Abstract
Translational control is crucial for proper timing of developmental events that take place in the absence of transcription, as in meiotic activation in oocytes, early embryogenesis in many organisms, and spermatogenesis. Here we show that a novel form of the translation initiation complex component eIF4G in Drosophila, eIF4G2, is required specifically for male germ cells to undergo meiotic division and proper spermatid differentiation. Flies mutant for eIF4G2 are viable and female fertile but male sterile. Spermatocytes form, but the germ cells in mutant males skip the major events of the meiotic divisions and form aberrant spermatids with large nuclei. Consistent with the failure to undergo the meiotic divisions, function of eIF4G2 is required post-transcriptionally for normal accumulation of the core cell cycle regulatory proteins Twine and CycB in mature spermatocytes. Loss of eIF4G2 function also causes widespread defects in spermatid differentiation. Although differentiation markers Dj and Fzo are expressed in late-stage eIF4G2 mutant germ cells, several key steps of spermatid differentiation fail, including formation of a compact mitochondrial derivative and full elongation. Our results suggest that an alternate form of the translation initiation machinery may be required for regulation and execution of key steps in male germ cell differentiation.
Keywords: Translational control, eIF4G, Cell cycle, Meiosis, Spermatocyte, Drosophila
INTRODUCTION
Translational control plays a key role in temporal regulation of developmental events that must be executed largely in the absence of transcription. For example, in metazoan organisms with large yolk-rich eggs, in which early stages of embryonic development commonly take place before onset of zygotic transcription, the order of events can be programmed by choreographed translation of maternally provided mRNAs (reviewed in Groisman et al., 2001; Kuersten and Goodwin, 2003). Likewise in spermiogenesis, in which drastic remodeling of the cell continues after transcription is largely shut down, key mRNAs are initially translationally repressed, then actively translated at specific times in the differentiation sequence (Giorgini et al., 2002; Hempel et al., 2006; Schafer et al., 1990). A third striking case is the specialized cell cycle of meiosis. In vertebrate oocytes arrested in meiotic prophase, large numbers of mRNAs required for subsequent oocyte maturation and early embryogenesis are present but translationally silent. Resumption of meiosis is triggered by physiological signals that initiate a cascade of stepwise translation of selected mRNAs that drives meiotic cell cycle progression. Emerging evidence suggests that distinct mechanisms of translational repression and activation act on specific mRNAs at different steps in the process (reviewed in Vasudevan et al., 2006).
A key regulatory point for translational control in eukaryotes is initiation, instigated by binding of the translational initiation complex to the 5′ cap of the mRNA, leading to recruitment of the ribosomal subunits. Components of the translation initiation complex are conserved across eukaryotes. The eIF4E subunit binds the 5′ cap, the RNA helicase eIF4A is believed to unwind secondary structure in the 5′ UTR, and eIF4G serves as a crucial scaffold to bring those two proteins together. eIF4G also binds an additional key regulator of translation initiation, poly(A)-binding protein (PABP) (Tarun and Sachs, 1996).
An important outstanding question for how sequential developmental events can be ordered by translational control is how the translation initiation machinery can become targeted to and activated at specific subsets of mRNAs, and how this machinery might change in different cell types and stages. Here we show that eIF4G2, a novel homolog of the core translation initiation complex scaffold protein eIF4G, is required for meiotic cell cycle progression and normal spermatid differentiation during male gamete differentiation in Drosophila. The requirement for eIF4G2 is cell type- and stage-specific: strong loss-of-function mutants are male sterile but viable and female fertile. We found that eIF4G2 is required for the normal stage-specific expression of cell cycle regulatory proteins in spermatocytes and for many aspects of spermatid differentiation.
MATERIALS AND METHODS
Fly husbandry, stocks and genetic mapping
All stocks and crosses were grown on cornmeal and dextrose media at 22°C. Wild-type flies were from the OregonR strain. Transgenic flies were generated by P-element-mediated transformation via embryo injection as in Rubin and Spradling (Rubin and Spradling, 1982).
The nc32 and XM alleles were identified in a screen for mutants that failed to complement β2-tubulin mutation(s) (Fuller, 1986). Z3-3283 and BR21-37 were identified in screens for viable but male sterile mutations by Wakimoto et al. (Wakimoto et al., 2004) and Elizabeth Raff (Indiana University, Bloomington, IN), respectively, and the four mutants were shown to be allelic by complementation. The following combinations of eIF4G2 alleles tested showed an equally severe mutant phenotype: nc32/Df(3R)mbc1, nc32/XM, XM/BR21-37, BR21-37/Z3-3283 and nc32/BR21-37. The nc32 allele was initially located by recombination mapping in the region between hh and Pr on 3R (Fuller, 1986). Df(3R)mbc1 and Df(3R)Exel9014 did not complement nc32, whereas Df(3R)Exel6194 and Df(3R)crb-F89-4 complemented nc32, placing the locus in polytene interval 95A5-7;95D6-11. For sequencing each of the four alleles, genomic DNA was extracted from flies carrying that allele over Df(3R)mbc1. In the text and figures, all eIF4G2 homozygous mutants shown were BR21-37/Z3-3283.
The twe-lacZ reporter is as previously described (White-Cooper et al., 1998). The original Dj-GFP reporter line (on chromosome 3) was generated in the Renkawitz-Pohl laboratory (Santel et al., 1997) and was obtained (for ease of shipping) from the laboratory of Craig Montell (Johns Hopkins University, Baltimore, MD). The transgene was hopped off of the third chromosome by crossing in transposase and selecting for transpositions off the third.
Histology
Phase-Hoechst staining was performed as described in Lin et al. (Lin et al., 1996), and in situ hybridization as in Hiller et al. (Hiller et al., 2001). Anti-Cyclin B (CycB) (1:10, F2F4; BD Biosciences) and anti-tubulin (1:200, DM1A; Sigma) immunostaining was performed using methanol/acetone fixation as in Glover and Gonzalez (Glover and Gonzalez, 1993). Anti-Fzo [1:700, rabbit polyclonal (Hales and Fuller, 1997)] and anti-HA (1:1000, 16B12; Covance) immunostaining was performed using ethanol/formaldehyde fixation as in Hime et al. (Hime et al., 1996). β-galactosidase activity assays and in situ hybridization were performed as in White-Cooper et al. (White-Cooper et al., 1998).
