Abstract
Bovine respiratory disease (BRD) is the most common economically important disease affecting cattle. For developing accurate diagnostics that can predict disease susceptibility/resistance and stratification, it is necessary to identify the molecular mechanisms that underlie BRD. To study the complex interactions among the bovine host and the multitude of viral and bacterial pathogens, as well as the environmental factors associated with BRD etiology, genome-scale high-throughput functional genomics methods such as microarrays, RNA-seq, and proteomics are helpful. In this review, we summarize the progress made in our understanding of BRD using functional genomics approaches. We also discuss some of the available bioinformatics resources for analyzing high-throughput data, in the context of biological pathways and molecular interactions. Although resources for studying host response to infection are avail-able, the corresponding information is lacking for majority of BRD pathogens, impeding progress in identifying diagnostic signatures for BRD using functional genomics approaches.
Keywords: bovine respiratory disease (BRD), functional genomics, bovine genome atlas, AgBase, host–pathogen interaction database (HPIDB)
Introduction
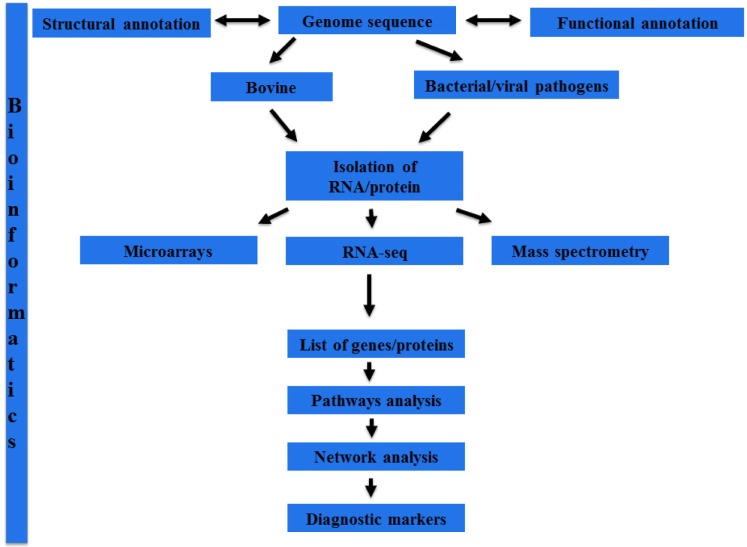
Despite research for decades and investments made in genomics to understand disease progression, bovine respiratory disease (BRD) continues to cause considerable economic losses for the cattle industry. BRD has a complex etiology that involves a multitude of interactions among the host, various pathogens, and the environment. Studying host–pathogen interaction (HPI) networks in BRD has the potential for identifying diagnostic markers or signatures that are indicative of disease and its severity. Functional genomics approaches (often referred to as “omics” approaches) generate genome-scale expression data for mRNA, protein, metabolites, and non-coding RNA, and applying these methods to BRD research can expedite the development of diagnostics. The list of genes/proteins identified by high-throughput approaches require analysis in a systems biology framework, ie, in the context of protein–protein interaction (PPI) networks, to decipher the underlying regulatory mechanisms that are responsible for a given phenotype or biological response. Studying gene/protein expression in a systems biology framework requires a comprehensive description of all parts of the system (structural annotation), identifying the interactions among the parts (protein–protein, protein–DNA interactions, etc), identifying regulatory mechanisms that govern these interactions, and finally developing descriptive mathematical models and iterative refinement of these mathematical models to generate predictive models of the system. At present, applying this entire workflow is not feasible for bovine–pathogen interactions that result in BRD. However, functional genomics approaches allow the generation of testable hypotheses from high-throughput data using gene ontology (GO) enrichment, pathways, and network analyses. Regardless of the data analysis strategy, bioinformatics tools, databases, and algorithms are required for leveraging functional genomics data for BRD diagnostics. The proposed functional genomics-based workflow is described in Figure 1.
Figure 1.
A flowchart describing functional genomics-based approach for BRD diagnostics. The availability of sequenced genomes for bovine and BRD pathogens enables the study of genome expression using various high-throughput approaches such as microarrays, RNA-seq, and proteomics, which often generate a list of genes/proteins. Continuous updates to existing structural and functional annotations enhance the quality of functional genomics findings. At the core of these annotation approaches as well as analysis of functional genomics data in the context of biological pathways and networks is the necessary bioinformatics analysis and resources. An integrated systems biology framework for analyzing functional genomics data is expected to identify accurate biomarkers for prediction of BRD resistance and susceptibility, as well as disease stratification.
For the purposes of this review, we used the following definition of functional genomics: “Functional genomics uses genomic data to study gene and protein expression and function on a global scale (genome-wide or system-wide), focusing on gene transcription, translation and protein-protein interactions, and often involving high-throughput methods” (http://www.nature.com/subjects/functional-genomics). Therefore, genome-wide association studies that identify genetic variations responsible for BRD susceptibility/resistance to aid beef cattle breeding programs and studies that focus on the role of small non-coding RNAs in post-transcriptional gene regulation are not discussed in this article. The reader is pointed to reviews in literature that focus on microarray and RNA-sequencing (RNA-seq)1–7 and proteomics8–11 methodologies and challenges associated with the data analysis.
In this review, we describe BRD, enumerate challenges associated with BRD diagnostics, and outline the bioinformatics methods/resources that enhance the structural and functional annotation of BRD genomes for analyzing gene/protein expression data for hypothesis generation. Progress made toward BRD diagnostics using proteomics, microarrays, and RNA-seq is summarized.
Bovine Respiratory Disease
BRD is the most common and economically important disease affecting cattle.12 BRD causes 70%–80% morbidity and 40–50% mortality in feedlot cattle.13 BRD is tempo rally associated with factors such as weaning, transportation, and nutritional and social alteration.14 These environmental factors, often referred to generically as stress, are host dependent, highly interconnected, largely immeasurable, and consequently poorly understood.15–17 The most common viruses involved in the BRD complex include bovine herpesvirus-1 (BHV-1), bovine parainfluenza virus type-3 (BPIV3), bovine respiratory syncytial virus (BRSV), and bovine viral diarrhea virus (BVDV).18 Typically, these viruses infect the upper respiratory tract, resulting in rhinitis, tracheitis, and bronchitis, and generally do not cause major lung damage but do predispose the lower airway to bacterial invasion.19 The primary bacterial agents, often obtained from BRD mortalities and associated with severe inflammation and lung damage, include Mannheimia haemolytica, Pasteurella multocida, Histophilus somni, and Mycoplasma bovis.20 The bacterial agents are commensals, found commonly in the respiratory tract or on other mucous membrane surfaces21–23 and cause lung damage only when host defenses are diminished because of increased stress and/or concurrent viral infection.24,25 Nearly all (99.8%) cattle identified with clinical BRD receive antimicrobial treatment, and antimicrobials are used extensively in disease prevention.26 In addition to the cost incurred in the production phase, evidence clearly indicates that BRD has negative effects on carcass weight, marbling, and other carcass value factors.27,28 Animals affected with BRD do not always exhibit clinical symptoms.29–31 Animal health products and management strategies to prevent infection and reduce the negative effects of BRDC pathogens over the past 50 years are yet to be conclusively linked to increased disease control.32 Thus, BRD diagnostics continues to be a problem to date.
Existing Measures for BRD Diagnosis
There are very limited reliable tools to identify animals affected with BRD.32,33 In vivo sampling techniques, including brochoalveolar lavage, transtracheal wash, and nasopharyngeal swab, have been described and used primarily to procure samples for agent identification.34–36 Clinical signs are used to determine disease onset and severity, but clinical signs are subjective, difficult to standardize, and nonspecific for BRD.27,31,37 However, observation of clinical signs remains the predominant tool for detection of BRD in vivo. Generally, cattle that become ill have moderate risk of developing pulmonary pathology and exhibiting lung lesions at harvest, but the majority of cattle with evidence of pulmonary pathology at harvest are not detected as being ill in life.27,29,31,38,39 Infrared thermography40 and ultrasonography41 are often used for diagnosis, along with lung biopsy,42 continually reporting rumen temperature,43 blood and breath biomarkers,44–46 haptoglobin measurement,30,47,48 and eating and drinking behaviors.38 However, none of these approaches has shown promise for in vivo field use. As such there is no “gold standard” for BRD diagnostics as noted in a recent review of laboratory tests for BRD.49
Thus, progress in BRD research is at an impasse and continued investigations focusing on independent elements (ie, pathogen or host) are unlikely to result in major breakthroughs in BRD research. This situation has been well recognized, and research focusing on identification of markers associated with genetic resistance to BRD has been initiated through the Bovine Respiratory Disease Coordinated Agricultural Project (BRD CAP),50 which will among other data generate gene expression profiles for single-agent challenge using the majority of BRD pathogens.51 For field application, diagnostic criteria that allow early disease identification and assessment of lung damage would allow treatment strategies to be tailored to the condition and help identify animals with poor predicted outcomes, similar to the way procalcitonin has been advocated for use in human bacterial lung infections.52 Such diagnostic criteria would enable more rational treatment decisions, limit antimicrobial use to those most likely to respond, and decrease suffering by identification of animals unlikely to survive.
