ABSTRACT
Burkholderia thailandensis has three acyl-homoserine lactone (AHL) LuxR-LuxI quorum-sensing circuits and two orphan LuxR homologs. Orphans are LuxR-type transcription factors that do not have cognate LuxI-type AHL synthases. One of the orphans, MalR, is genetically linked to the mal gene cluster, which encodes enzymes required for production of the cytotoxic polyketide malleilactone. Under normal laboratory conditions the mal gene cluster is silent; however, antibiotics like trimethoprim induce mal transcription. We show that trimethoprim-dependent induction of the mal genes requires MalR. MalR has all of the conserved amino acid residues characteristic of AHL-responsive LuxR homologs, but in B. thailandensis, MalR activation of malleilactone synthesis genes is not responsive to AHLs. MalR can activate transcription from the mal promoter in E. coli without addition of AHLs or trimethoprim. Expression of malR in B. thailandensis is induced by trimethoprim. Our data indicate that MalR binds to a lux box-like element in the mal promoter and activates transcription of the mal genes in an AHL-independent manner. Antibiotics like trimethoprim appear to activate mal gene expression indirectly by somehow activating malR expression. MalR activation of the mal genes represents an example of a LuxR homolog that is not a receptor for an AHL quorum-sensing signal. Our evidence is consistent with the idea that mal gene activation depends solely on sufficient transcription of the malR gene.
IMPORTANCE LuxR proteins are transcription factors that are typically activated by acyl-homoserine lactone (AHL) signals. We demonstrate that a conserved LuxR family protein, MalR, activates genes independently of AHLs. MalR is required for transcription of genes coding for synthesis of the cytotoxic polyketide malleilactone. These genes are not expressed when cells are grown under normal laboratory conditions. In laboratory culture, MalR induction of malleilactone requires certain antibiotics, such as trimethoprim, which increase malR expression by an unknown mechanism. At sufficient levels of malR expression, MalR functions independently of any external signal. Our findings show that MalR is an activator of the silent malleilactone biosynthesis genes and that MalR functions independently of AHLs.
INTRODUCTION
Acyl-homoserine lactone (AHL) quorum-sensing systems are widespread among the Proteobacteria. AHLs are generated by members of the LuxI family of AHL synthases. Members of the LuxR protein family serve as cognate AHL-dependent transcription factors. Often, the genes targeted by LuxR family members have an 18- to 20-base inverted repeat in their promoter regions that shows conservation across species. These repeat elements are called lux box-like sequences and serve as binding sites for the transcription factors. Sequence identity in pairwise comparisons of LuxR polypeptides is rather limited (18 to 35%), but there is a conserved N-terminal AHL binding region and a conserved C-terminal DNA binding region where sequence identity is higher and where there are highly conserved residues (see references 1 and 2 for reviews).
Burkholderia thailandensis has three pairs of luxI-luxR-type genes called btaI1-btaR1, btaI2-btaR2, and btaI3-btaR3. Each of the luxR homologs is linked to its cognate luxI homolog, as is the case for luxI-luxR-type gene pairs in many other bacteria. This species also has two additional luxR homologs, btaR4 (now called malR [see below]) and btaR5 (3). LuxR family members like MalR and BtaR5 are called orphans (4) or solos (5), because the genes encoding these polypeptides are not associated with a luxI homolog. Relatively few orphans have been characterized. Some orphans, like Pseudomonas aeruginosa QscR or Salmonella enterica serovar Typhimurium SdiA, show complete conservation of all of the invariant residues in the signal binding region of AHL-responsive LuxR family members and in fact respond to AHLs that are self-produced (6) or exogenously supplied (7, 8). Other LuxR family members, like Xanthomonas oryzae OryR and Photorhabdus asymbiotica PauR, show conservation in the AHL binding region but vary with respect to at least one of the conserved residues. OryR and PauR do not respond to AHLs. Rather, they respond to host-derived factors or an endogenously produced secondary metabolite, respectively (9, 10).
We are interested in the B. thailandensis MalR protein for several reasons. First, it shows complete identity in all of the residues conserved among AHL-responsive LuxR family members. Second, it is adjacent to and divergently transcribed from the mal gene cluster, which is required for the production of the B. thailandensis cytotoxic polyketide malleilactone. The genes for MalR and malleilactone biosynthesis are silent when B. thailandensis is grown in standard laboratory media but expressed in cells grown with certain antibiotics, such as trimethoprim (11). Third, malR and the mal genes are required for Caenorhabditis elegans infections (12). There is a lux box-like sequence in the promoter region of the mal operon. One might imagine that MalR is an AHL-responsive activator of the mal operon. However, the mal operon is silent under laboratory conditions where AHLs are produced. Perhaps MalR binds to a signal other than an AHL. This would be unusual, but perhaps not unique (13), for a LuxR family member showing complete identity with the conserved residues in the AHL binding and DNA binding regions. We seek to understand how the mal operon is regulated because its product, malleilactone, appears to be a virulence factor and because it may be regulated by MalR in an unusual manner.
Here, we show MalR is required for trimethoprim activation of the malleilactone genes and mal gene transcription is stimulated by several, but not all, antibiotics. Activation of the mal genes does not involve any of the AHLs tested in B. thailandensis or in recombinant Escherichia coli. We also show that induction is not the consequence of a general antibiotic stress response, nor does it involve a direct interaction of MalR with a malleilactone-inducing antibiotic. Our results are consistent with a model whereby certain antibiotics activate expression of the silent malR gene and the malR gene product in turn activates transcription of genes for malleilactone synthesis.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and reagents.
