ABSTRACT
GINS is a key component of the eukaryotic Cdc45-minichromosome maintenance (MCM)-GINS (CMG) complex, which unwinds duplex DNA at the moving replication fork. Archaeal GINS complexes have been shown to stimulate the helicase activity of their cognate MCM mainly by elevating its ATPase activity. Here, we report that GINS from the thermoacidophilic crenarchaeon Sulfolobus solfataricus (SsoGINS) is capable of DNA binding and binds preferentially to single-stranded DNA (ssDNA) over double-stranded DNA (dsDNA). Notably, SsoGINS binds more strongly to dsDNA with a 5′ ssDNA tail than to dsDNA with a 3′ tail and more strongly to an ssDNA fragment blocked at the 3′ end than to one at the 5′ end with a biotin-streptavidin (SA) complex, suggesting the ability of the protein complex to slide in a 5′-to-3′ direction along ssDNA. DNA-bound SsoGINS enhances DNA binding by SsoMCM. Furthermore, SsoGINS increases the helicase activity of SsoMCM. However, the ATPase activity of SsoMCM is not affected by SsoGINS. Our results suggest that SsoGINS facilitates processive DNA unwinding by SsoMCM by enhancing the binding of the helicase to DNA. We propose that SsoGINS stabilizes the interaction of SsoMCM with the replication fork and moves along with the helicase as the fork progresses.
IMPORTANCE GINS is a key component of the eukaryotic Cdc45-MCM-GINS complex, a molecular motor that drives the unwinding of DNA in front of the replication fork. Archaea also encode GINS, which interacts with MCM, the helicase. But how archaeal GINS serves its role remains to be understood. In this study, we show that GINS from the hyperthermophilic archaeon Sulfolobus solfataricus is able to bind to DNA and slide along ssDNA in a 5′-to-3′ direction. Furthermore, Sulfolobus GINS enhances DNA binding by MCM, which slides along ssDNA in a 3′-to-5′ direction. Taken together, these results suggest that Sulfolobus GINS may stabilize the interaction of MCM with the moving replication fork, facilitating processive DNA unwinding.
INTRODUCTION
Chromosomal DNA replication is a highly regulated process involving a large number of essential and optional protein factors (1, 2). Efficient DNA unwinding is an essential prerequisite for the duplication of the genetic material. In Eukarya, the minichromosome maintenance (MCM) complex comprising six subunits (MCM2 to MCM7) is the replicative helicase. However, the heterohexameric MCM itself shows little helicase activity (3, 4), and two accessory factors, i.e., the tetrameric GINS complex (Sld5, Pfs1, Psf2, and Psf3) and Cdc45, are required to activate MCM (5). The complex of MCM, Cdc45, and GINS, referred to as the CMG complex, is considered to be the active form of eukaryotic helicase (5, 6). The eukaryotic GINS complex is able to bind to the origin of replication and move along with the replication fork (6–11). Cdc45 is also required for the progression of the replication fork (12, 13). Three-dimensional reconstructions of the overall arrangement of the Drosophila CMG based on single-particle electron microscopy reveal that Cdc45 and GINS serve as a “latch” to bridge the MCM2/MCM5 gate, forming a topologically closed structure with a large interior channel (14). Biochemical evidence shows that the ATPase and the DNA binding activities of CMG are significantly higher than those of MCM2 to MCM7, suggesting that GINS, together with Cdc45, activates MCM2 to MCM7 mainly by stimulating the ATPase activity of the complex and promoting its binding to DNA (15). In addition to its ability to bind to MCM, GINS also interacts with polymerase α (Pol α)-primase and Pol ε (16, 17), indicating the important roles of GINS in both the initiation and the elongation phases of DNA replication. It has been suggested that GINS may act as a molecular linker that mediates the assembly of replication proteins around the MCM helicase during DNA replication (18).
The archaeal DNA replication machinery represents a simplified and ancestral version of the eukaryotic one (19). Unlike genomes of eukarya, most known archaeal genomes encode only one MCM homolog that forms a homohexamer. Notably, the archaeal MCM homohexamer alone exhibits robust helicase activity and unwinds DNA in a 3′-to-5′ direction (20–26). Unlike eukarya, in which the GINS complex comprises four distantly related subunits, archaea possess one or two GINS-encoding genes (27). While most crenarchaea encode two GINS proteins that are homologous to Sld2/Sld3 and Psf1/Sld5, respectively, the majority of euryarchaea have a single Psf1/Sld5 homolog (27, 28). Therefore, the archaeal GINS complex is either a tetramer of two different subunits assembled at a 2:2 ratio or a homotetramer of a single subunit (27). Structural studies show that the heterotetrameric GINS complex from the euryarchaeon Thermococcus kodakaraensis exhibits an overall similarity in architecture to that of human GINS (29). The physical and functional interactions between GINS and MCM have been well documented in archaea. GINS from the euryarchaeon Pyrococcus furiosus interacts with the P. furiosus MCM (PfuMCM) and increases the helicase activity of PfuMCM by stimulating its ATPase activity. It is worth noting that PfuGINS does not bind to DNA and is unable to assist the loading of PfuMCM (28). GINS complexes from the euryarchea Thermococcus kodakaraensis and Thermoplasma acidophilum also interact with their cognate MCMs and stimulate both the ATPase and the helicase activities of the MCMs (30, 31). It was noticed that although T. acidophilum GINS alone was unable to bind DNA, it was able to form a glutaraldehyde-cross-linkable complex with its cognate MCM on single-stranded DNA (ssDNA), suggesting the possibility that the GINS helps the MCM bind to DNA (31, 32). Like their eukaryotic counterparts, archaeal GINS complexes also interact with other replication proteins, in addition to their cognate MCMs. P. furiosus GINS binds to Orc1/Cdc6 (28), and T. kodakaraensis GINS interacts with DNA polymerase D, PCNA, replication factor C (RFC), and GINS-associated nuclease (GAN) (33). Intriguingly, GAN, a RecJ-like exonuclease, is suggested to be the archaeal counterpart of eukaryotic Cdc45 (34). However, the presence of an archaeal CMG complex has not been demonstrated (35).
The heterotetrameric GINS complex from the hyperthermophilic crenarchaeon Sulfolobus solfataricus (SsoGINS) contains two subunits of Gins23, a Psf2/Psf3 homolog, and two subunits of Gins51, a remote homolog of Psf1/Sld5 (36). Unlike GINS from euryarchaea investigated so far, SsoGINS was shown to be unable to affect the helicase activity of its cognate MCM (SsoMCM) despite the demonstrated physical interaction between the two protein complexes (36). Here, we report that SsoGINS was capable of binding to DNA and sliding in a 5′-to-3′ direction along ssDNA. The protein complex promoted DNA binding by SsoMCM and stimulated the helicase activity of SsoMCM. Our results suggest that SsoGINS enhances the unwinding ability of SsoMCM at the replication fork through its ability to interact and move along with the helicase.
MATERIALS AND METHODS
DNA substrates.
Unlabeled and biotin-, Cy3-, and Cy5-labeled oligonucleotides (see Table S1 in the supplemental material) were synthesized at Sangon BioTech (Shanghai, China). To prepare blunt-ended double-stranded DNA (dsDNA) or dsDNA with one or two single-stranded tails, an oligonucleotide was labeled with [γ-32P]ATP (PerklinElmer) at the 5′ end using T4 polynucleotide kinase (TaKaRa) and purified using a G-50 MicroSpin column (GE Healthcare). The 32P-labeled oligonucleotide was annealed to a complementary oligonucleotide (see Table S1) at a molar ratio of 1:1.2 in 20 mM Tris-HCl, pH 8.0, and 100 mM NaCl. A 3′ biotin-labeled oligonucleotide (S15) (see Table S1) was radiolabeled at the 5′ end as described above. To radiolabel a 5′ biotin-labeled oligonucleotide (S14) (see Table S1), the DNA fragment was annealed to oligonucleotide S20 and filled in with [α-32P]ATP using Klenow fragment (Thermo). After the sample was boiled for 3 min and subsequently cooled on ice, it was subjected to electrophoresis in a 15% polyacrylamide gel containing 8 M urea in 1× Tris-borate-EDTA (TBE) buffer. The band containing the desired ssDNA was sliced and eluted out of the gel slice in 0.5 M ammonium acetate, 100 mM magnesium acetate, and 1 mM EDTA, pH 8.0. Following ethanol precipitation, the DNA was dissolved in Tris-EDTA (TE) buffer. To prepare DNA fragments blocked at the end with a streptavidin (SA)-biotin complex, the biotin-labeled DNA was incubated for 30 min at 4°C with a 50-fold molar excess of streptavidin to ensure a 1:1 streptavidin/biotin stoichiometry before gel purification.
