Abstract
Immunoglobulin hypermutation provides the structural correlate for the affinity maturation of the antibody response. Characteristic modalities of this mechanism include a preponderance of point-mutations with prevalence of transitions over transversions, and the mutational hotspot RGYW sequence. Recent evidence suggests a mechanism whereby DNA-breaks induce error-prone DNA synthesis in immunoglobulin V(D)J regions by error-prone DNA polymerases. The nature of the targeting mechanism and the trans-factors effecting such breaks and their repair remain to be determined.
Introduction
The generation of B lymphocytes capable of producing high-affinity antibodies occurs through the accumulation of mutations in the variable (V) regions of rearranged immunoglobulin (Ig) genes [1] and the selection for B cells whose Ig antigen-receptors (i.e. BCRs) have accumulated mutations that increase affinity for the immunizing antigen [2]. Hypermutation of antigen-receptor genes has been described for a wide variety of vertebrates including sharks, frogs, sheep, mice and humans (reviewed in [3]). Although some differences among species do exist in the pattern of mutations effected by the Ig hypermutation machinery, many similarities reveal a common mechanism. Particularly striking similarities among species include a vast preponderance of base substitutions, identical mutational hotspots and, in the absence of antigenic-selection, a bias to generate transitions over transversions. In most species, somatic Ig hypermutation is triggered upon exposure to antigen, although sheep additionally utilize Ig hypermutation to generate the primary repertoire — as do chickens, pigs and rabbits, but in the context of gene conversion (reviewed in [3]).
Many aspects of the molecular machinery responsible for Ig hypermutation remain unknown, but some studies have revealed the cis-acting elements within the Ig locus that are required for hypermutation. These include the intronic enhancer for the κ chain and the heavy chain, and the V-region promoter, although the Ig promoter itself is not necessary as it can be replaced by a heterologous promoter (reviewed in [4]). The V region of the sequence is not necessary to target hypermutation as it too can be replaced by heterologous sequence. A putative role for the transcriptional machinery is likely, but it remains controversial whether transcription plays a direct role in hypermutation or whether it simply facilitates accessibility of the locus to the mutational machinery.
Recent advances have begun to reveal some of the molecular components and intermediate products of the Ig hypermutation molecular machinery. It now appears that DNA lesions, either double-strand breaks (DSBs) or single-strand breaks, are a feature of this mechanism [5,6••,7••,8]. Indeed, templated nucleotide additions in the IgV region were detected in the human B-cell line Ramos that hypermutates constitutively following transfection with a TdT (terminal deoxy-nucleotidyl transferase) transgene. The mutational rate and pattern suggest error-prone synthesis by one or more of the translesion synthesis polymerases typically involved in the mutagenic bypass of DNA lesions, and recent evidence strongly implicates their involvement [9•,10••,11••].
In addition, recent studies have revealed a mechanistic link between Ig hypermutation and Ig gene conversion [12••], as well as the existence of a germinal-center-restricted cytidine deaminase (activation-induced cytidine deaminase [AID]) that appears to play a critical role both in Ig hypermutation and class-switch recombination (CSR) [13••].
A critical but yet-unknown aspect of Ig hypermutation is the mechanism that ensures the targeting of lesions to the IgV region, while sparing the constant domains, which are only a few kilobases (kb) downstream from the V region. Addressing this feature of Ig hypermutation is likely to reveal molecules that are unique to this process.
Here, we will review these findings and integrate them into a multilayered model of Ig hypermutation that at least includes the following: targeting by Ig-hypermutation-specific elements; introduction of DNA lesions into IgV regions; and gap synthesis by error-prone DNA polymerases. We then examine a possible role for AID in the regulation of Ig hypermutation.
Targeting of hypermutation to the V(D)J regions of rearranged Ig loci
Ig hypermutation specifically targets a region of about 1.5 kb that includes the region containing the V, D (diversity) and J (joining) region — the V(D)J region — and part of the JC intron. The C (constant) domains of the heavy and the κ chains are largely spared, although some very low-grade mutation can be detected in the first C domain of the mouse λ chain, perhaps because of the short length of the JC intron.
