Abstract
A noncompetitive peak decay method was used with 1 mm × 4.6 mm i.d. silica monoliths to measure the dissociation rate constants (kd) for various drugs with human serum albumin (HSA) and α1-acid glycoprotein (AGP). Flow rates up to 9 mL/min were used in these experiments, resulting in analysis times of only 20-30 s. Using a silica monolith containing immobilized HSA, dissociation rate constants were measured for amitriptyline, carboplatin, cisplatin, chloramphenicol, nortriptyline, quinidine, and verapamil, giving values that ranged from 0.37 s−1 to 0.78 s−1. Similar work with an immobilized AGP silica monolith gave kd values for amitriptyline, nortriptyline, and lidocaine of 0.39 s−1 to 0.73 s−1. These kd values showed good agreement with values determined for drugs with similar structures and/or affinities for HSA or AGP. It was found that a kd of up to roughly 0.80 s−1 could be measured by this approach. This information made it possible to obtain a better understanding of the advantages and possible limitations of the noncompetitive peak decay method and in the use of affinity silica monoliths for the high-throughput analysis of drug-protein dissociation.
Keywords: Silica monolith, Affinity microcolumn, Drug-protein dissociation, Human serum albumin, α1-Acid glycoprotein
1. Introduction
The interactions between a drug and serum proteins are important in determining the activity of a drug once it is in the circulation. This information is useful for describing the absorption, metabolism, distribution, and excretion (ADME) of a drug within the body [1]. Two serum proteins that bind to various drugs are human serum albumin (HSA) and α1-acid glycoprotein (AGP) [1]. HSA is composed of a single polypeptide chain of 585 amino acids and has a molecular weight of 66.5 kDa [2]. AGP is composed of a single polypeptide chain with up to five carbohydrate groups and has a molecular weight of roughly 41 kDa [3]. Many acidic (anionic) drugs bind to HSA, whereas basic (cationic) drugs tend to bind to AGP [4].
Several techniques have been used to study the interactions and affinities of drugs with either HSA or AGP [5-15]. These methods include ultrafiltration [5, 9], equilibrium dialysis [6, 8], fluorescence assays [7, 12], capillary electrophoresis [10], UV-vis spectroscopy [11], and solid-phase microextraction [13-15]. Another technique used to study drug-protein interactions is high-performance affinity chromatography (HPAC) [1, 16]. In HPAC, a biologically-related ligand (e.g., a serum protein) is immobilized onto a support and used as the stationary phase. HPAC has been shown to give results comparable to those seen with soluble protein methods (e.g., equilibrium dialysis and ultrafiltration); however, this method is more easily automated, can reuse the same ligand for up to hundreds of experiments, and requires much shorter analysis times for binding studies [1, 16].
The use of monolithic supports along with affinity ligands in HPAC, or traditional affinity separations, is referred to as affinity monolith chromatography (AMC) [17]. A monolithic support consists of a continuous bed that has both large through-pores to permit solvent flow and smaller side pores to allow for analyte interactions with the stationary phase. The advantages of this type of support include its better mass transfer properties, higher permeability and lower back pressures than particulate supports. Several reports have demonstrated that monolithic supports can be employed in affinity chromatography (see review in Ref. [17]). For instance, it has been shown in prior work that these monolithic supports can be used with immobilized serum proteins such as bovine serum albumin [18] or HSA [17] and can be used in the study of various biological interactions [17,19-21].
HPAC and AMC can also be used to study the dissociation rates of drugs from serum proteins. Methods that have been employed for this purpose include band-broadening measurements and peak fitting methods. Band-broadening methods are typically performed within linear elution conditions and involve careful plate height measurements. The peak fitting method does not require linear elution conditions, but it does make the assumption that the rates of other kinetic processes (e.g., stagnant mobile phase mass transfer) are fast compared to the rate of analyte-ligand dissociation. As a result, dissociation rate constants that are determined by this method can be a function of more than one process if this assumption is not valid [22].
Another technique that has been used is the peak decay method [1, 22]. In this method, a small plug of analyte is injected onto a column, followed by the use of conditions that prevent re-association of the analyte as it is later released from the immobilized ligand. The resulting decay curve is then used to estimate the dissociation rate constant for the analyte from the ligand, as illustrated in Figure 1. This experiment can be carried out for systems that have high affinities by placing a displacing agent in the mobile phase that competes with the analyte for binding sites in the column and blocks re-association of the analyte [1]. A noncompetitive peak decay method can be used for systems with weak-to-moderate affinities by employing short high-performance affinity columns and fast flow rates to prevent analyte re-association [22].
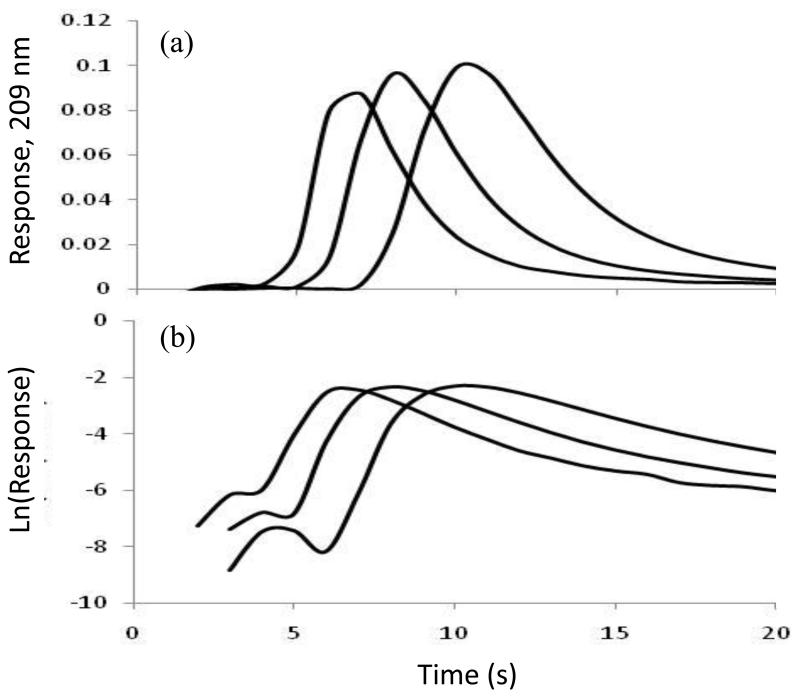
Figure 1.