An M13 ssDNA-based helicase substrate was prepared as described previously (37) with some modifications. Briefly, a 45-nucleotide (nt) oligonucleotide containing a run of 20 deoxyribosylthymine (dT) residues at the 5′ end (oligonucleotide S12; 2 pmol) (see Table S1 in the supplemental material) was annealed to M13 ssDNA (4 μg). The annealed primer was extended with Sequenase (26 U; United States Biochemicals) first in the presence of dCTP, dGTP, and dTTP (0.18 μM each) and [α-32P]dATP (40 μCi) for 5 min at 22°C and subsequently in the presence of the deoxynucleoside triphosphates (dNTPs; 80 μM each) and 4 μM ddCTP for 5 min at 37°C, according to the manufacturer's instructions. Reaction products were purified by using a G-50 MicroSpin column.
To prepare a 200-nt minicircular DNA substrate, the 5′-phosphorylated 100-nt strand A and 5′-phosphorylated 100-nt strand B (1 nmol each) (see Table S1 in the supplemental material) were mixed with the bridge strand AB (1.2 nmol) (see Table S1) in 50 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 10 mM dithiothreitol (DTT), 1 mM ATP, and 0.025 mg/ml bovine serum albumin (BSA) in a total volume of 200 μl. The mixture was incubated for 5 min at 65°C and cooled slowly to room temperature. T4 DNA ligase (10,000 U; New England BioLabs) was added, and the incubation was continued at 16°C for 12 h. The sample volume was then expanded to 12 ml with the addition of ligation buffer. Following the addition of the second oligonucleotide bridge strand BA (1.2 nmol) (see Table S1), annealing and ligation reactions were performed as described above. The sample was extracted with phenol-chloroform. The DNA was precipitated with ethanol, dissolved in Tris-EDTA (TE) buffer, and subjected to electrophoresis in an 8% polyacrylamide gel containing 8 M urea in 1× Tris-borate-EDTA (TBE) buffer. The gel was stained with ethidium bromide, and the band corresponding to the 200-nt minicircular DNA was sliced under UV light. The DNA was eluted out of the gel slice in 0.5 M ammonium acetate, 100 mM magnesium acetate, 1 mM EDTA, pH 8.0, and 0.1% sodium dodecyl sulfate (SDS). Following ethanol precipitation, the minicircular DNA was dissolved in TE buffer. The circular nature of the DNA was verified by treatment with ExoI and ExoIII (New England BioLabs).
Proteins.
Recombinant S. solfataricus MCM (SsoMCM), DNA polymerase B1 (SsoPolB1), Cren7, and single-stranded DNA binding protein (SsoSSB) were prepared as described previously (26, 38–40). To prepare recombinant S. solfataricus GINS (SsoGINS), genes encoding Gins23 (SSO0772) and Gins51 (SSO1049) were amplified from the genomic DNA of S. solfataricus P2 by PCR using Pfu DNA polymerase (see Table S1 in the supplemental material for primer sequences). The PCR products were inserted into NcoI/HindIII and NdeI/XhoI sites of plasmid pET-Duet-1 (Novagen). This introduced a hexahistidine tag at the C-terminal end of Gins23. The expression plasmid was transformed into Escherichia coli strain BL21 Rosetta. The cells were grown to an optical density at 600 nm of 0.4, and protein synthesis was induced by the addition of 0.2 mM isopropyl-β-d-thiogalactoside (IPTG). After incubation for 12 h at 15°C, the cells were harvested by centrifugation and suspended in 20 mM Tris-HCl, pH 8.0, 500 mM NaCl, and 0.2% Triton X-100. The cells were lysed by sonication, and the lysate was incubated for 30 min at 70°C. The sample was centrifuged, and the supernatant was purified by chromatography on a HiTrap chelating column (1 ml; GE Healthcare) and subsequently on a Mono Q 5/50 GL column (1 ml; GE Healthcare) as described previously (41).
Electrophoretic mobility shift assays (EMSAs).
The standard reaction mixture (10 μl) contained 25 mM HEPES-NaOH, pH 7.0, 50 mM sodium acetate, 5 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, 5% (vol/vol) glycerol, 2 nM 32P-labeled oligonucleotide substrate, and the amounts of protein indicated in the figure legends. After incubation for 10 min at 22°C, the samples were subjected to electrophoresis in a 5% polyacrylamide (79:1) gel in 0.1× TBE buffer. The gel was exposed to X-ray film and quantified by using a PhosphorImager (GE Healthcare).
For EMSAs with fluorescence-labeled protein and DNA, SsoGINS was labeled by incubation with a 5-fold molar excess of Cy5 succinimidyl ester in labeling buffer (25 mM HEPES-NaOH, pH 7.0, 100 mM NaCl, 5 mM MgCl2, and 10% [vol/vol] glycerol) for 4 h at 22°C, and excess dye was removed through extensive dialysis in labeling buffer. The labeling efficiency was determined by spectroscopy. A Cy3-labeled forked DNA fragment was prepared by annealing oligonucleotide L1 with oligonucleotide S5 (see Table S1 in the supplemental material). EMSAs were performed as described above, with the exception that Cy3-labeled DNA (0.1 μM), Cy5-labeled SsoGINS (1 μM), and SsoMCM (4 μM) were added as indicated (see the legend to Fig. 5B). The gel was imaged on a Typhoon scanner (GE Healthcare).
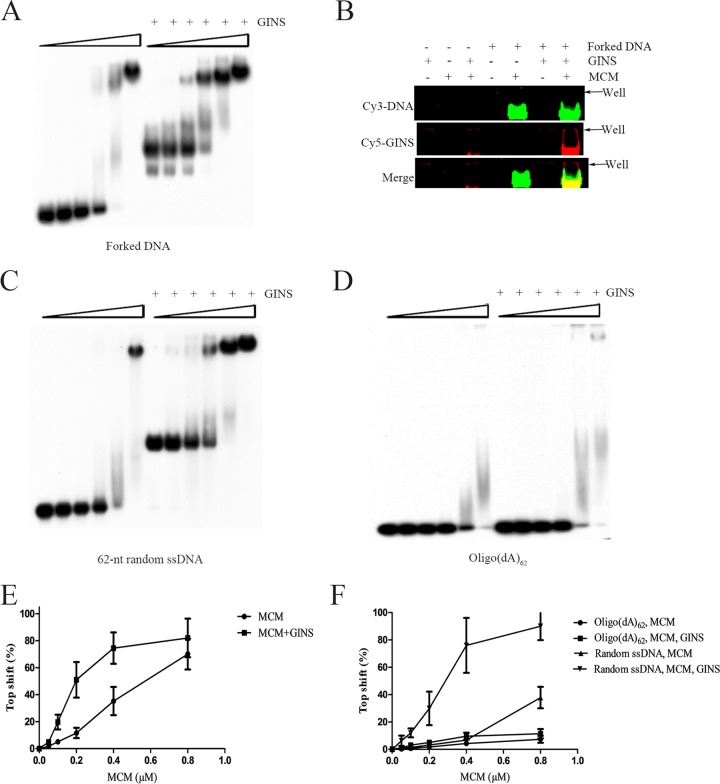
FIG 5.
Effect of SsoGINS on DNA binding by SsoMCM. (A, C, and D) Effect of SsoGINS on the gel retardation patterns of SsoMCM on various DNAs. SsoMCM was mixed with a 32P-labeled forked DNA (D4) (see Table S1 in the supplemental material) (A), a 62-nt random ssDNA (S1) (see Table S1) (C), or oligo(dA)62 (D) in the presence or absence of SsoGINS (1 μM). The protein-DNA complexes were subjected to polyacrylamide gel electrophoresis. The gel was exposed to X-ray film. Concentrations of SsoMCM were 0, 0.05, 0.1, 0.2, 0.4, and 0.8 μM, indicated by the triangles above the lanes. (B) EMSAs with fluorescence-labeled protein and DNA. Cy5 labeled SsoGINS (1 μM) and/or unlabeled SsoMCM (4 μM) were incubated with a Cy3-labeled forked DNA (LD2; 0.1 μM) (see Table S1). The protein-DNA complexes were subjected to polyacrylamide gel electrophoresis. The gel was imaged on a Typhoon scanner. (E and F) Quantitative analysis of gel retardation patterns shown in Fig. 3A, C, and D by phosphorimaging. Data shown represent an average of three independent measurements.