Cloning
Our cDNA clone for a portion of the 5′ half of the eIF4G2 coding sequence, amplified from testis RNA, represented a different splice isoform from the annotated sequence in FlyBase. In particular, the sequence corresponding to bases 1318 to 1434 of the original annotated coding sequence was spliced out; thus, the protein sequence encoded by this testis transcript did not include the 39 amino acids VCKR…NSTLTQ. All amino acid numberings in this paper reflect the sequence of this cDNA clone, which was used for making the tagged protein for expression in S2 cells. This splice form was also identified in the accompanying paper (Franklin-Dumont et al., 2007).
The rescue construct consisted of genomic DNA from 2221 bp 5′ of the ATG, 6718 bp of protein coding sequence plus introns, 703 bp downstream of the stop codon, and an SV40 terminator. The first three were PCR-amplified from genomic DNA, with the following restriction sites added in the primers: NotI/XbaI (5′ region), XbaI/EcoRV (coding sequence plus introns), EcoRV/XhoI (3′ region). All three were subcloned and sequenced for mutations. The SV40 terminator was PCR-amplified from pFAF (Chen and Fischer, 2002), with SalI/XhoI sites added to the ends of the primers. The transgene was assembled sequentially into pBS-KS+, then moved into pCASPER4 using NotI/XhoI. To generate an HA-tagged eIF4G2 reporter protein, we added 3xHA (PCR-amplified with SpeI/XbaI sites added in primers) to the N-terminus, by inserting the 3xHA fragment into the XbaI site of the rescue construct just upstream of and in frame with the AUG.
Templates for eIF4G2, eIF4G and twine in situ probes were generated by PCR from genomic DNA, then cloned into pCRII-TOPO (Invitrogen). The following primers were used: for eIF4G, 5′-GGATCAACACCGATATCCAGA-3′ and 5′-GGTTGTATACGTGAGGCCTT-3′; for eIF4G2, 5′-AACCGTTCCCGAGGTACATC-3′ and 5′-TCACCGTAGACTTGTGCA-3′; for twine, 5′-GCCAATAAAGTTGACCGCA-3′ and 5′-ATGCCGCTTCAGCATCCATT-3′. The cycB cDNA was used as a probe template, as in White-Cooper et al. (White-Cooper et al., 1998).
To generate the tagged protein constructs for expression in S2 cells, the eIF4G2 protein coding sequence was amplified in three sections from testis cDNA and genomic DNA, and the N-terminal portion of the coding sequence was assembled in pBS-KS using a three-part ligation and an internal DraIII site. The entire coding sequence was then inserted into the KpnI/NotI sites of pMT-HA (Bunch et al., 1988) by means of another three-part ligation, this time using an internal BamHI site. The eIF4G protein coding sequence was amplified from testis cDNA, cloned into pCRII-TOPO and inserted into the KpnI/SacI sites of pMT-HA (Bunch et al., 1988). The coding sequence of eIF4E1 was likewise amplified from testes cDNA, ligated into pCRII-TOPO and inserted into the SalI/SacI sites of pMT-Myc (Bunch et al., 1988). The eIF4E1 protein is one of the two polypeptides that are generated from the eIF-4E (FlyBase) locus; eIF-4E-RA and eIF-4E-RC encode eIF4E1 and eIF4E2, respectively (Hernandez et al., 2005).
Tissue culture and co-immunoprecipitations
S2 cells were grown in Schneider's S2 cell media (Gibco) supplemented with 12.5% fetal bovine calf serum and 0.1 mg/ml gentamycin. Transfections were performed (on cells split the same day) in six-well plates, using the FuGENE-HD transfection reagent (Roche) and leaving the cells in the transfection mix for 48 hours. All 2 ml of cells from each well was then transferred to a T-75 flask, to which 13 ml of fresh media was added. Copper sulfate was immediately added to a final concentration of 0.7 mM, and induction proceeded for 24 hours. One-tenth volume was saved for crude extract (+ 30 μl 2× protein loading buffer, boiled 5 minutes, frozen). The remainder was pelleted, washed once with phosphate-buffered saline (PBS) and resuspended in 500 μl cold lysis buffer [100 mM NaCl, 50 mM Tris, 2 mM EDTA, 2 mM EGTA, 1% NP-40 + protease inhibitor tablet (Roche)] in a 1.5 ml tube. The lysate was rocked for 20 minutes at 4°C and spun at full speed in a microcentrifuge for 5 minutes. The supernatant was transferred to a fresh tube, to which 20 μl anti-HA beads (3F10, Roche) was added. The anti-HA immunoprecipitation (IP) samples were incubated for 3 hours at 4°C, washed 3× with cold lysis buffer and 1× with cold PBS. 25 μl 2× loading buffer was added to the beads, and the samples were boiled for 5 minutes. Samples were run on a 4–20% gradient minigel (BioRad). Proteins were subsequently transferred to a BioRad Immunoblot PVDF membrane overnight at 25–30 V. The blot was then blocked in 5% milk in Tris-buffered saline (TBS) for 2 hours, incubated with primary antibody (anti-HA or anti-Myc, 1:5000) (16B12, Covance and 4A6, Upstate) for 1 hour (5% milk/TBS), and secondary antibody (HRP-anti-mouse, 1:5000; Jackson ImmunoResearch) for 1 hour, then washed in several changes of TBS over 1.5 hours. Detection was performed using Western Lightning ECL detection reagents (Amersham Biosciences).