Functional Genomics for Studying BRD
The complex interplay of bacteria, viruses, and environmental factors that results in BRD is poorly understood and is unlikely to be fully elucidated using reductionist strategies. Pathogens have evolved numerous strategies to successfully invade their hosts (acquire nutrients and evade immune system), and host defense systems are continually evolving to combat infections.53,54 The availability of genome sequences for bovine55,56 and BRD pathogens, including BHV-1,57–59 BVDV,60 BRSV,61 BPIV3,62 M. haemolytica,63–67 P. multocida subsp. multocida str. Anand1_cattle (contigs: GCA_000291645.1), H. somni,68 and M. bovis,69,70 facilitates transcriptomics and proteomics studies to identify molecular functions, pathways, and networks that are involved in BRD pathogenesis. To detect, manage, and treat BRD, it is critical to elucidate the key molecules in the host–pathogen interplay and generate systems biology models for emergent behavior. Systems biology focuses on the systematic study of complex interactions in/between biological systems and aims to produce quantitative models that are predictive.71 A number of biological processes, including inflammation, apoptosis, and thrombosis, are involved in the pathogenesis of BRD. Identification of these processes by the “omics” approaches, with subsequent evaluation in a systems biology framework, and linking the findings back to traditional end points will help develop diagnostic tools specific to BRD.
Improving Genome Sequence Quality: Structural Annotation
High-throughput technologies, such as microarrays, RNA-seq, and proteomics, provide readout of gene/protein expression on a genome scale. Studying the gene/protein expression in cattle or BRD pathogens could identify disease mechanisms for developing sound diagnostics. Involvement of genes/proteins in BRD pathology can only be determined provided they are annotated, ie, demarcated in the genomes. The process of identifying and defining all the functional elements within a genome sequence is defined as “structural annotation”. Structural annotation is the foundation for functional genomics approaches that focus on measuring the expression of all known genes/proteins in an organism. Genome structural annotation is concurrent with genome sequencing efforts. This initial annotation is based on in silico predictions that rely on sequence similarity. Although computational approaches have high accuracy of prediction, using experimental data to update existing in silico predictions72 is warranted. Experimental annotation at the RNA level using tiling arrays and single-nucleotide resolution RNA-seq results is expected to accurately identify transcriptional start sites, operon structures (in bacteria), exon–intron boundaries, and novel non-coding functional elements such as small RNAs (sRNAs) and long non-coding RNAs (lncRNAs). Of relevance to this review article, open reading frames with protein-coding potential that were missed in the initial annotation can be identified by experimental genome annotation methods. We reported RNA-seq based expression profiling studies for two bacterial BRD pathogens, H. somni73 and M. haemolytica.74 In H. somni strain 2336, we identified 94 sRNAs, of which 82 sRNAs were not reported elsewhere. We also identified 38 novel potential protein-coding open reading frames and 278 operon structures in the genome. Compared to a nonvirulent strain 129Pt, we found that ~30% of the identified sRNAs were unique to the virulent strain 2336, indicating that a number of the newly identified sRNAs in strain 2336 may be involved in strain-specific adaptations. Our RNA-seq based transcriptome map of M. haemolytica resulted in the identification of 14 novel protein-coding regions and 44 potential novel sRNAs. While both of these studies reported the identification of novel non-coding RNAs and protein-coding regions, confirmation of these findings by additional experimental approaches is necessary before this new knowledge can be incorporated into existing databases for functional genomics studies. Although the role of non-coding RNAs in regulating gene expression is well known, it still remains poorly studied in BRD. Using computational pipelines and nonprotein coding transcripts from expressed sequence tags (ESTs), thousands of non-coding RNAs (ncRNAs) were identified in the bovine genome.75 Inspection of the genomic distribution of the ncRNAs, majority of which were not reported previously, indicated their transcription adjacent to open reading frames with a possible involvement of ncRNA in cis-regulatory roles. These computational predictions for the existence of ncRNAs in bovine systems need confirmation by complementary experimental annotation approaches. The proposed whole-genome sequencing in the 1,000 Bull Genomes Consortium Project is expected to provide annotated sequence variants and genotypes of key ancestor bulls, which will enhance the quality of the genome enormously. Sequencing results of 23476 bulls has facilitated the mapping of monogenic as well as complex traits in cattle. Analysis of functional genomics data in the context of such information-rich bovine genome sequences will revolutionize our understanding of BRD for developing diagnostics.
Enhancing Genome Sequence Utility: Functional Annotation
Analysis of functional genomics data often generates a list of genes/proteins that are differentially expressed in response to a biological perturbation (biotic or abiotic). For understanding the mechanisms of BRD pathophysiology, it is important that all genes/proteins identified as being associated with BRD have functional information, ie, biological function. Bioontologies are controlled vocabularies for describing biological concepts such as gene functions, and the relationships that exist between concepts, to facilitate data sharing and computational analysis of high-throughput data sets.77–79 GO is one of the most commonly used bioontologies,80–82 which describes gene product functions. GO is the prevailing standard for functional annotation and consists of three separate ontologies: Biological Process, Cellular Component, and Molecular Function. BRD researchers can access GO for bovine gene products at AgBase.83 AgBase is a curated, open-source, Web-accessible resource for carrying out functional analysis of agricultural plant and animal gene products. AgBase provides GO for bovine gene products and also has tools for functional genomics data analysis. AgBase tools like GORetriever fetch available GO for an input gene list and GOslimviewer summarizes GO annotations at a higher level. For bovine gene products that lack GO, functional domains with GO annotations in other species are identified by GOAnna by a Basic Local Alignment Search Tool (BLAST) search, enabling the transfer of GO annotations from other species to bovine gene products, when appropriate. Recognizing that not all gene products have information that can be annotated to GO consortium standards, AgBase captures community annotations to provide information for any gene product.
Bioinformatics Resources for BRD Research
In the following sections, we summarize some of the bioinformatics resources available for analyzing functional genomics data in the bovine host and BRD pathogens. Although not comprehensive, this section describes some of the fundamental resources for conducting “omics”-based BRD research. Web links are provided for these resources in Table 1 for easy access.
Table 1.
Bioinformatics resources available for analyzing BRD functional genomics data.
| DESCRIPTION | WEB ADDRESS |
|---|---|
| The bovine genome database85 | http://bovinegenome.org/ |
| Microbial Gbrowsers83 | http://www.agbase.msstate.edu/cgi-bin/microbialGBrowsers.pl |
| The Bovine gene atlas87 | http://bovineatlas.arl.arizona.edu/ |
| InnateDB97 | http://www.innatedb.com/ |
| Animal TFDB99 | http://www.bioguo.org/AnimalTFDB/ |
| HPIDB103 | http://www.agbase.msstate.edu/hpi/main.html |
| KEGG89 | http://www.genome.jp/kegg/ |
| Unipathways92 | http://www.unipathway.org/ |
| Metacyc90,91 | http://metacyc.org/ |
| Reactome94,95 | http://www.reactome.org/ |
| Cytoscape104 | http://www.cytoscape.org |
| Pathguide96 | http://www.pathguide.org/ |
| AgBase83 | http://www.agbase.msstate.edu/ |
The bovine genome database
A genome sequence is not very useful as a linear string of ATCGs unless there is a way to visualize the sequence. The architecture of genome browsers such as GBrowse84 is a combination of database and query-based visualization interface that allows users to view the genome. This interface allows the users to add metadata, which are displayed as a separate track that can be turned on or off. The bovine genome database85 is a database that was developed to facilitate the visualization and annotation of the bovine genome sequence. This is a valuable resource that allows the bovine research community to utilize the existing two competing versions of genome assembly for research. As summarized by the bovine genome database, “the Btau_4.6.1 assembly includes the Y chromosome, while UMD3.1 version does not include the Y chromosome; however, bovine research community perceives UMD3.1 assembly as the more contiguous version”. This database displays both bovine genome assemblies in a GBrowse format and includes information regarding the official gene set, gene models from data repositories such as RefSeq and Ensembl, non-coding RNAs, repeats, pseudogenes, single-nucleotide polymorphisms, markers, quantitative trait loci (QTLs), and alignments to complementary DNAs, ESTs, and protein homologs. Because the bovine genome database is connected to the Bovine QTL viewer, it facilitates the identification of candidate genes underlying a quantitative trait loci.