FRET assays.
Steady-state fluorescence resonance energy transfer (FRET) assays were performed on a Hitachi Fluorescence Spectrophotometer (F-7000). The standard reaction mixture (200 μl) contained 50 mM HEPES-NaOH, pH 7.0, 5 mM MgCl2, 0.1 mg/ml BSA, 0.1 μM forked DNA with a 3′ oligo(dT)20 tail labeled at the 3′ end with Cy3 and a 5′ oligo(dT)20 tail labeled at the 5′ end with Cy5 (LD1) (see Table S1 in the supplemental material), and indicated amounts of SsoMCM and/or SsoGINS (see the legend to Fig. 4). The excitation and emission wavelengths were at 532 and 545 to 750 nm, respectively. The FRET intensity (I) was defined as follows: IFRET = ICy5/(ICy5 + ICy3).
FIG 4.
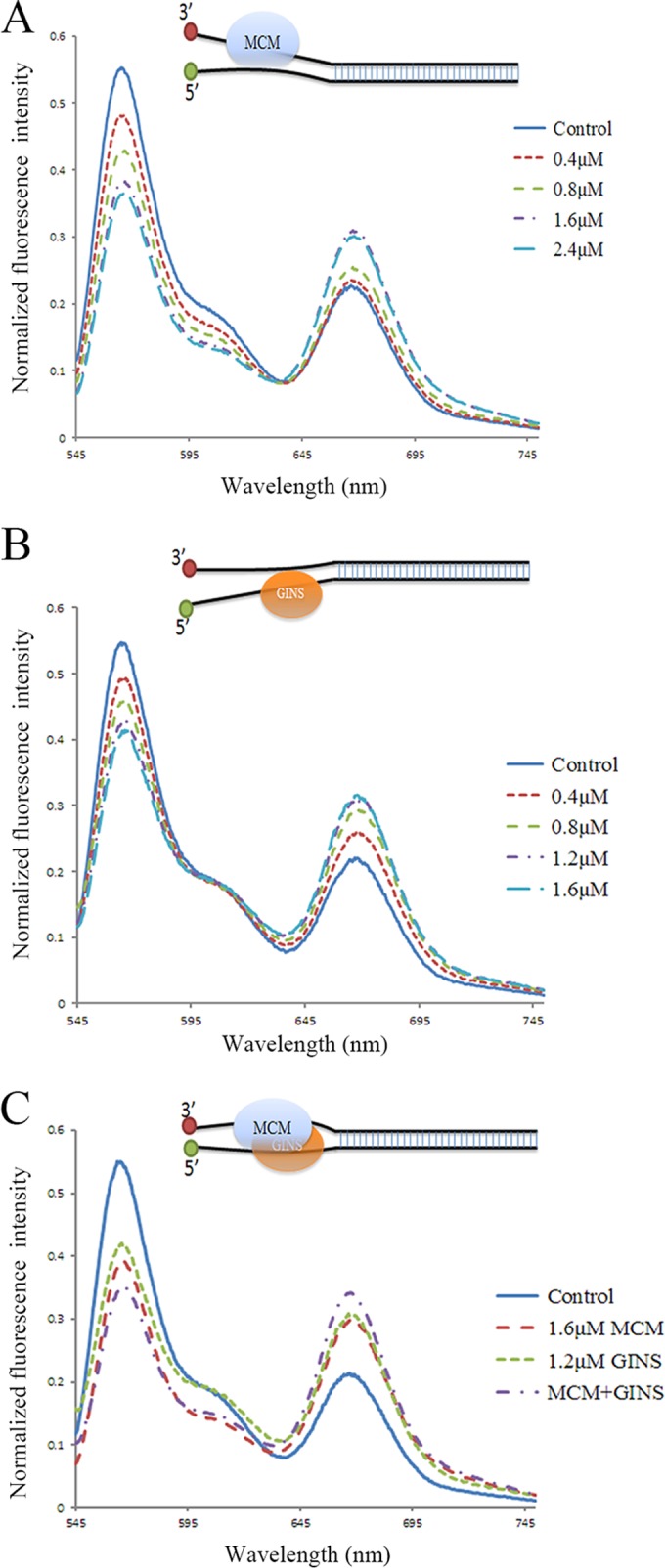
Effect of binding by SsoMCM (A) and/or SsoGINS (C and B) to forked DNA on the distance between the two single-stranded tails of the DNA. SsoMCM and/or SsoGINS were mixed with forked DNA with a 3′ oligo(dT)20 tail labeled at the 3′ end with Cy3 and a 5′ oligo(dT)20 tail labeled at the 5′ end with Cy5 (LD1) (see Table S1 in the supplemental material). Fluorescence intensity was recorded over a range of wavelengths from 545 to 750 nm. Cartoons show the proposed effect of the binding of the protein(s) on the distance between the two single-stranded tails of the forked DNA.
DNA helicase assays.
The standard reaction mixture (20 μl) contained 25 mM HEPES-NaOH, pH 7.0, 50 mM sodium acetate, 5 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, 2.5 mM ATP, 1 nM 32P-labeled oligonucleotide substrate (D4) (see Table S1 in the supplemental material) or M13 ssDNA-based substrate, and specified amounts of SsoMCM (see the legends to Fig. 6A, C, and D). SsoGINS was added when indicated. After incubation for 30 min at 55°C, reactions were stopped by the addition of 5 μl of a stop solution (50 mM EDTA, 0.5% SDS, 30% [vol/vol] glycerol, 0.1% bromophenol blue, and 0.1% xylene cyanol FF). The samples were subjected to electrophoresis in a 10% (for assays on the oligonucleotide substrate) or 5% (for M13-based assays) polyacrylamide (19:1) gel in 1× TBE buffer. The gel was exposed to X-ray film and quantitated by phosphorimaging.
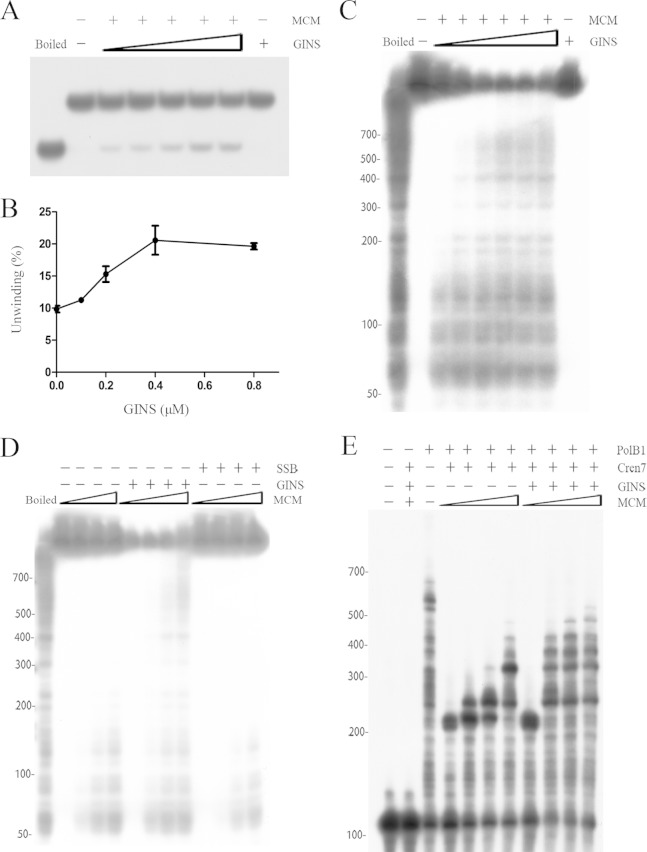
FIG 6.