The 5′ boundary of the mutational track is sharp and it is near the transcriptional initiation site whereas the 3′ end tapers throughout a region downstream of the J clusters, within 1.5–2.0 kb of the transcription initiation site [14,15]. Interestingly, the mutational track maps with the location of the promoter as placing DNA segments, as spacers just downstream of the promoter displaces the mutational track upstream [16–18] prompting many investigators to propose a direct role of transcription in Ig hypermutation. Mutations are rarely seen upstream of the leader intron, suggesting that, although the location of the promoter is important for the local distribution of mutations, there is a paucity of mutations for a short track of about 200 bps (basepairs) just downstream of the promoter [19].
Hypermutation of the targeted region appears to be under the direction of transcription-related elements including the promoter and enhancer regions whereas sequences upstream of the promoter are dispensable as their deletion or replacement does not impact hypermutation [16]. Storb and colleagues found that duplicating the V-gene promoter upstream of the C region and moving the intronic enhancer downstream of the C region in a κ transgene conferred hypermutability to the C region [20], further emphasizing the role of these elements in hypermutation. Interestingly, the IgV promoter, as well as the V sequence itself, can be replaced by a heterologous promoter or heterologous sequence, respectively, without impacting hypermutability [21–23]. The Ig enhancer regions are critical (reviewed in [19]), particularly the intronic enhancer in the case of the κ locus, and much research is currently under way to determine the region within the enhancers that confers hypermutability. Therefore it appears that whereas the Ig enhancers and a promoter are required for hypermutation, the IgV promoter is not, suggesting that the region that targets the hypermutation machinery to the Ig locus is not within the promoter.
Recently, it was demonstrated that, in addition to the Ig locus, at least one other gene in humans, encoding BCL-6, can be targeted by the hypermutation mechanism [24,25]. The targeting of hypermutation in human BCL-6 requires the application to the B cells of the same stimuli that are critical for the induction of Ig hypermutation: T-cell/B-cell contact, involving CD40/CD154, CD80/CD28 engagement and crosslinking to the BCR [26•]. Nevertheless, because BCL-6 hypermutation occurs in human B cells but not in mouse B cells [19], because mutations are restricted to untranslated regions and because there is no obvious biological reason for BCL-6 to be hypermutated, human BCL-6 hypermutation is probably the accidental acquisition in the evolution of the human BCL-6 gene of sequences that help target the Ig hypermutation machinery. It will be interesting to identify regions within the human BCL-6 gene that are different from the mouse ortholog and that resemble enhancer regions in the Ig loci.
Introduction of DNA lesions
The discovery of B-cell lines that can either be induced to hypermutate [27] or that hypermutate constitutively [5] has enabled researchers to probe for the presence of DNA lesions in the IgV regions that correlate with hypermutation. Utilizing the Ramos human B-cell line, which hypermutates spontaneously, Sale and Neuberger [5] unveiled evidence for either DSBs or staggered nicks, throughout the IgV region. These authors introduced a TdT-expressing transgene into the Ramos B-cell line and found nontemplated nucleotide additions in the IgV regions; these breaks tended to be at, or near, known mutational hotspots.
Recently, Papavasiliou and Schatz [6••], and Bross and colleagues [7••] found evidence for DNA DSBs using ligation-mediated PCR in Ramos B cells and/or in mouse germinal-center B cells. In these studies, the DSBs tended to occur near the mutational hotspot RGYW. In the study by Papavasiliou and Schatz, the DSBs were found in the late-S and G2 phases of the cell cycle, prompting these authors to suggest that a homologous-recombination mechanism of break repair, with the sister chromatid as template, is operative in Ig hypermutation. The resolution of these breaks, however, remains undefined but it does not require the nonhomologous DNA end-joining (NHEJ) mechanism of DSB repair [28••], strengthening the argument for homologous recombination. In addition, targeted deletion of molecules important to a variety of DNA-repair processes, such as nucleotide repair and base excision repair, did not impair hypermutation [29].
Further evidence for DNA breaks in hypermutation comes from a recent study where evidence for the long-suspected mechanistic link between Ig hypermutation and Ig gene conversion [3,30,31] was detected in a chicken B-cell line, which normally undergoes gene conversion in the IgV region. Strikingly, Sale and colleagues [12••] found that chicken DT40 cell subclones that were defective for various molecules involved in the repair of breaks via homologous recombination (XRCC2, XRCC3 and RAD51B) underwent significantly higher rates of somatic hypermutation than gene conversion. Considering that gene conversion, like hypermutation, requires a rearranged V(D)J (the acceptor), and that transcription or transcription-related sequences are necessary [32], it appears likely that Ig hypermutation and gene conversion share both the targeting phase and the introduction of DNA breaks. The mechanism utilized to process those breaks, however, probably differs between the two mechanisms — perhaps one (gene conversion) utilizing a pseudogene and the other (hypermutation) utilizing a sister chromatid as donors, during break repair via homologous recombination [12••].