RESULTS
CG10192 encodes an alternate eIF4G protein in Drosophila
The Drosophila CG10192 gene (henceforth renamed eIF4G2) encodes a novel homolog of the core translation initiation factor component eIF4G. Drosophila has three genes encoding eIF4G-like proteins. The canonical eIF4G (also known as eIF-4G – FlyBase), encoded by CG10811, was shown to be part of the cap-binding complex in embryo extracts and showed high-affinity interactions with cap-binding protein homologs eIF4E1, eIF4E2 and eIF4E4 in a yeast two-hybrid assay (Hernandez et al., 2005). The predicted protein encoded by CG10192 (eIF4G2) contained the conserved, signature middle domain [39% identical to the middle domain of human eIF4GI (EIF4G1 – HUGO), 42% identical to the fly eIF4G], which is known in human eIF4GI to bind eIF4A and eIF3 (Imataka and Sonenberg, 1997). The predicted eIF4G2 protein also contained a region resembling the alpha-helical MA3 domain found in other eIF4Gs and their homologs (Fig. 1A). However, the predicted eIF4G2 protein lacked the eIF5C domain [a domain of unknown function found in the C termini of eIF4Gs and some other translation factors (Marchler-Bauer et al., 2007)] and had a long N-terminal region (1124 amino acids, compared with 418 for eIF4G and 752 for human eIF4GI) with no homology to any known protein domains. The predicted eIF4G2 protein lacked a canonical eIF4E-binding site [YXXXXLΦ, where Φ is a hydrophobic residue (Mader et al., 1995)] at the stereotypical distance from the middle domain (roughly 100–150 residues N-terminal). However, epitope-tagged eIF4E1 co-immunoprecipitated with eIF4G2 when both were expressed in Drosophila tissue culture cells, indicating that eIF4G2 directly or indirectly associates with the cap-binding protein eIF4E1. Either HA-tagged eIF4G2 or HA-tagged canonical eIF4G was co-expressed in S2 cells with Myc-tagged eIF4E1. Immunoprecipitation of HA-eIF4G via the HA epitope tag resulted in co-immunoprecipitation of Myc-eIF4E1, as expected, visualized by western blot of the precipitate with anti-Myc (Fig. 1B, lane 2). Likewise, even though the HA-eIF4G2 was either less abundantly expressed or transferred to the blot less efficiently than HA-eIF4G (Fig. 1B, lane 6), when eIF4G2 was immunoprecipitated via the HA tag, Myc-eIF4E1 co-immunoprecipitated (Fig. 1B, lane 3). The co-immunoprecipitation of Myc-eIF4E1 with HA-eIF4G2 supports the idea, based on the conservation of the key middle domain, that eIF4G2 may also act in translation initiation. Consistent with this, eIF4G2 was shown in the accompanying paper to associate with 7-methyl-GTP-sepharose, a cap analog (Franklin-Dumont et al., 2007).
Fig. 1. eIF4G2 encodes a novel eIF4G homolog in Drosophila.
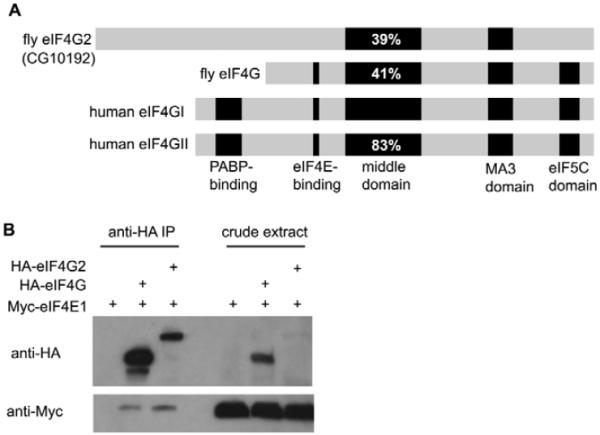
(A) Alignments of Drosophila eIF4G and eIF4G2 proteins with human eIF4GI (EIF4G1 – HUGO) and eIF4GII (EIF4G3 – HUGO). Black sections indicate recognized domains, with percentage identical to the eIF4GI middle domain marked. (B) Myc-eIF4E1 co-immunoprecipitates with HA-eIF4G2 from S2 cells. Immunoprecipitation with anti-HA. Western blot probed with anti-Myc and anti-HA, as indicated. Crude extract is 1/10 of each pre-immunoprecipitation (IP) sample.
eIF4G2 is required for male fertility
Several strong loss-of-function alleles of eIF4G2 were recovered in a variety of screens for mutants that affect male fertility (see Materials and methods). Trans-heteroallelic combinations of these alleles resulted in testes that contained plentiful spermatocytes but completely lacked elongated spermatids or mature sperm (Fig. 2C,D). Recombination mapping and deficiency complementation localized a representative allele to the region defined by deficiency Df(3R)mbc1, which failed to complement the male sterile phenotype. Sequence analysis of eIF4G2, which lies in this interval, revealed mutations in the eIF4G2 coding region for each of the four alleles tested. Two alleles had early stop codons and one a splice site mutation, all three resulting in partial or complete deletion of the signature conserved middle domain of the protein and therefore likely to be strong loss-of-function mutations (Fig. 2A). The fourth allele had a missense mutation in the conserved middle domain that changed a methionine to lysine in a position where the residue is invariably hydrophobic across species. The equivalent residue in human eIF4GII occurs in helix 5b (Marcotrigiano et al., 2001). Two nearby residues in the equivalent helix of human eIF4GI are required together for binding of both eIF4A and eIF3 (Imataka and Sonenberg, 1997).
Fig. 2. Loss-of-function mutations in eIF4G2 result in male sterility.