We used B. thailandensis strain E264 (14) and E. coli strain DH5α or MG4 for genetic manipulations and recombinant DNA expression, respectively (see Table S1 in the supplemental material). Our B. thailandensis malA-lacZ chromosomal insertion mutant (BT01447) was from a sequence-defined transposon mutant library (15). This mutant has a transposon insertion in the malA coding sequence after bp 5704 (out of 8,379) relative to the predicted malA translational start site. The other strains are listed in Table S1 in the supplemental material.
All E. coli growth was in Luria-Bertani (LB) broth at 37°C with shaking, and B. thailandensis was grown in morpholinepropanesulfonic acid (MOPS)-buffered LB broth (LB-MOPS) at 30°C with shaking. When appropriate, the following antibiotics were used (per milliliter): 100 μg trimethoprim (E. coli and B. thailandensis) and 15 μg gentamicin and 100 μg ampicillin (E. coli). We added IPTG (isopropyl-β-d-thiogalactopyranoside) as indicated.
We measured β-galactosidase activity with a Tropix Galacto-Light Plus chemiluminescence kit according to the manufacturer's protocol (Applied Biosystems, Foster City, CA). β-Galactosidase activity in B. thailandensis is reported as light units relative to the optical density at 600 nm (OD600). Genomic DNA, PCR and DNA fragments, and plasmid DNA were purified by using a DNeasy blood and tissue kit, PCR, plasmid purification kit, or gel extraction kit (Qiagen) according to the manufacturer's protocol.
Genetic manipulations.
We used standard procedures for DNA manipulations (16). To assess malA-lacZ expression in a malR-null background, an unmarked, in-frame malR deletion was introduced into the malA-lacZ reporter strain BT01447 by using the deletion construct pJRC115 btaR4, which we now call malR, and methods described previously (17). To assess malA-lacZ activity in an AHL synthase-deficient strain, we prepared genomic DNA from BT01447 and introduced it into the btaI1 btaI2 btaI3 triple mutant JBT112 (17) by using natural transformation as described previously (18). The pUC18-mini-Tn7T derivatives were introduced into B. thailandensis strains with the helper plasmid pTNS2 by electroporation, as previously described (19). We used PCR to verify insertion into the attn7 site near glmS1 as previously described (15). Where indicated, we removed the trimethoprim resistance marker dhfrIIb from pUC18-mini-Tn7T derivatives by using plasmid pFLPe2, as described previously (20).
The plasmids are listed in Table S1 in the supplemental material. To generate pJN105.malR for recombinant malR expression in E. coli, malR was amplified from genomic DNA isolated from B. thailandensis E264 by using primers that incorporated restriction sites (NheI and SpeI) into the product. The amplicon was cut with NheI and SpeI and ligated into NheI-SpeI-digested pJN105 to make pJN105.malR. To generate the E. coli malA-lacZ expression vector pQF50.malA, the region upstream of malA extending from positions −1 to −500 with respect to the translational start site was amplified by PCR by using primers that incorporated restriction sites (NcoI and HindIII) into the product. The amplicon was digested with NcoI and HindIII and ligated to NcoI-HindIII-digested pQF50 to make pQF50.PmalA. To make the malA-lacZ expression vector with mutations in the putative lux box, we used a synthetic DNA fragment (IDT gBlock) identical to the PCR amplicon used to make pQF50.PmalA, except with the base substitutions T4C and G5A in the putative lux box. This gBlock was digested with NcoI and HindIII and ligated to NcoI-HindIII-digested pQF50 to make pQF50.mutPmalA. To make the plac-malR expression cassette, we first constructed the IPTG-inducible expression vector pUC18-mini-Tn7T-LAC-Tp by digesting the fragment containing the lac promoter and lacI repressor gene from pUC18-mini-Tn7T-LAC with NsiI and SpeI and ligating it to NsiI-SpeI-digested pUC18-mini-Tn7T-Tp (21). We then cloned malR into this vector by cutting malR from pJN105.malR using the NheI and SpeI sites and ligating it to SpeI-cut pUC18-mini-Tn7T-LAC-Tp. We verified the orientation of malR with respect to the lac promoter in the resulting construct, pUC18-mini-Tn7T-LAC-Tp.malR, by PCR.
Transcription reporter assays.
To assess MalR activation of malA expression in recombinant E. coli, we used E. coli MG4 with arabinose-inducible malR (pJN105.malR) and either pQF50.PmalA with lacZ fused to the wild-type malA promoter or pQF50.mutPmalA with lacZ fused to the T4C and G5A mutant malA promoter. Overnight cultures were used as starters by diluting them to an OD600 of 0.05. When experimental cultures reached an OD600 of 0.5, l-arabinose was added at the concentrations indicated to induce MalR expression. These cultures were added to sterile 16-mm test tubes or test tubes containing dried AHLs, as indicated. The volume of culture in each tube was 0.5 ml. After 2 h at 37°C with shaking, β-galactosidase activity was measured as described above.