(a) Elution profiles and (b) logarithmic form of the elution peak profiles for 100 μL injections of 20 μM nortriptyline at 5, 7, or 9 mL/min (from right-to-left) on a 1 mm × 4.6 mm i.d. silica monolith containing immobilized AGP. Other conditions are given in the text.
Recently, the noncompetitive peak decay method has been used with small columns containing silica particles or silica monoliths to measure the dissociation rate constants of various model drugs from HSA [23, 24]. One goal of this study will be to explore the use of this approach with a broader range of drugs and to estimate the range of dissociation rate constants that can be measured with this method. A second goal will be to see if this approach can be used with AGP. Although 10 cm long affinity monolith columns containing AGP have been utilized for chiral separations [25], no prior work has been carried out with AGP in a microcolumn format or with the peak profiling method to study drug-protein dissociation rates. The data that will be obtained for HSA and AGP will be used to compare the reported association equilibrium constants and measured dissociation rate constants for these proteins with the tested drugs. These experiments should lead to a better understanding of the limitations and advantages of the noncompetitive peak decay method and in the use of affinity silica monoliths as tools for the high-throughput analysis of drug-protein interactions.
2. Experimental
2.1 Reagents
The HSA (Cohn fraction V, essentially fatty acid free, ≥ 96% pure) and AGP (99% pure) were from Sigma (St. Louis, MO, USA). The various drugs that were examined in this study (e.g., see Figure 2) were obtained from Sigma or Fluka (Milwaukee, WI, USA). All buffers and aqueous solutions were prepared using water from a Nanopure system (Barnstead, Dubuque, IA, USA) and filtered using Osmonics 0.22 μm nylon filters from Fisher (Pittsburgh, PA, USA).
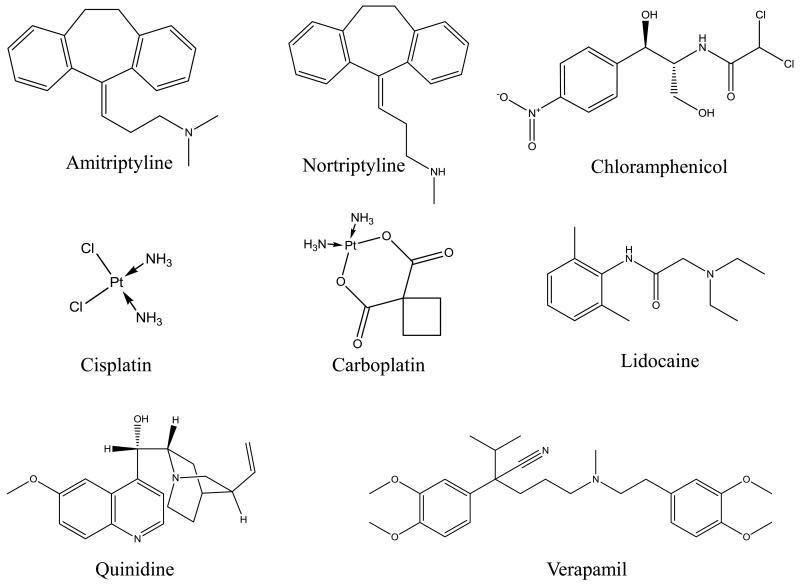
Figure 2.
Structures of the drugs examined in this study.
The drugs shown in Figure 2 all have known binding to HSA and/or AGP. For instance, amitriptyline and nortriptyline are antidepressants that bind to both HSA and AGP [11, 26, 27]. Carboplatin and cisplatin are anti-cancer drugs that bind to HSA [28, 29]. Chloramphenicol is an antibiotic and binds to HSA [12]. Lidocaine is used to treat ventricular cardiac arrythmias and binds to AGP with moderate-to-strong affinity and HSA with weak affinity [30]. Quinidine is an antiarrhythmic drug that binds to HSA [31, 32]. Verapamil is used to treat hypertension and binds to both HSA and AGP [5, 33].
2.2 Apparatus
A Chromolith Performance Si column (10 cm × 4.6 mm i.d.) was donated by Merck KGaA (Darmstadt, Germany). Using a lathe, this column was cut into 1 mm long pieces to make silica monolith microcolumns. Reagents to activate the silica monoliths and to immobilize HSA or AGP were applied using a Beckman System Gold 118 Solvent Module pump (Fullerton, CA, USA). The chromatographic system consisted of an isocratic HPLC PU-2080 Plus pump and a UV-2075 detector (Jasco, Easton, MD, USA). Injection onto this system was carried out by using a six-port Rheodyne Lab Pro valve (Cotati, CA, USA) and a 100 μL sample loop. An Alltech water jacket (Deerfield, IL, USA) and a circulating water bath from Fisher (Pittsburgh, PA, USA) were used to maintain a temperature of 37.0 (± 0.1) °C for the chromatographic system during all experiments described in this report. The chromatographic data were collected and processed using in-house programs written in LabView 5.1 (National Instruments, Austin, TX, USA).