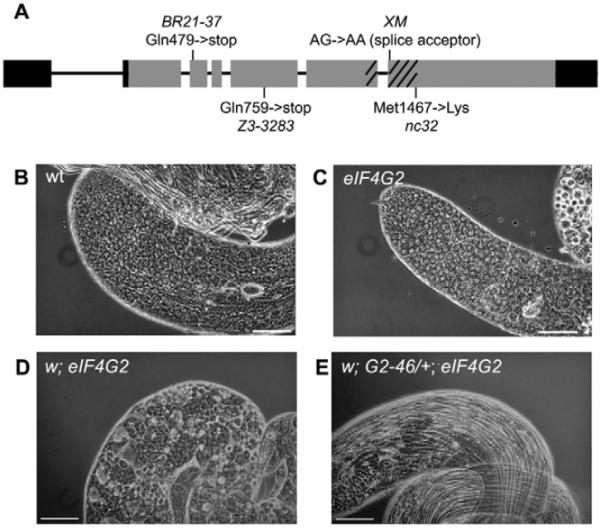
(A) Diagram of eIF4G2 intron/exon structure and four strong loss-of-function alleles. Gray, protein coding sequence; black, untranslated regions; diagonal lines, region encoding conserved middle domain. (B) Apical third of a wild-type testis. (C) Apical third of a testis from an eIF4G2 homozygote. (D) Part of testis from eIF4G2 homozygote. (E) Part of testis from eIF4G2 homozygote carrying a single copy of the genomic rescue transgene. eIF4G2 mutant flies were eIF4G2BR21-37/eIF4G2Z3-3283. Scale bars: 100 μm.
A 9.6 kb fragment of genomic DNA containing the eIF4G2 protein coding region plus 2.2 kb upstream and 0.7 kb downstream rescued the male sterile phenotype when introduced into flies as a transgene, confirming that the male sterility was due to loss-of-function of eIF4G2 (Fig. 2E).
eIF4G2 is expressed in a stage-specific pattern in the testis
In situ hybridization to testes revealed that eIF4G2 mRNA was strongly expressed in differentiating male germ cells from early spermatocytes to elongating spermatids (Fig. 3A). By contrast, eIF4G mRNA was present at the apical tip of the testis, in mitotic cells and early spermatocytes (Fig. 3C). The domains in which eIF4G and eIF4G2 transcripts were detected overlapped in early spermatocytes. Consistent with the distribution of eIF4G2 mRNA, epitope-tagged eIF4G2 protein expressed from the rescuing genomic transgene was detected in male germ cells starting from the early spermatocyte stage to elongating spermatids (Fig. 3E,F).
Fig. 3. eIF4G2 is expressed in a stage-specific pattern within the testis.
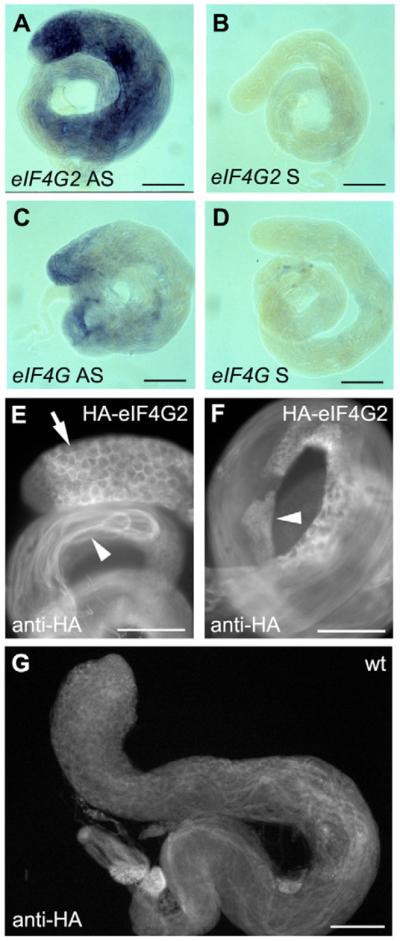
(A–D) In situ hybridization on wild-type testes; (A) eIF4G2 antisense, (B) eIF4G2 sense, (C) eIF4G antisense, (D) eIF4G sense. (E,F) Anti-HA immunostaining on testes from flies carrying the HA-eIF4G2-rescuing transgene. (E) Spermatocytes (arrow) and elongated spermatids (arrowhead). (F) Early post-meiotic spermatids (arrowhead). (G) Anti-HA staining on a non-transgenic testis (yw). Scale bars: 100 μm.
The eIF4G2 transcript was detected by RT-PCR in females and agametic males (data not shown). Flies mutant for the strong loss-of-function alleles of eIF4G2 were viable and female fertile, however, suggesting that function of eIF4G2 is required mainly for spermatogenesis.
eIF4G2 is required in males for meiotic division
Testes from eIF4G2 mutant males had early germ cells, including spermatogonia and spermatocytes (Fig. 4C,D). However, spermatocytes did not appear to properly execute the meiotic divisions. In wild type, entry into meiotic division is accompanied by condensation of the chromosomes, which subsequently move to the metaphase plate at the center of the nucleus (Fig. 4F; Fig. 4J, arrow), then separate in anaphase (Fig. 4J, arrowheads). Spermatocytes in eIF4G2 mutant males were formed, and meiotic chromosomes initiated condensation in preparation for the G2/M transition of meiosis I (Fig. 4H). However, meiotic chromosomes neither condensed completely nor moved to the center of the nucleus in eIF4G2 mutant spermatocytes, and no metaphase or anaphase figures were detected, indicating that spermatocytes in eIF4G2 males fail to undergo the major events of meiotic division.
Fig. 4. eIF4G2 is required in the testis for meiotic division.
