Abstract
MicroRNA-155 (miR-155) is a multifunctional molecule involved in both normal and malignant hematopoiesis. It has been found to be involved in the pathogenesis of many different hematological malignancies with either an oncogenic or a tumor-repressor effect, depending on the nature of the cell and the type of malignancy. In particular, it has been strongly implicated in the causation of diffuse large B-cell lymphomas. This review focuses on the molecular interactions of miR-155, its oncogenic mechanisms, and its potential as an effective therapeutic target for the associated malignancies.
Keywords: microRNA-155, hematological malignancies
Introduction
MicroRNAs (miRNAs) are a group of small nonprotein-coding RNAs (19–24 nucleotides in size), which act as sequence-specific regulators of posttranscriptional gene expression. They have been discovered to play important roles in many cellular processes, including cell proliferation, differentiation, and apoptosis in normal as well as malignant hematopoiesis.1 Different miRNAs have tumor-suppressor or oncogenic effects in the pathogenesis of various hematological malignancies and solid tumors.2 This review focuses on the association between miRNAs, especially microRNA-155 (miR-155), and hematological malignancies.
Biosynthesis of miRNAs
miRNAs are transcribed from different genomic locations, either from independent noncoding RNAs, from introns of protein-coding genes, or even from exons of long nonprotein-coding transcripts, as long polyadenylated primary precursors (pri-microRNA) by RNA polymerase II. miRNA-coding regions account for 1%–5% of all human genes.3 One pri-miRNA can contain up to six hairpin loop structures, each made up of around 70 nucleotides. Within the nucleus, pri-miRNA associates with a nuclear protein known as DiGeorge Syndrome Critical Region 8 (DGCR8 also known as “Pasha” in invertebrates) and an RNase III called Drosha to form the “microprocessor” complex. Drosha cleaves pri-miRNA into individual hairpin-shaped 70–100 nucleotide units that are called precursor miRNAs (pre-miRNA), which are exported to the cytoplasm. In the cytoplasm, another RNase III called Dicer processes pre-miRNAs into ~22-nucleotide-long miRNA duplexes by removing the terminal loop. Only one strand of this duplex structure acts as the mature miRNA. Mature miRNA associates with other proteins, such as the AGO (Argonaute) protein, TRBP (transactivation-responsive RNA-binding protein), and PACT (protein activator of the interferon-induced protein kinase), to form an effector complex called the miRNA-containing ribonucleoprotein particle (miRNP) or RNA-induced silencing complex, which then acts on the target messenger RNAs (mRNAs).4
Mechanism of Action of miRNAs
The miRNP complex is directed toward its target by complementary base-pairing of the miRNA with the target mRNA. Unlike plant miRNAs that bind to a single complementary site in the coding region of the target mRNA, animal miRNAs including human miRNAs usually do not have exact complementarity to the target mRNAs and bind to multiple partially complementary sites in the 3′-untranslated regions. This binding leads to either repression of translation of the target mRNA or its destabilization and degradation.5
Till date, more than 1000 pre-miRNAs have been reported to be present in the human genome. Based on bioinformatic predictions, at least 50% of all gene products in the human genome are thought to be regulated by miRNAs, with each miRNA regulating hundreds of genes.6
Role of miRNAs in Normal Hematopoiesis
Different miRNAs have been found to play an important role in different aspects of hematopoiesis. The actual effect of these miRNAs on the level of expression of genes involved in hematopoiesis is usually modest, causing not more than twofold reduction in the protein levels.7,8 However, some of these proteins such as the transcription factor PU.1 and the transcriptional activator MYB have to be expressed in precise dosages and even a modest reduction in their levels can have significant phenotypic effects.9 Additionally, each mRNA usually has more than one miRNA-binding site and each miRNA can target multiple genes in the same pathway, thereby leading to significant cumulative modulatory effects.10
Apart from the simple negative regulation of target mRNA, miRNAs act through regulatory circuits such as the “coherent feed-forward loop,” the “mutual negative feedback loop,” and the “positive feedback/feed-forward loop” to maintain inhibition/inactivation of specific proteins for lineage commitment during hematopoiesis.11 Some miRNAs also act as buffers for random changes in gene expression resulting from stochastic events in transcription and translation.11
Role of miRNAs in Hematological Malignancies
One of the earliest evidences for the role of miRNA deregulation in hematological malignancies was the finding that the 13q14 locus, which is deleted in many cases of chronic lymphocytic leukemia (CLL), includes the coding regions of two miRNAs miR-15a and miR-16–1. These two miRNAs are highly expressed in CD5+ B-lymphocytes, and if the loss of one of their alleles contributes to the pathogenesis of CLL, they must be acting as tumor suppressors.12
Further studies have suggested that deregulation of expression of different miRNAs is a common event in many hematological malignancies. Table 1 lists some of the known associations between miRNAs and hematological malignancies.13–37
Table 1.