2.3 Preparation of diol silica monolith
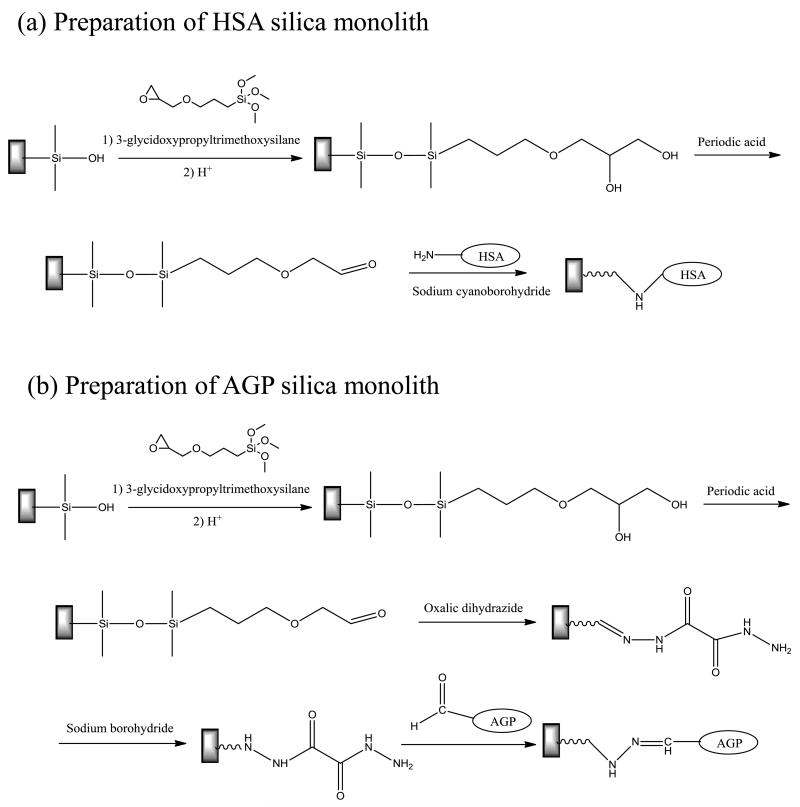
As shown in Figures 3(a)-(b), bare silica monoliths were first converted into a diol-bonded form, as described previously [25, 34, 35]. To do this, 1 mm long sections of the original silica monolith were cut and assembled into column housings made of Delrin. Each of these silica monoliths was washed with 0.10 M, pH 5.5 sodium acetate buffer for 40 min at 0.5 mL/min (Note: Unless otherwise indicated, this step and all following steps were conducted at room temperature). Pure 3-glycidoxypropyltrimethoxysilane was passed through the monolith for 50 min at 0.2 mL/min. After sealing both ends, the monolith column was placed in a water bath at 97 °C for 5 h. A solution of 0.10 M, pH 5.5 sodium acetate buffer was used to wash the column for 50 min at 0.1 mL/min and pure 3-glycidoxypropyltrimethoxysilane was again passed through the column for 50 min at 0.1 mL/min to ensure maximum diol coverage. The column ends were sealed and the column was placed in a water bath at 97 °C for 5 h. The column was removed from the water bath and washed with water for 4 h at 0.2 mL/min. A pH 3.0 solution of dilute sulfuric acid in water was passed through the column for 50 min at 0.2 mL/min. The column was then again sealed at both ends and placed in a water bath at 70 °C for 3 h. All of the resulting diol silica monolith columns were washed with water at 0.2 mL/min for over 5 h. Some of these columns were used for HSA or AGP immobilization while others were used as control columns in further studies.
Figure 3.
Reactions used in the preparation of (a) a HSA silica monolith and (b) a AGP silica monolith. The bonds shown above and below each silicon atom are to neighboring oxygen atoms in the support or to silanol groups.
2.4 Preparation of HSA silica monolith
As shown in Figure 3(a), HSA was immobilized onto a diol silica monolith by using the Schiff base method [34, 35]. In this method, a 90% (v/v) acetic acid solution in water was passed through each desired monolith for 4 h at 0.2 mL/min. A solution of 0.5 g/mL periodic acid in 90% acetic acid in water was then passed through the column in the dark for 7 h at 0.2 mL/min to oxidize the diol groups and form aldehyde groups. The column was washed with water for 8 h at 0.2 mL/min. A 10 mL solution containing 50 mg HSA and 25 mg sodium cyanoborohydride in 1.5 M, pH 6.0 potassium phosphate buffer was circulated through each column for 24 h at 0.5 mL/min. A second fresh 12 mL solution of 60 mg HSA and 30 mg sodium cyanoborohydride in the same pH 6.0 buffer was circulated through the column for 60 h at 0.5 mL/min. A 5 mL solution of 0.10 M, pH 8.0 potassium phosphate buffer containing 1 mg/mL sodium borohydride was applied to each column for 3 h at 0.1 mL/min, with this solution being used to reduce any remaining aldehyde groups on the support. The monolith columns were then washed with 0.10 M, pH 8.0 potassium phosphate buffer containing 0.5 M sodium chloride, which was passed through each column for 50 min at 0.2 mL/min, followed by an additional washing with 0.067 M, pH 7.4 potassium phosphate buffer for 1.5 h at 0.5 mL/min. The resulting HSA silica monolith was stored in this last buffer at 4 °C when not in use. The HSA silica monolith column was used within a period of 4 months; this type of support is known from prior studies to be stable for over one year under the storage and experimental conditions that were utilized in this study [24].
2.5 Preparation of AGP silica monolith
As illustrated in Figure 3(b), AGP was immobilized to a hydrazide-activated silica monolith [3, 25], using a diol-bonded silica monolith column as the starting material. To do this, a solution of a 90% (v/v) acetic acid solution in water was passed through the column for 4 h at 0.2 mL/min. A solution of 0.5 g/mL periodic acid in 90% acetic acid in water was then passed through the column in the dark for 6.7 h at 0.2 mL/min. The column was washed with water for 4.2 h at 0.2 mL/min. A 50 mL solution containing 0.13 g oxalic dihydrazide in 0.10 M, pH 5.0 potassium phosphate buffer was circulated through the column at 0.5 mL/min for 1.7 h. A solution of 1 mg/mL sodium borohydride in 0.10 M, pH 8.0 potassium phosphate buffer was passed through the column at 0.1 mL/min for 3 h. The column was washed with water for 1.7 h at 0.5 mL/min.