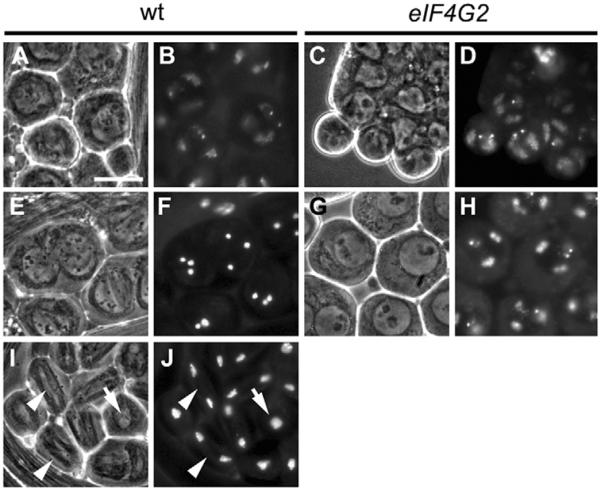
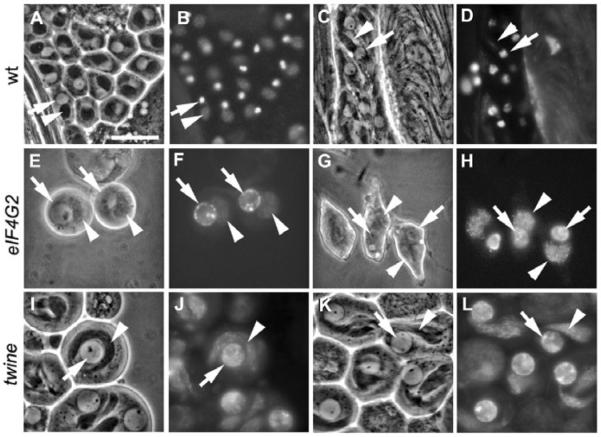
Live squashes of wild-type and mutant germ cells undergoing meiosis. (A,C,E,G,I) Phase contrast. (B,D,F,H,J) Hoechst staining. (A,B) Wild-type and (C,D) eIF4G2 mature spermatocytes. (E,F) Wild-type spermatocytes with condensed chromosomes. (G,H) eIF4G2 spermatocytes with partially condensed chromosomes. (I,J) Wild-type cells in metaphase (arrows) and anaphase (arrowheads). All images are at the same magnification. Scale bar: 20 μm. eIF4G2 mutant flies were eIF4G2BR21-37/eIF4G2Z3-3283.
Translation of two key meiotic cell cycle regulatory proteins requires eIF4G2
Function of eIF4G2 was required for the dramatic upregulation of the key cell cycle regulatory proteins Twine and CycB that normally takes place in mature spermatocytes. In wild type, the level of CycB protein is low in immature primary spermatocytes, rising in mature spermatocytes just before onset of the G2/M transition for meiosis I. CycB protein is then abruptly degraded at the metaphase to anaphase transition of meiosis I (White-Cooper et al., 1998) (Fig. 5A). The increase in expression of CycB protein was not detected in spermatocytes mutant for eIF4G2 (Fig. 5B), even though, based on chromatin condensation state (Fig. 4H), eIF4G2 mutant spermatocytes reached a stage in meiotic progression in which the increase of CycB levels would normally take place (White-Cooper et al., 1998). The meiotic Cdc25 phosphatase Twine triggers the G2/M transition of meiosis I. In wild-type mature spermatocytes, translation of twine in preparation for entry into the meiotic divisions (Alphey et al., 1992) can be visualized by expression of β-galactosidase from a twine-lacZ reporter transgene. For testes from males heterozygous for eIF4G2 and carrying one copy of the twine-lacZ reporter, each of the eight testes examined had cysts positive for β-galactosidase staining (Fig. 5C). In eIF4G2 mutant spermatocytes, however, we did not detect expression of β-galactosidase from the twine-lacZ reporter in any of the 12 testes examined (Fig. 5D), suggesting that expression of the twine reporter is much reduced in spermatocytes lacking eIF4G2 function. Both cycB and twine transcripts were expressed in eIF4G2 mutant spermatocytes, as in wild type (Fig. 5F,H), indicating that eIF4G2 is required for translation, not transcription or mRNA stability, of cycB and twine in spermatocytes. Lack of the normal increased expression levels of Twine and CycB proteins in eIF4G2 mutant spermatocytes was not likely to be because of arrest of spermatogenesis, as male germ cells proceeded to early spermatid stages in the mutant (Fig. 6).
Fig. 5. eIF4G2 is required for translation of twine and cycB in spermatocytes.
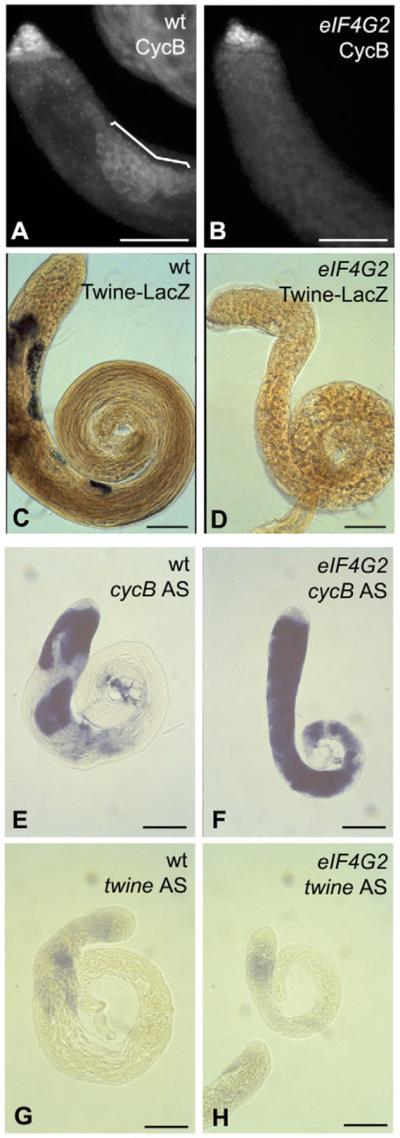
(A,B) Anti-CycB immunofluorescence on (A) wild-type and (B) eIF4G2 mutant testes. Bracket indicates expression of CycB in mature spermatocytes in wild type. (C,D) X-gal staining of (C) wild-type and (D) eIF4G2 mutant testes expressing a twine-lacZ reporter. (E,F) In situ hybridization on wild-type (E) and eIF4G2 (F) testes with cycB antisense probe. (G,H) In situ hybridization on wild-type (G) and eIF4G2 (H) testes with twine antisense probe. Scale bars: 100 μm. eIF4G2 mutant flies were eIF4G2BR21-37/eIF4G2Z3-3283.
Fig. 6. eIF4G2 is required for proper spermatid differentiation.