AgBase83 hosts GBrowse for three BRD bacterial pathogens, namely, M. haemolytica strain PHL213, H. somni strain 2336, and P. multocida strain 3480. The purpose of these microbial genome browsers is to primarily display proteogenomics data for structural annotation. The genome itself could be viewed independent of additional data such as peptide-level data to gain insights into the organization of functional elements in these bacterial genomes. The University of California Santa Cruz genome browser86 hosts H. somni 129Pt and M. succiniproducens, strains that do not cause BRD but can be used for comparative genomics.
Bovine gene atlas
A comprehensive description of the abundance of transcripts in all tissues is often referred to as a gene atlas, and availability of this information can help identify tissue-specific expression. The bovine gene atlas87 is available through the AgBase website. This resource was developed by applying next-generation sequencing to determine the RNA abundance in 92 adult, juvenile, and fetal cattle tissues and 3 cattle cell lines. The results are displayed in a GBrowse format, allowing the end user to navigate and access the project results. Bovine expression atlas results confirmed the expression of 16,517 annotated protein-coding loci in the genome. Analysis of BRD functional genomics data in the context of lung tissue expression atlas will help identify lung-specific gene/protein expression during BRD infection, when compared to expression in the nonrelevant tissues for disease onset and progression.
Resources for pathways analysis
Analysis of “omics” data sets for drawing biological conclusions often requires linking a gene/protein set to pathways. The HUGO Gene Nomenclature Committee Comparison of Orthology Predictions88 search tool helps identify bovine–human orthologs to tap into available functional annotation of human genes that can be transferred to bovine genes. Pathway resources for BRD pathogens are available through databases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG),89 Metacyc,90,91 and Unipathway,92 which have dedicated pages for the individual genomes also in addition to cataloging the annotated pathways. UniPathway is expected to be integrated with detailed UniProt93 information. Uniprot is a curated database resource for protein functions and pathways. Reactome94,95 is another good source of pathways information. For a comprehensive list of bioinformatics resources relevant to BRD, we suggest the use of Pathguide,96 a resource that lists all available PPI databases, as well as pathways and networks resources.
InnateDB
While it is important to interrogate the bovine and BRD pathogen genome sequences to identify the genomic context of functional genomics data, it is critical to identify a subset of host genes that constitute the immune response to infection with different BRD pathogens. Innate immune host responses are the first line of defense against pathogens and, when dysregulated, can cause damage to host tissues. To develop a diagnostic marker for BRD that is specific to a particular pathogen, it is important to characterize bovine innate immune responses to all BRD pathogens, in order to determine singe-agent specificities. InnateDB97 is a repository of >196,000 curated human, mouse, and bovine molecular interactions. There are 3,000 pathway annotations of relevance to cellular systems, including immune-relevant pathways. There are 46 bovine genes with documented roles in innate immune responses in this database. As more information regarding innate immune responses in bovine physiology becomes available from large-scale projects such as BRD CAP, results can be incorporated into InnateDB, which has evolved into a resource that can support systems biology framework for analyzing functional genomics data. Molecular interactions in InnateDB contain contextual information that meets the standards set forth by the minimum information required for reporting a molecular interaction experiment (MIMIx) standards.98 Because this information includes the evidence for each interaction, the tissue or cell type that the interaction was identified, and other related data, in conjunction with tissue expression atlases, InnateDB will be a powerful tool for identifying tissue-specific immune response networks for BRD diagnostics.
AnimalTFDB
To identify gene regulatory networks in BRD, it is important to analyze functional genomics data in the context of transcription factors (TFs) that are known to be involved in the regulation of gene transcription. TFs have different DNA-binding domains, which form the basis for their classification into different families, and up to 5% of genes in vertebrate genomes are predicted to be TFs.99 The animal TF database AnimalTFDB has information regarding bovine TFs, cofactors, and chromatin-remodeling factors.99 A recent update of the database includes evaluation of the expression of TFs from tissues using RNA-seq based expression data. This update to the database also includes a TF prediction server and allows for comparison of TFs from different species for the identification and prediction of this important class of proteins involved in the regulation of gene expression.
HPIDB
The study of HPIs to identify drug targets and develop vaccine strategies has a strong precedent in biomedical research. In infectious disease research, it is important to understand the commonalities between diseases for efficient development of therapeutics. Computational methods such as multitask learning are being applied to test this “commonality hypothesis” for multiple bacterial species, including Bacillus anthracis, Francisella tularensis, Yersinia pestis, and Salmonella typhi.100 With a multitude of pathogens being associated with BRD, identifying such a commonality in bovine response to pathogens will help provide specificity for BRD diagnostics. To study functional genomics data in the context of bovine–BRD pathogen interaction networks, availability of such HPI data is a prerequisite. Although a number of interaction databases for HPI exist, they are primarily focused on human pathogens. One such resource, VirHostNet,101,102 is a database of curated virus–host molecular interaction data. At present, VirHostNet has only seven HPIs for BVDV and three HPIs for BHV, with very limited number of HPIs. We developed host-pathogen interaction database (HPIDB),103 which focuses on integrating available HPIs from multiple databases and thus generates a set of nonredundant HPIs for multiple hosts and pathogens. At present, HPIDB contains 23,735 unique protein interactions between 68 host and 567 pathogen species. HPIDB provides data in a format compatible with Cytoscape104 for visualization of the interaction network. Similar to VirHostNet, HPIDB also allows the transfer of homologous HPIs to species of interest by conducting BLAST searches. Of the total species represented in the HPIDB, at least 20% are bacterial species, while 70% are viruses. Therefore, by using HPIDB, it is possible to get a first-pass computationally predicted HPI network for bovine BRD pathogens, which will help prioritize genes/proteins identified by functional genomics for BRD diagnostics.
In summary, bioinformatics resources available for bovine systems allow rapid analysis of functional genomics data for biological interpretation. Given a list of bovine genes, it is possible to retrieve the available functional GO information using the AgBase83 tool GORetriever and summarize the GO using GOSlimviewer. For a set of bovine genes that lack functional annotation, GOAnna at AgBase can add GOs based on sequence homology. Pathways resources such as Reactome,94,95 KEGG,89 Unipathways,92 InnateDB,97 TFDB,99 and HPIDB103 can each add additional bits of relevant information to the list of genes, such as pathways represented, involvement in host innate immune responses, activity as TFs, etc. Resources such as the bovine genome database facilitate the analysis of the genomic context of bovine genes, while tissue-specific expression patterns can be inferred from the bovine gene atlas. Utilizing resources such as HPIDB103 allows the visualization of bovine–pathogen interactions inferred from homology as networks in Cytoscape.104
Proteomics in BRD
Although not comprehensive, a few of the reported proteomics studies in BRD are summarized below. Bronchoalveolar lavage fluid (BALF), obtained by washing the epithelial lining of lungs, is often used to analyze proteins that are representative of pulmonary diseases.105 BALF is often used to understand respiratory disease mechanisms and to identify novel markers of pathogenicity.105 Characterization of the changes in protein expression in calves challenged with M. haemolytica by nano-liquid chromatography-tandem mass spectrometry (LC-MS/MS)42 identified ITIH4, an acute phase protein commonly associated with chronic obstructive pulmonary disease in humans, to be elevated, along with haptoglobin, a known acute phase marker for BRD. Interestingly, increase in the expression of the antimicrobial peptides (AMPs) cathelicidin-1 and -4 were also reported in this study. AMPs, also called as natural antibiotics or host defense peptides, are cationic peptides of the host innate immune response and are considered to be ideal candidates for novel peptide-based therapeutics.42,106 Efforts to catalog bovine AMPs based on prediction tools and databases such as AMPer107 are under way. A recent study used AMPer to search and identify novel AMPs in the bovine genome and sequence tags from the National Center for Biotechnology Information’s Database of Expressed Sequence Tags (NCBI dbEST) project. This study identified 27 AMPs with high confidence and 68 AMPs were predicted from the EST data.108
A major etiological factor of BRD is stress associated with transportation,32,109 weaning, commingling,110 preconditioning, sanitation, and air quality,111 which increases the susceptibility of cattle to bacterial pneumonia. In BRD, stress often correlates with immunodepression, and cattle succumb to bacterial infection by M. haemolytica.44,112 Although studies have identified stress as an etiological agent for BRD, molecular markers and mechanisms associated with stress response at the gene and protein levels remain elusive. Stress-associated changes in BALF were assessed by two-dimensional (2D) gel electrophoresis and mass spectroscopy by comparing nonstressed and stressed calves.113 In this study, expression of annexins A1 and A5, odorant-binding protein (OBP), isocitrate dehydrogenase, fibrinogen, heme-binding protein, alpha-2-Heremans-Schmid-glycoprotein (a-2-HSglycoprotein), alpha-1-antichymotrypsin, and albumin were significantly altered in response to stress. Similar studies were also performed with BALF to identify the effects of glucocorticoids in BRD by administering dexamethasone to calves.114 Increased levels of glucocorticoids suppress the immune system, rendering calves susceptible to bacterial and viral infections. This study identified several acute phase markers whose expression was elevated, such as alpha-1-acid glycoprotein (orosomucoid) and alpha-1-antitrypsin. Additionally, the amount of a-2-HSglycoprotein (fetuin) was reduced in BALF. Dexamethasone also increased the expression of two hydrophobic ligand-binding proteins, adipocyte-fatty acid binding protein, OBP, alpha-enolase, cofilin-1, and immunoglobulin J chain. Together, these studies evaluated the changes in the protein signature in response to stress associated with BRD,113,114 underpinning the probable mechanistic pathways. Consistent with these reports, Difference Gel Electrophoresis and MS analysis of BALF showed increased expression of annexins A1 and A2 in calves weaned and transported to the rearing site.115 Differentially expressed proteins in this study included three antioxidants such as dihydrodiol dehydrogenase-3, peroxiredoxin I, superoxide dismutase, calcyphosin, a calcium binding protein, and a macrophage capping protein.115 The authors recommend the use of annexin A1 as a potential stress-associated biomarker for BRD.