Role of microRNAs in haematological malignancies.
| HAEMATOLOGICAL MALIGNANCY | MicroRNAs REPORTED TO SHOW SIGNIFICANT ASSOCIATION WITH THE MALIGNANCY | PROPOSED ROLE OF ASSOCIATED MicroRNAs IN PATHOGENESIS |
|---|---|---|
| Leukemias | ||
| Acute myeloid leukemia (AML) | MiR-181a/b MiR-125b MiR-146a MiR-155 |
Over-expression of MiR-181a/b inhibits granulocytic and macrophage-like differentiation of HL-60 cells and CD34+ hematopoietic stem/progenitor cells by directly targeting and down-regulating the expression of PRKCD (which further affects the PRKCD-P38-C/EBPα pathway), CTDSPL (which further affects the phosphorylation of retinoblastoma protein) and CAMKK1.13,14 MiR-125b is significantly up-regulated in blast cells and promotes transformation of normal hematopoietic cells into malignant cells. Its anti-apoptotic and pro-proliferative effects are proposed to be mediated by targeting of ABTB1 and CBFB.15,16 MiR-146a is an inhibitor of TRAF6. As TRAF is involved in the activation of the canonical and noncanonical NF-kB signaling pathways, AML pathogenesis associated with mir-146a is believed to be mediated by NF-kB.15 The role of MiR-155 is discussed in the text. |
| Acute promyelocytic leukemia (APML) | MiR-181a/b | MiR-181a/b expression is activated by the PML/RARα oncogene and miR-181a/b in turn, targets the tumor suppressor gene RASSF1A by direct binding to its 3′-untranslated region.17 MiR-181a also decreases the expression of AC9 which is an enhancer of the trans-activity of the retinoic acid receptor; down-regulation of AC9 inhibits ATRA-induced differentiation.18 |
| Acute lymphoblastic leukemia (ALL) | MiR-125b MiR-155 MiR-21 MiR-196b MiR-17–92 |
MiR-125b facilitates the leukemogenic action of the BCR–ABL fusion gene, probably through down-regulation of IRF4, a transcription factor which inhibits the proto-oncogene BCL-6.19,20 The role of miR-155 is discussed in the text. MiR-21 has an oncogenic effect, through inhibition of pro-apoptotic and anti-proliferative genes.21 Over-expression of MiR-196b results in up-regulation of HOXA.22 Increased expression of the miR-17–92 cluster is associated with down-regulation of p21 which is an inhibitor of cell cycle progression.23 |
| Chronic lymphocytic leukemia (CLL) | MiR-15a, miR-16–1 MiR-34 MiR-29 MiR-181 MiR-155 |
MiR-15a and miR-16–1 act as tumor-suppressors and negatively regulate BCL2 at post-transcriptional level; down-regulation of these microRNAs results in increased Bcl2 expression with resultant inhibition of apoptosis.24 They also act in conjunction with DLEU7 to promote the activation of NF-kB and NFAT via TACI and BCMA.25,26 Over-expression of miR-34 results in p53-like effects on apoptosis or cell cycle arrest, as it is a downstream target of p53.27 MiR-29 and miR-181 act as tumor-suppressors by down-regulating TCL1 expression.28 The role of MiR-155 is discussed in the text. |
| Chronic myeloid leukemia (CML) | MiR-10a MiR-150 |
MiR-10a acts as tumor suppressor and down-regulates upstream stimulatory factor 2 (USF2). With inhibition of miR-10a, there is over-expression of USF2 which contributes to increased cell proliferation.29 MiR-150 acts as a tumor suppressor, probably by targeting c-Myb which is necessary for Bcr-Abl-mediated transformation.30 |
| Multiple myeloma (MM) | MiR-15a, miR-16–1 MiR-21 |
MiR-15a/16–1 acts as a tumor-suppressor through down-regulation of several target genes including WT1, FGFR1, PI3KCa, MDM4 and VEGFa.31,32 Over-expression of miR-21 leads to down-regulation of PTEN, Rho-B, and BTG2 and up-regulation of AKT and extracellular signal-regulated kinase signaling, which contribute to the pathogenesis of multiple myeloma.31 |
| Lymphomas | ||
| Hodgkin lymphoma (HL) | MiR-21 MiR-30d |
Over-expression of MiR-21 inhibits apoptosis by increasing the BCL2/BAX ratio.33 MiR-30d negatively regulates the TP53 pathway and also causes repression of CDKN1A expression.33 |
| Diffuse large B-cell lymphoma (DLBCL) | MiR-155 MiR-17–92 MiR-21 MiR-34a |
The role of miR-155 is discussed in the text. The MiR-17–92 cluster microRNAs act through repression of PTEN phosphatase and the pro-apoptotic BIM protein and down-regulation of the pro-apoptotic E2F1.34,35 MiR-21 acts by down-regulation of the tumour suppressor PTEN.36 MiR-34a has a tumour suppressor effect and its down-regulation promotes lympho-proliferation by allowing increased expression of GC transcription factors FOXP1 and BCL6.37 |
| Mantle cell lymphoma (MCL) | MiR-17–92 MiR-155 |
The MiR-17–92 cluster microRNAs act through repression of PTEN phosphatase and the pro-apoptotic BIM protein and down-regulation of the pro-apoptotic E2F1.34,35 The role of MiR-155 is discussed in the text. |
MicroRNA-155
