. Vision Res. 1998;38(10):1495–1504. doi: 10.1016/s0042-6989(97)00444-6. [DOI] [PubMed] [Google Scholar]
- 11.Koene S, Willems PH, Roestenberg P, Koopman WJ, Smeitink JA. Mouse models for nuclear DNA-encoded mitochondrial complex I deficiency. J Inherit Metab Dis. 2011;34(2):293–307. doi: 10.1007/s10545-009-9005-x. [DOI] [PubMed] [Google Scholar]
- 12.Cwerman-Thibault H, Sahel JA, Corral-Debrinski M. Mitochondrial medicine: To a new era of gene therapy for mitochondrial DNA mutations. J Inherit Metab Dis. 2011;34(2):327–344. doi: 10.1007/s10545-010-9131-5. [DOI] [PubMed] [Google Scholar]
- 13.Schon EA, DiMauro S, Hirano M, Gilkerson RW. Therapeutic prospects for mitochondrial disease. Trends Mol Med. 2010;16(6):268–276. doi: 10.1016/j.molmed.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin CS, et al. Mouse mtDNA mutant model of Leber hereditary optic neuropathy. Proc Natl Acad Sci USA. 2012;109(49):20065–20070. doi: 10.1073/pnas.1217113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maul GG, Rovera G, Vorbrodt A, Abramczuk J. Membrane fusion as a mechanism of simian virus 40 entry into different cellular compartments. J Virol. 1978;28(3):936–944. doi: 10.1128/jvi.28.3.936-944.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaeppel C, et al. A largely random AAV integration profile after LPLD gene therapy. Nat Med. 2013;19(7):889–891. doi: 10.1038/nm.3230. [DOI] [PubMed] [Google Scholar]
- 17.Yu H, et al. Gene delivery to mitochondria by targeting modified adenoassociated virus suppresses Leber’s hereditary optic neuropathy in a mouse model. Proc Natl Acad Sci USA. 2012;109(20):E1238–E1247. doi: 10.1073/pnas.1119577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu H, et al. Mutant NADH dehydrogenase subunit 4 gene delivery to mitochondria by targeting sequence-modified adeno-associated virus induces visual loss and optic atrophy in mice. Mol Vis. 2012;18:1668–1683. [PMC free article] [PubMed] [Google Scholar]
- 19.Sligh JE, et al. Maternal germ-line transmission of mutant mtDNAs from embryonic stem cell-derived chimeric mice. Proc Natl Acad Sci USA. 2000;97(26):14461–14466. doi: 10.1073/pnas.250491597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark MA, Shay JW. Mitochondrial transformation of mammalian cells. Nature. 1982;295:605–607. doi: 10.1038/295605a0. [DOI] [PubMed] [Google Scholar]
- 21.Vestweber D, Schatz G. DNA-protein conjugates can enter mitochondria via the protein import pathway. Nature. 1989;338(6211):170–172. doi: 10.1038/338170a0. [DOI] [PubMed] [Google Scholar]
- 22.Collombet JM, Wheeler VC, Vogel F, Coutelle C. Introduction of plasmid DNA into isolated mitochondria by electroporation. A novel approach toward gene correction for mitochondrial disorders. J Biol Chem. 1997;272(8):5342–5347. doi: 10.1074/jbc.272.8.5342. [DOI] [PubMed] [Google Scholar]
- 23.Yoon YG, Koob MD. Efficient cloning and engineering of entire mitochondrial genomes in Escherichia coli and transfer into transcriptionally active mitochondria. Nucleic Acids Res. 2003;31(5):1407–1415. doi: 10.1093/nar/gkg228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan SM, Bennett JP., Jr Development of mitochondrial gene replacement therapy. J Bioenerg Biomembr. 2004;36(4):387–393. doi: 10.1023/B:JOBB.0000041773.20072.9e. [DOI] [PubMed] [Google Scholar]
- 25.Ibrahim N, et al. DNA delivery to mitochondria: Sequence specificity and energy enhancement. Pharm Res. 2011;28(11):2871–2882. doi: 10.1007/s11095-011-0516-4. [DOI] [PubMed] [Google Scholar]
- 26.Mileshina D, et al. Mitochondrial transfection for studying organellar DNA repair, genome maintenance and aging. Mech Ageing Dev. 2011;132(8-9):412–423. doi: 10.1016/j.mad.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Kanamori A, et al. Superoxide is an associated signal for apoptosis in axonal injury. Brain. 2010;133(9):2612–2625. doi: 10.1093/brain/awq105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cogné B, et al. NGS library preparation may generate artifactual integration sites of AAV vectors. Nat Med. 2014;20(6):577–578. doi: 10.1038/nm.3578. [DOI] [PubMed] [Google Scholar]