M. haemolytica can produce membrane vesicles that are made of outer membrane proteins (OMPs). Fifty-eight OMPs from M. haemolytica vesicles were identified by LC-MS/MS116 and these are potential targets for vaccine development.115 Cattle vaccinated with OMPs showed remarkably lower lung lesion scores compared to nonvaccinated calves. The identification of these novel OMPs was possible because of the availability of the M. haemolytica genome sequence.65,66 These studies demonstrate the importance of proteomics in identifying novel candidates for developing diagnostics and therapeutics that target either the host or the pathogen.
Proteomics of cytopathic (cp) BVDV identified bovine proteins involved in the immune function of professional antigen-presenting cells during BVDV infection.117,118 Proteins of significant interest included proteins involved in cell adhesion, apoptosis, antigen uptake, processing, and presentation such as the transporter associated with antigen processing, acute phase response proteins (fetuins, serum amyloid A, and apolipoprotein A-II), and major histocompatibility complex class I- and II-related proteins. Proteins involved in professional antigen presentation were significantly altered after BVDV infection. This analysis provided mechanistic insights into BVDV infection and showed that cp BVDV, while promoting monocyte activation and differentiation, inhibits antigen presentation to immunocompetent T-cells and subsequent infection.117 Follow-up proteomic studies using 2D-LC-electrospray ionization (ESI)-MS/MS characterized the changes in protein expression in bovine monocytes by both cp and noncytopathic (ncp) BVDV infections119 and revealed signaling pathways that were differentially regulated in cp and ncp BVDV infections. Macropinocytosis signaling, virus entry via the endocytic pathway, integrin signaling, and primary immunodeficiency signaling were activated by ncp BVDV-infected monocytes, while pathways of actin cytoskeleton signaling, RhoA signaling, clathrin-mediated endocytosis signaling, and interferon signaling were identified only in cp BVDV-infected cells. Among the common pathways involved in cp and ncp BVDV infection, acute phase response signaling was the most significant for both BVDV biotypes, while integrin alpha 2b (ITGA2B) and integrin beta 3 (ITGB3) were downregulated by ncp BVDV and upregulated by cp BVDV infection.119 A proteomics based study reported the characterization of differentially regulated protein kinases in response to cp and ncp BVDV infection in bovine monocytes.120 Using differential detergent fractionation and 2D-LC-ESI-MS, 378 proteins with homology to known protein kinases or related proteins were identified. This approach found upregulation of three kinases that mediate cellular signaling in response to BVDV, such as urokinase-type plasminogen activator receptor (u-PAR), myristoilated alanine-rich C kinase substrate (MARCKS), and nucleoside diphosphate kinase B (NDPKB). Rho-associated protein kinase 1 (ROK1) GTPase involved in cell differentiation and hexokinase type 3 (HK3), a kinase involved in glucose utilization, were downregulated in bovine monocytes in response to BVDV infection. This provides a molecular basis for sugar starvation in BVDV pathogenesis. The expression of the receptor of activated C kinase, pyridoxal kinase, diacylglycerol kinase, and Bruton’s tyrosine kinase was reduced in monocytes infected with cp BVDV compared to those with ncp BVDV, suggesting a role for these proteins in the differential pathogenicity of cp and ncp BVDV viruses.
Transcriptomics in BRD
The past decade has seen several technologies designed to analyze gene expression changes in an organism, a tissue, and individual cells. Techniques to profile gene expression include microarrays, RNA-seq based on next-generation sequencing, custom quantitative reverse transcription polymerase chain reaction arrays, and multiplex branched DNA assays. These techniques have enabled profiling of the gene expression of both the host and the pathogen, as well as identifying critical players that are differentially expressed under different pathogenic conditions associated with BRD. A few transcriptomics studies in BRD are summarized below.
Fatal cases of BRD are because of viral–bacterial synergy and are often associated with BHV-1 and M. haemolytica infections in the lower respiratory tract.121 Approximately 44,000 transcripts were evaluated in bovine bronchial epithelial cell (BBEC) following exposure to BHV-1 alone, M. haemolytica alone, or both BHV-1 and M. haemolytica.121 This analysis revealed that M. haemolytica exposure led to a significant increase in the production of inflammatory molecules such as chemokine (C-X-C motif) ligand 2 (CXCL2), interleukin (IL)-6, IL-1α, E-selectin, and IL-8, compared to animals challenged with BHV-1 alone in BBEC. However, in the coinfection model, differential regulation of several markers of inflammation, such as tumor necrosis factor-α, IL-8, toll-like receptor-2 (TLR2), IL-1, CXCL2, and colony stimulating factor 2 (CSF-2), were observed along with vascular endothelial growth factor (VEGF), endothelin 2 (EDN2), intercellular adhesion molecule 1 (ICAM1), and IL-16. Taken together, gene expression profiling of BBE cells by microarrays revealed that the viral bacterial synergy could compound the pathogenesis of BRD. Although follow-up studies confirming these observations are warranted, transcriptomics has identified pathways that go awry during the pathogenesis of BRD.
In another study, gene expression profiling of M. haemolytica A1strain was conducted 6 days postchallenge in pneumonic lungs,122 after establishment of infection. The in vivo gene expression signature was compared to an in vitro-cultured sample. Forty-four genes were differentially expressed by more than eight-fold. Interestingly, 17 genes were upregulated in vivo, while 27 genes were downregulated. The genes that were downregulated included virulence-associated genes encoding leukotoxin, a capsule biosynthetic enzyme, and a serotype-specific antigen, Ssa, suggesting that expression of these genes is not critical once infection is established.122
To understand the mechanism by which M. haemolytica adapts to low-iron environments, changes in gene expression under iron-limiting growth conditions were assessed at 15 minutes, 30 minutes, and 60 minutes following iron deprivation.123 Upregulation of several ATP-binding cassette transporters and receptors was found under iron-limiting conditions, suggesting new mechanisms by which M. haemolytica can compensate for lack of iron. Although limited in number, these studies clearly demonstrate the potential for knowledge discovery for BRD diagnostics utilizing transcriptomics.