MiR-155 is processed from a noncoding RNA transcribed from the B-cell integration cluster (BIC) gene located on chromosome 21. Depending upon the arm of the pre-miRNA-155 hairpin structure from which the mature miRNA-155 is derived, it is denoted as miR-155-5p (from the 5′ arm) and miR-155-3p (from the 3′ arm).38,39 MiR-155 is a multifunctional miRNA molecule that has been found to be involved in various biological processes. The list of target genes of miR-155 includes around 140 genes, which include genes encoding for regulatory proteins for myelopoiesis and erythropoiesis, tumor-suppressor genes, and genes encoding for inflammatory proteins.40
Apart from its role in normal and malignant hematopoiesis that is described in detail in this review, miR-155 has been found to have important roles in immune pathways, inflammatory processes, and cardiovascular pathophysiology.40 It has been shown to be an important component of the primary macrophage response to different inflammatory mediators such as bacterial lipopolysaccharide (LPS), interferon beta, poly IC (polyriboinosinic–polyribocytidylic acid), and tumor necrosis factor alpha.41 It has been found to be important in lymphocyte development and generation of B-cell- and T-cell-mediated immune responses.42 It has also been implicated in the causation of hypertension and cardiovascular diseases through repression of angiotensin II type I receptor.43
Role of miR-155 in Normal Hematopoiesis
MiR-155, like many of the other miRNAs involved in hematopoiesis, is expressed in high levels in normal hematopoietic stem-progenitor cells (HSPCs) and in low levels in mature hematopoietic cells. These miRNAs keep the genes specifying hematopoietic differentiation in check, until differentiation occurs. MiR-155 has been found to control both myelopoiesis and erythropoiesis.44 Masaki et al.45 demonstrated a 200-fold reduction in miR-155 expression during the differentiation of purified normal human erythroid progenitors in a liquid culture system, thereby confirming its role in erythroid differentiation.
However, the exact mechanisms by which miR-155 regulates normal myeloid lineage commitment are not clearly understood yet. It is likely to be involved in negative regulation of apoptosis, in increasing the rate of proliferation among myeloid progenitors, or in promoting commitment of HSPCs to the common myeloid progenitor lineage.15
Role of miR-155 in Hematological Malignancies
MiR-155 is one of the most frequently overexpressed miRNAs in solid as well as hematological malignancies.46 Aberrant expression of miR-155 has been found to be associated with various types of hematological malignancies. It can have either an oncogenic or a tumor-repressor effect, depending on the nature of the tissue and the type of malignancy. Over-expression of miR-155 has been found to be associated with many hematological malignancies including diffuse large B-cell lymphoma (DLBCL), Hodgkin’s lymphoma, follicular lymphoma, primary mediastinal B-cell lymphoma, chronic lymphoid leukemia, and acute myeloid leukemia (AML) with McDonough feline sarcoma viral oncogene homolog (FMS)-like tyrosine kinase 3 (FLT3)-internal tandem duplication mutations, suggesting its oncogenic role in their pathogenesis. On the other hand, it has been found to be downregulated in certain other hematological cancers such as Burkitt’s lymphoma (BL), mantle cell lymphoma (MCL), chronic myeloid leukemia, and AML with inv (16) and 3q26 cytogenetic abnormalities, suggesting a tumor-repressor role in these malignancies.7,38,47,48
Role of miR-155 in the pathogenesis of lymphomas
Different mechanisms have been postulated for the miR-155-mediated pathogenesis of hematological malignancies. One of these proposed pathogenic mechanisms, especially for lymphomas such as DLBCL, is the downregulation of B-cell lymphoma 6 protein (BCL6) and histone deacetylase 4 (HDAC4) by miR−155.38 BCL6 is an evolutionarily conserved zinc finger transcription factor, belonging to the family of POK (Pox viruses and Zinc-finger and Kruppel) factors, which normally has an inhibitory effect on transcription. BCL6 mediates transcriptional repression in processes such as hematopoietic cell differentiation, leukemogenesis, and inflammation by recruiting HDACs like HDAC4. Sandhu et al.49 studied the micro array data of 84 patients with DLBCL from two gene expression omnibus microarray data sets and found that miR-155 expression inversely correlated with BCL6 and HDAC4 expression. Further, they performed a genome-wide transcriptome analysis of naïve B cells in Eµ-miR-155-transgenic mice that overexpress miR-155. They found that BCL6 is significantly downregulated in Eµ-miR-155 mice, which in turn leads to de-repression of its targets like inhibitor of differentiation (Id2), interleukin-6 (IL-6), cMyc, Cyclin D1, and Mip1α/ccl3, which together promote cell survival and proliferation. BCL6 was found to be indirectly targeted by miR-155 through Mxd1/Mad1 upregulation, while its co-repressor partner HDAC4 was found to be directly targeted (Fig. 1). They also found that increased ectopic expression of HDAC4 in human activated B-cell (ABC)-type DLBCL cells reduces miR-155-induced proliferation and clonogenic potential and increases apoptosis (Fig. 1).49
Figure 1.