Error-prone synthesis
The frequency of mutations generated by the Ig hypermutation mechanism is estimated to be about one-million-fold over background with an estimated rate of 10−3/bp/generation [2,33]. Furthermore, the generation of a mismatched pair during synthesis requires that a putative hypermutation polymerase be capable of commonly inserting an incorrect base and also of extending from the newly generated mismatched terminus. It is likely that such putative error-prone synthesis during somatic hyper-mutation occurs outside of global replication and in the context of short patch error-prone repair [34].
Interestingly, in the hypermutation of the novel antigen-receptor (NAR) in sharks — where the mutational pattern is otherwise very similar to mammalian hypermutation —and in a shark Ig light-chain locus (NS3), many mutations occur in doublets, suggesting two misincorporation and mismatch-extension events [35,36].
The unprecedented mutation frequency and the likely requirement for mismatch extension is very difficult, if not impossible, to attain if the involved polymerases synthesize DNA with high fidelity and have proofreading capabilities. It is instead much more likely that the hypermutation polymerase is one (or several) of the growing family of DNA polymerases that have evolved to allow continued synthesis opposite DNA lesions that would otherwise stall a replication fork: the translesion synthesis polymerases. These polymerases — which among others include DNA polymerases η, ι and ζ — are error-prone and capable of extending, with various efficiencies, from a mismatched terminus [37•,38,39].
Whereas DNA polymerases η and ι are highly error-prone when acting on undamaged DNA, DNA polymerase ζ may not be as error-prone but is very efficient at extending from a mismatched terminus. In fact, recent evidence suggests that these polymerases can act in concert, where either η or ι inserts an incorrect base, and the ubiquitous polymerase ζ extends from the mismatched pair [40,41], thus stabilizing the mutation. Given the recent data suggesting a role for DSBs in Ig hypermutation, the finding in yeast that increased incidences of base-substitutions near DSBs are entirely dependent on DNA polymerase ζ [42] makes this polymerase a natural candidate for Ig hypermutation [3].
Several recent studies have provided evidence for at least two of these polymerases in Ig hypermutation. Zeng and colleagues [9•] found that in patients with the variant form of Xeroderma pigmentosum, who have been shown to have mutations in the DNA polymerase η gene, Ig mutations from A and T nucleotides were greatly reduced, prompting the investigators to suggest that polymerase η is an AT mutator in Ig hypermutation. Rogozin and colleagues [43•] further found that polymerase η tended to be more likely than other polymerases to generate mutations at Ig hyper-mutation hotspot dinucleotides. Zan and colleagues [10••] obtained a dramatic reduction in Ig and BCL-6 mutations when the hypermutation-inducible human B-cell line CL-01 was exposed to antisense oligonucleotides that target the RNA for the catalytic subunit of DNA polymerase ζ (Rev3). Such a reduction in Ig hypermutation was associated with a comparable reduction in UV-damaged induced mutagenesis, as typically seen for cells with low levels of this polymerase.
Furthermore, Diaz and colleagues [11••] generated a mouse transgenic model where antisense RNA is made to the RNA encoding part of the REV3 gene. In this in vivo study, transgenic mice generated vigorous immune responses, and large and abundant germinal centers, but most failed to generate high-affinity antibodies; the accumulation of mutations in IgV regions was reduced, with a pronounced reduction in mutations that enhance affinity to antigen. In contrast to the polymerase η study, no appreciable alteration in the targeting of nucleotides was detected in the studies where the role for polymerase ζ was examined.
Perhaps this difference is explained by a mechanism whereby both polymerases act together, one (polymerase η) as a mispair inserter and the other (polymerase ζ) as a mispair extender. It is also possible that polymerase ι is involved in Ig hypermutation because the pattern of mutations it generates in undamaged DNA is strikingly similar to Ig hypermutation, particularly in its asymmetrical targeting of A rather than T nucleotides depending on the DNA strand that it is acting upon [44]. However, to date, no direct analysis of a putative role of this polymerase in Ig hypermutation has been examined. Finally, other error-prone DNA polymerases such as β, μ and λ have been shown not to be required for Ig hypermutation ([45]; C-A Reynaud, J-C Weill, personal communication).