The BRD CAP team analyzed pathogen-specific host response in the bronchial lymph nodes of steers recently.51 In brief, single-pathogen challenges were performed in steers for three different viral pathogens, such as BHV-1, BRSV, and BVDV, and three bacterial agents, M. haemolytica, P. multocida, and M. bovis. Transcriptome profiling by RNA-seq identified differential expressions that were common and unique to each challenge agent. Among the differentially expressed genes, a cumulative total of 5,757 genes were identified as unique to single challenges, among which 200 genes that showed high variance were common to all pathogen challenges. Apart from identifying 25 common genes that were differentially expressed in all the infections, the study reported unique genes that were specific to individual challenges. Differentially expressed genes specific to BRSV infection included FRZB, MT1E, NPTX1, TSPAN18, and CA4, while ALB, ITIH2, KRT24, MMP7, and TTR were unique to BVDV. Infection with M. haemolytica resulted in the differential expression of BMPR1B, COL6A6, and KIR3DL2. EBD and TSPAN1 were differentially regulated in response to M. bovis. Not surprisingly, the panel of genes common to all pathogens mostly comprised genes of the host innate immune response, such as MMP9, S100A8, and S100A9. S100A8 and S100A9 are expressed in neutrophils and monocytes124 and are known danger-associated molecular patterns that bind pattern recognition receptors in response to inflammation.125 This study exemplifies the value of functional genomics to decipher pathogen-specific response in host tissue, enabling the development of specific gene panels that could be evaluated for developing BRD.
Concluding Remarks
Current therapeutics and accurate diagnostics for human diseases is possible as the human genome is sequenced and well annotated, enabling the study of gene functions and disease mechanisms, making subsequent development of diagnostics and drug targets126 easier to accomplish. Unlike human diseases, most veterinary diseases are challenged with lack of complete genomic information and the availability of species-specific research tools.127 This is true for bovine respiratory disease research aimed at developing diagnostics tools. Although there is some information regarding the genes/proteins in the bovine genome that are implicated in BRD, the information is not comprehensive. Availability of annotated bovine–pathogen interspecies interaction is very rudimentary. On the pathogen side of the equation, knowledge pertaining to intraspecies interactions is lacking or minimal. The ongoing human and mouse ENCyclopedia Of DNA Elements projects126,128 highlight the importance of identifying functional elements in the genome at high resolution. However, such an effort is marginal in veterinary diseases. Although parallels can be drawn from human diseases, the differences in causative agents and interspecies differences has often resulted in failure to identify similar disease mechanisms127 in veterinary clinical science. Ongoing international effort to conduct whole-genome sequencing of 1,000 bulls76 will address the deficit in the availability of a high-resolution genome sequence that is well annotated on the host side. The recent formation of an organization of an international scale, the Functional Annotation of Animal Genomes (FAANG) project, aims to generate comprehensive maps of functional elements in the genomes of domesticated animal species.129 There is a large deficit in our knowledge of BRD mostly because of lack of species-specific gene sequences from the host and pathogen, as well as lack of species-specific immunological reagents to perform molecular analyses. The FAANG project should help bridge this gap in veterinary research and generate central repositories of data, tools, and reagents. In multifactorial diseases such as BRD, high-throughput omics approaches such as transcriptomics and proteomics offer a convenient alternative than the traditional hypothesis-driven research to explore disease complexity. These technologies help compare and contrast disease states, tissue-specific biomarkers, and the micro and macroenvironments in pathogenesis that could affect diagnosis, prognosis, and therapy. In human diseases, considerable progress has been made in the identification of biomarkers using functional genomics technologies with the integration of bioinformatics and systems biology.130 To maximize the impact of future BRD research, particularly when targeting naturally occurring disease in a field environment, a diagnostic strategy with high sensitivity, specificity, and reproducibility across time and geographical sites is essential. Continued investment in deciphering molecular mechanisms of BRD by utilizing functional genomics, with concomitant investments in improving the genomics infrastructure, ie, bovine and BRD pathogen genome sequences and bioinformatics, and computational biology resources is mandatory for making progress toward developing such BRD diagnostics.
Acknowledgments
We thank Leslie A. Shack for assistance with this article.
Footnotes
ACADEMIC EDITOR: J.T Efird, Associate Editor
PEER REVIEW: Seven peer reviewers contributed to the peer review report. Reviewers’ reports totaled 820 words, excluding any confidential comments to the academic editor.
FUNDING: This work was partially supported by the National Institutes of Health Centers of Biomedical Research Excellence, under the award number P20GM103646-01A1, and by the National Science Foundation, under award number EPS 0903787. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Wrote the first draft of the manuscript: ANR, BN. Contributed to the final manuscript: ANR, WBE, BN. Jointly developed the structure and arguments for the paper: ANR, BN. Made critical revisions and approved the final version: ANR, WBE, BN. All the authors reviewed and approved the final manuscript.
REFERENCES
- 1.Evans AC, Forde N, O’Gorman GM, Zielak AE, Lonergan P, Fair T. Use of microarray technology to profile gene expression patterns important for reproduction in cattle. Reprod Domest Anim. 2008;43(suppl 2):359–67. doi: 10.1111/j.1439-0531.2008.01185.x. [DOI] [PubMed] [Google Scholar]
- 2.Finotello F, Di Camillo B. Measuring differential gene expression with RNA-seq: challenges and strategies for data analysis. Brief Funct Genomics. 2015;14(2):130–42. doi: 10.1093/bfgp/elu035. [DOI] [PubMed] [Google Scholar]
- 3.Friedman BA, Maniatis T. ExpressionPlot: a web-based framework for analysis of RNA-Seq and microarray gene expression data. Genome Biol. 2011;12(7):R69. doi: 10.1186/gb-2011-12-7-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantione KJ, Kream RM, Kuzelova H, et al. Comparing bioinformatic gene expression profiling methods: microarray and RNA-Seq. Med Sci Monit Basic Res. 2014;20:138–42. doi: 10.12659/MSMBR.892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGettigan PA. Transcriptomics in the RNA-seq era. Curr Opin Chem Biol. 2013;17(1):4–11. doi: 10.1016/j.cbpa.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Ramskold D, Kavak E, Sandberg R. How to analyze gene expression using RNA-sequencing data. Methods Mol Biol. 2012;802:259–74. doi: 10.1007/978-1-61779-400-1_17. [DOI] [PubMed] [Google Scholar]
- 7.Wilson HL, Aich P, Roche FM, et al. Molecular analyses of disease pathogenesis: application of bovine microarrays. Vet Immunol Immunopathol. 2005;105(3–4):277–87. doi: 10.1016/j.vetimm.2005.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregorich ZR, Ge Y. Top-down proteomics in health and disease: challenges and opportunities. Proteomics. 2014;14(10):1195–210. doi: 10.1002/pmic.201300432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Washburn MP, Wolters D, Yates JR., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 10.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR., III Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 2013;113(4):2343–94. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith RA. Impact of disease on feedlot performance: a review. J Anim Sci. 1998;76(1):272–4. doi: 10.2527/1998.761272x. [DOI] [PubMed] [Google Scholar]
- 13.Hilton WM. BRD in 2014: where have we been, where are we now, and where do we want to go? Anim Health Res Rev. 2014;15(2):120–2. doi: 10.1017/S1466252314000115. [DOI] [PubMed] [Google Scholar]
- 14.McVey DS. BRD research needs in the next 10–20 years. Anim Health Res Rev. 2009;10(2):165–7. doi: 10.1017/S1466252309990247. [DOI] [PubMed] [Google Scholar]
