Abstract
We have produced three antitoxins consisting of the variable domains of camelid heavy chain-only antibodies (VHH) by expressing the genes in the chloroplast of green algae. These antitoxins accumulate as soluble proteins capable of binding and neutralizing botulinum neurotoxin. Furthermore, they accumulate at up to 5% total soluble protein, sufficient expression to easily produce these antitoxins at scale from algae. The genes for the three different antitoxins were transformed into Chlamydomonas reinhardtii chloroplasts and their products purified from algae lysates and assayed for in vitro biological activity using toxin protection assays. The produced antibody domains bind to botulinum neurotoxin serotype A (BoNT/A) with similar affinities as camelid antibodies produced in Escherichia coli, and they are similarly able to protect primary rat neurons from intoxication by BoNT/A. Furthermore, the camelid antibodies were produced in algae without the use of solubilization tags commonly employed in E. coli. These camelid antibody domains are potent antigen binding proteins and the heterodimer fusion protein containing two VHH domains was capable of neutralizing BoNT/A at near equimolar concentrations with the toxin. Intact antibody domains were detected in the gastrointestinal (GI) tract of mice treated orally with antitoxin producing microalgae. These findings support the use of orally delivered antitoxins produced in green algae as a novel treatment for botulism.
Introduction
Antibody-based antitoxins are a therapeutic option for neutralizing toxins in subjects that have been exposed to the toxin or infected with a toxin-producing microorganism. Historically, antitoxins have been produced by generating antisera in large animals immunized with inactive forms of the toxin of interest. As recombinant DNA technologies and transformation techniques have advanced, toxin-specific recombinant monoclonal antibodies (mAbs) have been produced which are also capable of neutralizing toxins by binding to them in sera (Cheng et al., 2009). Antisera and mAbs are laborious to produce and difficult to stably maintain and distribute at scale, thus necessitating either different forms of treatment or more efficient antitoxins.
Camelid VHH domains are the smallest of the antibody-derived biomolecules currently employed in medical and industrial biotechnology applications (Saerens et al., 2008). At 12-15 kDa, these so-called ‘nanobodies’ are half the size of single chain variable fragment antibodies, and contain only a single domain instead of both a light and heavy chain domain. Meanwhile, full-sized common antibodies are 150-160 kDa and require the assembly of multiple heavy and light chain fragments and proper linkage of disulfide bonds. VHH domains are water-soluble, heat-stable, pepsin-resistant, and have a high binding capacity, making them an ideal candidate for a preparative medicine against intoxication from organisms like Clostridium botulinum (Dolk et al., 2005; van der Linden et al., 1999).
Botulinum neurotoxin (BoNT) from Clostridium botulinum is the most potent biological toxin known to vertebrates, and has an intravenous LD50 of 1.3 to 2.1 ng/kg in primates (Arnon SS et al., 2001). BoNTs and other Clostridial toxins are metallo-proteases that target the neuroexocytosis apparatus, with variants of BoNT targeting different individual subunits of the complex. Loss of neurotransmission capabilities in the intoxicated nerve cells leads to a potentially lethal flaccid paralysis, which currently can only be treated by artificial ventilation and feeding. There is no way to speed recovery once intoxicated, and the condition must be endured until the toxin is cleared naturally. In the blood, BoNT that has not yet entered neurons may be neutralized by antisera, but there is currently no way to remove ingested toxin that has entered the small intestine. This is especially relevant in infant botulism, where the gut is colonized by C. botulinum before normal flora is present and exposure to toxin is prolonged.
Algae have already been identified as a potential platform for the large-scale and inexpensive production of medicinal recombinant proteins like anti-cancer immunotoxins and vaccine antigens (Gregory et al., 2012; Tran et al., 2013). To assess whether the green alga C. reinhardtii would also be able to produce a functional single-chain or multivalent BoNT/A antitoxins, we expressed three previously characterized VHH domains derived from alpacas immunized with BoNT/A as recombinant proteins in algal chloroplasts. All three proteins were expressed as soluble, apparently correctly folded molecules, and were purified by affinity chromatography. All three proteins bound to the target BoNT/A toxin in ELISA assays and all three algae produced antitoxin VHHs were capable of protecting rat primary cerebellar neurons from BoNT/A inactivation. A multivalent nanobody containing two genetically linked binding domains had significantly greater binding avidity and higher efficacy in the protection assays than VHH monomers, as observed previously for these agents produced in E. coli (Mukherjee et al., 2012). Furthermore, the antitoxin-producing microalgae were able to deliver and maintain intact neutralizing antibodies inside the stomach and small intestine of algae-fed mice.
Results
Vector design and genetic transformation
Genes for each recombinant anti-BoNT/A VHH domain were taken from a previously characterized phage display library derived from immunized alpacas, and were chosen based on their potency in neutralizing BoNT/A (Mukherjee et al., 2012). Two monomer VHH domains (C2 and H7) and a heterodimer of two VHH domains separated by a flexible spacer (H7-fs-B5) were cloned by PCR and ligated to a psbA replacement vector. The H7 domain in the heterodimer is the same domain as the H7 monomer. All VHH domains were tagged with a FLAG peptide at the C-terminal end for detection. Each gene was codon optimized for expression in the C. reinhardtii chloroplast. The VHH genes were ligated into a psbA replacement vector as diagrammed in Figure 1. This vector utilizes 5′ and 3′ homologous regions outside of the psbA gene to direct homologous recombination between the recombinant DNA plasmid and the chloroplast genome upon transformation. To select for propagation of transformants, a Kanamycin resistance gene cassette was placed downstream of the chimeric VHH gene. Transformed algae were obtained using biolistic methods with DNA-coated gold particles accelerated onto a mat of algae cells (Rasala et al., 2010).