Live squashes of wild-type and mutant germ cells initiating spermatid differentiation. (A,C,E,G,I,K) Phase contrast. (B,D,F,H,J,L) Hoechst staining. (A,B) Wild-type early round spermatids with haploid nuclei (arrow) and mitochondrial derivative (arrowhead). (C,D) Elongating spermatids in wild type. (E,F) Late-stage eIF4G2 cells with large nuclei (arrow) and aggregating mitochondria (arrowhead). (G,H) Terminal cells in eIF4G2 with aggregated mitochondria (arrowhead), large nuclei (arrow) and aberrant partial cellular elongation. (I,J) twine early spermatids. (K,L) twine elongating spermatids. All images are at the same magnification. Scale bar: 20 μm. eIF4G2 mutant flies were eIF4G2BR21-37/eIF4G2Z3-3283. twine mutant flies were tweHB5/tweK08310.
Loss of eIF4G2 causes defects in differentiation
Loss of eIF4G2 function also caused profound defects in spermatid differentiation. In wild type, early spermatids form after the two meiotic divisions (Fig. 6A,B). Each haploid round onion-stage spermatid has a phase-bright nucleus (arrow) and a phase-dark mitochondrial derivative (arrowhead). Further differentiation includes elongation of the mitochondrial derivative (Fig. 6C,D), growth of flagella, and dramatic cell elongation. In eIF4G2 males, although spermatocytes did not execute the major events of meiotic division, male germ cells appeared to proceed to early stages of spermatid differentiation, producing large aberrant cells resembling abnormal early spermatids with a large nucleus (arrow) and a partially aggregated mitochondrial cloud (arrowhead) (Fig. 6E,F). This was the most common aberrant cell type seen in the lower part of eIF4G2 mutant testes. The nuclear morphology in these late-stage eIF4G2 mutant germ cells appeared similar to that of early spermatids in twine mutant testes (Fig. 6I–L), in which the cells fail to undergo meiotic divisions but form spermatids that differentiate (Alphey et al., 1992). This is consistent with the model that the failure to execute major events of meiosis observed in eIF4G2 mutant male germ cells may in part reflect lack of key cell cycle regulators for the G2/M transition of meiosis I.
A fraction of late-stage eIF4G2 mutant germ cells appeared to initiate spermatid elongation (Fig. 6G,H). To examine the extent to which eIF4G2 mutant germ cells are capable of differentiating, we examined the morphology of late-stage and terminal germ cells stained with anti-tubulin, as well as the expression of two spermatid differentiation markers, Fzo and Dj, by immunofluorescence microscopy. In wild-type elongating spermatids, microtubules aligned along the length of the cell are easily visible by anti-tubulin immunofluorescence (Fig. 7B, arrowheads). In terminal-stage eIF4G2 mutant germ cells, arrays of parallel microtubules were detected in the cells that had initiated (defective) elongation (Fig. 7D, arrowheads).
Fig. 7. eIF4G2 is not required for the translation of the spermatid differentiation markers fzo and dj.
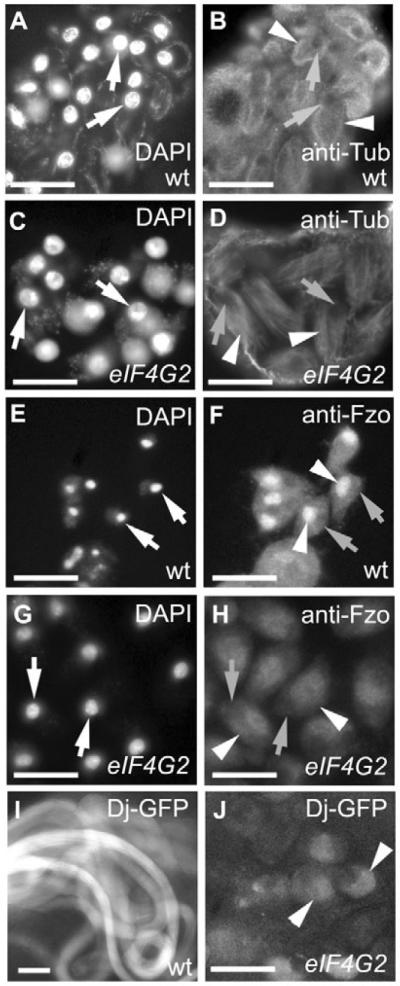
(A–D) Anti-tubulin immunostaining of squashed preparations of (A,B) wild-type and (C,D) eIF4G2 testes. (A,C) DAPI; (B,D) anti-Tub. (E–H) Anti-Fzo immunostaining (F,H) and DAPI (E,G) on squashed preparations of (E,F) wild-type and (G,H) eIF4G2 testes. Arrows indicate location of nuclei; arrowheads mark mitochondrial derivatives (F) or mitochondrial aggregates (H). (I,J) Dj-GFP reporter in (I) wild-type and (J) eIF4G2 testes. Arrowheads indicate Dj-GFP in J. Scale bars: 20 μm. eIF4G2 mutant flies were eIF4G2BR21-37/eIF4G2Z3-3283.
Consistent with progression of eIF4G2 mutant germ cells into spermatid differentiation stages, two proteins involved in spermatid differentiation, Fzo and Dj, were expressed in eIF4G2 mutant germ cells. Expression of fzo mRNA initiates in early spermatocytes, but the Fzo protein normally only accumulates to detectable levels by late in anaphase II, reaching a peak in haploid round spermatids (Hales and Fuller, 1997). Fzo protein localizes to the mitochondrial derivative in wild-type spermatids (Fig. 7F, arrowheads). In the eIF4G2 mutant, Fzo protein was detected in late-stage germ cells in a cloud next to the nucleus (Fig. 7H, arrowheads), resembling the unfused mitochondrial aggregates observed by phase-contrast microscopy in mutant spermatids. Likewise, although expression of dj mRNA initiates in early spermatocytes, translation of dj is normally delayed until well after completion of the meiotic divisions (Santel et al., 1997). The Dj-green fluorescent protein (GFP) fusion protein marks elongated spermatids in wild type (Fig. 7I), localizing to the mitochondrial derivative within each flagellum (Santel et al., 1998; Santel et al., 1997). In eIF4G2 mutant testes, Dj-GFP colocalized with the aggregated mitochondria in late-stage cells (Fig. 7J, arrowheads).