To assess MalR activation of malA in B. thailandensis, we used a B. thailandensis malA-lacZ reporter strain (BT01447) (15). Logarithmic-phase cultures were diluted to an OD600 of 0.05 in growth medium supplemented with antibiotics or IPTG at the concentrations indicated. When used, AHLs were added to the culture tube and dried prior to adding inoculated LB broth. The final AHL concentrations were 5 μM. After 24 h at 30°C with shaking, β-galactosidase activity was measured as described above.
Purification of malleilactone.
Malleilactone was purified from B. thailandensis as follows. A stationary-phase culture of B. thailandensis E264 was used as the inoculum for 8 640-ml cultures in 4-liter Erlenmeyer flasks. The growth medium was LB-MOPS plus 9 μg/ml trimethoprim. Cells were inoculated to an OD600 of 0.05. After 32 h at 30°C with shaking (200 rpm), the cultures were extracted with 1 volume of ethyl acetate, and the extract was dried over Na2SO4 and evaporated completely in a vacuum. The remaining residue was resolved through a Kupchan liquid extraction scheme. Briefly, the dried extract was dissolved in 200 ml of methanol (MeOH)-H2O (9:1) and extracted four times with 200 ml hexanes. The remaining methanolic extract was diluted in 800 ml of MeOH-H2O (6:4), which was extracted four times with an equal volume of CH2Cl2. High-performance liquid chromatography–mass spectrometry (HPLC-MS) analysis of the three resulting fractions (MeOH-H2O, CH2Cl2, and hexanes) showed that malleilactone was in the CH2Cl2 fraction. This fraction was dried, suspended in a small volume (2 to 3 ml) of CH2Cl2, and resolved on a silica gel column (16 g; diameter, 1.25 cm; length, 20 cm). The column was equilibrated in CH2Cl2, loaded with the malleilactone-containing mixture, and eluted with 3 column volumes (CV) of CH2Cl2, followed by 3 CV (each) of 2.5%, 5%, 7.5%, and 10% MeOH in CH2Cl2. Fractions containing malleilactone, as judged by thin-layer chromatography (TLC) with 5% MeOH in CH2Cl2 as a solvent, were pooled; the pooled fractions consisted of a number of malleilactone analogs. Pure malleilactone A was obtained by a final HPLC step consisting of a preparative Eclipse XCB-C8 column (Agilent; 7 μm; 21.2 by 250 mm) operating at 12 ml/min with an elution program that contained an isocratic step (6 min; 28% acetonitrile [MeCN] in H2O) followed by a gradient from 28 to 100% MeCN over 36 min. Both MeCN and H2O contained 0.1% NH4OH, which helped stabilize the malleilactone. This procedure yielded 4 mg of pure malleilactone, which was verified by UV-visible (λmax = 370 nm), 1H nuclear magnetic resonance (NMR), and high-resolution electrospray ionization (ESI)-MS ([M+H]+ observed, 307.1907; [M+H]+ calculated, 307.1909).
RESULTS
MalR is required for trimethoprim induction of mal biosynthesis genes.
MalR is a LuxR orphan with all of the LuxR family conserved amino acids and 35% amino acid sequence identity to the P. aeruginosa AHL-responsive orphan QscR. MalR is genetically linked to a 13-gene cluster, the malleilactone (mal)-biosynthetic genes. The mal genes are transcriptionally activated by trimethoprim (11). To address the question of whether MalR is required for trimethoprim to induce expression of the mal genes, we used a B. thailandensis strain with a chromosomal lacZ fusion to malA, the first gene in the mal cluster (15). We compared malA-lacZ activation in a malR mutant to that in wild-type malR. The lacZ reporter was activated by trimethoprim in the wild type, as previously reported (11), but not in the malR mutant (Fig. 1). These results show that MalR is required for trimethoprim activation of the mal biosynthesis genes.
FIG 1.

Trimethoprim-dependent expression of malA requires MalR. Shown is growth-adjusted activity of a malA-lacZ chromosomal reporter in the malR mutant (TT04) (diamonds) or its isogenic parent (BT01447) (circles) grown with trimethoprim (solid symbols) or without trimethoprim (open symbols). The data are the means ± ranges from the results of two independent experiments.
Influence of antibiotics on mal gene expression and the B. thailandensis growth rate.
Trimethoprim is a dihydrofolate reductase inhibitor. In addition to trimethoprim, several other antibiotics activate the mal operon. They include fluoroquinolone DNA gyrase inhibitors and the cell wall biosynthesis inhibitors piperacillin, ceftazidime, and cefotaxime (11). The fact that these antibiotics are structurally distinct and target different activities suggests that they might not interact with MalR directly. An alternative explanation is that they affect the growth rate of B. thailandensis at the sublethal concentrations tested, and this might correlate with activation of the mal genes. We first tested the influence of trimethoprim, the strongest mal gene inducer (11), on growth and mal gene induction by using our B. thailandensis malA-lacZ reporter. At concentrations that induced mal expression, trimethoprim also inhibited bacterial growth (Table 1 and Fig. 2), and in fact, the level of mal gene expression was inversely correlated with growth over a range of trimethoprim concentrations (Fig. 2). We also tested other antibiotics at sub-MIC but growth-slowing concentrations. Not all of the antibiotics we tested served to activate malA-lacZ. Kanamycin, for example, did not activate lacZ expression, but it did inhibit growth (Table 1 and Fig. 2). Thus, slow growth is not sufficient for activation of the mal genes, but it may be necessary.