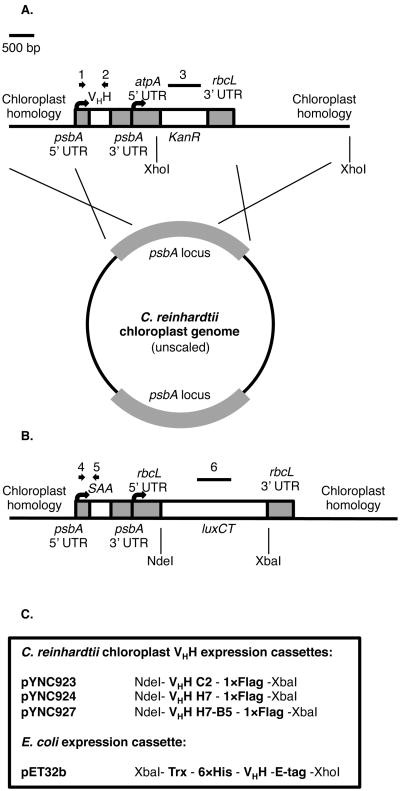
Figure 1. Camelid derived VHH domains and VHH expression vectors.
(A) Chloroplast codon-optimized VHH genes were ligated to the backbone of a psbA replacement vector. The coding sequences are governed by psbA elements including the promoter, 5′ UTR, and 3′ UTR. Downstream of the VHH cassette is a kanamycin resistance coding sequence (AphVI) governed by the elements of two other chloroplast genes, atpA and rbcL. The VHH cassette and the antibiotic resistance cassette are both flanked by sequence homology to the 5′ and 3′ regions of the psbA locus, directing the replacement of the psbA gene by homologous recombination. Primers from PCR analysis of gene integration in Figure 2 are indicated by 1 (psbA UTR forward) and 2 (VHH reverse). The probe from Figure 2C is indicated by 3. (B) Schematic of the psbA locus of recipient strain W1.1 used to generate VHH producing strains. Primers used in PCR homoplasmy screens in Figure 2B for the loss of mSAA amplicon are indicated by 4 (psbA UTR forward) and 5 (mSAA reverse). The probe for the Southern blot used in Figure 2D is indicated by 6. (C) The construction of each VHH chimera for C. reinhardtii chloroplast and E. coli vectors are shown. In the chloroplast vectors, both unique domains and the heterodimer contain a C-terminal FLAG peptide for purification purposes. In the bacterial vectors, each VHH has an N-terminal Trx domain for solubility and a 6×His tag for purification. On the C-terminal end, there is an E-tag, which is also for purification.
Identification of transgenic C. reinhardtii
Several C. reinhardtii colonies resistant to Kanamycin were selected for each construct transformed into recipient strain W1.1. Transgenic lines were propagated onto tris-acetate-phosphate (TAP) plates with 150 ug/mL Kanamycin, and positive transformants were identified by PCR analysis. Figure 2A shows PCR amplification products specific to each nanobody transgene. The primers used are indicated in Figure 1 and described in the methods. The forward primer is the same for all three reactions (psbA untranslated region), but the reverse were designed as sequences complementary to the 3′ end of each VHH gene, which is why the band for H7-fs-B5 is larger. Figure 2B shows the loss of an amplicon specific to the recipient strain for all transformed strains, demonstrating that all copies of the chloroplast genome have been transformed. Southern blot analyses were also performed to support data obtained from PCR screening. Figure 2C shows the presence of the Kanamycin resistance gene in all transformed strains of C. reinhardtii, but not in the recipient strain used for transformation. Figure 2D shows the presence of the luxCT gene in the recipient strain used for transformation, but no detectable copies of luxCT in the VHH-producing strains, thus demonstrating homoplasmy of the chloroplast genomes. C. reinhardtii contains 80 copies of its chloroplast genome, and even a single copy of the replaced locus would be apparent as an amplification product of the PCR screen. These screens are required because chloroplasts which are heteroplasmic for a transgene may not accumulate the desired protein well, due to competition for translation factors from the wild type gene. A control amplification product for the 16S rRNA locus is present in all samples demonstrating PCR efficacy.
Figure 2. Amplification of PCR products specific to VHH domains from C. reinhardtii chloroplasts and demonstration of homoplasmic chloroplast genomes.
(A) PCR analysis using primers specific to the psbA 5′ UTR primer and VHH gene demonstrate that the algal strain was successfully transformed. The image depicts an amplicon in each lane containing transformed algae, and no amplicon in adjacent lanes containing lysates from the recipient strain (W1.1, negative control). Primer locations and indicated in Figure 1. (B) PCR analysis using primers specific to the psbA 5′ UTR and the mSAA gene in the recipient strain shows that our strains are homoplasmic for the transformed locus. The PCRs also contain primers specific to the 16S rRNA locus to control for efficacy (upper band) because we are screening for a loss of the mSAA amplicon (lower band) in the recipient strain for transformation. (C) Southern blot analysis demonstrating gene integration into transformed strains relative to the recipient strain (W1.1). Chloroplast DNA was digested with XhoI and probed with a DIG-labeled fragment of DNA complementary to the Kanamycin resistance gene shown in Figure 1. (D) Southern blot analysis demonstrating homoplasmy of the chloroplast genomes of the transformed strains. Chloroplast DNA was digested with NdeI and XbaI and probed with a DIG-labeled fragment of luxCT. There are no detectable copies of the W1.1 chloroplast genome, indicating that all copies of the psbA locus have been replaced with VHH and antibiotic resistance genes.