Processivity of SsoMCM. (A and B) DNA unwinding by SsoMCM. SsoMCM (0.2 μM) was incubated with a radiolabeled oligonucleotide substrate (D4) (see Table S1 in the supplemental material) in the presence of increasing amounts of SsoGINS. Reaction products were subjected to electrophoresis in a polyacrylamide gel in 1× TBE buffer. The gel was exposed to X-ray film (A) and quantitated by phosphorimaging (B). Data shown in panel B represent an average of three independent measurements. Left lane, boiled substrate; second lane from left, no protein added; far-right lane, 0.8 μM SsoGINS. Concentrations of SsoGINS were 0, 0.1, 0.2, 0.4, and 0.8 μM, indicated by the triangles above the lanes. (C) Effect of SsoGINS on the processivity of SsoMCM. SsoMCM (0.2 μM) was incubated with a radiolabeled M13-based substrate in the presence of increasing amounts of SsoGINS. Reaction products were subjected to electrophoresis in 5% polyacrylamide gel in 1× TBE buffer. Left lane, boiled substrate; second lane from left, no protein added; far-right lane, 1.2 μM SsoGINS. Concentrations of SsoGINS were 0, 0.1, 0.2, 0.4, 0.8, and 1.2 μM, indicated by the triangles above the lanes. (D) Effect of SsoSSB on the processivity of SsoMCM. Various amounts of SsoMCM were incubated with the radiolabeled M13-based substrate in the presence of SsoGINS (1 μM) or SsoSSB (1.6 μM). Reaction products were subjected to electrophoresis in 5% polyacrylamide gel in 1× TBE buffer. The gel was exposed to X-ray film. Left lane, boiled substrate. Concentrations of SsoMCM were 0, 0.1, 0.2, and 0.4 μM, indicated by the triangles above the lanes, for each set of conditions. (E) Coupled DNA helicase and polymerase assays. SsoMCM was preincubated for 5 min with a 32P-labeled oligonucleotide (S13) (see Table S1) annealed to a 200-nt circular ssDNA in the presence or absence of SsoGINS (1 μM). Cren7 (1 μM) was added, and the incubation was continued for an additional 5 min. SsoPolB1 (25 nM) was then added. After further incubation, reaction products were processed for electrophoresis in polyacrylamide gel containing 7 M urea in 1× TBE buffer. The gel was exposed to X-ray film. Concentrations of SsoMCM were 0, 0.1, 0.2, and 0.4 μM, respectively, as indicated by the triangles above the lanes, for the conditions with SsoGINS and without SsoGINS. Values at the left of panels C to E are nucleotides.
Coupled DNA helicase and polymerase assays.
The standard reaction mixture (20 μl) contained 50 mM Tris-HCl, pH 8.0, 1 mM DTT, 0.1 mg/ml BSA, 5 mM MgCl2, 2 nM 200-nt circular ssDNA annealed to a 32P-labeled oligonucleotide (S13) (see Table S1 in the supplemental material), and indicated amounts of SsoMCM (see the legend to Fig. 6E). After incubation for 5 min at 55°C in the presence or absence of SsoGINS (1 μM), Cren7 (1 μM) was added, and the incubation was continued for an additional 5 min. SsoPolB1 (25 nM) was then added. The reaction was started by the addition of 2.5 mM ATP and 1 mM dNTPs (Roche). After incubation for 30 min at 55°C, the reaction was extracted with phenol-chloroform and mixed with an equal volume of 2× loading buffer (95% deionized formamide, 100 mM EDTA, 0.025% bromphenol blue, and 0.025% xylene cyanol FF). After samples were boiled for 3 min and subsequently cooled on ice, they were subjected to electrophoresis in a 5% polyacrylamide (19:1) gel containing 7 M urea in 1× TBE buffer. The gel was exposed to X-ray film.
RESULTS
DNA binding by SsoGINS.
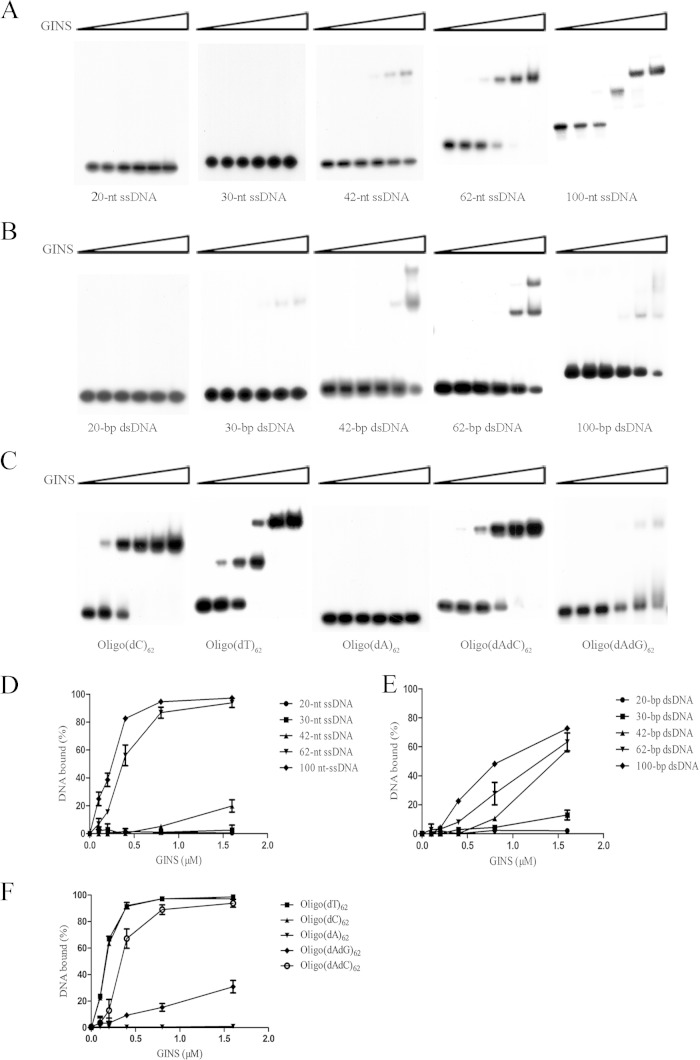
It has been demonstrated that human GINS interacts strongly with DNA, binding preferentially to ssDNA or to dsDNA containing ssDNA stretches over dsDNA (42). However, the GINS complexes from the euryarchaea P. furiosus and T. acidophilum do not bind DNA (28, 32). To determine if GINS from the crenarchaeon S. solfataricus (SsoGINS) was able to bind to DNA, we first prepared ssDNAs and dsDNAs of different lengths and with random sequences and tested the ability of SsoGINS to interact with these DNAs by an electrophoretic mobility shift assay (EMSA). As shown in Fig. 1A and D, SsoGINS failed to bind to a 20-nt or 30-nt ssDNA, bound weakly to a 42-nt ssDNA, but bound strongly to a 62-nt ssDNA, as indicated by the appearance of a shifted band. When a 100-nt ssDNA was used, the second shift was generated, suggesting the binding of two molecules of SsoGINS to the DNA. On the other hand, SsoGINS was unable to bind a 20-bp dsDNA but bound weakly to a 30-bp dsDNA. Furthermore, the protein complex generated two retarded shifts on a dsDNA fragment of 42-bp or longer (Fig. 1B and E). These observations suggest that SsoGINS was able to bind shorter dsDNA fragments than ssDNA fragments. However, it is worth noting that SsoGINS showed greater affinity for ssDNA fragments than for dsDNA fragments when the sizes of the fragments were no shorter than those required for stable binding. For example, SsoGINS bound to the 62-nt ssDNA considerably more strongly than to a 62-bp dsDNA, prepared by annealing the 62-nt ssDNA with its complementary strand, with apparent dissociation constants (Kds) of 0.33 ± 0.13 μM for the former and 1.56 ± 0.3 μM for the latter, as measured on the basis of the amount of the protein required for the retardation of one-half of the amount of the labeled input DNA. Therefore, SsoGINS is similar to human GINS in binding preferentially to ssDNA. In addition, the binding of SsoGINS to ssDNA appears to be as strong as that of the human protein (42).
FIG 1.
Binding of SsoGINS to ssDNA and dsDNA fragments. (A, B, D, and E) Binding of SsoGINS to ssDNA or dsDNA of various lengths. SsoGINS was mixed with 32P-labeled 20-, 30-, 42-, 62-, and 100-nt ssDNA fragments (S1, S3, S7, S17, and OligoA) (see Table S1 in the supplemental material) or with 20-, 30-, 42-, 62-, and 100-bp dsDNA fragments (D1, D8, D9, D7, and D10) (see Table S1). The protein-DNA complexes were subjected to polyacrylamide gel electrophoresis. The gel was exposed to X-ray film (A and B) and quantified by phosphorimaging (D and E). Concentrations of SsoGINS were 0, 0.1, 0.2, 0.4, 0.8, and 1.6 μM, indicated by the triangles above the lanes. (C and F) Sequence preference of SsoGINS in ssDNA binding. SsoGINS was mixed with a 32P-labeled 62-nt homopolymeric ssDNA or a 62-nt ssDNA with alternating A and G or A and C (see Table S1). The protein-DNA complexes were subjected to polyacrylamide gel electrophoresis. The gel was exposed to X-ray film (C) and quantified by phosphorimaging (F). Concentrations of SsoGINS used in the assays were 0, 0.1, 0.2, 0.4, 0.8, and 1.6 μM, in respective order, as indicated by the triangle above the lanes. Data shown in panels D to F represent an average of three independent measurements.