DISCUSSION
We have identified a novel form of the core translational initiation factor component eIF4G that is required only for male gamete differentiation. Although precedent for developmentally regulated translation initiation factor components comes from data on the cap binding protein eIF4E, such as Caenorhabditis elegans IFE-1 and IFE-4, and various eIF4Es from Drosophila, zebrafish and mammals (Amiri et al., 2001; Cho et al., 2006; Cho et al., 2005; Dinkova et al., 2005; Hernandez et al., 2005; Joshi et al., 2004; Robalino et al., 2004), less is known about the potential for the core eIF4G subunit to show such tissue specificity. In a human hematopoetic stem cell line, eIF4GII is specifically recruited to 5′ cap structures of mRNAs upon thrombopoietin-mediated induction of megakaryocyte differentiation, whereas levels of eIF4GI at the cap remain constant (Caron et al., 2004). However, the authors acknowledge that this recruitment of eIF4GII could represent an overall increase in active initiation factor complex within differentiating megakaryocytes, rather than intrinsic transcript specificity on the part of eIF4GII.
Function of Drosophila eIF4G2 is required for both meiotic cell cycle progression and for many aspects of spermatid differentiation. However, loss of eIF4G2 does not cause meiotic arrest. The eIF4G2 loss-of-function phenotype in testes is different from the phenotype of mutations in the testis TAFs (tTAFs). In tTAF mutant males, spermatocytes arrest at the G2/M transition, fail to undergo meiotic division and show a complete absence of spermatid differentiation (Hiller et al., 2004; Hiller et al., 2001; Lin et al., 1996). By contrast, in eIF4G2 mutant males, germ cells appear to skip the major events of meiotic division but initiate spermatid differentiation. Germ cells in males mutant for the cell cycle phosphatase Twine, or cdc2ts mutant males shifted to the non-permissive temperature, also skip the major events of meiotic division but proceed to execute spermatid differentiation (Alphey et al., 1992; Sigrist et al., 1995). These data show that initiation and execution of the spermatid differentiation program can proceed even when male germ cells fail to execute the meiotic divisions.
The failure to undergo the meiotic divisions in eIF4G2 is likely to be due, at least in part, to failure to upregulate twine and cycB translation as spermatocytes mature. Although eIF4G2 is a homolog of a known translation initiation factor, and eIF4G2 mutant spermatocytes have defects in translation of cycB and twine, it is formally possible that eIF4G2 does not act directly on these transcripts, but rather on an upstream regulator of their translation. Future experiments will address whether eIF4G2 binds these two mRNAs, to determine whether its effect on their translation is likely to be direct or indirect.
Function of eIF4G2 also appears to be required for many aspects of spermatid differentiation. Although early spermatids form in eIF4G2 mutant males, the mitochondrial cloud fails to condense and form a compact mitochondrial derivative, and very little spermatid elongation takes place. The defects in spermatid differentiation in eIF4G2 mutant males are more severe than the defects observed in males mutant for the RNA-binding protein Boule, homolog of human BOULE and DAZL (Eberhart et al., 1996). These observations suggest that although both Boule and eIF4G2 are required for normal translation of twine (Maines and Wasserman, 1999), the requirement for eIF4G2 is more widespread. A broad requirement for eIF4G2 for timing or execution of many events during male germ cell differentiation is reflected in the pleiotropic nature of the eIF4G2 mutant phenotype in testes. As shown in the accompanying paper (Franklin-Dumont et al., 2007), loss-of-function of eIF4G2 also affects spermatocyte growth as well as timing of events of the meiotic program in primary spermatocytes.
Given the broad defects observed in male germ cells, the predicted role of eIF4G2 in translation initiation, and the apparent reduction in transcript levels for the canonical eIF4G, it was surprising that Fzo and Dj proteins were expressed in spermatids from eIF4G2 mutant males. These findings suggest that eIF4G2 is not required (directly or indirectly) for translation of all mRNAs in mature spermatocytes and post-meiotic germ cells. It is possible that some of the canonical eIF4G protein persists from earlier germ cell stages, sufficient for translation of fzo and dj. However, if so, this is not sufficient for robust translation of cell cycle regulators twine and cycB in late spermatocytes, or for sufficient translation of additional mRNAs required for proper spermatid differentiation.
Acknowledgments
The authors would like to thank Elizabeth Raff for the BR21-37 allele, the Bloomington Drosophila Stock Center for fly stocks, and members of the Fuller laboratory for helpful discussions and feedback. C.C.B. was supported by an NRSA postdoctoral fellowship (1 F32 GM071260) and a Neonatology and Developmental Training Grant (NIH, T32 HD007249). This research was supported by NICHD/NIH through cooperative agreement U54-HD31398 as part of the Specialized Cooperative Centers Program in Reproduction and Fertility Research.