Accumulation of recombinant nanobodies in C. reinhardtii chloroplast
Transgenic cell lines containing the C2, H7, and B5-fs-H7 VHH genes were tested for their ability to accumulate their respective recombinant nanobodies in the chloroplast. Immunobloting was performed on the soluble fraction of cell lysates for each nanobody-producing strain using an anti-Flag antibody directed against the C-terminal Flag-tag contained in each protein. Figure 3A demonstrates accumulation of both 14 kDa anti-BoNT VHH domains (C2 and H7) as well as the 28 kDa chimeric protein containing two unique anti-BoNT/A nanobodies genetically fused with a flexible spacer (H7-fs-B5). All recombinant proteins migrated according to their predicted sizes. Below each lane is the percent total soluble protein of each VHH domain that accumulated in the different transgenic lines, as determined by direct ELISA.
Figure 3. Accumulation of VHH domains in C. reinhardtii chloroplasts and purification by the FLAG tag.
(A) The soluble fraction of transformed algae lysates were separated by SDS-PAGE under reducing conditions (10 μg), transferred to a nitrocellulose membrane, and decorated with an anti-FLAG antibody conjugated to an alkaline phosphatase for detection. The image shows accumulation of each VHH domain (C2, H7, H7-fs-B5), and no accumulation of any FLAG peptides in the recipient strain W1.1. In a separate experiment, the % total soluble protein of each VHH domain was determined by direct ELISA. (B) Accumulation of the heterodimer is high enough to be visualized by Coomassie stain. The arrow points to the heterodimer band which is not present in W1.1 lysate. (C) Each VHH domain was purified using the FLAG peptide, and 20 μL of each concentrated, buffer-exchanged (PBS) protein was separated by reducing SDS-PAGE and stained with Coomassie to show purity.
To support ELISA data, a Coomassie stain gel was performed to show accumulation of the heterodimer (Figure 3B). The heterodimer band can be seen in the lysate from the algae strain expressing H7-fs-B5, while this band is absent in the lysate from the parental strain W1.1, as expected. All three recombinant nanobodies were then purified from the soluble fraction of algae lysates using an anti-Flag conjugated agarose resin. A Coomassie stain of the purified proteins resolved by SDS-PAGE showed that they had been separated from other C. reinhardtii protein present in the soluble fraction of the lysates (Figure 3C), and each was purified sufficiently for comparison with previously described VHH domains purified from E. coli.
Antigen binding capacity of chloroplast-produced anti-BoNT/A nanobodies
To determine whether algae chloroplast produced VHH domains would be capable of binding their target antigen, an enzyme-linked immunosorbent assay (ELISAs) was performed using BoNT/A coated plates (Figure 4). The E. coli expressed and purified proteins previously described by Mukherjee et al. (2012) exhibited binding to BoNT/A in the subnanomolar range (Figure 4A). The same proteins purified from C. reinhardtii chloroplasts also bound to BoNT/A with comparable affinities (Figure 4B). Protein B11, another VHH specific to the cell wall of C. reinhardtii, was used as a negative control as it does not bind to BoNT/A (Jiang et al., 2013).
Figure 4. C. reinhardtii chloroplast produced VHH domains bind BoNT/A.
(A) Recombinant VHH domains purified from E. coli were subjected to ELISA using anti-E-tag antibodies. All three domains that were also made in algae demonstrated binding to BoNT/A. A fourth VHH (B11), produced in E. coli, binds to a surface antigen of C. reinhardtii and served as a negative control (–) for BoNT/A binding. (B) All three binding domains produced from algae were capable of binding BoNT/A with similar affinity to those produced from bacteria. The heterodimer produced from algae had an EC50 in the picomolar range, while the same one produced from bacteria had an EC50 in the nanomolar range.
BoNT/A neutralization and cell survival assays
To assess the efficacy of algae-derived VHH domains in neutralizing BoNT/A toxicity, rat cerebellar primary neurons were isolated and subjected to BoNT/A intoxication. The quantity of toxin required to induce complete cleavage of its target SNAP-25 was determined by titration to be 50 pM. Primary neurons were pretreated with log dilutions on the nanomolar to picomolar scale of each unique VHH protein purified from either C. reinhardtii or E. coli before intoxication with 50 pM BoNT/A. After 24 hours of incubation, cells were harvested and their lysates were subjected to SDS-PAGE and immunoblotted for SNAP-25 (Figure 5). An inhibition of BoNT/A was demonstrated by a reduction in cleavage of SNAP-25 while in the presence of each chimeric VHH purified from either E. coli (Figure 5A) or C. reinhardtii (Figure 5B).
Figure 5. C. reinhardtii chloroplast produced VHH domains neutralize BoNT/A.