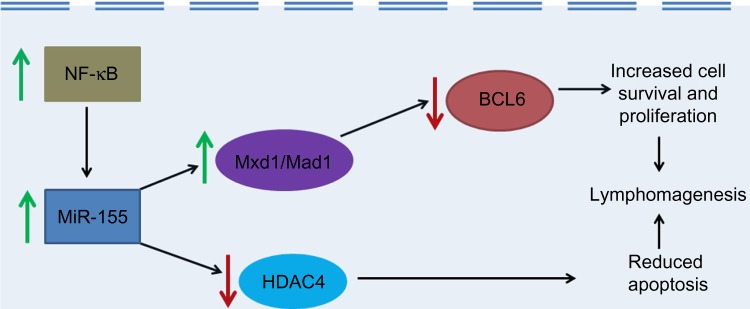
The role of MiR-155 in lymphomagenesis. Green arrows indicate increased activity and red arrows indicate decreased activity.
Thompson et al.50 studied the role of nuclear factor kappa B (NF-κB) in the miR-155-mediated pathogenesis of DLBCL. They found that tumor cells of the ABC subtype of DLBCL, which has low expression of the cell surface protein CD10 and poorer prognosis, had higher NF-κB activity when compared to tumor cells of the germinal center B-cell subtype of DLBCL, which has higher CD10 expression and a better prognosis. Higher activity of NF-κB correlated with increased expression of miR-155 and reduced expression of the transcription factor PU.1 and CD10 in many B-lymphoma cell lines. Both overexpression of miR-155 and treatment with the NF-κB inducer LPS were found to cause reduced expression of CD10 in the BJAB B-lymphoma cell line. Based on these findings, it was concluded that increased NF-κB activity results in increased miR-155, which in turn leads to decrease in PU.1 and consequent reduction in CD10 mRNA and protein, leading to a more aggressive form of DLBCL.50
MiR-155 has also been proposed to contribute to the pathogenesis of DLBCL by targeting the human germinal center associated lymphoma (HGAL) protein, a lymphocyte motility inhibitor.51 Dagan et al studied HGAL expression in DLBCL cell lines transfected with hsa-miR-155 and found that miR-155 directly downregulates HGAL expression, which in turn leads to decreased RhoA activation and increased lymphoma cell motility. This effect was proposed to contribute to lymphoma cell dissemination and aggressiveness in DLBCL.51
SMA and MAD-related protein 5 (SMAD5) is another miR-155 target considered to be involved in the pathogenesis of DLBCL. It normally plays a very important role in the signaling pathway by which transforming growth factor-beta (TGF-β) inhibits the proliferation of human hematopoietic progenitor cells. A study by Rai et al.52 found that miR-155 overexpression, through inhibition of SMAD5 activity, made DLBCLs resistant to the growth-inhibitory effects of both TGF-β1 and bone morphogenetic proteins, through defective induction of p21 and impaired cell cycle arrest.
MiR-155 has also been found to have an oncogenic role in the causation of anaplastic large-cell lymphoma (ALCL) lacking the t(2;5) translocation (which lacks the nucleophosmin-anaplastic lymphoma tyrosine kinase fusion protein), that is, the ALCL ALK(−) subtype. In a recently published study by Merkel et al.53, it was found that ALCL ALK(−) show reduced miR-155 promoter methylation and high miR-155 expression. Murine engraftment models of ALCL ALK(−) treated with antisense miR-155 mimics showed increased levels of CCAAT-enhancer binding protein beta (C/EBPβ) and of SOCS1 (Suppressor of cytokine signaling 1), which correlated with reduced tumor growth.53
As opposed to its oncogenic effects mentioned above, miR-155 has been found to have tumor-suppressor effects in Burkitt’s lymphoma (BL). Kluiver et al.54 demonstrated the absence or very low expression of miR-155 in Ebstein–Barr virus (EBV)-positive BL cells, EBV-negative BL cells, and EBV latency type I BL cells. However, they reported higher levels of BIC and miR-155 in in vitro transformed lymphoblastoid EBV latency type III BL cell lines. In a subsequent study, they demonstrated that even ectopic expression of BIC in the BL-derived Ramos cell line as well as other BL-derived cell lines did not result in miR-155 expression, indicating a specific block in the processing of miR-155 from BIC in BL. This was attributed to the regulation of BIC expression at the transcriptional level by protein kinase C and NF-κB and at the processing level by an unknown mechanism in BL cells.55 A study by Dorsett et al.56 showed that knockdown mouse models with mutation in the miR-155 binding site in the 3′-untranslated region of activation-induced cytidine deaminase (AID) had increase in steady-state AID mRNA and protein amounts, which resulted in a high degree of Myc-Igh translocations, which are the key transforming events in BL.