Ig hypermutation regulation — a possible role for AID
Recently an RNA-editing enzyme that is specific to activated germinal-center B cells and/or cells undergoing CSR was discovered by Muramatsu and colleagues [13••]. Inactivation of this gene in mice resulted in abrogation of CSR and somatic hypermutation, strongly suggesting a mechanistic and/or regulatory link between Ig hypermutation and CSR [46]. Humans with mutations in the AID gene develop a type of hyper-IgM syndrome with absolute impairment in CSR and significant but not complete impairment in Ig hypermutation [47••]. Given its potential function as an RNA-editing enzyme, it is very likely that AID plays a role in the modification of RNA transcripts that encode molecules critical to Ig hypermutation and CSR [46,48,49] (and perhaps Ig gene conversion?). Whether this molecule is involved in the regulation of hypermutation targeting, lesion introduction or error-prone repair remains a fascinating question that is likely to unveil novel molecules important for these mechanisms.
Conclusions
An emerging model of somatic hypermutation based on the most recent data from different laboratories, including ours, incorporates the targeted introduction of DNA breaks into Ig V(D)J regions, followed by error-prone repair, perhaps via homologous recombination using a sister chromatid as a template (Figure 1). It is likely that the mutational hotspots are the sites where the breaks occur, although one cannot rule out that they are a signature of the error-prone DNA polymerases involved. DSBs are likely to be necessary but not sufficient for the introduction of somatic mutations. However, a direct causal relationship between DNA breaks and mutation induction has not been determined and it is possible that these breaks are the by-products rather than the cause of hypermutation. The nature of the molecules responsible for effecting the DNA breaks is undefined but Rag-1 does not appear to be involved [28••].
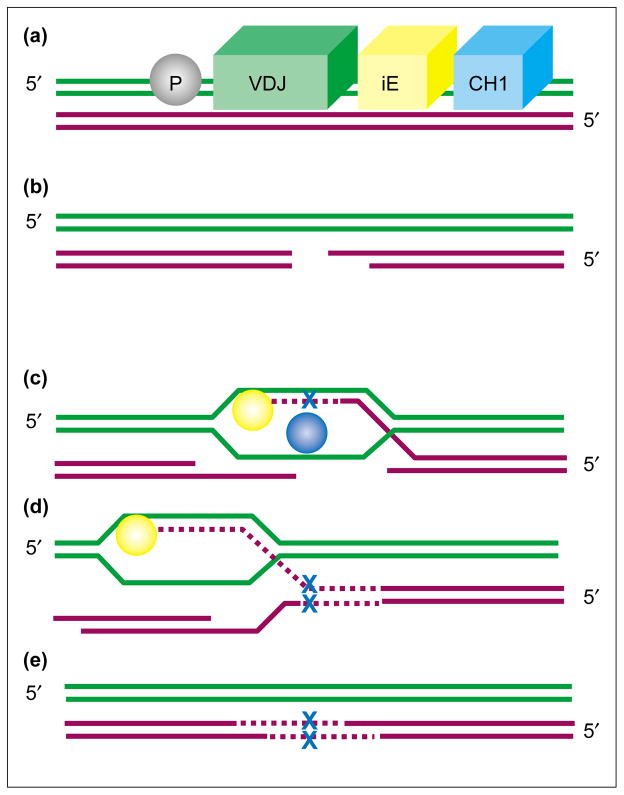
Figure 1.
The emerging model of somatic hypermutation. (a) Specific elements in the intronic enhancer (iE) target hypermutation to the V(D)J region (the heavy-chain is used as an example; CH1 is its first constant region). A promoter (P) (although not necessarily the Ig promoter) is required for hypermutation. (b) The introduction of DNA-breaks in the V(D)J region leads to the opening of a gap. (c) Homologous recombination is initiated and uses the sister chromatid as a template. BCR cross-linking regulates the expression of translesion DNA polymerases, including ζ and η. This results in error-prone gap DNA synthesis, including mispair insertion (blue cross) by one of several translesion polymerases (blue sphere). (d) Mispair extension by polymerase ζ (yellow sphere) will then occur. (e) The resultant hypermutation allows for high-affinity antibodies to be produced.