To gain some clues about the striking difference between SsoGINS and the GINS complexes from the euryarchaea P. furiosus and T. acidophilum in DNA binding properties, we performed a sequence analysis on archaeal GINS. All known crenarchaeal GINS proteins except for Sld5/Psf1 homologs from Pyrobaculum species are separated phylogenetically from their homologs in euryarchaea (see Fig. S6 in the supplemental material). The Psf1/Sld5 homolog (SSO1049) from S. solfataricus is distantly related to homologs from P. furiosus (17% similarity at the amino acid sequence level) and T. acidophilum (13% sequence similarity), the latter of which encodes a single GINS protein. S. solfataricus Psf2/Psf3 (SSO0772) is only 24% similar at the amino acid sequence level to Psf2/Psf3 from P. furiosus. Therefore, it is possible that GINS complexes from crenarchaea, such as S. solfataricus, differ from those from euryarchaea in properties such as DNA binding ability.
We then sought to determine if SsoGINS exhibited sequence preference in DNA binding. As shown in Fig. 1C and F, both oligo(dT)62 and an oligonucleotide containing a run of 62 deoxyribosylcytosine residues [oligo(dC)62] were tightly bound by the protein, whereas an oligonucleotide containing a series of 62 deoxyribosyladenine residues [oligo(dA)62] was not measurably bound. Unexpectedly, SsoGINS was able to form two shifts on oligo(dT)62 but only one shift on oligo(dC)62, although it showed similar affinities for the two oligonucleotides. It is possible that the size of oligo(dT) efficiently bound by the protein was shorter than that of oligo(dC). This possibility is supported by the finding that SsoGINS bound well to oligo(dT)42 but could hardly bind to oligo(dC)42 (see Fig. S1 in the supplemental material). Since an oligonucleotide with a run of 62 deoxyribosylguanine residues [oligo(dG)62] could not be readily synthesized, we tested the binding of SsoGINS to an oligonucleotide comprising alternating dA and dG [oligo(dAdG)62] or alternating dA and dC [oligo(dAdC)62] (Fig. 1C and F) residues. SsoGINS bound oligo(dAdC)62 well but was barely able to bind oligo(dAdG)62. Based on these results, we conclude that SsoGINS preferentially binds to pyrimidine-rich ssDNA over purine-rich ssDNA.
SsoGINS slides along ssDNA in a 5′-to-3′ direction.
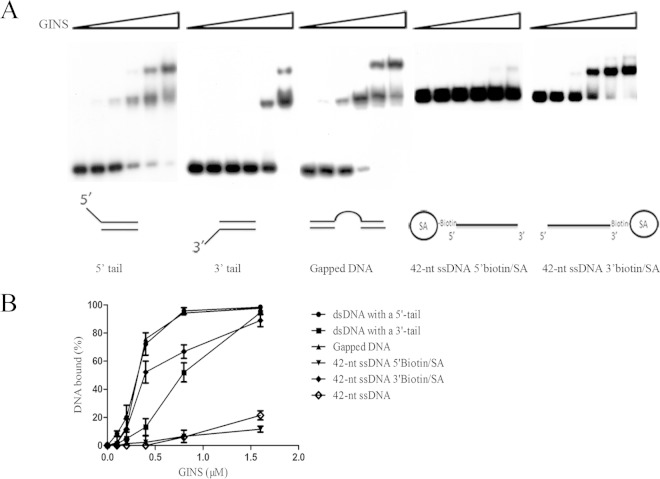
To further investigate the nature of DNA binding by SsoGINS, we prepared DNA fragments comprising both double-stranded and single-stranded regions in various arrangements (Fig. 2). Interestingly, SsoGINS bound more tightly to a dsDNA with a 5′ ssDNA tail (Kd, 0.28 ± 0.06 μM) than to a dsDNA with a 3′ ssDNA tail (Kd, 0.84 ± 0.22 μM). A possible interpretation for this observation is that SsoGINS was able to slide in a 5′-to-3′ direction along the ssDNA upon binding to the DNA but could hardly move laterally on dsDNA. Therefore, the protein complex was retained more readily on the dsDNA with a 5′ ssDNA tail than on the dsDNA with a 3′ ssDNA tail. To exclude the possibility that the observed polarity in binding by SsoGINS resulted from the difference of the protein in affinities for the two termini, we performed an EMSA on a DNA fragment containing a 20-dT single-stranded region flanked by two 21-bp dsDNA stretches (Fig. 2A). The binding was as strong as that observed on the dsDNA with a 5′ ssDNA tail (Kd, 0.30 ± 0.09 μM), indicating that a single-stranded terminus did not play an important role in DNA binding by the protein.
FIG 2.
Binding of SsoGINS to DNAs of different structures. SsoGINS was mixed with a 32P-labeled DNA fragment containing a 42-bp dsDNA region with a 5′ tail (D2) or a 3′ tail (D3) (see Table S1 in the supplemental material), a 32P-labeled partial dsDNA fragment containing a 20-dT region in the middle (D6) (see Table S1), a 42-nt ssDNA with 5′ biotin-SA, or a 42-nt ssDNA with 3′ biotin-SA. The protein-DNA complexes were subjected to polyacrylamide gel electrophoresis. The gel was exposed to X-ray film (A) and quantified by phosphorimaging (B). Concentrations of SsoGINS were 0, 0.1, 0.2, 0.4, 0.8, and 1.6 μM, indicated by the triangles above the lanes. Data shown in panel B represent an average of three independent measurements.
To further verify the suggestion that GINS is able to track unidirectionally on ssDNA, we prepared a 42-nt ssDNA blocked at either the 5′ or the 3′ end with a biotin-streptavidin (SA) complex. The biotin-SA complex served as a physical block, which would presumably prevent SsoGINS from sliding off the end of the DNA fragment. As shown in Fig. 1A and 2, SsoGINS bound approximately 20-fold more strongly to the 42-nt ssDNA containing a 3′ biotin-SA block (Kd, 0.36 ± 0.06 μM) than to the 42-nt ssDNA without the biotin-streptavidin block (Kd, 7.74 ± 0.95 μM). In contrast, binding of the protein complex to the 42-nt ssDNA with a 5′ biotin-SA block (Kd, 8.27 ± 0.92 μM) was similar to that to the unblocked 42-nt ssDNA. Based on these results, we conclude that SsoGINS is able to slide along ssDNA in a 5′-to-3′ direction.
SsoGINS interacts with both ssDNA tails of a forked DNA.
Notably, binding of SsoGINS to a dsDNA fragment with both 5′ and 3′ tails at one end (forked DNA; Kd, 0.13 ± 0.02 μM) or a dsDNA containing a single-stranded bubble in the middle (bubbled DNA; Kd, 0.17 ± 0.06 μM) was even stronger than that to the dsDNA with a 5′ ssDNA tail (Kd, 0.28 ± 0.06 μM) or to a dsDNA containing a single-stranded region in the middle (gapped DNA; Kd, 0.30 ± 0.09 μM) (Fig. 2 and 3). It appears that although SsoGINS bound preferentially to the 5′ tail of the forked DNA, the 3′ tail of the DNA was likely involved in stabilizing the binding. To look further into the potential role of the 3′ tail in the binding of SsoGINS to the forked DNA, we performed a fluorescence resonance energy transfer (FRET) assay using a forked DNA molecule labeled with Cy3 (donor) on the 3′ tail and with Cy5 (acceptor) on the 5′ tail. As shown in Fig. 4B, the labeled DNA molecule displayed a FRET value of 33.63% ± 0.55% in the absence of SsoGINS. As an increasing amount of SsoGINS was added, the FRET efficiency increased, with a maximum of 44.96% ± 0.28% at 1.2 μM SsoGINS, suggesting that the 5′ and the 3′ tails were drawn closer as a result of the binding by the protein complex (Fig. 4B). As a control, the addition of the single-stranded DNA binding protein from S. solfataricus (SsoSSB) to the assay system resulted in a slight decrease in FRET efficiency (see Fig. S5 in the supplemental material), presumably due to the reduced flexibility of the SsoSSB-bound single-stranded tails. These data support the contention that although SsoGINS bound preferentially to the 5′ tail of the forked DNA, it was also in contact with the 3′ tail in such a fashion that the binding of the complex to the DNA was stabilized.