- 29.Ni TH, et al. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J Virol. 1998;72(4):2777–2787. doi: 10.1128/jvi.72.4.2777-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290(5806):470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 31.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 32.Facucho-Oliveira JM, St John JC. The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev. 2009;5(2):140–158. doi: 10.1007/s12015-009-9058-0. [DOI] [PubMed] [Google Scholar]
- 33.Montoya J, Gaines GL, Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34(1):151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 34.Chou TH, et al. Postnatal elongation of eye size in DBA/2J mice compared with C57BL/6J mice: In vivo analysis with whole-eye OCT. Invest Ophthalmol Vis Sci. 2011;52(6):3604–3612. doi: 10.1167/iovs.10-6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi X, Sun L, Hauswirth WW, Lewin AS, Guy J. Use of mitochondrial antioxidant defenses for rescue of cells with a Leber hereditary optic neuropathy-causing mutation. Arch Ophthalmol. 2007;125(2):268–272. doi: 10.1001/archopht.125.2.268. [DOI] [PubMed] [Google Scholar]
- 36.Qi X, Lewin AS, Sun L, Hauswirth WW, Guy J. SOD2 gene transfer protects against optic neuropathy induced by deficiency of complex I. Ann Neurol. 2004;56(2):182–191. doi: 10.1002/ana.20175. [DOI] [PubMed] [Google Scholar]
- 37.Guy J, et al. Rescue of a mitochondrial deficiency causing Leber Hereditary Optic Neuropathy. Ann Neurol. 2002;52(5):534–542. doi: 10.1002/ana.10354. [DOI] [PubMed] [Google Scholar]
- 38.Qi X, Sun L, Lewin AS, Hauswirth WW, Guy J. The mutant human ND4 subunit of complex I induces optic neuropathy in the mouse. Invest Ophthalmol Vis Sci. 2007;48(1):1–10. doi: 10.1167/iovs.06-0789. [DOI] [PubMed] [Google Scholar]
- 39.Ellouze S, et al. Optimized allotopic expression of the human mitochondrial ND4 prevents blindness in a rat model of mitochondrial dysfunction. Am J Hum Genet. 2008;83(3):373–387. doi: 10.1016/j.ajhg.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi X, Lewin AS, Sun L, Hauswirth WW, Guy J. Mitochondrial protein nitration primes neurodegeneration in experimental autoimmune encephalomyelitis. J Biol Chem. 2006;281(42):31950–31962. doi: 10.1074/jbc.M603717200. [DOI] [PubMed] [Google Scholar]
- 41.Cideciyan AV, et al. Human retinal gene therapy for Leber congenital amaurosis shows advancing retinal degeneration despite enduring visual improvement. Proc Natl Acad Sci USA. 2013;110(6):E517–E525. doi: 10.1073/pnas.1218933110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman NJ, Lott MT, Wallace DC. The clinical characteristics of pedigrees of Leber’s hereditary optic neuropathy with the 11778 mutation. Am J Ophthalmol. 1991;111(6):750–762. doi: 10.1016/s0002-9394(14)76784-4. [DOI] [PubMed] [Google Scholar]
- 43.Smith KH, Johns DR, Heher KL, Miller NR. Heteroplasmy in Leber’s hereditary optic neuropathy. Arch Ophthalmol. 1993;111(11):1486–1490. doi: 10.1001/archopht.1993.01090110052022. [DOI] [PubMed] [Google Scholar]
- 44.Howell N, Xu M, Halvorson S, Bodis-Wollner I, Sherman J. A heteroplasmic LHON family: Tissue distribution and transmission of the 11778 mutation. Am J Hum Genet. 1994;55(1):203–206. [PMC free article] [PubMed] [Google Scholar]
- 45.Chinnery PF, Andrews RM, Turnbull DM, Howell NN. Leber hereditary optic neuropathy: Does heteroplasmy influence the inheritance and expression of the G11778A mitochondrial DNA mutation? Am J Med Genet. 2001;98(3):235–243. doi: 10.1002/1096-8628(20010122)98:3<235::aid-ajmg1086>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 46.Sacconi S, et al. A functionally dominant mitochondrial DNA mutation. Hum Mol Genet. 2008;17(12):1814–1820. doi: 10.1093/hmg/ddn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yuan H, Guy J (2014) Mutant human T8993G ATP6 expression by a mitochondrial targeting sequence modification of AAV capsid VP2 in transgenic mice. ARVO Supplement Invest Ophthalmol and Vis Sci 55:3311.
- 48. Committee on Care and Use of Laboratory Animals (1996) Guide for the Care and Use of Laboratory Animals (Natl Inst Health, Bethesda), DHHS Publ No (NIH) 85–23.
- 49.Porciatti V. The mouse pattern electroretinogram. Doc Ophthalmol. 2007;115(3):145–153. doi: 10.1007/s10633-007-9059-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.