In addition, the role of AID and the importance of mismatch-repair proteins in hypermutation remain unclear. Mismatch-repair proteins have been implicated in Ig hypermutation [49–51], but the potential for indirect effects, such as genomic instability and reduced proliferative potential from mismatch repair deficiency [52,53], has obscured their importance in Ig hypermutation. The fact that mismatch-repair-deficient mice display a significant alteration in the Ig mutational pattern (namely a bias for targeting of GC nucleotides) suggests a direct role in Ig hypermutation, because it is difficult to explain how a defect in proliferation would yield an alteration in the pattern of hypermutation. DNA polymerase ζ seems to play a critical role in Ig and BCL-6 hypermutation, but the nature of the break-repair mechanism and how high-fidelity polymerases are excluded from it remain to be better defined. DNA polymerase η may also play a role, although it may be minor ([54]; but see [55,56]).
Finally, the mechanism that targets hypermutation to the Ig locus and human BCL-6 remains unknown, although there is strong evidence for an important role of the Ig enhancers and perhaps transcription in this process [57•]. The spontaneously hypermutating and inducible mono-clonal B-cell lines that have become recently available should prove to be important transfectable vehicles for the definition of the role of different cis elements in hyper-mutation of the Ig and BCL-6 loci, as well as for the identification of the trans-factors and DNA polymerases that effect and mediate the DNA lesions and their repair.
Acknowledgments
We are grateful to Norman Klinman and Hong Zan for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health (NIH) grants AR 40908, AI 45011, AG 13910 and AI 07621 to PC; MD is currently supported by NIH Training Grant T32 A1-07244.
Abbreviations
- AID
activation-induced cytidine deaminase
- C
constant
- CSR
class-switch recombination
- D
diversity
- DSB
double-strand break
- Ig
immunoglobulin
- J
joining
- kb
kilobases
- TdT
terminal deoxy-nucleotidyl transferase
- V
variable
Contributor Information
Marilyn Diaz, Email: mdiaz@scripps.edu.
Paolo Casali, Email: pcasali@med.cornell.edu.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Weigert MG, Cesari IM, Yonkovich SJ, Cohn M. Variability in the lambda light chain sequences of mouse antibody. Nature. 1970;228:1045–1047. doi: 10.1038/2281045a0. [DOI] [PubMed] [Google Scholar]
- 2.Clarke SH, Huppi K, Ruezinsky D, Staudt L, Gerhard W, Weigert M. Inter- and intraclonal diversity in the antibody response to influenza hemagglutinin. J Exp Med. 1985;161:687–704. doi: 10.1084/jem.161.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diaz M, Flajnik MF. Evolution of somatic hypermutation and gene conversion in adaptive immunity. Immunol Rev. 1998;162:13–24. doi: 10.1111/j.1600-065x.1998.tb01425.x. [DOI] [PubMed] [Google Scholar]
- 4.Storb U, Peters A, Klotz E, Kim N, Shen HM, Hackett J, Rogerson B, Martin TE. Cis-acting sequences that affect somatic hypermutation of Ig genes. Immunol Rev. 1998;162:153–160. doi: 10.1111/j.1600-065x.1998.tb01438.x. [DOI] [PubMed] [Google Scholar]
- 5.Sale JE, Neuberger MS. TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity. 1998;9:859–869. doi: 10.1016/s1074-7613(00)80651-2. [DOI] [PubMed] [Google Scholar]
- 6••.Papavasiliou FN, Schatz DG. Cell-cycle-regulated DNA double stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 2000;408:216–221. doi: 10.1038/35041599. Strong evidence for DNA DSBs is provided in this study utilizing both Ramos B cells and mouse germinal-center B cells. The breaks were found in the late-S and G2 phases of the cell cycle and correlated with transcription. [DOI] [PubMed] [Google Scholar]
- 7••.Bross L, Fukita Y, McBlane F, Demolliere C, Rajewsky K, Jacobs H. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity. 2000;13:589–597. doi: 10.1016/s1074-7613(00)00059-5. Further evidence for DSBs is provided in this study with germinal-center B cells. [DOI] [PubMed] [Google Scholar]