A study by Yim et al.57 has also suggested a tumor-suppressor effect of miR-155 in the pathogenesis of MCL. Complete methylation of miR-155-3p was documented in one MCL cell line (REC-1), and demethylation with 5-aza-2′-deoxycytidine treatment of REC-1 led to re-expression of miR-155-3p with consequent increased apoptosis and decreased cellular viability. Lymphotoxin-beta (LT-β), which is an upstream activator of the noncanonical NF-κB signaling pathway, was established to be the target of miR-155-3p by luciferase assay. Further, miR-155-3p was found to be hypermethylated in a significant proportion of primary MCL as well as in B-cell, T-cell, and Natural Killer cell (NK-cell) non-Hodgkin’s lymphomas (NHLs). As miR-155-3p methylation correlated with miR-155-3p downregulation and LT-β upregulation, it was concluded to be a potential tumor-suppressive miRNA for MCL and other NHL subtypes.57
Role of miR-155 in the pathogenesis of leukemias
MiR-155-associated pathogenesis of acute myeloid and lymphoblastic leukemias has been proposed to be mediated through SHIP1 (Src homology 2 domain-containing inositol phosphatase) and C/EBPβ, two important regulators of B-cell maturation.15 Studies have shown that miR-155 directly inhibits SHIP1 as well as C/EBPβ.58,59 SHIP1 mediates the conversion of phosphatidylinositol triphosphate (PIP3) to phosphatidylinositol diphosphate (PIP2). PIP3 facilitates the Phosphoinositide 3-kinase (PI3K)–Akt pathway by functioning as a docking site for signaling molecules in the pathway. By promoting the conversion of PIP3 to PIP2, SHIP1 blocks the activation of the PI3K–Akt pathway and probably thereby suppresses the development of AML.60 MiR-155 is believed to promote the pathogenesis of AML by downregulating SHIP1, and thereby reversing SHIP1-mediated PI3K–Akt pathway suppression (Fig. 2). O’Connell et al.58 found that overexpression of miR-155 in hematopoietic cells both in vitro and in vivo studies resulted in repression of endogenous SHIP1 and increased activation of the kinase AKT. Further, they also found that knocking down SHIP1 or overexpressing miR-155 in HSPCs produced similar myeloproliferative phenotypes, with an increased number of CD11b+ myeloid cells in the bone marrow and spleen, decreased marrow erythropoiesis, and splenomegaly.58 C/EBPβ is a transcription factor involved in negative regulation of the IL-6 signaling pathway in B-cells and also plays an important role in myeloid and lymphoid maturation.61 A study by Costinean et al.59 showed that miR-155-mediated downregulation of SHIP1 and C/EBPβ is the most likely mechanism for the pathogenesis of acute lymphoblastic leukemia/high-grade lymphoma in Eµ-MiR-155 transgenic mice. Both SHIP1 and C/EBPβ protein levels were found to be markedly diminished in leukemic pre-B-cells in Eµ-miR-155-transgenic mice. Mir-155-induced downregulation of both SHIP1 and C/EBPβ was proposed to cause a block in B-cell differentiation through de-repression of the IL-6 signaling pathway and to induce a reactive proliferation of the relatively apoptosis-resistant myeloid precursor cells (Fig. 2).59
Figure 2.
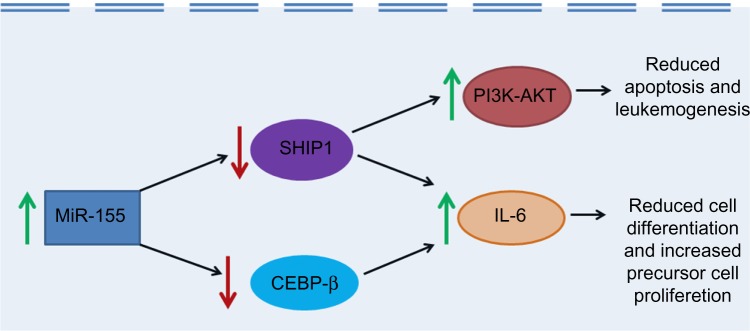
The role of MiR-155 in leukemogenesis. Green arrows indicate increased activity and red arrows indicate decreased activity.