FIG 3.
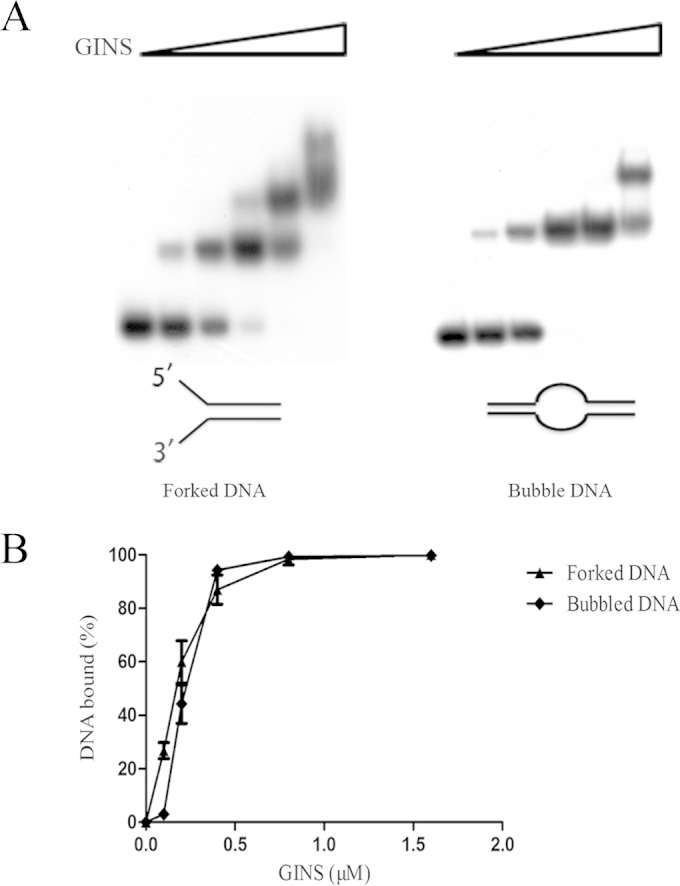
Binding of SsoGINS to forked and bubbled DNA. SsoGINS was mixed with a 32P-labeled DNA fragment containing a 42-bp dsDNA region with both 5′ and 3′ tails at one end (D4; forked DNA) or a 32P-labeled partial dsDNA fragment containing a 20-T bubble in the middle (D5; bubbled DNA) (see Table S1 in the supplemental material). The protein-DNA complexes were subjected to polyacrylamide gel electrophoresis. The gel was exposed to X-ray film (A) and quantified by phosphorimaging (B). Concentrations of SsoGINS were 0, 0.1, 0.2, 0.4, 0.8, and 1.6 μM, in respective order, as indicated by the triangles above the lanes. Data shown represent an average of three independent measurements.
SsoGINS stabilizes the binding of SsoMCM to DNA.
SsoGINS is able to interact with SsoMCM (36). It has been reported that SsoMCM loads on the 3′ tail of a forked DNA while externally grabbing the 5′ tail (24, 25). In the present study, we show that SsoGINS preferentially bound to the 5′ tail of a forked DNA while interacting with the 3′ tail. Therefore, it is of interest to learn if SsoGINS would influence the binding of SsoMCM to DNA. In the EMSAs, SsoMCM alone was able to generate a slow-migrating shift on the forked DNA (Fig. 5A). In the presence of 1 μM SsoGINS, which by itself produced a fast-migrating shift, the addition of an increasing amount of SsoMCM was accompanied by a decrease in the amount of the SsoGINS-DNA shift and an increase in the amount of the SsoMCM-DNA shift. The drastic difference between the SsoGINS-DNA and the SsoMCM-DNA complexes in mobility is presumably due to the difference between the molecular masses of the two DNA binding protein complexes. Notably, the formation of the SsoMCM-DNA complex occurred at significantly lower SsoMCM concentrations in the presence of SsoGINS (Kd, 0.18 ± 0.03 μM by monomer) than in the absence of the protein (Kd, 0.56 ± 0.09 μM) (Fig. 5A and E). A similar observation was made when an ssDNA with a random sequence instead of the forked DNA was used in the assays (Fig. 5C and F). Furthermore, the slow-migrating band was slightly upshifted in the presence of SsoGINS, suggesting the formation of an SsoMCM-SsoGINS-DNA complex. Given the large size difference between the SsoMCM hexamer and the SsoGINS complex, the difference between the SsoMCM-DNA and the SsoMCM-SsoGINS-DNA in gel mobility was expected to be small. Therefore, to confirm the association of SsoGINS with the slow-migrating shift, we performed an EMSA on the same DNA fragment as that used in the experiment shown in Fig. 5A, except that the DNA was labeled with Cy3 instead of 32P. SsoGINS was labeled with Cy5. SsoGINS clearly comigrated with the band containing the DNA and SsoMCM, demonstrating the formation of the SsoGINS-SsoMCM-DNA complex (Fig. 5B). Taking advantage of the finding that SsoGINS bound very poorly to oligo(dA)62 (Fig. 1C), we also determined if the effect of SsoGINS on DNA binding by SsoMCM depended on the binding of SsoGINS to the DNA. While SsoMCM bound less strongly to oligo(dA)62 than to the 62-nt ssDNA with a random sequence (see Fig. S2 in the supplemental material), the binding of the helicase to oligo(dA)62 was barely enhanced by SsoGINS (Fig. 5D and F). Taken together, these results suggest that SsoGINS was able to promote the binding of SsoMCM to DNA, and the ability of SsoGINS to bind the DNA was required for the stimulatory effect.
To gain further insights into the effect of SsoGINS on the interaction of SsoMCM with DNA, we performed the following FRET assays. As predicted from the previous studies (24), FRET increased as an increasing amount of SsoMCM bound to the fluorescence-labeled forked DNA, and the highest FRET value (45.98% ± 0.65%) was obtained at a protein concentration of 1.6 μM (Fig. 4A). Intriguingly, when SsoGINS and SsoMCM were added to the binding mixture at 1.2 and 1.6 μM, respectively, levels that would allow each of them to produce the highest FRET efficiency or to generate the largest gel shift in the EMSAs, a FRET value higher than the maximum FRET obtained with either protein was recorded (50.74% ± 0.45%) (Fig. 4C). It appears that SsoGINS did not compete with SsoMCM for binding to the DNA, and, instead, the two proteins bound simultaneously to the DNA in a manner that would bring the two single-stranded tails closer than each of the proteins alone could. Based on these and previous results, we speculate that SsoMCM and SsoGINS bind to the 3′ tail and 5′ tail, respectively, of the forked DNA and contribute to the stability of the SsoMCM-SsoGINS-DNA complex by interacting externally with their respective unbound tail as well as by protein-protein interactions.
SsoGINS enhances processive DNA unwinding by SsoMCM.
SsoGINS showed no significant effect on the helicase activity of SsoMCM in a previous study (36). Since we found that SsoGINS enhanced DNA binding by SsoMCM, we decided to redetermine if the former would affect the helicase activity of the latter. The forked DNA substrate (D4) (see Table S1 in the supplemental material) was used in the helicase activity assays. As shown in Fig. 6A, when SsoGINS was added to the helicase reaction, an ∼2-fold increase in the helicase activity of SsoMCM was observed (Fig. 6B). It was shown that PfuGINS stimulated the helicase activity of PfuMCM by increasing its ATPase activity. By comparison, SsoGINS did not affect the ATPase activity of SsoMCM (see Fig. S4 in the supplemental material).