- 15.Duff GC, Galyean ML. Board-invited review: recent advances in management of highly stressed, newly received feedlot cattle. J Anim Sci. 2007;85(3):823–40. doi: 10.2527/jas.2006-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falkenberg SM, Carroll JA, Keiser DH, et al. Evaluation of the endocrine response of cattle during the relocation process. Livest Sci. 2012;151:203–12. [Google Scholar]
- 17.Galyean ML, Perino LJ, Duff GC. Interaction of cattle health/immunity and nutrition. J Anim Sci. 1999;77(5):1120–34. doi: 10.2527/1999.7751120x. [DOI] [PubMed] [Google Scholar]
- 18.Ellis JA. Update on viral pathogenesis in BRD. Anim Health Res Rev. 2009;10(2):149–53. doi: 10.1017/S146625230999020X. [DOI] [PubMed] [Google Scholar]
- 19.Edwards TA. Control methods for bovine respiratory disease for feedlot cattle. Vet Clin North Am Food Anim Pract. 2010;26(2):273–84. doi: 10.1016/j.cvfa.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 20.Booker CW, Abutarbush SM, Morley PS, et al. Microbiological and histopathological findings in cases of fatal bovine respiratory disease of feedlot cattle in Western Canada. Can Vet J. 2008;49(5):473–81. [PMC free article] [PubMed] [Google Scholar]
- 21.Frank GH. Pasteurellosis in cattle. In: Adlam C, Rutter JM, editors. Pasteurella and Pasteurellosis. London: Academic Press; 1989. pp. 197–222. [Google Scholar]
- 22.Miller RB, Barnum DA, McEntee KE. Hemophilus somnus in the reproductive tracts of slaughtered cows: location and frequency of isolations and lesions. Vet Pathol. 1983;20(5):515–21. doi: 10.1177/030098588302000502. [DOI] [PubMed] [Google Scholar]
- 23.Soehnlen MK, Aydin A, Murthy KS, et al. Epidemiology of Mycoplasma bovis in Pennsylvania veal calves. J Dairy Sci. 2012;95(1):247–54. doi: 10.3168/jds.2011-4309. [DOI] [PubMed] [Google Scholar]
- 24.Aich P, Jalal S, Czuba C, et al. Comparative approaches to the investigation of responses to stress and viral infection in cattle. OMICS. 2007;11(4):413–34. doi: 10.1089/omi.2007.0023. [DOI] [PubMed] [Google Scholar]
- 25.Griffin D. Bovine pasteurellosis and other bacterial infections of the respiratory tract. Vet Clin North Am Food Anim Pract. 2010;26(1):57–71. doi: 10.1016/j.cvfa.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Wilkinson A. Future of BRD research: an animal health industry perspective. Anim Health Res Rev. 2009;10(2):163–4. doi: 10.1017/S1466252309990235. [DOI] [PubMed] [Google Scholar]
- 27.Gardner BA, Dolezal HG, Bryant LK, Owens FN, Smith RA. Health of finishing steers: effects on performance, carcass traits, and meat tenderness. J Anim Sci. 1999;77(12):3168–75. doi: 10.2527/1999.77123168x. [DOI] [PubMed] [Google Scholar]
- 28.Larson RL. The effect of cattle health on performance, production costs, and carcass value. J Anim Sci. 2005;83(E. suppl):E37–43. [Google Scholar]
- 29.Bryant LK, Perino LJ, Griffin DD. Lung lesions in feeder cattle at salughter: a proposed method for lesion recording, and lesion effects on calf growth and carcass traits; Paper presented at: Proc. of the Am. Assoc. Bovine Pract. Ann. Mtg; San Diego, CA. 1996. [Google Scholar]
- 30.Holland BP, Step DL, Burciaga-Robles LO, et al. Effectiveness of sorting calves with high risk of developing bovine respiratory disease on the basis of serum haptoglobin concentration at the time of arrival at a feedlot. Am J Vet Res. 2011;72(10):1349–60. doi: 10.2460/ajvr.72.10.1349. [DOI] [PubMed] [Google Scholar]
- 31.Wittum TE, Woollen NE, Perino LJ, Littledike ET. Relationships among treatment for respiratory tract disease, pulmonary lesions evident at slaughter, and rate of weight gain in feedlot cattle. J Am Vet Med Assoc. 1996;209(4):814–8. [PubMed] [Google Scholar]
- 32.Taylor JD, Fulton RW, Lehenbauer TW, Step DL, Confer AW. The epidemiology of bovine respiratory disease: what is the evidence for preventive measures? Can Vet J. 2010;51(12):1351–9. [PMC free article] [PubMed] [Google Scholar]
- 33.Ribble C, Stitt T, Iwasawa S, Toews L, Stephen C. A review of alternative practices to antimicrobial use for disease control in the commercial feedlot - executive summary. Can J Infect Dis Med Microbiol. 2010;21(3):128–32. doi: 10.1155/2010/798465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angen O, Thomsen J, Larsen LE, et al. Respiratory disease in calves: microbiological investigations on trans-tracheally aspirated bronchoalveolar fluid and acute phase protein response. Vet Microbiol. 2009;137(1–2):165–71. doi: 10.1016/j.vetmic.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Godinho KS, Sarasola P, Renoult E, et al. Use of deep nasopharyngeal swabs as a predictive diagnostic method for natural respiratory infections in calves. Vet Rec. 2007;160(1):22–5. doi: 10.1136/vr.160.1.22. [DOI] [PubMed] [Google Scholar]
- 36.Poulsen KP, McGuirk SM. Respiratory disease of the bovine neonate. Vet Clin North Am Food Anim Pract. 2009;25(1):121–37. vi–vii. doi: 10.1016/j.cvfa.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 37.White BJ, Renter DG. Bayesian estimation of the performance of using clinical observations and harvest lung lesions for diagnosing bovine respiratory disease in post-weaned beef calves. J Vet Diagn Invest. 2009;21(4):446–53. doi: 10.1177/104063870902100405. [DOI] [PubMed] [Google Scholar]
- 38.Buhman MJ, Perino LJ, Galyean ML, Wittum TE, Montgomery TH, Swingle RS. Association between changes in eating and drinking behaviors and respiratory tract disease in newly arrived calves at a feedlot. Am J Vet Res. 2000;61(10):1163–8. doi: 10.2460/ajvr.2000.61.1163. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt TB. Survey of Carcass and Offal Characteristics of the Fed Cattle in the Texas Panhandle. West Texas A&M University; Canyon, TX: 2000. [Google Scholar]
- 40.Schaefer AL, Cook NJ, Church JS, et al. The use of infrared thermography as an early indicator of bovine respiratory disease complex in calves. Res Vet Sci. 2007;83(3):376–84. doi: 10.1016/j.rvsc.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flock M. Diagnostic ultrasonography in cattle with thoracic disease. Vet J. 2004;167(3):272–80. doi: 10.1016/S1090-0233(03)00110-2. [DOI] [PubMed] [Google Scholar]
- 42.Boehmer JL, Degrasse JA, Lancaster VA, McFarland MA, Callahan JH, Ward JL. Evaluation of protein expression in bovine bronchoalveolar fluid following challenge with Mannheimia haemolytica. Proteomics. 2011;11(18):3685–97. doi: 10.1002/pmic.201000710. [DOI] [PubMed] [Google Scholar]
- 43.Timsit E, Assie S, Quiniou R, Seegers H, Bareille N. Early detection of bovine respiratory disease in young bulls using reticulo-rumen temperature boluses. Vet J. 2010 Oct;190:136–42. doi: 10.1016/j.tvjl.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 44.Aich P, Babiuk LA, Potter AA, Griebel P. Biomarkers for prediction of bovine respiratory disease outcome. OMICS. 2009;13(3):199–209. doi: 10.1089/omi.2009.0012. [DOI] [PubMed] [Google Scholar]
- 45.Burciaga-Robles LO, Holland BP, Step DL, et al. Evaluation of breath biomarkers and serum haptoglobin concentration for diagnosis of bovine respiratory disease in heifers newly arrived at a feedlot. Am J Vet Res. 2009;70(10):1291–8. doi: 10.2460/ajvr.70.10.1291. [DOI] [PubMed] [Google Scholar]
- 46.Reinhold P, Becher G, Rothe M. Evaluation of the measurement of leukotriene B4 concentrations in exhaled condensate as a noninvasive method for assessing mediators of inflammation in the lungs of calves. Am J Vet Res. 2000;61(7):742–9. doi: 10.2460/ajvr.2000.61.742. [DOI] [PubMed] [Google Scholar]
- 47.Berry BA, Confer AW, Krehbiel CR, Gill DR, Smith RA, Montelongo M. Effects of dietary energy and starch concentrations for newly received feedlot calves: II. Acute-phase protein response. J Anim Sci. 2004;82(3):845–50. doi: 10.2527/2004.823845x. [DOI] [PubMed] [Google Scholar]
- 48.Young CR, Wittum TE, Stanker LH, Perino LJ, Griffin DD, Littledike ET. Serum haptoglobin concentrations in a population of feedlot cattle. Am J Vet Res. 1996;57(2):138–41. [PubMed] [Google Scholar]
- 49.Fulton RW, Confer AW. Laboratory test descriptions for bovine respiratory disease diagnosis and their strengths and weaknesses: Gold standards for diagnosis, do they exist? Can Vet J. 2012;53(7):754–61. [PMC free article] [PubMed] [Google Scholar]
- 50.Van Eenennaam A, Neibergs H, Seabury C, et al. Results of the BRD CAP project: progress toward identifying genetic markers associated with BRD susceptibility. Anim Health Res Rev. 2014;15(2):157–60. doi: 10.1017/S1466252314000231. [DOI] [PubMed] [Google Scholar]