Overexpression of miR-155 has also been found to be associated with the more aggressive and poorer prognosis type of CLL. Cui et al.62 reported that transfection of CLL cells with miR-155 reduced SHIP1 expression and enhanced responsiveness to B-cell receptor (BCR) ligation, whereas transfection with a miR-155 inhibitor had the opposite effect. Treatment of CLL or normal B cells with the CD40-ligand or B-cell-activating factor upregulated miR-155 and increased sensitivity to BCR ligation, but these effects got blocked by miR-155 inhibitors.62
In contrast to this, miR-155 has been found to have tumor-repressor effects in AML with FLT3-wild type, by inducing cell apoptosis through caspase-3 activation. In a study by Palma et al.47, knockdown of miR-155 by locked nucleic acid antisense oligonucleotides in the FLT3-wildtype AML cells was shown to lead to resistance to cytarabine arabinoside-induced apoptosis and to suppression of cell differentiation. Ectopic expression of miR-155 in FLT3-wildtype AML cells resulted in the gain of myelomonocytic markers (CD11b, CD14, and CD15) and increase in the expression of cleaved caspase-3 with a concomitant increase in apoptosis, reduced cell growth, and decreased clonogenic capacity.47
Role of miR-155 in the pathogenesis of solid tumors
In addition to its role in hematological malignancies, miR-155 also has complex gene regulatory effects on oncogenic and tumor-suppressor genes involved in solid tumors. By targeting numerous molecules in key signaling pathways such as glutathione metabolism, Stress-activated protein kinase (SAPK)/c-Jun N-terminal kinase (JNK), Toll-like receptor (TLR), and Extracellular signal-regulated kinase (ERK)/Mitogen-activated protein kinase (MAPK) pathways, it plays an important role in the pathogenesis of cancers of the breast, lung, stomach, and mesenchymal malignancies such as liposarcomas.38
MiR-155-Related Therapeutic Targets for Hematological Malignancies
As miR-155 plays an important role in oncogenesis, especially for malignancies such as DLBCL, anti-miRs targeting it might prove to be of significant therapeutic benefit. However, limitations to this treatment modality include the instability of free-floating anti-miRs in the plasma and their vulnerability to breakdown by nucleases, nonspecific tissue uptake, and renal clearance. These limitations can be overcome by nanoparticle-based delivery of the anti-miRs to target tissues. In a study by Babar et al.63, anti-miR-155 was loaded into poly (lactic-co-glycolic acid) nanoparticles and administered to mice with disseminated lymphoma due to miR-155 over-expression. To further facilitate intracellular delivery, these nanoparticles were “decorated” with a cell-penetrating peptide, penetratin. The pre-B-cells were found to favor uptake of nanoparticles coated with penetratin when compared to other cell-penetrating peptides such as Trans-activating transcriptional activator from human immunodeficiency virus 1 (TAT) and polyarginine. This nanoparticle-mediated anti-miR-155 administration was found to result in significant slowing of growth of the pre-B-cell tumor in vivo.63
Zhang et al.64 found that cell proliferation associated with B-cell lymphoproliferative disorders such as Walden-strom macroglobulinemia (WM) and CLL could be blocked in vitro with an anti-miR-155 oligonucleotide targeting the seed region of miR-155. When the anti-miR-155 was delivered systemically, it was taken up in the CD19+ cells in the bone marrow of WM-engrafted mice and led to the upregulation of miR-155 target mRNAs in these cells, which in turn resulted in significant reduction in tumour growth in vivo.64
For malignancies such as AML where mir-155 has a repressor effect, therapeutic benefits might be obtained by induction of miR-155 expression. Either way, miR-155 is a promising therapeutic target for various hematological malignancies and solid tumors, and studies into both aspects of miR-155-mediated therapy are ongoing.65
Conclusion
Various miRNAs play critical roles in the causation of different hematological malignancies. Among them, miR-155 is one of the important miRNAs that contributes to the pathogenesis of diverse hematological malignancies and solid tumors with complex oncogenic as well as tumor-repressor roles depending on the disease context and tissue type. Due to its important role in cancer pathogenesis, it is a promising therapeutic target for these cancers. Further clear elucidation of the role and pathogenic mechanisms of miR-155 in hematological malignancies may prove to be of great help in the development of effective treatment modalities for these conditions.
Footnotes
ACADEMIC EDITOR: Karen Pulford, Editor in Chief
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,092 words, excluding any confidential comments to the academic editor.
FUNDING: Author discloses no funding sources.
COMPETING INTERESTS: Author discloses no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
PR conceived and drafted the manuscript, and reviewed and approved the final manuscript.