Given the proposed possibility that SsoGINS and SsoMCM are able to slide along ssDNA in opposing directions, thus allowing the former to stabilize the interaction of the latter with a progressing fork, we then investigated if SsoGINS would influence the processivity of SsoMCM in DNA unwinding by using a modification of a well-established assay (5, 20, 37, 43). In the assay, an oligonucleotide primer annealed to M13 ssDNA was extended with Sequenase first in the presence of radiolabeled dNTPs and subsequently with a mixture of dNTPs and ddCTP to yield radiolabeled extension products of various sizes. As shown in Fig. 6C, SsoMCM was able to unwind dsDNA of no longer than 200 bp under the assay conditions. However, the addition of SsoGINS allowed up to ∼700 bp of dsDNA to be unwound by SsoMCM. It was noticed that more unwinding of dsDNA longer than ∼150 bp occurred with increasing amounts of SsoGINS. In contrast, unwinding of dsDNA shorter than ∼150 bp was not significantly enhanced by SsoGINS (Fig. 6C and D). It was reported that human replication protein A (RPA) stimulated the processivity of human CMG helicase by sequestering the emerging ssDNA (37). In our assays, however, SsoSSB displayed no stimulatory effect on the unwinding activity of SsoMCM, eliminating the possibility that SsoGINS promoted the processivity of SsoMCM simply by binding to the ssDNA generated in the unwinding reaction (Fig. 6D).
We then determined if SsoGINS would help SsoMCM function in a coordinated fashion with S. solfataricus DNA polymerase B1 (SsoPolB1). A rolling-circle assay was established, in which a 200-nt circular ssDNA was annealed to a radiolabeled 100-nt complementary ssDNA with a 5′ tail of 20 dT residues. Cren7 (1 μM), a chromatin protein highly conserved among crenarchaea, was included in the assay to inhibit the potent strand displacement activity of SsoPolB1 (44). The helicase activity of SsoMCM was not affected by Cren7 (1 μM) (see Fig. S3 in the supplemental material). Given the design of the minicircular template, extension products of >220 nt in length would have been synthesized by SsoPolB1 using as the template the ssDNA unwound by SsoMCM. As shown in Fig. 6E, SsoMCM (0.1 μM) was able to unwind ∼30 bp of dsDNA in the absence of SsoGINS under the assay conditions. SsoMCM became significantly more efficient in unwinding reactions in the presence of SsoGINS than in its absence, permitting SsoPolB1 to synthesize products of up to ∼500 bp (Fig. 6E). The stimulatory effect of SsoGINS was most pronounced at lower SsoMCM concentrations. It was noticed that hot spots existed for the termination of chain elongation by SsoPolB1. By contrast, termination of unwinding by SsoMCM occurred more randomly in the M13 ssDNA-based assays (Fig. 6D and E). Presumably, the presence of the strong termination signals in the minicircle assays was attributed to steric hindrance, resulting from the synthesis of the new DNA strand by SsoPolB1, to the movement of SsoMCM or SsoPolB1 along the template DNA. Taken together, our results suggest that SsoGINS increased the efficiency of DNA unwinding by SsoMCM.
DISCUSSION
Unlike all known archaeal GINS (28, 31, 32), SsoGINS bound strongly to DNA. Strong DNA binding was previously observed only with human GINS (42). Drosophila GINS exhibited rather weak affinity for forked DNA (15). In Archaea, GINS from neither P. furiosus nor T. acidophilum bound DNA in EMSAs (28, 31, 32). Like human GINS, SsoGINS preferentially bound to single-stranded DNA over double-stranded DNA. Structural studies show that the four subunits of human GINS form a ring-like tetramer with a central channel, which is proposed to be able to accommodate at least ssDNA (45). Since archaeal GINS complexes resemble their eukaryotic counterparts in architecture (29), the possibility exists that SsoGINS binds DNA in the same manner. Crystallographic and electron microscopic studies on eukaryotic GINS have revealed various sizes for the central channel, raising the possibility that the protein complex is flexible in structure to permit changes under various conditions (8, 42, 45–47). Therefore, it may be speculated that SsoGINS is capable of undergoing conformational changes upon binding to DNA or when passing DNA into its central channel.
Intriguingly, SsoGINS bound more tightly to dsDNA with a 5′ tail than to dsDNA with a 3′ tail. Furthermore, the affinity of the protein complex for an ssDNA fragment was substantially higher when the fragment contained a biotin-SA block at the 3′ end than when it had the block at the 5′ end. These results indicate that SsoGINS was able to slide along ssDNA in a 5′-to-3′ direction. It is worth noting that human Cdc45, a key factor in the human CMG complex, slides along ssDNA in a 3′-to-5′ direction (48). It would be of interest to determine if a eukaryotic GINS complex resembles SsoGINS in ssDNA tracking. If both GINS and Cdc45 are able to slide along ssDNA in the absence of ATP hydrolysis or any other forms of energy input, they would fit well with their roles as components of a CMG complex at a progressing replication fork. MCM serves as a motor for the moving CMG complex by tracking along the leading strand in a 3′-to-5′ direction at the expense of ATP. Cdc45 may migrate in front of the MCM on the same strand, acting as a molecular “wedge” (48). Given the polarity of its motion, GINS would presumably slide with the MCM along the lagging strand.
Like other archaeal GINS complexes, SsoGINS promoted DNA unwinding by SsoMCM. This observation contrasts with a previous report, in which no significant effects of S. solfataricus GINS on the helicase activity of MCM were detected (36). Since no experimental details are provided in the report, we speculate that the discrepancy results from the differences in the experimental conditions used in the two studies. We found that the SsoGINS-mediated stimulation was more pronounced with unwinding of longer dsDNA than with that of shorter dsDNA by SsoMCM. While unwinding of short oligonucleotide substrates by the helicase was moderately facilitated by SsoGINS, the sizes of dsDNAs unwound by SsoMCM in the presence of SsoGINS were significantly longer than those in the absence of SsoGINS. This appears to be consistent with the observation that SsoGINS did not stimulate the ATPase activity of SsoMCM. Stimulation of the ATPase activity of MCM by GINS has been widely observed. In Drosophila, the rate of ATP hydrolysis by CMG was about 300-fold higher than that by MCM2 to MCM7 (15). GINS from P. furiosus, T. kodakaraensis, and T. acidophilum were also shown to enhance the ATPase and thus the helicase activity of their cognate MCM (28, 30, 31). The lack of the drastic stimulation of unwinding of the oligonucleotide substrates may be attributed to the inability of SsoGINS to stimulate the ATPase of SsoMCM. How would SsoGINS affect the processivity of SsoMCM then? SsoGINS bound to the 5′ single-stranded tail of a forked DNA appeared to be in contact with the 3′ single-stranded tail, as demonstrated in our FRET assays. This agrees with the higher affinity of SsoGINS for the forked DNA substrate than for the same DNA except for the lack of the 3′ single-stranded tail. On the other hand, SsoMCM bound to the 3′ single-stranded tail of a forked substrate interacting with the 5′ single-stranded tail via the surface of the complex (24, 25). Indeed, the FRET efficiency was significantly higher when both protein complexes were allowed to bind to a forked DNA with two tails fluorescently labeled with donor and acceptor fluorophores, respectively, than when either alone was. Given the protein-protein interaction between SsoGINS and SsoMCM and the opposite directions in which they slide along ssDNA, SsoGINS probably serves to stabilize DNA binding by SsoMCM at a progressing fork, thereby enhancing the processivity of the helicase. The stimulatory effect of SsoGINS on the processivity of SsoMCM was also observed when DNA unwinding was coupled with DNA synthesis by SsoPolB1 on a circular template bound by Cren7, a chromatin protein, pointing to the physiological relevance of the functional interaction between the two protein complexes. In conclusion, our results suggest that the ability of SsoGINS and SsoMCM to bind to and migrate with a progressing replication fork in a coordinated and interactive fashion may provide a mechanistic basis for the promotion of the processivity of SsoMCM by SsoGINS.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by National Natural Science Foundation of China grant 31130003.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00496-15.