(A) VHH domains produced from E. coli were capable of preventing BoNT/A from cleaving its target SNAP-25 when co-administered with toxin to rat primary neuron cells. A western blot of SNAP-25 integrity is shown for harvested primary neuron cell cultures treated with each VHH. Domains C2 and H7 were capable of preventing cleavage, although not entirely, at 1 nM antitoxin. The heterodimer B5-fs-H7 was able to completely neutralize BoNT/A at 1 nM, and had some antitoxin activity at 100 pM. The lane indicating “no toxin” contained cells incubated without an antitoxin and serves as a negative control. (B) VHH domains produced from C. reinhardtii chloroplasts also demonstrated antitoxin activity. Domain C2 was capable of preventing SNAP-25 cleavage at 10 nM. Domain H7 offered complete neutralization at 1 nM and partial neutralization at 100 pM. The heterodimer B5-fs-H7 had potent antitoxin activity, completely neutralizing BoNT/A at 100 pM and mostly neutralizing BoNT/A at 10 pM.
The proteins produced from algae were clearly able to neutralize their toxin target, and demonstrated similar potency to those produced from bacteria. VHH C2 was able to neutralize BoNT/A at 10 nM and partially at 1 nM. H7 fully neutralized BoNT/A at 1 nM, and partially at 100 pM. The H7/B5 heterodimer proved to be 10-fold better at neutralizing toxin, as it completely neutralized BoNT/A at 100 pM, and partially at 10 pM. This large difference in efficacy is likely due to an increased avidity from having two target binding domains per molecule, which affords a neutralization concentration at near equimolar concentration with the toxin.
Detection of intact antitoxin in mice treated with H7-fs-B5 producing microalgae
To determine whether microalgae could serve as a vehicle for delivering recombinant antitoxin, transgenic algae producing the antitoxin was administered orally to mice and their GI tracts were examined for the presence of intact H7-fs-B5. Three time points were taken using two mice for each time point (45, 75, and 120 minutes), and the contents of the sectioned GI tract was extracted for western blot. One mouse was given wild type algae (cc1690) and its GI tract processed in the same manner after 45 minutes. The contents of three sections (stomach, jejunum and ileum) from each mouse were analyzed by western blot (Figure 6). Although the location and amount of antitoxin in the GI tract was variable, likely due to inter-mouse differences in water intake and activity that could influence algae transit speed, orally administered antitoxin was clearly detected intact in both the stomach and the small intestine. These results demonstrate that antitoxin-producing microalgae are able to deliver and maintain intact neutralizing antibodies inside the GI tract of mice treated orally with algae.
Figure 6. Orally administered microalgae producing H7-fs-B5 can deliver intact antitoxin to mouse stomach and small intestine.
Whole cell fresh algae was administered to mice by oral gavage, and sections of the gastrointestinal tract were examined 45, 75 or 120 minutes later. Two mice (#1 and #2) were examined per time point for antitoxin (H7-fs-B5) by blotting for Flag-tag of H7-fs-B5, and one mouse given wild type algae (WT) was examined after 45 minutes. The last lane on the right is control lysate derived from the the same preparation of antitoxin-producing algae orally administered to mice. Antitoxin was detected in the stomach at 45 minutes post-feeding, but moved to jejunum and ileum by 75 minutes and was primarily found in the ileum (or passed into colon) by 120 minutes.
Discussion
We have generated functional single chain antitoxin VHH domains in algae, and shown that these proteins are produced as soluble correctly folded proteins capable of neutralizing BoNT/A and protecting rat primary neuron cells from intoxication. We also show that these antitoxins can be orally delivered intact to the stomach and small intenstine of mice. Furthermore, these antitoxin molecules were produced in algae chloroplasts without extraneous solubilization tags (thioredoxin) that are commonly employed for production in bacteria. Finally, because algae are photosynthetic, they can be highly scalable and economically produced. Green algae are also edible making them an ideal platform for producing large, inexpensive quantities of antitoxin for oral delivery. If antitoxins are to be used to treat bacterial intoxication, it is best that they can be produced and stored inexpensively at scale.
The most common human form of BoNT intoxication in the U.S. is infant botulism, where the intestines are colonized by C. botulinum in the absence of normal gut flora. As an orphan disease, it has received little attention from pharmaceutical companies although it is massively expensive to treat as it requires weeks of intensive care in a hospital. One important advancement in treatment was the development of Botulism Immunoglobulin Intravenous (BIG-IV), a circulating antitoxin used to treat infant botulism. This development reduced the mean duration of intensive care by 3.2 weeks, of mechanical ventilation by 2.6 weeks, of tube or intravenous feeding by 6.4 weeks, and of the mean hospital charges per patient by $88,000 USD (Arnon et al., 2006). While an enormous achievement, this human serum-derived antitoxin does nothing to remove BoNT produced in the gut before it gets into circulation. This is why our algae-produced and orally delivered antitoxin is novel and has potential to vastly reduce duration and cost of hospitalization due to botulism.
C. botulinum, can also be found as a contaminant of both household food preservation methods such as canning as well as in industrial food processing. Recently, China issued a ban on imported dry milk from New Zealand when C. botulinum was found in dairy products made from the imported material (Reuters, 2013). Furthermore, C. botulinum is generally easy to cultivate and distribute, making this organism a serious bioterrorism threat. For these reasons it is very promising to see an antitoxin candidate that can be delivered orally and neutralizes BoNT/A at near equimolar quantities.