TABLE 1.
Abilities of antibiotics to activate malA-lacZ in B. thailandensis
| Antibiotic (μg/ml)a | Antibiotic class | Relative malA-lacZ activityb |
|---|---|---|
| None | 1 | |
| Trimethoprim (15)c | THF pathway inhibitord | 9.7 (0.4) |
| Kanamycin (115) | Ribosomal inhibitor | 0.5 (0.1) |
| Zeocin (1,200) | DNA intercalator | 1.5 (0) |
| Mitomycin C (10) | DNA cross-linker | 4.3 (0.1) |
| Sulfamethoxazole (3,000) | THF pathway inhibitor | 10 (0.7) |
| Malleilactone (15) | Unknown | 12.3 (1.5) |
The concentration of antibiotic used is indicated in parentheses, and in each case, the concentration resulted in a 70 to 80% reduction in the growth yield, as determined by the OD600 taken at the time of β-galactosidase measurement.
Growth-adjusted β-galactosidase activity relative to that of the untreated control. The β-galactosidase activity and OD600 were determined after 24 h. The values are the means of the results of two independent experiments, with the range indicated in parentheses.
Trimethoprim was previously shown to activate malA-lacZ (11).
THF pathway, tetrahydrofolate reductase pathway.
FIG 2.
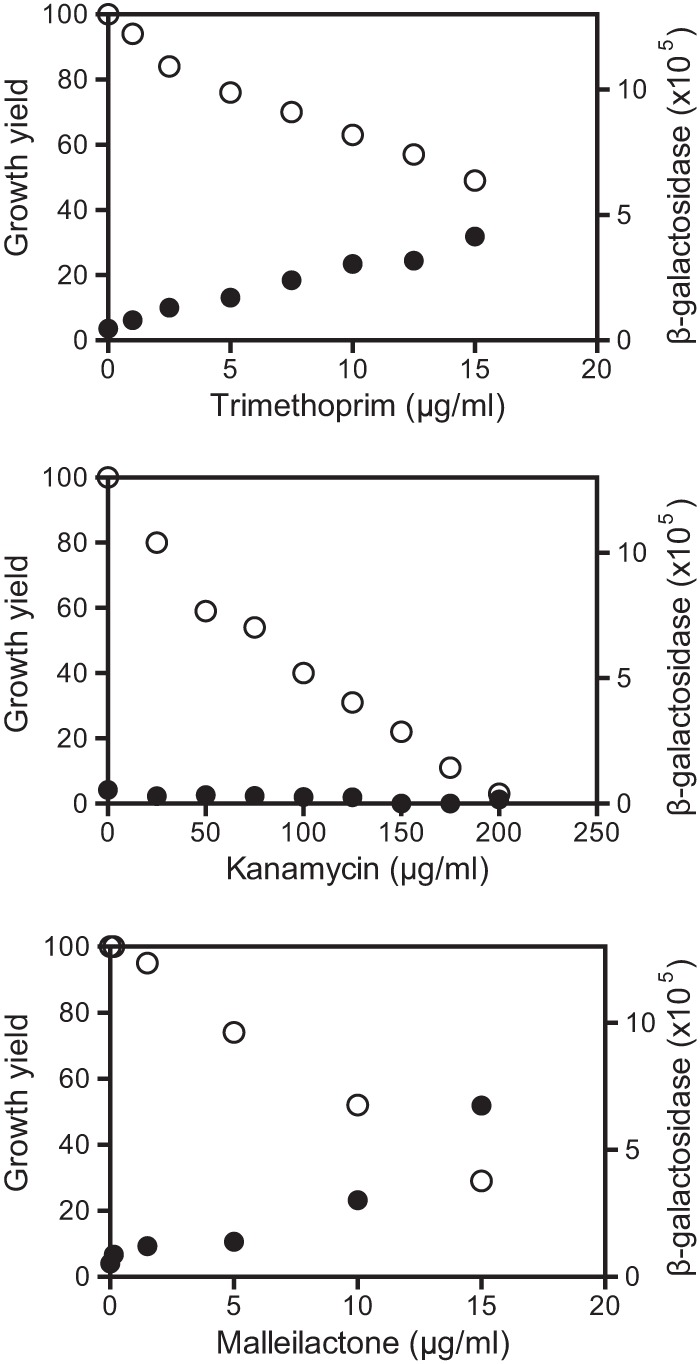
Effect of antibiotics on growth (open circles) and activation of a chromosomal malA-lacZ reporter (solid circles). The data are from the B. thailandensis malA-lacZ reporter BT01447 grown 24 h with antibiotics as indicated. The final growth yield is a percentage of that without added antibiotic measured as the OD600. β-Galactosidase activity is reported as relative light units normalized to the OD600.
Our malA-lacZ reporter disrupts malA, which is a predicted polyketide synthase thought to be critical for malleilactone synthesis (12). Thus, malleilactone itself is likely not required for trimethoprim and other antibiotics to activate the mal genes. Nevertheless, malleilactone might influence mal gene expression. To address this possibility, we purified malleilactone from cultures of trimethoprim-treated B. thailandensis cells, where it was estimated to be at a concentration of 0.8 μg/ml (see Materials and Methods). At this concentration, our purified malleilactone had a minimal effect on growth or mal gene expression in the B. thailandensis reporter strain (Fig. 2). However, at a higher concentration (15 μg/ml), malleilactone reduced growth by 70 to 80% and showed potency as a malA activator comparable to that of trimethoprim (Fig. 2).