REFERENCES
- 1.Bell SP, Dutta A. 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Masai H, Matsumoto S, You ZY, Yoshizawa-Sugata N, Oda M. 2010. Eukaryotic chromosome DNA replication: where, when and how? Annu Rev Biochem 79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- 3.Lee JK, Hurwitz J. 2000. Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J Biol Chem 275:18871–18878. doi: 10.1074/jbc.M001118200. [DOI] [PubMed] [Google Scholar]
- 4.Bochman ML, Schwacha A. 2007. Differences in the single-stranded DNA binding activities of MCM2-7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J Biol Chem 282:33795–33804. doi: 10.1074/jbc.M703824200. [DOI] [PubMed] [Google Scholar]
- 5.Moyer SE, Lewis PW, Botchan MR. 2006. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A 103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. 2006. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 7.Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K. 2003. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423:720–724. doi: 10.1038/nature01692. [DOI] [PubMed] [Google Scholar]
- 8.Kubota Y, Takase Y, Komori Y, Hashimoto Y, Arata T, Kamimura Y, Araki H, Takisawa H. 2003. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Gene Dev 17:1141–1152. doi: 10.1101/gad.1070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H. 2003. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev 17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. 2005. Molecular anatomy and regulation of a stable replisome eukaryotic DNA at a paused replication fork. Gene Dev 19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 12.Tercero JA, Labib K, Diffley JF. 2000. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J 19:2082–2093. doi: 10.1093/emboj/19.9.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pacek M, Walter JC. 2004. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J 23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. 2011. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. 2010. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell 37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shikata K, Sasa-Masuda T, Okuno Y, Waga S, Sugino A. 2006. The DNA polymerase activity of Pol epsilon holoenzyme is required for rapid and efficient chromosomal DNA replication in Xenopus egg extracts. BMC Biochem 7:21. doi: 10.1186/1471-2091-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Falco M, Ferrari E, De Felice M, Rossi M, Hubscher U, Pisani FM. 2007. The human GINS complex binds to and specifically stimulates human DNA polymerase alpha-primase. EMBO Rep 8:99–103. doi: 10.1038/sj.embor.7400870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labib K, Gambus A. 2007. A key role for the GINS complex at DNA replication forks. Trends Cell Biol 17:271–278. doi: 10.1016/j.tcb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Barry ER, Bell SD. 2006. DNA replication in the archaea. Microbiol Mol Biol Rev 70:876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chong JP, Hayashi MK, Simon MN, Xu RM, Stillman B. 2000. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc Natl Acad Sci U S A 97:1530–1535. doi: 10.1073/pnas.030539597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelman Z, Lee JK, Hurwitz J. 1999. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum ΔH contains DNA helicase activity. Proc Natl Acad Sci U S A 96:14783–14788. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shechter DF, Ying CY, Gautier J. 2000. The intrinsic DNA helicase activity of Methanobacterium thermoautotrophicum ΔH minichromosome maintenance protein. J Biol Chem 275:15049–15059. doi: 10.1074/jbc.M000398200. [DOI] [PubMed] [Google Scholar]
- 23.Carpentieri F, De Felice M, De Falco M, Rossi M, Pisani FM. 2002. Physical and functional interaction between the mini-chromosome maintenance-like DNA helicase and the single-stranded DNA binding protein from the crenarchaeon Sulfolobus solfataricus. J Biol Chem 277:12118–12127. doi: 10.1074/jbc.M200091200. [DOI] [PubMed] [Google Scholar]
- 24.Rothenberg E, Trakselis MA, Bell SD, Ha T. 2007. MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J Biol Chem 282:34229–34234. doi: 10.1074/jbc.M706300200. [DOI] [PubMed] [Google Scholar]
- 25.Graham BW, Schauer GD, Leuba SH, Trakselis MA. 2011. Steric exclusion and wrapping of the excluded DNA strand occurs along discrete external binding paths during MCM helicase unwinding. Nucleic Acids Res 39:6585–6595. doi: 10.1093/nar/gkr345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. 2005. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol 12:756–762. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 27.Makarova KS, Wolf YI, Mekhedov SL, Mirkin BG, Koonin EV. 2005. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res 33:4626–4638. doi: 10.1093/nar/gki775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshimochi T, Fujikane R, Kawanami M, Matsunaga F, Ishino Y. 2008. The GINS complex from Pyrococcus furiosus stimulates the MCM helicase activity. J Biol Chem 283:1601–1609. doi: 10.1074/jbc.M707654200. [DOI] [PubMed] [Google Scholar]
- 29.Oyama T, Ishino S, Fujino S, Ogino H, Shirai T, Mayanagi K, Saito M, Nagasawa N, Ishino Y, Morikawa K. 2011. Architectures of archaeal GINS complexes, essential DNA replication initiation factors. BMC Biol 9:28. doi: 10.1186/1741-7007-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishino S, Fujino S, Tomita H, Ogino H, Takao K, Daiyasu H, Kanai T, Atomi H, Ishino Y. 2011. Biochemical and genetical analyses of the three mcm genes from the hyperthermophilic archaeon, Thermococcus kodakaraensis. Genes Cells 16:1176–1189. doi: 10.1111/j.1365-2443.2011.01562.x. [DOI] [PubMed] [Google Scholar]
- 31.Ogino H, Ishino S, Haugland GT, Birkeland NK, Kohda D, Ishino Y. 2014. Activation of the MCM helicase from the thermophilic archaeon, Thermoplasma acidophilum by interactions with GINS and Cdc6-2. Extremophiles 18:915–924. doi: 10.1007/s00792-014-0673-6. [DOI] [PubMed] [Google Scholar]
- 32.Ogino H, Ishino S, Mayanagi K, Haugland GT, Birkeland NK, Yamagishi A, Ishino Y. 2011. The GINS complex from the thermophilic archaeon, Thermoplasma acidophilum may function as a homotetramer in DNA replication. Extremophiles 15:529–539. doi: 10.1007/s00792-011-0383-2. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Santangelo TJ, Cubonova L, Reeve JN, Kelman Z. 2010. Affinity purification of an archaeal DNA replication protein network. mBio 1(5):e00221-10. doi: 10.1128/mBio.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makarova KS, Koonin EV, Kelman Z. 2012. The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biol Direct 7:7. doi: 10.1186/1745-6150-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onesti S, MacNeill SA. 2013. Structure and evolutionary origins of the CMG complex. Chromosoma 122:47–53. doi: 10.1007/s00412-013-0397-x. [DOI] [PubMed] [Google Scholar]
- 36.Marinsek N, Barry ER, Makarova KS, Dionne I, Koonin EV, Bell SD. 2006. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep 7:539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. 2012. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc Natl Acad Sci U S A 109:6042–6047. doi: 10.1073/pnas.1203734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wadsworth RI, White MF. 2001. Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res 29:914–920. doi: 10.1093/nar/29.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo L, Feng YG, Zhang ZF, Yao HW, Luo YM, Wang JF, Huang L. 2008. Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among Crenarchaea. Nucleic Acids Res 36:1129–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lou HQ, Duan ZH, Huo XF, Huang L. 2004. Modulation of hyperthermophilic DNA polymerase activity by archaeal chromatin proteins. J Biol Chem 279:127–132. doi: 10.1074/jbc.M309860200. [DOI] [PubMed] [Google Scholar]
- 41.Guo X, Huang L. 2010. A superfamily 3 DNA helicase encoded by plasmid pSSVi from the hyperthermophilic archaeon Sulfolobus solfataricus unwinds DNA as a higher-order oligomer and interacts with host primase. J Bacteriol 192:1853–1864. doi: 10.1128/JB.01300-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boskovic J, Coloma J, Aparicio T, Zhou M, Robinson CV, Mendez J, Montoya G. 2007. Molecular architecture of the human GINS complex. EMBO Rep 8:678–684. doi: 10.1038/sj.embor.7401002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barry ER, McGeoch AT, Kelman Z, Bell SD. 2007. Archaeal MCM has separable processivity, substrate choice and helicase domains. Nucleic Acids Res 35:988–998. doi: 10.1093/nar/gkl1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun F, Huang L. 2013. Sulfolobus chromatin proteins modulate strand displacement by DNA polymerase B1. Nucleic Acids Res 41:8182–8195. doi: 10.1093/nar/gkt588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang YP, Wang G, Bermudez V, Hurwitz J, Chen XS. 2007. Crystal structure of the GINS complex and functional insights into its role in DNA replication. Proc Natl Acad Sci U S A 104:12685–12690. doi: 10.1073/pnas.0705558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi JM, Lim HS, Kim JJ, Song OK, Cho Y. 2007. Crystal structure of the human GINS complex. Genes Dev 21:1316–1321. doi: 10.1101/gad.1548107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamada K, Kubota Y, Arata T, Shindo Y, Hanaoka F. 2007. Structure of the human GINS complex and its assembly and functional interface in replication initiation. Nat Struct Mol Biol 14:388–396. doi: 10.1038/nsmb1231. [DOI] [PubMed] [Google Scholar]
- 48.Szambowska A, Tessmer I, Kursula P, Usskilat C, Prus P, Pospiech H, Grosse F. 2014. DNA binding properties of human Cdc45 suggest a function as molecular wedge for DNA unwinding. Nucleic Acids Res 42:2308–2319. doi: 10.1093/nar/gkt1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.