The platforms for producing many diverse therapeutic proteins are limited due to the complex nature of their domain structures, as well as the logistics of mass production, purification, distribution, and administration. Algae are a promising alternative for production of these agents because of their demonstrated ability to fold complex proteins, resulting in the production of soluble active proteins for many complex recombinant proteins (Barrera and Mayfield, 2013). Many important therapeutic proteins require complex folding and formation of disulfide bonds, which often cannot be done in bacterial platforms like E. coli. Additionally, there is no glycosylation of proteins in C. reinhardtii chloroplasts, which can cause problems with immunogenic responses and half-life when produced from yeasts (Gerngross, 2004). The camelid antitoxins characterized in this study accumulate at up to 4.6% total soluble protein, which is more than sufficient to afford commercial scale production (Manuell et al., 2007). In addition to forming soluble active proteins, algae are also edible making them a useful vehicle for the delivery of therapeutic proteins orally. For example, recently an oral vaccine against malaria was produced in algae and shown to remain active after several months storage at room temperature (Gregory et al., 2013).
There are other plant chloroplast-based platforms capable of orally delivering recombinant therapeutic proteins. In tobacco chloroplasts, a peptide based therapy for type II diabetes was developed and delivered orally in mice fed tobacco accumulating extendin-4 (Kwon et al., 2013). Also, VHH domains against rotavirus were produced in tobacco chloroplasts at 2-3% total soluble protein, although they required translational fusions to stability domains or redirection to the lumen in order to accumulate to detectable levels (Lentz et al., 2012). In our study the transgenic microalgae was non-photosynthetic due to the higher accumulation levels afforded by psbA knockouts in C. reinhardtii. Current levels of accumulation in photosynthetic microalgae can be quite low at 0.25% for m-SAA under the regulation of psbD elements (Manuell et al., 2007), so we opted to use non-photosynthetic strains better suited for in vivo studies with mice. However, there are several avenues of research currently being pursued toward improving recombinant protein accumulation in photosynthetic-competent microalgae (Rasala et al., 2011)(Specht and Mayfield, 2013).
Microalgae have several advantages over other platforms, like food crop species, for oral delivery of recombinant therapeutic proteins. First, C. reinhardtii is not a food crop, so production of algae would not displace land area suitable for food production. Second, there is not a risk of transgene transfer from algae into food crops, which is a primary concern of many people for GMO organisms (Davison, 2010). Also, as a single-celled plant, more energy can be utilized toward producing therapeutic proteins instead of diverting it to higher order structures like roots, leaves, stalks, and flowers. Finally, algae can be grown in containment inside bioreactors, further protecting natural populations of algae from accidental transfer of modified genetic material. Additional work will be needed to determine the efficacy of algae-derived antitoxins delivered orally in BoNT-intoxicated mouse models using whole-cell and lyophilized algae.
Materials and methods
Design and cloning of VHH transgenes for chloroplast expression
Genes encoding VHH monomers (C2, H7) and the H7-B5 heterodimer generated by Mukherjee et al. (2012) were adapted to the chloroplast codon usage of C. reinhardtii (KDRI species number 3055) according to the table in Kazusa DNA Research Institute's Database (http://www.kazusa.or.jp/codon). Further gene optimization and restriction site mapping were performed in silico using the Gene Designer bioinformatics software suite prior to synthesis (DNA2.0, Menola Park, CA). An additional protein domain fused to the VHHs include a single C-terminal Flag tag. Each cassette was flanked by NdeI and XbaI restriction sites upstream and downstream, respectively, for ligation in-frame with the chloroplast transformation vector driven by the psbA promoter (Manuell et al., 2007). Schematic diagrams of the C. reinhardtii chloroplast VHH expression cassettes related to the present study can be found in Figure 1. All bacterial proteins were expressed from pET32b plasmid described by Mukherjee et al. (2012) with an N-terminal thioredoxin (Trx), internal 6×His, and C-terminal E-tag.
Transformation of C. reinhardtii chloroplasts
Transformation of algae chloroplasts was performed using the biolistic method as described previously (Ramesh et al., 2004). Gold microprojectiles were coated with undigested plasmid DNA and accelerated onto a mat of 15 million C. reinhardtii cells plated onto TAP with 100 μg/ml Kanamycin. These cells were maintained under low light conditions until un-tranformed cells died and Kanamycin resistant transformants grew into visible colonies.
PCR screening of transgenic algae
To ensure transgene integration into the chloroplast genome and loss of all copies of the transformation locus, PCR screens were performed on lysates of the transformed algae. To test transgenic algae for the presence of the recombinant nanobody genes a primer to the psbA 5′ UTR, 5′-GTGCTAGGTAACTAACGTTTGATTTTT-3′ was combined with a reverse nanobody coding sequence primer specific to the C-terminal FLAG tag of each construct. The sequence for this primer is , 5′- GTCGACTTATTTATCGTCATCATCTTTGTAATC-3′. To test for a chloroplast homoplasmic for the transgenes, the same psbA primer was combined with a primer for mSAA, 5′-CTTGAATAGTTTCTCTAGCGTTAC-3′ which was the transformation locus replaced in these strains. Since we were looking for the loss of an amplicon, control primers for the 16S rRNA locus, 5′-CCGAACTGAGGTTGGGTTTA-3′ and reverse primer, 5′-GGGGGAGCGAATAGGATTAG-3′ were used as a control for PCR efficacy.