Transcription from the malA promoter in recombinant E. coli requires MalR and the lux box-like sequence.
We engineered a plasmid with a 500-bp fragment containing the presumed malA promoter fused to a promoterless lacZ (from positions −1 to −500 with respect to the malA translation start codon) and a plasmid with an arabinose-inducible malR. In E. coli, lacZ expression was dependent on the presence of both plasmids and on arabinose (Fig. 3A).
FIG 3.
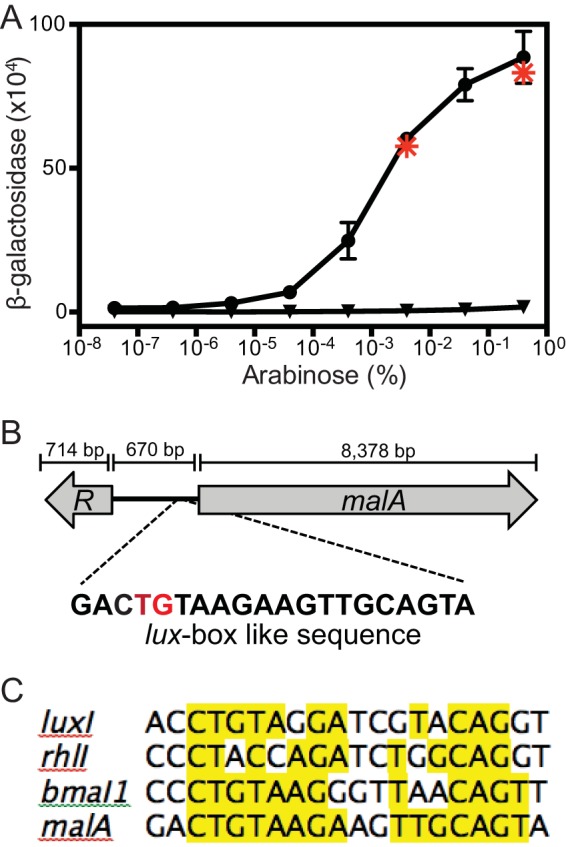
Expression of malA requires MalR and an intact lux box-like sequence in recombinant E. coli. (A) Activation of the malA promoter in E. coli containing an arabinose-inducible malR expression vector (pJN105.malR) and a lacZ reporter with either the wild-type malA promoter (pQF50.PmalA) (circles) or the malA promoter with the T4C and G5A base substitutions (pQF50.mutPmalA) (triangles). The values are the means ± ranges of the results of two independent experiments. Asterisks, E. coli containing pJN105.malR and pQF50.PmalA grown with 0.4 or 0.004% arabinose and 5 μM (each) C8-HSL, 3OHC8-HSL, and 3OHC10-HSL (the B. thailandensis AHLs). Similar results were obtained with 5 μM C4-, C6-, C10-, C12-, C14-, 3OHC6-, and p-coumaroyl-HSL. (B) malR-malA B. thailandensis genomic region. The putative lux box-like element is centered 63.5 bp upstream of the malA translation start site. The red letters indicate the T4C and G5A base substitutions in pQF50.PmalA. (C) Alignment of the malA lux box-like sequence with the lux boxes from Vibrio fischeri luxI, P. aeruginosa rhlI, and B. mallei bmaI1.
The lux box-like sequence is centered 63.5 bp upstream of the malA translational start site (Fig. 3B and C). We tested the hypothesis that MalR requires this lux box-like sequence to activate malA by introducing the base substitutions T4C and G5A (Fig. 3B), which are critical for transcription factor binding in LuxR-LuxI-type systems (22). In E. coli containing this construct, the basal level of lacZ expression was similar to that in E. coli containing the full-length malA-lacZ fusion; however, the T4C and G5A substitutions abolished MalR-dependent activation of malA-lacZ (Fig. 3A). Our results are consistent with the idea that MalR binds to the lux box-like element upstream of malA and that this binding leads to transcriptional activation of malA. Activation of reporter transcription in E. coli did not require AHLs or antibiotics.
MalR-dependent activation of the mal genes is not influenced by AHLs.
Although activation of the malA-lacZ reporter in E. coli does not require an AHL, it might nevertheless be influenced by AHLs either positively or negatively. Thus, we measured lacZ expression in our E. coli reporter containing both the malA-lacZ expression vector and the arabinose-dependent malR expression vector as follows. We grew the E. coli reporter strain in the presence of arabinose at a concentration that elicits maximal or half-maximal MalR-dependent malA-lacZ activation (0.4 or 0.004% arabinose, respectively) plus the following AHLs (5 μM each): octanoyl-homoserine lactone (HSL), 3-hydroxy-octanoyl-HSL, and 3-hydroxy-decanoyl-HSL (the B. thailandensis AHLs); butyryl-HSL, hexanoyl-HSL, decanoyl-HSL, dodecanoyl-HSL, tetradecanoyl-HSL, and 3-hydroxy-hexanoyl-HSL (C4-, C6-, C10-, C12-, C14-, and 3OHC6-HSL); and p-coumaroyl-HSL. Addition of any of the above-listed AHLs to the growth medium did not positively or negatively affect lacZ transcription (Fig. 3A and data not shown).