Prior to AGP immobilization, a solution containing 10 mL of 5 mg/mL AGP in 20 mM, pH 7.0 sodium acetate buffer and containing 0.15 M sodium chloride plus 10 mL of 20 mM periodic acid in the same buffer was prepared and allowed to stir for 30 min at 4 °C. This step was performed to oxidize the carbohydrate residues on AGP under mild conditions to give aldehyde groups. The oxidation reaction was quenched by adding 5 mL of ethylene glycol. Using 0.10 M, pH 7.0 potassium phosphate buffer as the elution buffer, the oxidized AGP was purified by collecting it as the reaction mixture was applied to a Econo-Pac 10DG desalting column (6 kDa exclusion limit, 10 mL volume, BioRad, Hercules, CA, USA).
The hydrazide-activated silica monolith was washed with 0.10 M, pH 7.0 potassium phosphate buffer for 1 h at 0.5 mL/min. A 15 mL portion of the oxidized AGP solution in 0.10 M, pH 7.0 potassium phosphate buffer was circulated through the monolith at 0.5 mL/min for 48 h. Another 15 mL portion of the oxidized AGP solution and circulated through the monolith for 48 h for 0.5 mL/min. The monolith was then washed with 0.067 M, pH 7.4 potassium phosphate buffer for 2 h at 0.5 mL/min. The resulting AGP silica monolith was stored in this last buffer at 4 °C when not in use. The AGP silica monolith column was used within a period of 1 month; this type of support is known to be stable for at least 2-3 months under the storage and experimental conditions that were utilized in this study [3, 25].
2.6 Chromatographic studies
All solutions used in this report for chromatographic studies were made in 0.067 M, pH 7.4 potassium phosphate buffer. These solutions were used within one week of preparation and stored at 4 °C when not in use. All mobile phases were prepared from this buffer and were degassed for 25 min prior to use. A sample concentration of 20 μM was used for all samples. The following detection wavelengths were employed: amitriptyline and nortriptyline, 209 nm; carboplatin, cisplatin, and chloramphenicol, 204 nm; lidocaine, 207 nm; quinidine, 234 nm; and verapamil, 230 nm. Samples were injected at flow rates ranging from 4 to 9 mL/min in the chromatographic studies. A baseline correction was performed on the collected data, and these corrected results were then used to prepare a plot of the natural logarithm of the response versus time, as shown in Figure 1. The slope of the linear range of natural logarithm of the elution profile was used to determine the dissociation rate constant for the drug-protein interaction. The linear range was selected by comparing the profiles for the control column and HSA column and choosing a region that did not include any significant overlap in their elution profiles for a given analyte, according to methods described in Refs. [23] and [24].
3. Results and discussion
3.1 General results of noncompetitive peak decay method
The noncompetitive peak decay method was performed at 37 °C by injecting each drug sample onto silica monoliths containing immobilized HSA, immobilized AGP, or control supports at various flow rates. In this method, some of the injected analyte was first allowed to bind to the immobilized protein as the sample plug entered the column. After the excess sample had been washed through the column, any retained analyte that then dissociated from the protein and entered the mobile phase was quickly eluted from the column.
Along with the presence of negligible re-association, it is also necessary in the peak decay method to have a rate of mass transfer for the analyte from the stagnant mobile phase to the flowing mobile phase that is faster than the rate of analyte-ligand dissociation. If these conditions are met, the following equation can be used to determine the dissociation rate constant for the analyte from the immobilized protein or binding agent in the column [22, 23].
| (1) |
In this equation, mAe is the moles of analyte that elutes from the column at time t, mA0 represents the moles of analyte that were initially bound to the column, and kd is the dissociation rate constant for the analyte-ligand interaction. According to Eq. (1), a plot of the natural logarithm of the elution profile (or ) versus time should give a linear relationship with a slope for the tailing portion of the profile that is equal to −kd, as illustrated in Figure 1(b). This relationship means it is possible from the slope of such a plot to obtain an estimate of the dissociation rate constant for the release of the analyte from the immobilized ligand [22, 23].
Elution profiles like those shown in Figure 1 were obtained at several flow rates for each drug on silica monoliths that contained immobilized HSA and AGP. Identical studies were also carried out on silica monoliths that contained a control support but no immobilized protein. In most cases, the decay profile for the immobilized HSA and AGP columns showed a much slower release of the drug from the immobilized protein compared to that of the drug washing off from the control column (see Section 3.4 for a discussion of exceptions). These same decay profiles each gave a linear relationship for their tailing portion when plotted in a logarithmic format, which was then used with Eq. (1) to estimate kd. This type of behavior has been noted before in the use of HSA columns for peak decay studies, which have previously been validated with model drugs for use in the estimation of drug-protein dissociation rates [23, 24]. However, this current study is the first report indicating that the same method can be used to examine the dissociation rates of drugs from AGP.