Southern blot analysis of transgenic algae
Genomic DNA was extracted from algae biomass by phenol-chloroform extraction. An inoculating loop was used to scrape cells from TAP agar plates into 1.5 mL Eppendorf tubes containing TEN buffer (10 mM Tris-HCl, 10 mM EDTA, 150 mM NaCl). The cells were resuspended by vortexing and then centrifuged for 1 minute at 14,000 RPM in a tabletop centrifuge. The supernatant was aspirated and the cells were resuspended in 150 uL water and 300 uL SDS-EB buffer (2% SDS, 400 mM NaCl, 40 mM EDTA, 100 mM Tris-HCl, pH 8.0) by vortexing. The DNA was extracted once with 350 uL of phenol/chloroform/isoamyl alcohol (25:24:1 ratio), and then again with 300 uL chloroform/isoamyl alcohol (24:1 ratio). Aqueous and organic layers were separated by centrifuging at 14,000 RPM in a tabletop centrifuge. The DNA was precipitated by adding 2 volumes of absolute ethanol to the aqueous layer, allowing 30 minutes of incubation on ice, and centrifuging for 10 minutes at 14,000 RPM. The resulting pellet was washed with 70% ethanol, air dried, and resuspended in 40 uL of water.
For Southern blot analysis, 3 ug of genomic DNA was digested overnight with either XhoI (Figure 2C) or a combination of NdeI and XbaI (Figure 2D). Digested DNA fragments were separated by gel electrophoresis and then transferred to nylon membranes by capillary transfer overnight. DNA was fixed to the membranes by UV-crosslinking, and then pre-hybridized with DIG Easy Hyb granules (Roche) for 30 minutes. The membranes were probed overnight in the same hybridization solution with a DIG-labeled PCR product of either a 754 bp fragment of the Kanamycin resistance gene (Figure 2C), or a 547 bp fragment of luxCT (Figure 2D), which is present only in the recipient strain (W1.1). Stringency washes were performed at room temperature and at 68 degrees Celsius using 2X SSC with SDS (0.3 M NaCl, 30 mM sodium citrate, 0.1% SDS, pH 7.0). The membranes were blocked for 30 minutes in blocking solution (Roche) and then decorated with an anti-DIG antibody conjugated to an alkaline phosphatase (Roche) for 30 minutes. The membranes were washed twice for 15 minutes each with Washing Buffer (0.1 M Maleic acid, 0.15 M NaCl, 0.3% Tween 20, pH 7.5), and band signal was developed with nitro-blue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) in AP buffer. The DIG-labeled PCR products were made with forward primer 5′-GAGCCAAATAAAATTGGTCAGTCG-3′ and reverse primer 5′- CGCAGTTTCCTCCGATGC-3′ (Kanamycin resistance gene), or forward primer 5′- GAGGTTCTTCAGGTGGTGGA-3′ and reverse primer 5′- AATTGTGCAGGACCACCTTC-3′ (luxCT).
Evaluation of recombinant nanobody accumulation
PCR-confirmed transgenic and homoplasmic algae were grown on TAP agar plates containing 150 μg/ml Kanamycin under constant illumination (100 μE m-2 s-1) for 5 days. When the plates had thick, healthy, green mats of algae a disposable inoculating loop was used to collect cells from each strain. Approximately one loop full of cells were resuspended in 500 μL TBST and sonicated on ice for two 10-second intervals at 25% amplitude (Fisher Scientific Sonic Dismembrator Model 500). Soluble protein was separated from the lysates by centrifugation in a tabletop centrifuge at 20,000 × g for 30 minutes at 4° C. SDS-PAGE was performed using Bio-Rad mini-protean TGX 10% gels and the gels were analyzed by immunoblot against the Flag tag. To detect the recombinant nanobodies the gels were transferred to a nitrocellulose membrane, decorated with a Sigma rabbit anti-Flag antibody conjugated to an alkaline phosphatase (AP) at 4° C overnight, and then developed with NBT and BCIP in AP buffer. To show accumulation of the heterodimer by stain gel, samples were processed as described above and the PAGE gel was stained overnight using Coomassie, and then destained with 25% methanol in water for 8 hours. Molecular masses of proteins were indicated with AccuRuler RGB protein ladder (BioPioneer).
To assess % total soluble protein production of each VHH domain, direct ELISA was employed using Nunc 96 well plates coated with 1 μg of total soluble protein in triplicate. The quantity of VHH in each microgram of soluble protein was determined by titrating purified VHH proteins into soluble protein from the parental recipient strain (W1.1). Wells were coated with protein diluted into coating buffer (100 mM bicarbonate/carbonate pH 9.6) overnight, washed five times with TBST, blocked with SuperBlock blocking buffer (Thermo) for one hour, and then incubated with an anti-Flag HRP antibody produced in mouse (Sigma) at 1:10,000 in blocking buffer for one hour. Each well was rinsed five times with TBST and then incubated with TMB substrate kit (Thermo) until signal developed. Signal was read using a Tecan microplate reader measuring absorbance at 450 nm.
Purification on the basis of the Flag-tag
Each strain was grown to a density of 1 × 106 cells/ml in 250 ml TAP flasks at 100 μE, used to inoculate 20 L carboys also containing TAP, and then grown for 3-5 days at 300 μE until mid log-phase growth or about 5 × 106 cells/ml. Cells were collected in a continuous flow centrifuge, pelleted and resuspended in lysis buffer [50 mM Tris pH 8.0, 500 mM NaCl, 0.1% Tween-20, complete protease inhibitor (Roche)], and sonicated for 7.5 minutes at 25% amplitude on ice to break open the cells. The lysates were separated into soluble and insoluble fractions by centrifugation at 20,000 × g. The soluble fractions were applied to 1 ml of equilibrated anti-Flag conjugated agarose resin (Sigma) and allowed to tumble for 1 hour at 4° C. The resins were washed with 50 column volumes of lysis buffer twice, three times with lysis buffer without tween-20, and then eluted six times with 1 column volume of elution buffer (100 mM glycine pH 3.5, 500 mM NaCl). Proteins were buffer exchanged into PBS with Vivaspin 6 concentrator columns (Sartorius Stedim). SDS-PAGE and immunoblotting were performed as described above. To assess purity, buffer exchanged protein samples were subjected to SDS-PAGE and subsequently stained by Coomassie.