It is possible that AHLs do not affect MalR or transcription from the malA promoter in recombinant E. coli but that they do have an influence on malA expression in B. thailandensis. To test this possibility, we measured lacZ expression from the malA-lacZ reporter in the parent B. thailandensis strain, a malR mutant, and a mutant incapable of producing any AHLs. Cells were grown both with and without added AHLs (Fig. 4). As previously shown (Fig. 1), trimethoprim induced lacZ expression in the malR wild type but not in the malR mutant. Trimethoprim also induced malA-lacZ activity in the AHL synthesis mutant (Fig. 4). In the absence of trimethoprim, self-produced or exogenously added AHLs modestly repressed malA-lacZ reporter activity by about 2-fold (Fig. 4). Because malR is not expressed under these conditions (11), we posit that this may be due to cross-regulation by one of the other LuxR homologs encoded in the B. thailandensis chromosome. Our results with both recombinant E. coli and B. thailandensis are consistent with our conclusion that trimethoprim activates malA-lacZ expression in a MalR-dependent fashion and show that the AHLs we tested, including the B. thailandensis AHLs, are not required for MalR-mediated activation of the mal operon.
FIG 4.
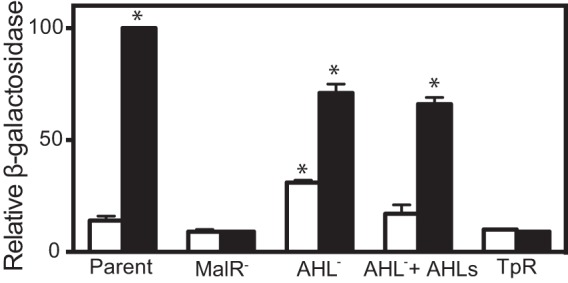
Transcriptional activation of a malA-lacZ chromosomal fusion in B. thailandensis. Growth-adjusted β-galactosidase activity is shown as a percentage of wild-type control values. The open bars indicate activity in the absence of trimethoprim, and the solid bars indicate activity in the presence of 15 μg/ml trimethoprim. Parent, B. thailandensis with the malA-lacZ chromosomal reporter (BT01447); MalR−, BT01447 with a malR deletion (TT04); AHL−, BT01447 with deletions of all three of the AHL synthase genes (ΔbtaI1-3) (TT10); AHLs, 5 μM (each) C8-HSL, 3OHC8-HSL, and 3OHC10-HSL; TpR, BT01447 with the dhfrII gene conferring trimethoprim resistance inserted in the neutral chromosomal attn7 site (TT05). The values are the means and ranges of the results from two independent experiments. Statistical analysis was done with a t test compared to the untreated parent; *, P ≤ 0.05.
MalR-dependent activation of malA transcription correlates with malR transcription.
Our results with recombinant E. coli (Fig. 3) suggest the possibility that MalR does not require a ligand to activate mal gene expression. It may be that a sufficient level of MalR is the only requirement for activation of mal gene expression and that trimethoprim and certain other antibiotics somehow activate malR transcription. In fact, trimethoprim, piperacillin, and malleilactone all induce malR expression in B. thailandensis (11) (see Fig. S1 in the supplemental material). Furthermore, we constructed a strain with plac-malR in a neutral site (glmS1) on the chromosome of a B. thailandensis malR mutant containing the malA-lacZ reporter. We monitored malA expression in this strain, grown with or without trimethoprim and increasing concentrations of IPTG to induce plac-malR expression. We found that activation of malA-lacZ required IPTG but not trimethoprim (Fig. 5).
FIG 5.
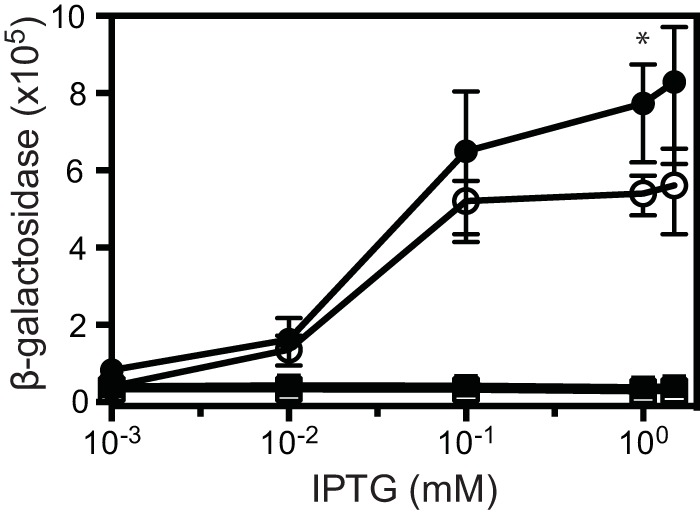
IPTG-dependent activation of malA-lacZ by a lac promoter-controlled malR in B. thailandensis. Cells were grown with 15 μg/ml trimethoprim (solid symbols) or without trimethoprim (open symbols) and IPTG as indicated. Shown is the growth-adjusted activity of a malA-lacZ chromosomal reporter in the native malR deletion with a lac promoter-controlled malR strain (TT08) (circles) and an isogenic strain with the lac promoter but no malR (TT07) (squares). The values are the means ± ranges from the results of two independent experiments. Statistical analysis was done by t test of the trimethoprim-treated strain compared to the identical strain grown without trimethoprim; *, P ≤ 0.05.