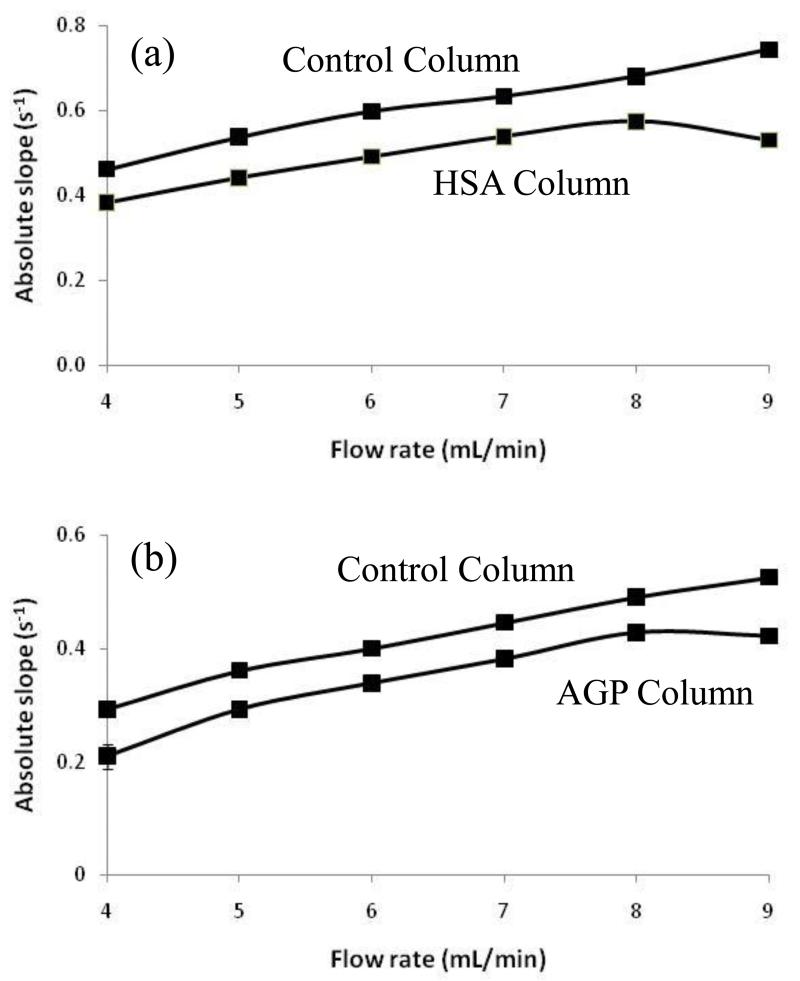
Figure 1(b) shows the logarithmic form of elution profiles that were obtained at flow rates of 5, 7, and 9 mL/min for nortriptyline on an AGP silica monolith. It can be seen that the use of flow rates up to 9 mL/min allowed for analysis times of only 20 s in this type of experiment. Similar analysis times (i.e., 20-30 s) were observed for the other drugs that were tested on the HSA and AGP silica monoliths. Besides decreasing the analysis time, the use of high flow rates in these experiments helped to prevent movement of analyte from the flowing mobile phase and back into the stagnant mobile phase, where the analyte could then re-associate with the stationary phase. Figure 4 shows how the measured slopes and apparent kd values changed with flow rate for some representative drugs that were applied to the HSA and AGP monoliths and their corresponding control columns. As noted in previous work with other model drugs and HSA [23, 24], the slope and apparent value of kd approached a constant value on the immobilized protein columns as the contribution due to stagnant mobile phase mass transfer became small compared to the rate of analyte-protein dissociation. However, the slopes measured on the control columns showed a steady increase in value even at the highest flow rates. This type of behavior was expected in the peak decay method and made it desirable to use high flow rates to obtain the most accurate estimates of drug-protein dissociation rates [22-24].
Figure 4.
Absolute value of the slopes measured at various flow rates for the logarithmic form of elution profiles obtained for injections of (a) quinidine on monoliths containing immobilized HSA or a control support and (b) nortriptyline on monoliths containing immobilized AGP or a control support. All injections were made at 37 °C onto 1 mm × 4.6 mm i.d. silica monoliths using 100 μL of a sample containing 20 μM of the drug being examined. Other conditions are given in the text.
A useful feature of silica monoliths in this type of work is their better mass transfer properties compared to silica particles, which makes it easier to obtain conditions in which the rate of stagnant mobile phase mass transfer is small compared to the rate of drug dissociation from an immobilized protein such as HSA [24]. The lower backpressures of silica monoliths versus silica particle-based columns, and the use of small monoliths in the peak decay method, were also valuable features in allowing the use of high flow rates in these experiments. For instance, even at 9 mL/min, the backpressures observed for the 1 mm × 4.6 mm i.d. silica monoliths used in this study were 5.3-14.7 MPa (767-2132 psi). All of these features make this approach of potential interest for the high-throughput screening or analysis of drug-protein dissociation rates [24].
3.2 Measurement of drug dissociation rates from HSA
The silica monolith columns containing immobilized HSA and a control support were used to obtain the dissociation rates of various drugs from HSA. Table 1 shows a summary of the results that were obtained in these experiments. Although no previous dissociation rate constants have been reported for these particular interactions, it was possible to compare these results with reported kd values for other drugs that have similar structures and/or binding affinities for HSA. For amitriptyline and nortriptyline, which have reported association equilibrium constants in the range of 1.4-4.7 × 102 M−1 [27, 31, 36], the estimated kd values of 0.39 s−1 (amitriptyline) and 0.36 s−1 (nortriptyline) were in good agreement with each other. Similar agreement was seen in the kd values that were measured for cisplatin and carboplatin (i.e., two structurally similar and platinum-containing anticancer drugs) which gave kd values of 0.65 s−1 and 0.70 s−1, respectively.
Table 1. Dissociation rate constants measured for various drugs with HSA.
| Drug | Association Equilibrium Constant, Ka (M−1) [Ref.] |
Dissociation Rate Constant, kd (s−1)a |
|---|---|---|
| Amitriptyline | 3.7-4.7 × 102 [27, 36] | 0.39 (± 0.02) |
| Carboplatin | Not reported | 0.70 (± 0.03) |
| Chloramphenicol | 0.82-2.5 × 104 [12, 37] | 0.78 (± 0.01) |
| Cisplatin | 8.5 × 102 [28] | 0.65 (± 0.14) |
| Nortriptyline | 1.41-4.4 × 102 [27, 31] | 0.37 (± 0.01) |
| Quinidine | 0.16-4.78 × 104 [36, 38] | 0.53 (± 0.01) |
| Verapamil | 0.11-1.4 × 104 [33, 39] | 0.38 (± 0.05) |
All of the kd values were measured at pH 7.4 and at 37 °C. The values in parentheses represent a range of ± 1 S.D., as determined for the slopes of the best-fit lines for the logarithmic elution profiles.
Chloramphenicol, quinidine, and verapamil all have association equilibrium constants that have been determined to be in the range of 103-104 M−1 for HSA [12, 33, 36-39]. A drug with a similar affinity for HSA and a previously-measured kd value for this interaction is tolbutamide. Tolbutamide has an association equilibrium with HSA of approximately 5 × 104 M−1 [40] and a kd of 0.49 s−1, as determined earlier by using the noncompetitive peak decay method [24]. The measured kd values for quinidine and verapamil were 0.53 s−1 and 0.38 s−1, respectively, and differed by only 8-22% with the kd value reported for tolbutamide from HSA. The measured kd for chloramphenicol with HSA was 0.78 s−1, which was 1.6-fold higher than kd for tolbutamide with the same protein.