REFERENCES
- 1.Vasilatou D, Papageorgiou S, Pappa V, et al. The role of microRNAs in normal and malignant hematopoiesis. Eur J Haematol. 2010;84(1):1–16. doi: 10.1111/j.1600-0609.2009.01348.x. [DOI] [PubMed] [Google Scholar]
- 2.Lawrie CH. MicroRNAs and haematology: small molecules, big function. Br J Haematol. 2007;137(6):503–12. doi: 10.1111/j.1365-2141.2007.06611.x. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez A, Griffiths-Jones S, Ashurst JL, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol Biol. 2006;342:33–47. doi: 10.1385/1-59745-123-1:33. [DOI] [PubMed] [Google Scholar]
- 5.Williams AE. Functional aspects of animal microRNAs. Cell Mol Life Sci. 2008;65(4):545–62. doi: 10.1007/s00018-007-7355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen TB, Kjems J, Bramsen JB. Enhnacing miRNA annotation confidence in miRBase by continuous cross dataset analysis. RNA Biol. 2011;8:378–83. doi: 10.4161/rna.8.3.14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musilova K, Mraz M. MicroRNAs in B-cell lymphomas: how a complex biology gets more complex. Leukemia. 2015;29(5):1004–17. doi: 10.1038/leu.2014.351. [DOI] [PubMed] [Google Scholar]
- 8.Baek D, Villen J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Çonnell RM, Baltimore D. MicroRNAs and hematopoietic cell development. Curr Top Dev Biol. 2012;99:145–74. doi: 10.1016/B978-0-12-387038-4.00006-9. [DOI] [PubMed] [Google Scholar]
- 10.Akbari Moqadam F, Pieters R, den Boer ML. The hunting of targets: challenge in miRNA research. Leukemia. 2013;27(1):16–23. doi: 10.1038/leu.2012.179. [DOI] [PubMed] [Google Scholar]
- 11.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149(3):515–24. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weng H, Lal K, Yang FF, et al. The pathological role and prognostic impact of miR-181 in acute myeloid leukemia. Cancer Genet. 2015;208(5):225–9. doi: 10.1016/j.cancergen.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su R, Lin HS, Zhang XH, et al. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene. 2015;34(25):3226–39. doi: 10.1038/onc.2014.274. [DOI] [PubMed] [Google Scholar]
- 15.Khalaj M, Tavakkoli M, Stranahan AW, et al. Pathogenic microRNAs in myeloid malignancies. Front Genet. 2014;5:361. doi: 10.3389/fgene.2014.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bousquet M, Nguyen D, Chen C, et al. MicroRNA-125b transforms myeloid cell lines by repressing multiple mRNA. Haematologica. 2012;97:1713–21. doi: 10.3324/haematol.2011.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bräuer-Hartmann D, Hartmann JU, Wurm AA, et al. PML/RARα-Regulated miR-181a/b Cluster Targets the Tumor Suppressor RASSF1A in Acute Promyelocytic Leukemia. Cancer Res. 2015;75(16):3411–24. doi: 10.1158/0008-5472.CAN-14-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhuang LK, Xu GP, Pan XR, et al. MicroRNA-181a-mediated downregulation of AC9 protein decreases intracellular cAMP level and inhibits ATRA-induced APL cell differentiation. Cell Death Dis. 2014;5:e1161. doi: 10.1038/cddis.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schotte D, Pieters R, Den Boer ML. MicroRNAs in acute leukemia: from biological players to clinical contributors. Leukemia. 2012;26:1–12. doi: 10.1038/leu.2011.151. [DOI] [PubMed] [Google Scholar]
- 20.Bousquet M, Harris MH, Zhou B, et al. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA. 2010;107:21558–63. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 22.Schotte D, Lange-Turenhout EA, Stumpel DJ, et al. Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica. 2010;95:1675–82. doi: 10.3324/haematol.2010.023481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong P, Iwasaki M, Somervaille TC, et al. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 2010;70:3833–42. doi: 10.1158/0008-5472.CAN-09-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palamarchuk A, Efanov A, Nazaryan N, et al. 13q14 deletions in CLL involve cooperating tumor suppressors. Blood. 2010;115(19):3916–22. doi: 10.1182/blood-2009-10-249367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balatti V, Pekarky Y, Rizzotto L, et al. miR deregulation in CLL. Adv Exp Med Biol. 2013;792:309–25. doi: 10.1007/978-1-4614-8051-8_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabbri M, Bottoni A, Shimizu M, et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. Jama. 2011;305(1):59–67. doi: 10.1001/jama.2010.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pekarsky Y, Santanam U, Cimmino A, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66(24):11590–3. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 29.Agirre X, Jiménez-Velasco A, José-Enériz ES, et al. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol Cancer Res. 2008;6:1830–40. doi: 10.1158/1541-7786.MCR-08-0167. [DOI] [PubMed] [Google Scholar]
- 30.Flamant S, Ritchie W, Guilhot J, et al. Micro-RNA response to imatinib mesylate in patients with chronic myeloid leukemia. Haematologica. 2010;95:1325–33. doi: 10.3324/haematol.2009.020636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdi J, Qiu L, Chang H. Micro-RNAs Newperformers in multiple myeloma bone marrow microenvironment. Biomark Res. 2014;2:10. doi: 10.1186/2050-7771-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao SM, Xing CY, Chen CQ, et al. miR-15a and miR-16-1 inhibit the proliferation of leukemic cells by down-regulating WT1 protein level. J Exp Clin Cancer Res. 2011;30:110. doi: 10.1186/1756-9966-30-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sánchez-Espiridión B, Martín-Moreno AM, Montalbán C, et al. MicroRNA signatures and treatment response in patients with advanced classical Hodgkin lymphoma. Br J Haematol. 