- 51.Tizioto PC, Kim J, Seabury CM, et al. Bovine Respiratory Disease Complex Coordinated Agricultural Project Research Team Immunological response to single pathogen challenge with agents of the bovine respiratory disease complex: an RNA-sequence analysis of the bronchial lymph node transcriptome. PLoS One. 2015;10(6):e0131459. doi: 10.1371/journal.pone.0131459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ham SW, Lew WK, Weaver FA. Thrombin use in surgery: an evidence-based review of its clinical use. J Blood Med. 2010;1:135–42. doi: 10.2147/JBM.S6622. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Dyer MD, Murali TM, Sobral BW. Computational prediction of host-pathogen protein-protein interactions. Bioinformatics. 2007;23(13):i159–66. doi: 10.1093/bioinformatics/btm208. [DOI] [PubMed] [Google Scholar]
- 54.Forst CV. Host-pathogen systems biology. Drug Discov Today. 2006;11(5–6):220–7. doi: 10.1016/S1359-6446(05)03735-9. [DOI] [PubMed] [Google Scholar]
- 55.Bovine Genome Sequencing and Analysis Consortium. Elsik CG, Tellam RL, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324(5926):522–8. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zimin AV, Delcher AL, Florea L, et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009;10(4):R42. doi: 10.1186/gb-2009-10-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.d’Offay JM, Fulton RW, Eberle R. Complete genome sequence of the NVSL BoHV-1.1 Cooper reference strain. Arch Virol. 2013;158(5):1109–1113. doi: 10.1007/s00705-012-1574-6. [DOI] [PubMed] [Google Scholar]
- 58.Robinson KE, Meers J, Gravel JL, McCarthy FM, Mahony TJ. The essential and non-essential genes of Bovine herpesvirus 1. J Gen Virol. 2008;89(pt 11):2851–63. doi: 10.1099/vir.0.2008/002501-0. [DOI] [PubMed] [Google Scholar]
- 59.Schwyzer M, Ackermann M. Molecular virology of ruminant herpesviruses. Vet Microbiol. 1996;53(1–2):17–29. doi: 10.1016/s0378-1135(96)01231-x. [DOI] [PubMed] [Google Scholar]
- 60.Colett MS, Larson R, Gold C, Strick D, Anderson DK, Purchio AF. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988;165(1):191–9. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- 61.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73(1):251–9. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailly JE, McAuliffe JM, Skiadopoulos MH, Collins PL, Murphy BR. Sequence determination and molecular analysis of two strains of bovine parainfluenza virus type 3 that are attenuated for primates. Virus Genes. 2000;20(2):173–82. doi: 10.1023/a:1008130917204. [DOI] [PubMed] [Google Scholar]
- 63.Eidam C, Poehlein A, Brenner Michael G, et al. Complete genome sequence of Mannheimia haemolytica strain 42548 from a case of bovine respiratory disease. Genome Announc. 2013;1(3):e00318–13. doi: 10.1128/genomeA.00318-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gioia J, Qin X, Jiang H, et al. The genome sequence of Mannheimia haemolytica A1: insights into virulence, natural competence, and Pasteurellaceae phylogeny. J Bacteriol. 2006;188(20):7257–66. doi: 10.1128/JB.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harhay GP, Koren S, Phillippy AM, et al. Complete closed genome sequences of Mannheimia haemolytica serotypes A1 and A6, isolated from cattle. Genome Announc. 2013;1(3):e00188–13. doi: 10.1128/genomeA.00188-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hauglund MJ, Tatum FM, Bayles DO, Maheswaran SK, Briggs RE. Genome sequences of Mannheimia haemolytica serotype A1 Strains D153 and D193 from bovine pneumonia. Genome Announc. 2013;1(5):e00848–13. doi: 10.1128/genomeA.00848-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klima CL, Cook SR, Hahn KR, et al. Draft genome sequence of a Mannheimia haemolytica serotype 6 isolate collected from the nasopharynx of a beef calf with bovine respiratory disease. Genome Announc. 2013;1(2):e0005113. doi: 10.1128/genomeA.00051-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Challacombe JF, Duncan AJ, Brettin TS, et al. Complete genome sequence of Haemophilus somnus (Histophilus somni) strain 129Pt and comparison to Haemophilus ducreyi 35000HP and Haemophilus influenzae Rd. J Bacteriol. 2007;189(5):1890–8. doi: 10.1128/JB.01422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Zheng H, Liu Y, et al. The complete genome sequence of Mycoplasma bovis strain Hubei-1. PLoS One. 2011;6(6):e20999. doi: 10.1371/journal.pone.0020999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wise KS, Calcutt MJ, Foecking MF, Roske K, Madupu R, Methe BA. Complete genome sequence of Mycoplasma bovis type strain PG45 (ATCC 25523) Infect Immun. 2011;79(2):982–3. doi: 10.1128/IAI.00726-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hood L. Systems biology: integrating technology, biology, and computation. Mech Ageing Dev. 2003;124(1):9–16. doi: 10.1016/s0047-6374(02)00164-1. [DOI] [PubMed] [Google Scholar]
- 72.Kucharova V, Wiker HG. Proteogenomics in microbiology: taking the right turn at the junction of genomics and proteomics. Proteomics. 2014;14(23–24):2360–675. doi: 10.1002/pmic.201400168. [DOI] [PubMed] [Google Scholar]
- 73.Kumar R, Lawrence ML, Watt J, Cooksey AM, Burgess SC, Nanduri B. RNA-seq based transcriptional map of bovine respiratory disease pathogen “Histophilus somni 2336”. PLoS One. 2012;7(1):e29435. doi: 10.1371/journal.pone.0029435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reddy JS, Kumar R, Watt JM, Lawrence ML, Burgess SC, Nanduri B. Tran-scriptome profile of a bovine respiratory disease pathogen: Mannheimia haemolytica PHL213. BMC Bioinformatics. 2012;13(suppl 15):S4. doi: 10.1186/1471-2105-13-S15-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qu Z, Adelson DL. Bovine ncRNAs are abundant, primarily intergenic, conserved and associated with regulatory genes. PLoS One. 2012;7(8):e42638. doi: 10.1371/journal.pone.0042638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Daetwyler HD, Capitan A, Pausch H, et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat Genet. 2014;46(8):858–65. doi: 10.1038/ng.3034. [DOI] [PubMed] [Google Scholar]
- 77.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform. 2008:67–79. [PMC free article] [PubMed] [Google Scholar]
- 79.Rubin DL, Shah NH, Noy NF. Biomedical ontologies: a functional perspective. Brief Bioinform. 2008;9(1):75–90. doi: 10.1093/bib/bbm059. [DOI] [PubMed] [Google Scholar]
- 80.Lewis SE. Gene ontology: looking backwards and forwards. Genome Biol. 2005;6(1):103. doi: 10.1186/gb-2004-6-1-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gene Ontology C Gene ontology consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–56. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gene Ontology Consortium. Blake JA, Dolan M, et al. Gene ontology annotations and resources. Nucleic Acids Res. 2013;41(Database issue):D530–5. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McCarthy FM, Gresham CR, Buza TJ, et al. AgBase: supporting functional modeling in agricultural organisms. Nucleic Acids Res. 2010;39(Database issue):D497–506. doi: 10.1093/nar/gkq1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stein LD, Mungall C, Shu S, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12(10):1599–610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Childers CP, Reese JT, Sundaram JP, et al. Bovine genome database: integrated tools for genome annotation and discovery. Nucleic Acids Res. 2011;39(Database issue):D830–4. doi: 10.1093/nar/gkq1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Harhay GP, Smith TP, Alexander LJ, et al. An atlas of bovine gene expression reveals novel distinctive tissue characteristics and evidence for improving genome annotation. Genome Biol. 2010;11(10):R102. doi: 10.1186/gb-2010-11-10-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eyre TA, Wright MW, Lush MJ, Bruford EA. HCOP: a searchable database of human orthology predictions. Brief Bioinform. 2007;8(1):2–5. doi: 10.1093/bib/bbl030. [DOI] [PubMed] [Google Scholar]