3.3 Measurement of drug dissociation rates from AGP
The dissociation rate constants that were measured for various drugs with AGP by the peak decay method are summarized in Table 2. There were again no previous dissociation rate constants reported for these drugs with AGP, so the results were compared with drugs that had similar structures and/or affinities for AGP or HSA. The related drugs amitriptyline and nortriptyline both have association equilibrium constants for AGP in the range of 0.13–3.4 × 105 M−1. The measured kd values for these two drugs with AGP were also found to be similar, with values of 0.39 s−1 and 0.42 s−1, respectively. In addition, these values agreed with results that have been reported for drugs that have the same types of affinities for HSA. For example, warfarin has an association equilibrium constant of 2.1-2.6 × 105 M−1 for HSA and has reported kd values of 0.35-0.66 s−1 for this protein [23]. In addition, the kd of diazepam (i.e., a drug with an association equilibrium constant of 2.2 × 105 M−1 for HSA) has been reported to be 0.44 s−1 [24]. Lidocaine has association equilibrium constants in the range of 1.1-1.7 × 105 M−1 for AGP [30, 41]. The dissociation rate constant for this drug with AGP was 0.73 s−1. When comparing this result with those for drugs that have similar binding strengths to HSA, this value was comparable to the upper value of 0.66 s−1 [23] that has been reported for warfarin with HSA.
Table 2. Dissociation rate constants measured for various drugs with AGP.
| Drug | Association Equilibrium Constant, Ka (M−1) [Ref.] |
Dissociation Rate Constant, kd (s−1)a |
|---|---|---|
| Amitriptyline | 0.13–3.4 × 105 [27, 44] | 0.39 (± 0.01) |
| Nortriptyline | 0.2-1.2 × 105 [27] | 0.42 (± 0.01) |
| Lidocaine | 1.1-1.7 × 105 [30, 41] | 0.73 (± 0.07) |
All of the kd values were measured at pH 7.4 and at 37 °C. The values in parentheses represent a range of ± 1 S.D., as determined for the slopes of the best-fit lines for the logarithmic elution profiles.
3.4 Comparison of dissociation rate constants of drugs with HSA or AGP
The results obtained for HSA and AGP in Tables 1-2 were examined more closely to see what range of kd could be determined by the noncompetitive peak decay method. The overall ranges that were measured by this approach for drugs with the immobilized HSA and AGP silica monoliths were 0.37-0.78 s−1 and 0.39-0.73 s−1, respectively. A previous report using the same approach with an HSA silica monolith also resulted in a kd value for imipramine of 0.29 s−1 [24]. Thus, this information indicates that kd of at least 0.29-0.80 s−1 could be measured by the noncompetitive peak decay method. It is also known from previous work that systems with slower rates of dissociation can also be examined by the peak decay method when using the competitive detection mode and an appropriate labeled probe that binds to the same sites on the immobilized ligand as the analyte of interest [1].
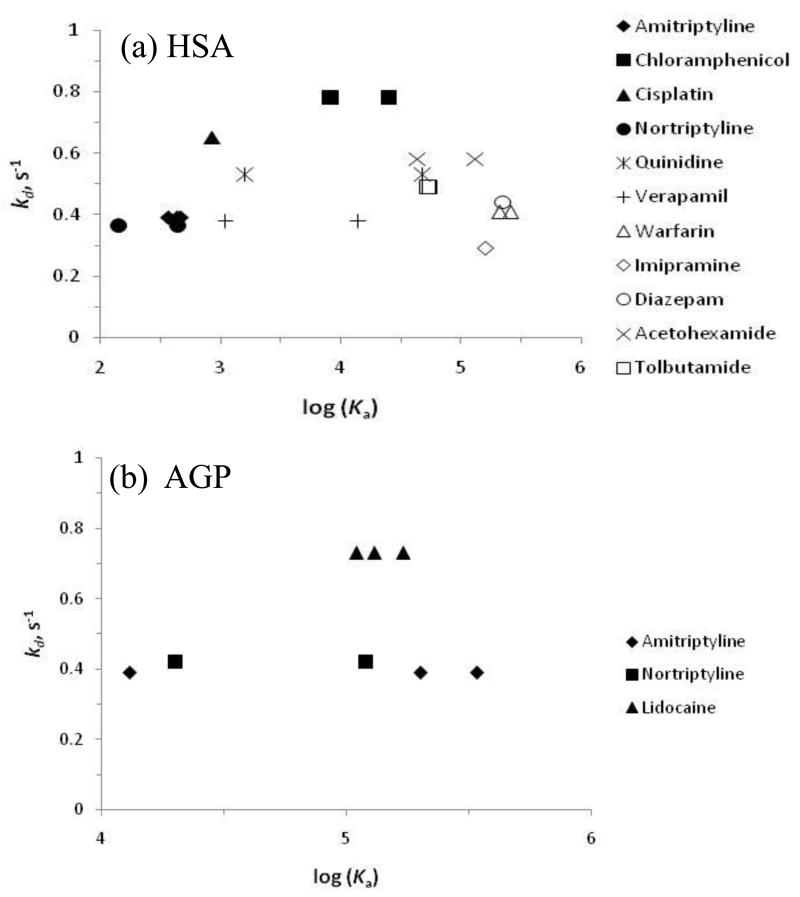
The kd values that were measured by the peak decay method, both in this current study and in prior work [24], were also compared to the association equilibrium constants (Ka) that have been reported for the same drugs with HSA and AGP. The results of this comparison are shown in Figure 5. As indicated earlier, the overall ranges of dissociation rate constants seen for HSA and AGP were similar. There was also no statistically significant trend apparent in the value of kd with the value of log(Ka) as indicated by the best-fit lines in Figure 5. Although an initial glance of these results suggests that there was slight increase in kd with log(Ka), this trend was not significant at the 90% confidence level, with correlation coefficients of only 0.1520 and 0.2571 being obtained for Figure 5(a) and 5(b), respectively. This type of behavior fits a model in which the rate of association is more important than the rate of dissociation in determining the overall affinity of a drug for these proteins. Further work with a broader set of drugs that bind to both HSA and/or AGP will be needed in future work to more fully explore this relationship.