Although trimethoprim was not required for malA-lacZ activation, it had a modest stimulatory influence on IPTG-dependent activation of malA-lacZ. This raised the possibility that trimethoprim does interact with MalR directly to affect its ability to activate mal gene expression. We addressed the possibility that trimethoprim is a ligand for MalR by introducing a gene coding for a trimethoprim-resistant dihydrofolate reductase (dhfrIIb) into our reporter strain. We found that dhfrIIb completely abolished trimethoprim-dependent activation of malA-lacZ (Fig. 5). Because the dhfrIIB product changes the target of trimethoprim and does not affect trimethoprim itself, we conclude that although trimethoprim activation of the mal operon is MalR dependent, the trimethoprim effect does not result from a direct interaction of trimethoprim and MalR. This is consistent with the hypothesis that trimethoprim serves to activate malR transcription and that elevated levels of MalR are sufficient to activate the otherwise silent mal genes in laboratory cultures of B. thailandensis.
DISCUSSION
Here, we describe an unusual LuxR family protein, MalR, which retains all of the conserved amino acid residues in the AHL binding domains of LuxR family members involved in AHL quorum sensing but does not require an AHL for activity. Generally, LuxR family members that do not respond to AHLs have one or more substitutions in the conserved residues of the AHL binding domain. For example, X. oryzae OryR responds to a plant-derived signal (9), P. asymbiotica PauR responds to dialkylresorcinols (9, 10), and Serratia sp. CarR appears to be like MalR in that it does not seem to require an AHL or any exogenously added ligand for activity (23). CarR activity may also correspond to its level of transcription (24, 25). Although CarR has a change in one of the conserved AHL binding residues, genetic evidence supports the idea that the conversion to AHL independence occurred through mutations in the C terminus (23). The C-terminal mutations in CarR (23) are in a region that is not present in MalR.
The B. thailandensis mal gene cluster is silent in laboratory-grown cultures, but mal gene expression can be activated by certain antibiotics. We have shown that antibiotic activation of the mal genes requires MalR. Our results also indicate that antibiotics activate mal gene expression indirectly by driving malR expression. This conclusion is based on our results with B. thailandensis (Fig. 1 and 4) and a B. thailandensis AHL signal synthesis mutant and a trimethoprim-resistant mutant (Fig. 4), as well as with E. coli (Fig. 3), showing that sufficient levels of malR expression are all that is needed to activate mal gene expression. In our experiments, MalR does not appear to require an AHL or a ligand added to the culture medium to influence mal gene expression. It is possible that a common metabolite in B. thailandensis and E. coli serves as a ligand. Recently, the E. coli LuxR orphan SdiA was shown to bind a conserved metabolite (1-octanoyl-rac-glycerol) in the absence of its cognate AHL (13). SdiA has some activity in the absence of AHLs, as do a few other AHL-responsive LuxR homologs (e.g., Burkholderia cenocepacia CepR2) (26, 27). It is not known if any other LuxR proteins bind common metabolites, like SdiA. In ways other than the AHL requirement, MalR is similar to LuxR quorum-sensing signal receptors. For example, MalR requires an intact lux box-like sequence to activate the promoter of the malleilactone-biosynthetic genes.
Transcription of malR is activated by several, but not all, of the antibiotics we tested. In every case, antibiotics that activated mal gene expression also slowed the growth of B. thailandensis, with growth and mal activation inversely correlated. However, the influence of an antibiotic on growth was not in itself sufficient for mal gene activation. Some antibiotics slowed growth but did not affect mal gene activation. We imagine that the presence of certain antibiotics activates a specific cellular response pathway, which in turn activates expression of malR. Malleilactone may activate the mal genes through a similar pathway, although the cellular target of malleilactone is unknown. We find it interesting that antibiotic modulation of AHL-responsive LuxR family members has been reported elsewhere (28, 29). In fact, several classes of antibiotics activate transcription of the P. aeruginosa orphan QscR (29). MalR might be a degenerate AHL quorum-sensing signal receptor. There is increasing evidence that AHL receptors play an important role in interspecies competition (30, 31). Antibiotics and cytotoxic factors are often activated by AHL quorum sensing (31–33), and as discussed above, MalR (11) and QscR (29) are antibiotic activated. The mal genes code for synthesis of malleilactone, which is a cytotoxin that also has antibiotic activity against several Gram-positive bacteria (12), and as we show here (Fig. 2), malleilactone even slows the growth of B. thailandensis at high concentrations. Activation of the mal genes by low, nonlethal concentrations of antibiotics might be an alarm response whereby B. thailandensis reacts to danger by mounting an attack against competitors.