Quantification of VHH binding affinity to BoNT/A by ELISA
Binding assays comparing the affinity of E. coli and C. reinhardtii expressed anti-BoNT/A VHH proteins were performed in 96-well plates pre-coated with BoNT/A and blocked of non-specific binding sites (Metabiologics, Inc., Madison, WI). E. coli expressed VHH B11 is a single-domain camelid VHH that binds to the cell wall of C. reinhardtii and served as a negative control since it does not bind to BoNT/A (Jiang et al. 2013, The Plant Journal, In Press). The ELISA procedure employed a dilution series of each VHH ranging from 1 μM to 10 pM in phosphate buffered saline (PBS) solution containing 0.1% tween-20 and 4% non-fat dry milk. Each dilution was administered in 100 μl volumes to duplicate wells and incubated at room temperature for 1 hour in rotary shaker at 40 RPM. All wells were then washed three times with 200 μl of phosphate buffered solution with 0.1% tween (PBST) followed by three washes with PBS. The extent of binding was determined by probing with a rabbit anti-E-tag antibody (Bethyl Labs, 1:5,000 in PBST) for the bacterially produced VHHs and a mouse anti-Flag-tag antibody (Sigma-Aldrich, 1:5,000 in PBST) for the VHHs derived from algae. For eventual colorimetric detection, both antibodies are conjugated with horseradish peroxidase (HRP). After incubation with the respective antibodies for 1 hour at room temperature, each well was again washed 3× with 200 μl of PBST and 3× with 200 μl of PBS. The ELISA was developed with 100 μl of 3,3′,5,5′-tetramethylbenzidine (TMB) liquid substrate per well (Sigma-Aldrich) and incubated at room temperature for 2-5 minutes until suitable colorimetric change occurred. The reaction was ceased by adding 100 μl of 1 N HCl and absorbance readings were measured at 450 nm using automated plate reader (Molecular Devices, MicroStation).
Cell survival assays
Primary neuron assays to assess BoNT/A neutralization were performed as previously described (Mukherjee et al., 2012). Briefly, rat cerebellar primary neuron cells were isolated from rat brains and cultured for 24 hours in the presence of 50 pM BoNT/A and a dilution series of each antitoxin. The cell cultures were pre-treated with separate dilutions of each VHH immediately before administering BoNT/A. After overnight culture, cells were harvested by centrifugation, lysed, and proteins were separated by SDS-PAGE. Western blots were performed and probed for SNAP-25. Neutralization of BoNT/A by test agents was assessed by comparing the extent of SNAP25 cleavage as compared to cells treated with toxin alone.
Oral administration of antitoxin producing microalgae to mice
All animal work was done in accordance with Institutional Animal Care and Use Guidelines of the University of California, San Diego. All mice were housed in specific pathogen-free conditions prior to use. Wild-type C57/BL6J mice were purchased from the Jackson Laboratory. Mice were gavaged with 150 mg of fresh, wet algae resuspended in 200 uL sterile PBS and sacrificed 45, 75, or 120 minutes later. Luminal contents were harvested from stomach, jejunum, and ileum and processed for immunoblotting as described below.
Digesta samples were resuspended in 400 uL TBST + protease inhibitor (Roche) and boiled for 4 minutes at 95 degrees while shaking at 1400 rpm. Total protein was analyzed (40 uL) by SDS-PAGE under reducing conditions using BioRad mini-protean TGX 12% PAGE gels. Gels were transferred to nitrocellulose membranes for immunoblotting. After blocking membranes for 30 minutes with 5% dry milk solubilized in TBST, membranes were decorated with an anti-FLAG antibody produced in mouse (Sigma). After washing the membrane 3× for 10 minutes with TBST, the alkaline phosphatase conjugated to the anti-FLAG antibody was used to detect H7-fs-B5 using NBT and BCIP.
Acknowledgments
This work was supported in part by U.S. Department of Energy grant DE-EE0003373 (SPM), U.S. National Institute of Health grant DK093507 (JTC), U.S. National Institute of Health grant U54-AI057159 (CBS) and a contract from the California Energy Commission (CILMSF #500-10-039). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Conflict of Interest: Stephen P. Mayfield is a co-founder of Sapphire Energy and Triton Algal Innovations, both of which may benefit from the commercialization of green algae for the production of recombinant proteins.
George A. Oyler is the president of Synaptic Research, LLC.