Figure 5.
Relationship between the dissociation rate constant (kd, s−1) and association equilibrium constant (Ka, M−1) for various drugs with (a) HSA and (b) AGP. These tables include previously-determined values for kd for some drugs with HSA, which were determined in Ref. 20 by also using affinity silica monoliths and the peak decay method. The Ka values shown were obtained from the literature, as listed in Tables 1-2 and provided in Ref. 20). For drugs in which several Ka values have been reported, a point is given for each pair of kd and Ka values. The best-fit lines for the data in these plots were as follows: (a) y = 0.0184 x + 0.4077, with a correlation coefficient = 0.1520; and (b) y = 0.0885 x + 0.0856, with a correlation coefficient of 0.2571.
It was noted in this study that there were some drugs that could not be easily examined by the noncompetitive peak decay method. For example, the elution profiles for carbamazepine were similar on the HSA, AGP and control columns, which all gave apparent kd values of around 0.8 s−1. It was also found that the slopes for verapamil on the AGP column and its control column were similar, with apparent kd values of roughly 0.7 s−1. There were two reasons for the similarity in these results for the protein columns and control columns. First, carbamazepine and verapamil both had significant non-specific interactions with the modified supports that were used in these particular columns. Although the effects of weak and fast dissociation due to nonspecific interactions can be minimized in peak decay studies by using longer times to determine the slopes of elution profiles [24], this correction becomes more difficult if strong and slow dissociation for these secondary interactions is instead present (i.e., as was the case for verapamil and carbamazepine). In addition, carbamazepine has been determined previously to have a dissociation rate constant of 1.7 s−1 from HSA [42], which was above the range of values that were found in this current report to be easily measurable by the peak decay method. In this situation, an alternative method that could be used is the peak profiling method, as has been employed with carbamazepine in prior work [42]. This later approach is complementary to the noncompetitive peak decay method in that it can measure dissociation rate constants in the range of at least 0.67-2.7 s−1 [42, 43] and can be used to correct for even strong nonspecific analyte interactions with the support [42].
4. Concluding remarks
This report examined the use of the noncompetitive peak decay method with 1 mm × 4.6 mm i.d. silica monoliths in the measurement of drug dissociation rates from HSA and AGP. It was determined that the range of kd values that could be reliably obtained for drugs with these columns and by this method extended down to at least 0.29 s−1 up to about 0.8 s−1. This technique allowed the measurement of a large number of drugs with HSA and AGP, with values for kd that ranged from 0.37-0.78 s−1 and 0.39-0.73 s−1, respectively. Although this method worked well for many of the tested drugs, it was determined that an alternative method (i.e., peak profiling) was needed for analytes with kd values above 0.8 s−1 and/or that had slow dissociation due to non-specific binding to the support. These results should be useful in the extension of the peak decay method to other drugs and in providing a better understanding of drug-protein interactions in the circulation.
Acknowledgements
This work was supported by the National Institutes of Health under grant R01 GM044931 and was conducted in facilities that were renovated under NIH grant RR015468.
References
- [1].Hage DS, Jackson A, Sobansky MR, Schiel JE, Yoo MJ, Joseph KS. J. Sep. Sci. 2009;32:835–853. doi: 10.1002/jssc.200800640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kragh-Hansen U. Pharmacol Rev. 1981;33:17–53. [PubMed] [Google Scholar]
- [3].Xuan H, Hage DS. Anal. Biochem. 2005;346:300–310. doi: 10.1016/j.ab.2005.08.025. [DOI] [PubMed] [Google Scholar]
- [4].Bertucci C, Domenici E. Curr. Med. Chem. 2002;9:1463–1481. doi: 10.2174/0929867023369673. [DOI] [PubMed] [Google Scholar]
- [5].Hanada K, Ohta T, Hirai M, Arai M, Ogata H. J. Pharm. Sci. 2000;89:751–757. doi: 10.1002/(SICI)1520-6017(200006)89:6<751::AID-JPS6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- [6].Gillis AM, Yee Y-G, Kates RE. Biochem. Pharmacol. 1985;34:4279–4282. doi: 10.1016/0006-2952(85)90285-0. [DOI] [PubMed] [Google Scholar]
- [7].Taheri S, Cogswell LP, III, Gent A, Strichartz GR. J. Pharmacol. Exp. Ther. 2003;304:71–80. doi: 10.1124/jpet.102.042028. [DOI] [PubMed] [Google Scholar]
- [8].Herve F, Gomas E, Duche J-C, Tillement J-P. Br. J. Clin. Pharmacol. 1993;36:241–249. doi: 10.1111/j.1365-2125.1993.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hong Y, Tang Y, Zeng S. Chirality. 2009;21:692–698. doi: 10.1002/chir.20666. [DOI] [PubMed] [Google Scholar]