References
- Alphey L, Jimenez J, White-Cooper H, Dawson I, Nurse P, Glover DM. Twine, a CDC25 homolog that functions in the male and female germline of Drosophila. Cell. 1992;69:977–988. doi: 10.1016/0092-8674(92)90616-k. [DOI] [PubMed] [Google Scholar]
- Amiri A, Keiper BD, Kawasaki I, Fan Y, Kohara Y, Rhoads RE. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development. 2001;128:3899–3912. doi: 10.1242/dev.128.20.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch TA, Grinblat Y, Goldstein LS. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron S, Charon M, Cramer E, Sonenberg N, Dusanter-Fourt I. Selective modification of eukaryotic initiation factor 4F (eIF4F) at the onset of cell differentiation: recruitment of eIF4GII and long-lasting phosphorylation of eIF4E. Mol. Cell. Biol. 2004;24:4920–4928. doi: 10.1128/MCB.24.11.4920-4928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Fischer JA. A P element transformation vector for high levels of gene expression in germ-line cells of the ovary and undifferentiated cells in the developing eye of Drosophila. Plasmid. 2002;47:61–65. doi: 10.1006/plas.2001.1546. [DOI] [PubMed] [Google Scholar]
- Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N. A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005;121:411–423. doi: 10.1016/j.cell.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, Sonenberg N. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr. Biol. 2006;16:2035–2041. doi: 10.1016/j.cub.2006.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova TD, Keiper BD, Korneeva NL, Aamodt EJ, Rhoads RE. Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Mol. Cell. Biol. 2005;25:100–113. doi: 10.1128/MCB.25.1.100-113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart CG, Maines JZ, Wasserman SA. Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature. 1996;381:783–785. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- Franklin-Dumont TM, Chatterjee C, Wasserman SA, DiNardo S. A novel eIF4G homolog, Off-schedule, couples translational control to meiosis and differentiation in Drosophila spermatocytes. Development. 2007;134:2851–2861. doi: 10.1242/dev.003517. [DOI] [PubMed] [Google Scholar]
- Fuller MT. Genetic analysis of spermatogenesis in Drosophila: the role of the testis-specific beta-tubulin and interacting genes in cellular morphogenesis. In: Gall JG, editor. Gametogenesis and the Early Embryo. Alan Liss; New York: 1986. pp. 19–41. [Google Scholar]
- Giorgini F, Davies HG, Braun RE. Translational repression by MSY4 inhibits spermatid differentiation in mice. Development. 2002;129:3669–3679. doi: 10.1242/dev.129.15.3669. [DOI] [PubMed] [Google Scholar]
- Glover DM, Gonzalez C. Techniques for studying mitosis in Drosophila. In: Brookes R, Fantes P, editors. The Cell Cycle: A Practical Approach. IRL Press; Oxford: 1993. pp. 163–168. [Google Scholar]
- Groisman I, Huang YS, Mendez R, Cao Q, Richter JD. Translational control of embryonic cell division by CPEB and maskin. Cold Spring Harb. Symp. Quant. Biol. 2001;66:345–351. doi: 10.1101/sqb.2001.66.345. [DOI] [PubMed] [Google Scholar]
- Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Hempel LU, Rathke C, Raja SJ, Renkawitz-Pohl R. In Drosophila, don juan and don juan like encode proteins of the spermatid nucleus and the flagellum and both are regulated at the transcriptional level by the TAF II80 cannonball while translational repression is achieved by distinct elements. Dev. Dyn. 2006;235:1053–1064. doi: 10.1002/dvdy.20698. [DOI] [PubMed] [Google Scholar]
- Hernandez G, Altmann M, Sierra JM, Urlaub H, Diez del Corral R, Schwartz P, Rivera-Pomar R. Functional analysis of seven genes encoding eight translation initiation factor 4E (eIF4E) isoforms in Drosophila. Mech. Dev. 2005;122:529–543. doi: 10.1016/j.mod.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- Hime GR, Brill JA, Fuller MT. Assembly of ring canals in the male germ line from structural components of the contractile ring. J. Cell Sci. 1996;109:2779–2788. doi: 10.1242/jcs.109.12.2779. [DOI] [PubMed] [Google Scholar]
- Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi B, Cameron A, Jagus R. Characterization of mammalian eIF4E-family members. Eur. J. Biochem. 2004;271:2189–2203. doi: 10.1111/j.1432-1033.2004.04149.x. [DOI] [PubMed] [Google Scholar]
- Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Lin TY, Viswanathan S, Wood C, Wilson PG, Wolf N, Fuller MT. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development. 1996;122:1331–1341. doi: 10.1242/dev.122.4.1331. [DOI] [PubMed] [Google Scholar]
- Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines JZ, Wasserman SA. Post-transcriptional regulation of the meiotic Cdc25 protein Twine by the Dazl orthologue Boule. Nat. Cell Biol. 1999;1:171–174. doi: 10.1038/11091. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Lomakin IB, Sonenberg N, Pestova TV, Hellen CU, Burley SK. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- Robalino J, Bhavesh J, Fahrenkrug SC, Jagus R. Two zebrafish eIF4E family members are differentially expressed and functionally divergent. J. Biol. Chem. 2004;279:10532–10541. doi: 10.1074/jbc.M313688200. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Santel A, Winhauer T, Blumer N, Renkawitz-Pohl R. The Drosophila don juan (dj) gene encodes a novel sperm specific protein component characterized by an unusual domain of a repetitive amino acid motif. Mech. Dev. 1997;64:19–30. doi: 10.1016/s0925-4773(97)00031-2. [DOI] [PubMed] [Google Scholar]
- Santel A, Blumer N, Kampfer M, Renkawitz-Pohl R. Flagellar mitochondrial association of the male-specific Don Juan protein in Drosophila spermatozoa. J. Cell Sci. 1998;111:3299–3309. doi: 10.1242/jcs.111.22.3299. [DOI] [PubMed] [Google Scholar]
- Schafer M, Kuhn R, Bosse F, Schafer U. A conserved element in the leader mediates post-meiotic translation as well as cytoplasmic polyadenylation of a Drosophila spermatocyte mRNA. EMBO J. 1990;9:4519–4525. doi: 10.1002/j.1460-2075.1990.tb07903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist S, Ried G, Lehner CF. Dmcdc2 kinase is required for both meiotic divisions during Drosophila spermatogenesis and is activated by the Twine/cdc25 phosphatase. Mech. Dev. 1995;53:247–260. doi: 10.1016/0925-4773(95)00441-3. [DOI] [PubMed] [Google Scholar]
- Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Seli E, Steitz JA. Metazoan oocyte and early embryo development program: a progression through translation regulatory cascades. Genes Dev. 2006;20:138–146. doi: 10.1101/gad.1398906. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT, Lindsley DL, Herrera C. Toward a comprehensive genetic analysis of male fertility in Drosophila melanogaster. Genetics. 2004;167:207–216. doi: 10.1534/genetics.167.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H, Schafer MA, Alphey LS, Fuller MT. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development. 1998;125:125–134. doi: 10.1242/dev.125.1.125. [DOI] [PubMed] [Google Scholar]