2013;162(3):336–47. doi: 10.1111/bjh.12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806–11. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamanaka Y, Tagawa H, Takahashi N, et al. Aberrant over-expression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009;114(15):3265–75. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- 37.Craig VJ, Cogliatti SB, Imig J, et al. Myc-mediated repression of microRNA-34a promotes high-grade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood. 2011;117(23):6227–36. doi: 10.1182/blood-2010-10-312231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higgs G, Slack F. The multiple roles of microRNA-155 in oncogenesis. J Clin Bioinforma. 2013;3(1):17. doi: 10.1186/2043-9113-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eis PS, Tam W, Sun L, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–32. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faraoni I, Antonetti FR, Cardone J, et al. MiR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792(6):497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 41.O’Connell RM, Taganov KD, Boldin MP, et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–9. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin MM, Lee EJ, Buckenberger JA, et al. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J Biol Chem. 2006;281:18277–84. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 44.Georgantas RW, 3rd, Hildreth R, Morisot S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104(8):2750–5. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masaki S, Ohtsuka R, Abe Y, et al. Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis. Biochem Biophys Res Commun. 2007;364(3):509–14. doi: 10.1016/j.bbrc.2007.10.077. [DOI] [PubMed] [Google Scholar]
- 46.Calin GA, Croce CM. C. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 47.Palma CA, Al Sheikha D, Lim TK, et al. MicroRNA-155 as an inducer of apoptosis and cell differentiation in Acute Myeloid Leukaemia. Mol Cancer. 2014;13:79. doi: 10.1186/1476-4598-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrie CH. MicroRNAs and lymphomagenesis: a functional review. Br J Haematol. 2012;160(5):571–81. doi: 10.1111/bjh.12157. [DOI] [PubMed] [Google Scholar]
- 49.Sandhu SK, Volinia S, Costinean S, et al. MiR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Emu-miR-155 transgenic mouse model. Proc Nat Acad Sci USA. 2012;109(49):20047–52. doi: 10.1073/pnas.1213764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson RC, Herscovitch M, Zhao I, et al. NF-kappaB down-regulates expression of the B-lymphoma marker CD10 through a miR-155/PU.1 pathway. J Biol Chem. 2011;286(3):1675–82. doi: 10.1074/jbc.M110.177063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dagan LN, Jiang X, Bhatt S, et al. MiR-155 regulates HGAL expression and increases lymphoma cell motility. Blood. 2012;119(2):513–20. doi: 10.1182/blood-2011-08-370536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rai D, Kim SW, McKeller MR, et al. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc Natl Acad Sci U S A. 2010;107(7):3111–6. doi: 10.1073/pnas.0910667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merkel O, Hamacher F, Griessl R, et al. Oncogenic role of miR-155 in anaplastic large cell lymphoma lacking the t(2;5) translocation. J Pathol. 2015;236(4):445–56. doi: 10.1002/path.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kluiver J, Haralambieva E, de Jong D, et al. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45:147–53. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- 55.Kluiver J, van den Berg A, de Jong D, et al. Regulation of primicroRNA BIC transcription and processing in Burkitt lymphoma. Oncogene. 2007;26(26):3769–76. doi: 10.1038/sj.onc.1210147. [DOI] [PubMed] [Google Scholar]
- 56.Dorsett Y, McBride KM, Jankovic M, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–8. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yim RL, Wong KY, Kwong YL, et al. Methylation of miR-155-3p in mantle cell lymphoma and other non-Hodgkin’s lymphomas. Oncotarget. 2014;5(20):9770–82. doi: 10.18632/oncotarget.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Connell RM, Chaudhuri AA, Rao DS, et al. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106(17):7113–8. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Costinean S, Sandhu S, Pedersen IM, et al. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. 2009;114(7):1374–82. doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo JM, Yoshida H, Komura S, et al. Possible dominant-negative mutation of the SHIP gene in acute myeloid leukemia. Leukemia. 2003;17(1):1–8. doi: 10.1038/sj.leu.2402725. [DOI] [PubMed] [Google Scholar]
- 61.Scott LM, Civin CI, Rorth P, et al. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725–35. [PubMed] [Google Scholar]
- 62.Cui B, Chen L, Zhang S, et al. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood. 2014;124(4):546–54. doi: 10.1182/blood-2014-03-559690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Babar IA, Cheng CJ, Booth CJ, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Nat Acad Sci USA. 2012;109(26):E1695–704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Roccaro AM, Rombaoa C, et al. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood. 2012;120(8):1678–86. doi: 10.1182/blood-2012-02-410647. [DOI] [PubMed] [Google Scholar]
- 65.Zhang M, Zhou X, Wang B, et al. Lactosylated gramicidin-based lipid nanoparticles (Lac-GLN) for targeted delivery of anti-miR-155 to hepatocellular carcinoma. J Cont Release. 2013;168(3):251–61. doi: 10.1016/j.jconrel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]