- 8.Kong Q, Maizels N. DNA breaks in hypermutating immunoglobulin genes: evidence for a break-and-repair pathway of somatic hypermutation. Genetics. 2001;158:369–378. doi: 10.1093/genetics/158.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol. 2001;2:537–541. doi: 10.1038/88740. In this study, Ig mutations from A and T nucleotides were greatly reduced in Xeroderma pigmentosum patients with a defect in the DNA polymerase η gene. A large increase in expression of this polymerase in mouse germinal-center B cells was found, but this finding is at odds with a dramatic down-regulation of DNA polymerase η expression in human B cells that are induced to hypermutate in vitro. [DOI] [PubMed] [Google Scholar]
- 10••.Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P. The translesion DNA polymerase zeta plays a major role in Ig and BCL-6 somatic hypermutation. Immunity. 2001;14:643–653. doi: 10.1016/s1074-7613(01)00142-x. This study provides strong evidence for a role of DNA polymerase ζ in somatic hypermutation because Ig mutations were severely reduced when a hypermutation-inducible human B-cell line underwent targeted reduction of REV3 transcripts. The Ig mutation reduction was paralleled by a similar reduction in UV-induced DNA mutagenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Diaz M, Verkoczy LK, Flajnik MF, Klinman NR. Decreased frequency of somatic hypermutation and impaired affinity maturation but intact germinal center formation in mice expressing antisense RNA to DNA polymerase zeta. J Immunol. 2001;167:327–335. doi: 10.4049/jimmunol.167.1.327. This study provides in vivo evidence for an involvement of DNA polymerase ζ in hypermutation. Mice expressing antisense transcripts to the RNA encoding mouse Rev3 mounted vigorous immune responses with class-switched antibodies and large germinal centers, but failed to generate high-affinity antibodies and had a reduction in the accumulation of Ig mutations. [DOI] [PubMed] [Google Scholar]
- 12••.Sale JE, Calandrini DM, Takata M, Takeda S, Neuberger MS. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–926. doi: 10.1038/35091100. In this study, the deletion of various RAD51 paralogues in the chicken B-cell line DT40 that normally undergoes gene conversion resulted in a dramatic increase in somatic hypermutation at the expense of gene conversion. This is the first evidence for a mechanistic link between Ig gene conversion and somatic hypermutation. [DOI] [PubMed] [Google Scholar]
- 13••.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. The discovery that a novel cytidine deaminase is highly expressed in germinal-center B cells, and the ablation of somatic hypermutation and CSR by its targeted deletion, strongly implicate this novel molecule in both mechanisms. [DOI] [PubMed] [Google Scholar]
- 14.Lebecque SG, Gearhart PJ. Boundaries of somatic mutation in rearranged immunoglobulin genes: 5′ boundary is near the promoter, and 3′ boundary is approximately 1 kb from V(D)J gene. J Exp Med. 1990;172:1717–1727. doi: 10.1084/jem.172.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogerson BJ. Mapping the upstream boundary of somatic mutations in rearranged immunoglobulin transgenes and endogenous genes. Mol Immunol. 1994;31:83–98. doi: 10.1016/0161-5890(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 16.Tumas-Brundage K, Vora KA, Giusti AM, Manser T. Characterization of the cis-acting elements required for somatic hypermutation of murine antibody V genes using conventional transgenic and transgene homologous recombination approaches. Semin Immunol. 1996;8:141–150. doi: 10.1006/smim.1996.0018. [DOI] [PubMed] [Google Scholar]
- 17.Tumas-Brundage K, Manser T. The transcriptional promoter regulates hypermutation of the antibody heavy chain locus. J Exp Med. 1997;185:239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber JS, Berry J, Manser T, Claflin JL. Mutations in Ig V(D)J genes are distributed asymmetrically and independently of the position of V(D)J. J Immunol. 1994;153:3594–3602. [PubMed] [Google Scholar]
- 19.Neuberger MS, Ehrenstein MR, Klix N, Jolly CJ, Yelamos J, Rada C, Milstein C. Monitoring and interpreting the intrinsic features of somatic hypermutation. Immunol Rev. 1998;162:107–116. doi: 10.1111/j.1600-065x.1998.tb01434.x. [DOI] [PubMed] [Google Scholar]