- 89.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Caspi R, Altman T, Billington R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome databases. Nucleic Acids Res. 2014;42(Database issue):D459–71. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karp PD, Riley M, Saier M, Paulsen IT, Paley SM, Pellegrini-Toole A. The EcoCyc and MetaCyc databases. Nucleic Acids Res. 2000;28(1):56–9. doi: 10.1093/nar/28.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morgat A, Coissac E, Coudert E, et al. UniPathway: a resource for the exploration and annotation of metabolic pathways. Nucleic Acids Res. 2012;40(Database issue):D761–9. doi: 10.1093/nar/gkr1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(Database issue):D204–12. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Croft D, Mundo AF, Haw R, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(Database issue):D472–7. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jupe S, Fabregat A, Hermjakob H. Expression data analysis with reactome. Curr Protoc Bioinformatics. 2015;49:8.20.1–8.20.9. doi: 10.1002/0471250953.bi0820s49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34(Database issue):D504–6. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Breuer K, Foroushani AK, Laird MR, et al. InnateDB: systems biology of innate immunity and beyond – recent updates and continuing curation. Nucleic Acids Res. 2013;41(Database issue):D1228–33. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Orchard S, Salwinski L, Kerrien S, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat Biotechnol. 2007;25(8):894–8. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 99.Zhang HM, Chen H, Liu W, et al. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 2012;40(Database issue):D144–9. doi: 10.1093/nar/gkr965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kshirsagar M, Carbonell J, Klein-Seetharaman J. Multitask learning for host-pathogen protein interactions. Bioinformatics. 2013;29(13):i217–26. doi: 10.1093/bioinformatics/btt245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guirimand T, Delmotte S, Navratil V. VirHostNet 2.0: surfing on the web of virus/host molecular interactions data. Nucleic Acids Res. 2015;43(Database issue):D583–7. doi: 10.1093/nar/gku1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Navratil V, de Chassey B, Meyniel L, et al. VirHostNet: a knowledge base for the management and the analysis of proteome-wide virus-host interaction networks. Nucleic Acids Res. 2009;37(Database issue):D661–8. doi: 10.1093/nar/gkn794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kumar R, Nanduri B. HPIDB – a unified resource for host-pathogen interactions. BMC Bioinformatics. 2010;11(suppl 6):S16. doi: 10.1186/1471-2105-11-S6-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Magi B, Bargagli E, Bini L, Rottoli P. Proteome analysis of bronchoalveolar lavage in lung diseases. Proteomics. 2006;6(23):6354–69. doi: 10.1002/pmic.200600303. [DOI] [PubMed] [Google Scholar]
- 106.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24(12):1551–7. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 107.Fjell CD, Hancock RE, Cherkasov A. AMPer: a database and an automated discovery tool for antimicrobial peptides. Bioinformatics. 2007;23(9):1148–55. doi: 10.1093/bioinformatics/btm068. [DOI] [PubMed] [Google Scholar]
- 108.Fjell CD, Jenssen H, Fries P, et al. Identification of novel host defense peptides and the absence of alpha-defensins in the bovine genome. Proteins. 2008;73(2):420–30. doi: 10.1002/prot.22059. [DOI] [PubMed] [Google Scholar]
- 109.Cernicchiaro N, White BJ, Renter DG, Babcock AH, Kelly L, Slattery R. Associations between the distance traveled from sale barns to commercial feedlots in the United States and overall performance, risk of respiratory disease, and cumulative mortality in feeder cattle during 1997 to 2009S16. J Anim Sci. 2012;90(6):1929–39. doi: 10.2527/jas.2011-4599. [DOI] [PubMed] [Google Scholar]
- 110.Ribble CS, Meek AH, Shewen PE, Guichon PT, Jim GK. Effect of pretransit mixing on fatal fibrinous pneumonia in calves. J Am Vet Med Assoc. 1995;207(5):616–9. [PubMed] [Google Scholar]
- 111.Lago A, McGuirk SM, Bennett TB, Cook NB, Nordlund KV. Calf respiratory disease and pen microenvironments in naturally ventilated calf barns in winter. J Dairy Sci. 2006;89(10):4014–25. doi: 10.3168/jds.S0022-0302(06)72445-6. [DOI] [PubMed] [Google Scholar]
- 112.Emau P, Giri SN, Bruss ML. Viral-bacterial pneumonia in calves: effects on plasma eicosanoids and long chain fatty acids. Int J Tissue React. 1987;9(3):199–214. [PubMed] [Google Scholar]
- 113.Mitchell GB, Clark ME, Siwicky M, Caswell JL. Stress alters the cellular and proteomic compartments of bovine bronchoalveolar lavage fluid. Vet Immunol Immunopathol. 2008;125(1–2):111–25. doi: 10.1016/j.vetimm.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 114.Mitchell GB, Clark ME, Caswell JL. Alterations in the bovine bronchoalveolar lavage proteome induced by dexamethasone. Vet Immunol Immunopathol. 2007;118(3–4):283–93. doi: 10.1016/j.vetimm.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 115.Senthilkumaran C, Clark ME, Abdelaziz K, et al. Increased annexin A1 and A2 levels in bronchoalveolar lavage fluid are associated with resistance to respiratory disease in beef calves. Vet Res. 2013;44:24. doi: 10.1186/1297-9716-44-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ayalew S, Confer AW, Shrestha B, Wilson AE, Montelongo M. Proteomic analysis and immunogenicity of Mannheimia haemolytica vesicles. Clin Vaccine Immunol. 2013;20(2):191–6. doi: 10.1128/CVI.00622-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee SR, Nanduri B, Pharr GT, Stokes JV, Pinchuk LM. Bovine viral diarrhea virus infection affects the expression of proteins related to professional antigen presentation in bovine monocytes. Biochim Biophys Acta. 2009;1794(1):14–22. doi: 10.1016/j.bbapap.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 118.Lee SR, Pharr GT, Cooksey AM, McCarthy FM, Boyd BL, Pinchuk LM. Differential detergent fractionation for non-electrophoretic bovine peripheral blood monocyte proteomics reveals proteins involved in professional antigen presentation. Dev Comp Immunol. 2006;30(11):1070–83. doi: 10.1016/j.dci.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 119.Ammari M, Mccarthy FM, Nanduri B, Pinchuk LM. Analysis of bovine viral diarrhea viruses-infected monocytes: identification of cytopathic and non-cytopathic biotype differences. BMC Bioinformatics. 2010;11(suppl 6):S9. doi: 10.1186/1471-2105-11-S6-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pinchuk GV, Lee SR, Nanduri B, Honsinger KL, Stokes JV, Pinchuk LM. Bovine viral diarrhea viruses differentially alter the expression of the protein kinases and related proteins affecting the development of infection and antiviral mechanisms in bovine monocytes. Biochim Biophys Acta. 2008;1784(9):1234–47. doi: 10.1016/j.bbapap.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 121.N’Jai AU, Rivera J, Atapattu DN, Owusu-Ofori K, Czuprynski CJ. Gene expression profiling of bovine bronchial epithelial cells exposed in vitro to bovine herpesvirus 1 and Mannheimia haemolytica. Vet Immunol Immunopathol. 2013;155(3):182–9. doi: 10.1016/j.vetimm.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sathiamoorthy S, Hodgins DC, Shewen PE, Highlander SK, Lo RY. A snapshot of Mannheimia hemolytica A1 gene expression during infection in the bovine host. FEMS Microbiol Lett. 2011;325(2):148–54. doi: 10.1111/j.1574-6968.2011.02422.x. [DOI] [PubMed] [Google Scholar]
- 123.Roehrig SC, Tran HQ, Spehr V, Gunkel N, Selzer PM, Ullrich HJ. The response of Mannheimia haemolytica to iron limitation: implications for the acquisition of iron in the bovine lung. Vet Microbiol. 2007;121(3–4):316–29. doi: 10.1016/j.vetmic.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 124.Edgeworth J, Gorman M, Bennett R, Freemont P, Hogg N. Identification of p8,14 as a highly abundant heterodimeric calcium binding protein complex of myeloid cells. J Biol Chem. 1991;266(12):7706–13. [PubMed] [Google Scholar]
- 125.Schiopu A, Cotoi OS. S100A8 and S100A9: DAMPs at the crossroads between innate immunity, traditional risk factors, and cardiovascular disease. Mediators Inflamm. 2013;2013:828354. doi: 10.1155/2013/828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carninci P. Genomics: mice in the ENCODE spotlight. Nature. 2014;515(7527):346–7. doi: 10.1038/515346a. [DOI] [PubMed] [Google Scholar]
- 127.Klopfleisch R, Gruber AD. Transcriptome and proteome research in veterinary science: what is possible and what questions can be asked? ScientificWorldJournal. 2012;2012:254962. doi: 10.1100/2012/254962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kellis M, Wold B, Snyder MP, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A. 2014;111(17):6131–8. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Andersson L, Archibald AL, Bottema CD, et al. FAANG Consortium Coordinated international action to accelerate genome-to-phenome with FAANG, the functional annotation of animal genomes project. Genome Biol. 2015;16:57. doi: 10.1186/s13059-015-0622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hu ZZ, Huang H, Wu CH, et al. Omics-based molecular target and biomarker identification. Methods Mol Biol. 2011;719:547–71. doi: 10.1007/978-1-61779-027-0_26. [DOI] [PMC free article] [PubMed] [Google Scholar]