- [10].Timerbaev AR, Aleksenko SS, Polec-Pawlak K, Ruzik R, Semenova O, Hartinger CG, Oszwaldowski S, Galanski M, Jarosz M, Keppler BK. Electrophoresis. 2004;25:1988–1995. doi: 10.1002/elps.200305984. [DOI] [PubMed] [Google Scholar]
- [11].Leis D, Barbosa S, Attwood D, Taboada P, Mosquera V. Langmuir. 2002;18:8178–8185. [Google Scholar]
- [12].Ding F, Zhao G, Chen S, Liu F, Sun Y, Zhang L. J. Mol. Struct. 2009;929:159–166. [Google Scholar]
- [13].Vuckovic D, Pawliszyn J. J. Pharm. Biomed. Anal. 2009;50:550–555. doi: 10.1016/j.jpba.2008.08.023. [DOI] [PubMed] [Google Scholar]
- [14].Koster EHM, Wemes C, Morsink JB, de Jong GJ. J. Chromatogr. B. 2000;739:175–182. doi: 10.1016/s0378-4347(99)00344-8. [DOI] [PubMed] [Google Scholar]
- [15].Yuan H, Pawliszyn J. Anal. Chem. 2001;73:4410–4416. doi: 10.1021/ac010227s. [DOI] [PubMed] [Google Scholar]
- [16].Hage DS. J. Chromatogr. B. 2002;768:3–30. doi: 10.1016/s0378-4347(01)00482-0. [DOI] [PubMed] [Google Scholar]
- [17].Mallik R, Hage DS. J. Sep. Sci. 2006;29:1686–1704. doi: 10.1002/jssc.200600152. [DOI] [PubMed] [Google Scholar]
- [18].Zacharis CK, Kalaitzantonakis EA, Podgornik A, Theodoridis GA. J. Chromatogr. A. 2007;1144:126–134. doi: 10.1016/j.chroma.2006.12.081. [DOI] [PubMed] [Google Scholar]
- [19].Platonova G, Tennikova TB. J. Chromatogr. A. 2005;1065:75–81. doi: 10.1016/j.chroma.2004.10.073. [DOI] [PubMed] [Google Scholar]
- [20].Kalashnikova IV, Ivanova ND, Tennikova TB. Russ. J. Appl. Chem. 2008;81:867–873. [Google Scholar]
- [21].Gupalova TV, Lojkina OV, Palagnuk VG, Totolian AA, Tennikova TB. J. Chromatogr. A. 2002;949:185–193. doi: 10.1016/s0021-9673(02)00032-8. [DOI] [PubMed] [Google Scholar]
- [22].Schiel JE, Hage DS. J. Sep. Sci. 2009;32:1507–1522. doi: 10.1002/jssc.200800685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen J, Schiel JE, Hage DS. J. Sep. Sci. 2009;32:1632–1641. doi: 10.1002/jssc.200900074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yoo MJ, Hage DS. J. Chromatogr. A. 2011;1218:2072–2078. doi: 10.1016/j.chroma.2010.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mallik R, Xuan H, Hage DS. J. Chromatogr. A. 2007;1149:294–304. doi: 10.1016/j.chroma.2007.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Leis D, Barbosa S, Attwood D, Taboada P, Mosquera V. J. Phys. Chem. 2002;106:9143–9150. [Google Scholar]
- [27].Brinkschulte M, Breyer-Pfaff U. Naunyn-Schmiedeberg’s Arch. Pharmacol. 1980;314:61–66. doi: 10.1007/BF00498432. [DOI] [PubMed] [Google Scholar]
- [28].Neault JF, Tajmir-Riahi HA. Biochim. Biophys. Acta. 1998;1384:153–159. doi: 10.1016/s0167-4838(98)00011-9. [DOI] [PubMed] [Google Scholar]
- [29].Xie R, Johnson W, Rodriguez L, Gounder M, Hall GS, Buckley B. Anal. Bioanal. Chem. 2007;387:2815–2822. doi: 10.1007/s00216-007-1147-9. [DOI] [PubMed] [Google Scholar]
- [30].Soman S, Yoo MJ, Jang YJ, Hage DS. J. Chromatogr. B. 2010;878:705–708. doi: 10.1016/j.jchromb.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hollosy F, Valko K, Hersey A, Nunhuck S, Keri G, Beran C. J. Med. Chem. 2006;49:6958–6971. doi: 10.1021/jm050957i. [DOI] [PubMed] [Google Scholar]
- [32].Leahey EB, Reiffel JA, Giardina E-GV, Bigger T., Jr. Ann. Int. Med. 1980;92:605–608. doi: 10.7326/0003-4819-92-5-605. [DOI] [PubMed] [Google Scholar]
- [33].Mallik R, Yoo MJ, Chen S, Hage DS. J. Chromatogr. B. 2008;876:69–75. doi: 10.1016/j.jchromb.2008.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mallik R, Hage DS. J. Pharm. Biomed. Anal. 2008;46:820–830. doi: 10.1016/j.jpba.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yoo MJ, Hage DS. J. Sep. Sci. 2009;32:2776–2785. doi: 10.1002/jssc.200900346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Peters T., Jr. All About Albumin: Biochemistry, Genetics, and Medical Applications. Academic Press; San Diego, CA: 1996. [Google Scholar]
- [37].Vodrazka Z, Jandova D, Grafnetterova J, Schuck O, Kalousek I, Tomasek R, Lachmanov J. Biochem. Pharmacol. 1978;27:1717–1720. doi: 10.1016/0006-2952(78)90545-2. [DOI] [PubMed] [Google Scholar]
- [38].Ueda CT, Makoid MC. J. Pharm. Sci. 1979;68:448–450. doi: 10.1002/jps.2600680414. [DOI] [PubMed] [Google Scholar]
- [39].Jia Z, Ramstad T, Zhong M. J. Pharm. Biomed. Anal. 2002;30:405–413. doi: 10.1016/s0731-7085(02)00223-6. [DOI] [PubMed] [Google Scholar]
- [40].Joseph KS, Hage DS. J. Chromatogr. B. 2010;878:1590–1598. doi: 10.1016/j.jchromb.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].McNamara PJ, Slaughter RL, Pieper JA, Wyman MG, Lalka D. Anesth. Analg. 1981;60:395–400. [PubMed] [Google Scholar]
- [42].Tong Z, Schiel JE, Papastavros E, Ohnmacht CM, Smith QR, Hage DS. J. Chromatogr. A. 2011;1218:2065–2071. doi: 10.1016/j.chroma.2010.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Schiel JE, Ohnmacht CM, Hage DS. Anal. Chem. 2009;81:4320–4333. doi: 10.1021/ac9000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Schley J, Muller-Oerlinghausen B. J. Pharm. Pharmacol. 1986;38:102–106. doi: 10.1111/j.2042-7158.1986.tb04520.x. [DOI] [PubMed] [Google Scholar]