- 20.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 21.Azuma T, Motoyama N, Fields LE, Loh DY. Mutations of the chloramphenicol acetyl transferase transgene driven by the immunoglobulin promoter and intron enhancer. Int Immunol. 1993;5:121–130. doi: 10.1093/intimm/5.2.121. [DOI] [PubMed] [Google Scholar]
- 22.Betz AG, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger MS. Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 23.Klix N, Jolly CJ, Davies SL, Bruggemann M, Williams GT, Neuberger MS. Multiple sequences from downstream of the J kappa cluster can combine to recruit somatic hypermutation to a heterologous, upstream mutation domain. Eur J Immunol. 1998;28:317–326. doi: 10.1002/(SICI)1521-4141(199801)28:01<317::AID-IMMU317>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 24.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 25.Migliazza A, Martinotti S, Chen W, Fusco C, Ye BH, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R. Frequent somatic hypermutation of the 5′ noncoding region of the BCL-6 gene in B-cell lymphoma. Proc Natl Acad Sci USA. 1995;92:12520–12524. doi: 10.1073/pnas.92.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Zan H, Li Z, Yamaji K, Dramitinos P, Cerutti A, Casali P. B cell receptor engagement and T cell contact induce BCL-6 somatic hypermutation in human B cells: identity with Ig hypermutation. J Immunol. 2000;165:830–839. doi: 10.4049/jimmunol.165.2.830. In this study, it was demonstrated that BCL-6 can also be induced to hypermutate in a human B-cell line and its induction requires the same stimuli as Ig hypermutation. [DOI] [PubMed] [Google Scholar]
- 27.Zan H, Cerutti A, Dramitinos P, Schaffer A, Li Z, Casali P. Induction of Ig somatic hypermutation and class switching in a human monoclonal IgM+ IgD+ B cell line in vitro: definition of the requirements and modalities of hypermutation. J Immunol. 1999;162:3437–3447. [PMC free article] [PubMed] [Google Scholar]
- 28••.Bemark M, Sale JE, Kim HJ, Berek C, Cosgrove RA, Neuberger MS. Somatic hypermutation in the absence of DNA-dependent protein kinase catalytic subunit (DNA-PK(cs)) or recombination-activating gene (RAG)1 activity. J Exp Med. 2000;192:1509–1514. doi: 10.1084/jem.192.10.1509. This study provides strong evidence that neither Rag-1 nor DNApK is required for somatic hypermutation, distinguishing hypermutation further from V(D)J recombination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs H, Fukita Y, van der Horst GT, de Boer J, Weeda G, Essers J, de Wind N, Engelward BP, Samson L, Verbeek S, et al. Hypermutation of immunoglobulin genes in memory B cells of DNA repair-deficient mice. J Exp Med. 1998;187:1735–1743. doi: 10.1084/jem.187.11.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maizels N. Might gene conversion be the mechanism of somatic hypermutation of mammalian immunoglobulin genes? Trends Genet. 1989;5:4–8. doi: 10.1016/0168-9525(89)90004-8. [DOI] [PubMed] [Google Scholar]
- 31.Weill JC, Reynaud CA. Rearrangement/hypermutation/gene conversion: when, where and why? Immunol Today. 1996;17:92–97. doi: 10.1016/0167-5699(96)80586-x. [DOI] [PubMed] [Google Scholar]
- 32.Carlson LM, McCormack WT, Postema CE, Humphries EH, Thompson CB. Templated insertions in the rearranged chicken IgL V gene segment arise by intrachromosomal gene conversion. Genes Dev. 1990;4:536–547. doi: 10.1101/gad.4.4.536. [DOI] [PubMed] [Google Scholar]
- 33.Chien NC, Pollock RR, Desaymard C, Scharff MD. Point mutations cause the somatic diversification of IgM and IgG2a antiphosphorylcholine antibodies. J Exp Med. 1988;167:954–973. doi: 10.1084/jem.167.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertocci B, Quint L, Delbos F, Garcia C, Reynaud CA, Weill JC. Probing immunoglobulin gene mutation with microsatellites suggests a nonreplicative short patch DNA synthesis process. Immunity. 1998;9:257–265. doi: 10.1016/s1074-7613(00)80608-1. [DOI] [PubMed] [Google Scholar]
- 35.Diaz M, Velez J, Singh M, Cerny J, Flajnik MF. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: the translesion synthesis model of somatic hypermutation. Int Immunol. 1999;11:825–833. doi: 10.1093/intimm/11.5.825. [DOI] [PubMed] [Google Scholar]
- 36.Lee SS, Greenberg A, Hsu E. Evolution and somatic diversification of immunoglobulin light chains. Curr Top Microbiol Immunol. 2000;248:285–300. doi: 10.1007/978-3-642-59674-2_13. [DOI] [PubMed] [Google Scholar]