References
- Arnon SS, Schechter R, Maslanka SE, Jewell NP, Hatheway CL. Human Botulism Immune Globulin for the Treatment of Infant Botulism. N Engl J Med. 2006;354:462–471. doi: 10.1056/NEJMoa051926. [DOI] [PubMed] [Google Scholar]
- Arnon SS, Schechter R, Inglesby TV, et al. Botulinum toxin as a biological weapon: Medical and public health management. JAMA. 2001;285:1059–1070. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- Barrera DJ, Mayfield SP. High-value Recombinant Protein Production in Microalgae. In: Emeritus ARPD, Q H Ph D, editors. Handbook of Microalgal Culture. John Wiley & Sons, Ltd; 2013. pp. 532–544. [Google Scholar]
- Cheng LW, Stanker LH, Henderson TD, Lou J, Marks JD. Antibody Protection against Botulinum Neurotoxin Intoxication in Mice. Infect Immun. 2009;77:4305–4313. doi: 10.1128/IAI.00405-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J. GM plants: Science, politics and EC regulations. Plant Sci. 2010;178:94–98. [Google Scholar]
- Dolk E, van Vliet C, Perez JMJ, Vriend G, Darbon H, Ferrat G, Cambillau C, Frenken LGJ, Verrips T. Induced refolding of a temperature denatured llama heavy-chain antibody fragment by its antigen. Proteins Struct Funct Bioinforma. 2005;59:555–564. doi: 10.1002/prot.20378. [DOI] [PubMed] [Google Scholar]
- Gerngross TU. Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat Biotechnol. 2004;22:1409–1414. doi: 10.1038/nbt1028. [DOI] [PubMed] [Google Scholar]
- Gregory JA, Li F, Tomosada LM, Cox CJ, Topol AB, Vinetz JM, Mayfield S. Algae-Produced Pfs25 Elicits Antibodies That Inhibit Malaria Transmission. PLoS ONE. 2012;7:e37179. doi: 10.1371/journal.pone.0037179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory JA, Topol AB, Doerner DZ, Mayfield S. Alga-Produced Cholera Toxin-Pfs25 Fusion Proteins as Oral Vaccines. Appl Environ Microbiol. 2013;79:3917–3925. doi: 10.1128/AEM.00714-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Rosenberg JN, Wauchope AD, Tremblay JM, Shoemaker CB, Weeks DP, Oyler GA. Generation of a phage display library of single-domain camelid VHH antibodies directed against Chlamydomonas reinhardtii antigens and characterization of VHHs binding cell surface antigens. Plant J. 2013:n/a–n/a. doi: 10.1111/tpj.12316. [DOI] [PubMed] [Google Scholar]
- Kwon KC, Nityanandam R, New JS, Daniell H. Oral delivery of bioencapsulated exendin-4 expressed in chloroplasts lowers blood glucose level in mice and stimulates insulin secretion in beta-TC6 cells. Plant Biotechnol J. 2013;11:77–86. doi: 10.1111/pbi.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz EM, Garaicoechea L, Alfano EF, Parreño V, Wigdorovitz A, Bravo-Almonacid FF. Translational fusion and redirection to thylakoid lumen as strategies to improve the accumulation of a camelid antibody fragment in transplastomic tobacco. Planta. 2012;236:703–714. doi: 10.1007/s00425-012-1642-x. [DOI] [PubMed] [Google Scholar]
- Van der Linden RHJ, Frenken LGJ, de Geus B, Harmsen MM, Ruuls RC, Stok W, de Ron L, Wilson S, Davis P, Verrips CT. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim Biophys Acta BBA - Protein Struct Mol Enzym. 1999;1431:37–46. doi: 10.1016/s0167-4838(99)00030-8. [DOI] [PubMed] [Google Scholar]
- Manuell AL, Beligni MV, Elder JH, Siefker DT, Tran M, Weber A, McDonald TL, Mayfield SP. Robust expression of a bioactive mammalian protein in Chlamydomonas chloroplast. Plant Biotechnol J. 2007;5:402–412. doi: 10.1111/j.1467-7652.2007.00249.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee J, Tremblay JM, Leysath CE, Ofori K, Baldwin K, Feng X, Bedenice D, Webb RP, Wright PM, Smith LA, et al. A Novel Strategy for Development of Recombinant Antitoxin Therapeutics Tested in a Mouse Botulism Model. PLoS ONE. 2012;7:e29941. doi: 10.1371/journal.pone.0029941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh VM, Bingham SE, Webber AN. A simple method for chloroplast transformation in Chlamydomonas reinhardtii. Methods Mol Biol Clifton NJ. 2004;274:301–307. doi: 10.1385/1-59259-799-8:301. [DOI] [PubMed] [Google Scholar]
- Rasala BA, Muto M, Lee PA, Jager M, Cardoso RMF, Behnke CA, Kirk P, Hokanson CA, Crea R, Mendez M, et al. Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol J. 2010;8:719–733. doi: 10.1111/j.1467-7652.2010.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Muto M, Sullivan J, Mayfield SP. Improved heterologous protein expression in the chloroplast of Chlamydomonas reinhardtii through promoter and 5′ untranslated region optimization. Plant Biotechnol J. 2011;9:674–683. doi: 10.1111/j.1467-7652.2011.00620.x. [DOI] [PubMed] [Google Scholar]
- Reuters. China Bans Milk Powder From New Zealand Over Botulism Fears. New York: 2013. [Google Scholar]
- Saerens D, Ghassabeh GH, Muyldermans S. Single-domain antibodies as building blocks for novel therapeutics. Curr Opin Pharmacol. 2008;8:600–608. doi: 10.1016/j.coph.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Specht EA, Mayfield SP. Synthetic oligonucleotide libraries reveal novel regulatory elements in Chlamydomonas chloroplast mRNAs. ACS Synth Biol. 2013;2:34–46. doi: 10.1021/sb300069k. [DOI] [PubMed] [Google Scholar]
- Tran M, Van C, Barrera DJ, Pettersson PL, Peinado CD, Bui J, Mayfield SP. Production of unique immunotoxin cancer therapeutics in algal chloroplasts. Proc Natl Acad Sci U S A. 2013;110:E15–22. doi: 10.1073/pnas.1214638110. [DOI] [PMC free article] [PubMed] [Google Scholar]