- 37•.Lawrence CW, Maher VM. Mutagenesis in eukaryotes dependent on DNA polymerase zeta and Rev1p. Philos Trans R Soc London B Biol Sci. 2001;356:41–46. doi: 10.1098/rstb.2000.0746. Informative review of the translesion synthesis polymerases, particularly of DNA polymerase ζ, both in humans and in yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase eta. J Mol Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 39.McDonald JP, Tissier A, Frank EG, Iwai S, Hanaoka F, Woodgate R. DNA polymerase iota and related rad30-like enzymes. Philos Trans R Soc London B Biol Sci. 2001;356:53–60. doi: 10.1098/rstb.2000.0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 41.Yuan F, Zhang Y, Rajpal DK, Wu X, Guo D, Wang M, Taylor JS, Wang Z. Specificity of DNA lesion bypass by the yeast DNA polymerase eta. J Biol Chem. 2000;275:8233–8239. doi: 10.1074/jbc.275.11.8233. [DOI] [PubMed] [Google Scholar]
- 42.Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147:1017–1024. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat Immunol. 2001;2:530–536. doi: 10.1038/88732. Mutational hotspots generated by DNA polymerase η in undamaged DNA are similar to those of known hotspots for Ig hypermutation. [DOI] [PubMed] [Google Scholar]
- 44.Frank EG, Tissier A, McDonald JP, Rapic-Otrin V, Zeng X, Gearhart PJ, Woodgate R. Altered nucleotide misinsertion fidelity associated with poliota-dependent replication at the end of a DNA template. EMBO J. 2001;20:2914–2922. doi: 10.1093/emboj/20.11.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esposito G, Texido G, Betz UA, Gu H, Muller W, Klein U, Rajewsky K. Mice reconstituted with DNA polymerase beta-deficient fetal liver cells are able to mount a T cell-dependent immune response and mutate their Ig genes normally. Proc Natl Acad Sci USA. 2000;97:1166–1171. doi: 10.1073/pnas.97.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durandy A, Honjo T. Human genetic defects in class-switch recombination (hyper-IgM syndromes) Curr Opin Immunol. 2001;13:543–548. doi: 10.1016/s0952-7915(00)00256-9. [DOI] [PubMed] [Google Scholar]
- 47••.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. This study demonstrates that a subset of a hyper-IgM-syndrome patients have a defect in the human AID gene. [DOI] [PubMed] [Google Scholar]
- 48.Tian M, Alt FW. RNA editing meets DNA shuffling. Nature. 2000;407:31–33. doi: 10.1038/35024189. [DOI] [PubMed] [Google Scholar]
- 49.Phung QH, Winter DB, Cranston A, Tarone RE, Bohr VA, Fishel R, Gearhart PJ. Increased hypermutation at G and C nucleotides in immunoglobulin variable genes from mice deficient in the MSH2 mismatch repair protein. J Exp Med. 1998;187:1745–1751. doi: 10.1084/jem.187.11.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rada C, Ehrenstein MR, Neuberger MS, Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- 51.Wiesendanger M, Kneitz B, Edelmann W, Scharff MD. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J Exp Med. 2000;191:579–584. doi: 10.1084/jem.191.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frey S, Bertocci B, Delbos F, Quint L, Weill JC, Reynaud CA. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity. 1998;9:127–134. doi: 10.1016/s1074-7613(00)80594-4. [DOI] [PubMed] [Google Scholar]
- 53.Vora KA, Tumas-Brundage KM, Lentz VM, Cranston A, Fishel R, Manser T. Severe attenuation of the B cell immune response in Msh2-deficient mice. J Exp Med. 1999;189:471–482. doi: 10.1084/jem.189.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dorner T, Lipsky PE. Smaller role for pol η? Nat Immunol. 2001;2:982–983. doi: 10.1038/ni1101-982. [DOI] [PubMed] [Google Scholar]
- 55.Rogozin IB, Pavlov YI, Kunkel TA. Response 1. Nat Immunol. 2001;2:983–984. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 56.Gearhart PJ, Zeng X. Response 2. Nat Immunol. 2001;2:984. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 57•.Storb U, Shen HM, Michael N, Kim N. Somatic hypermutation of immunoglobulin and non-immunoglobulin genes. Philos Trans R Soc London B Biol Sci. 2001;356:13–19. doi: 10.1098/rstb.2000.0743. An informative and stimulating review of the findings regarding the role of cis-acting elements and transcription in Ig and BCL-6 hypermutation. [DOI] [PMC free article] [PubMed] [Google Scholar]