B. thailandensis is closely related to two human pathogens, Burkholderia pseudomallei and Burkholderia mallei. The malleilactone-biosynthetic gene clusters are conserved (>80% identity at the amino acid sequence level), and MalR shows 100% identity across these three species. B. pseudomallei is the causative agent of melioidosis, an often fatal emerging infectious disease that is prevalent in northeast Thailand (34). One of the difficulties in treatment is the high level of antibiotic resistance in B. pseudomallei. Trimethoprim and sulfamethoxazole are two of the few clinically useful antibiotics (35). Both of these antibiotics activate malleilactone production in B. thailandensis (Table 1). These findings, together with the finding that malleilactone is important for B. thailandensis virulence in C. elegans (12), suggest the need to evaluate how antibiotics regulate virulence and the gene homologs for malleilactone and MalR in B. pseudomallei. Finally, it is interesting that at sufficiently high concentrations (15 μg/ml) malleilactone itself inhibits the growth of B. thailandensis. Members of the genus Burkholderia are quite insensitive to most antibiotics, and this creates clinical difficulties (36). It is not unreasonable to think that malleilactone might provide a scaffold for anti-Burkholderia agents or that discovery of the malleilactone target might be helpful in drug discovery.
Supplementary Material
ACKNOWLEDGMENTS
We thank Amy Schaefer and Charlotte Majerczyk for helpful discussions and technical advice about the project.
This work was supported by the National Science Foundation under Cooperative Agreement no. DBI-0939454 and by National Institutes of Health grant GM-59026.
Any opinions, findings, and conclusions or recommendations expressed in this material are ours and do not necessarily reflect the views of the granting agencies.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00425-15.
REFERENCES
- 1.Fuqua C, Parsek MR, Greenberg EP. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 2.Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev 25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 3.Ulrich RL, Hines HB, Parthasarathy N, Jeddeloh JA. 2004. Mutational analysis and biochemical characterization of the Burkholderia thailandensis DW503 quorum-sensing network. J Bacteriol 186:4350–4360. doi: 10.1128/JB.186.13.4350-4360.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuqua C. 2006. The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J Bacteriol 188:3169–3171. doi: 10.1128/JB.188.9.3169-3171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramoni S, Venturi V. 2009. PpoR is a conserved unpaired LuxR solo of Pseudomonas putida which binds N-acyl homoserine lactones. BMC Microbiol 9:125. doi: 10.1186/1471-2180-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chugani SA, Whiteley M, Lee KM, D'Argenio D, Manoil C, Greenberg EP. 2001. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. 2001. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J Bacteriol 183:5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith JN, Ahmer BM. 2003. Detection of other microbial species by Salmonella: expression of the SdiA regulon. J Bacteriol 185:1357–1366. doi: 10.1128/JB.185.4.1357-1366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferluga S, Bigirimana J, Hofte M, Venturi V. 2007. A LuxR homologue of Xanthomonas oryzae pv. oryzae is required for optimal rice virulence. Mol Plant Pathol 8:529–538. doi: 10.1111/j.1364-3703.2007.00415.x. [DOI] [PubMed] [Google Scholar]
- 10.Brachmann AO, Brameyer S, Kresovic D, Hitkova I, Kopp Y, Manske C, Schubert K, Bode HB, Heermann R. 2013. Pyrones as bacterial signaling molecules. Nat Chem Biol 9:573–578. doi: 10.1038/nchembio.1295. [DOI] [PubMed] [Google Scholar]
- 11.Seyedsayamdost MR. 2014. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci U S A 111:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biggins JB, Ternei MA, Brady SF. 2012. Malleilactone, a polyketide synthase-derived virulence factor encoded by the cryptic secondary metabolome of Burkholderia pseudomallei group pathogens. J Am Chem Soc 134:13192–13195. doi: 10.1021/ja3052156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen Y, Nguyen NX, Rogers JL, Liao J, MacMillan JB, Jiang Y, Sperandio V. 2015. Structural and mechanistic roles of novel chemical ligands on the SdiA quorum-sensing transcription regulator. mBio 6:e02429–14. doi: 10.1128/mBio.02429-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brett PJ, DeShazer D, Woods DE. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol 48:317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 15.Gallagher LA, Ramage E, Patrapuvich R, Weiss E, Brittnacher M, Manoil C. 2013. Sequence-defined transposon mutant library of Burkholderia thailandensis. mBio 4:e00604–13. doi: 10.1128/mBio.00604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 17.Chandler JR, Duerkop BA, Hinz A, West TE, Herman JP, Churchill ME, Skerrett SJ, Greenberg EP. 2009. Mutational analysis of Burkholderia thailandensis quorum sensing and self-aggregation. J Bacteriol 191:5901–5909. doi: 10.1128/JB.00591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thongdee M, Gallagher LA, Schell M, Dharakul T, Songsivilai S, Manoil C. 2008. Targeted mutagenesis of Burkholderia thailandensis and Burkholderia pseudomallei through natural transformation of PCR fragments. Appl Environ Microbiol 74:2985–2989. doi: 10.1128/AEM.00030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majerczyk C, Brittnacher M, Jacobs M, Armour CD, Radey M, Schneider E, Phattarasokul S, Bunt R, Greenberg EP. 2014. Global analysis of the Burkholderia thailandensis quorum sensing-controlled regulon. J Bacteriol 196:1412–1424. doi: 10.1128/JB.01405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi KH, Mima T, Casart Y, Rholl D, Kumar A, Beacham IR, Schweizer HP. 2008. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl Environ Microbiol 74:1064–1075. doi: 10.1128/AEM.02430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi KH, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 22.Antunes LC, Ferreira RB, Lostroh CP, Greenberg EP. 2008. A mutational analysis defines Vibrio fischeri LuxR binding sites. J Bacteriol 190:4392–4397. doi: 10.1128/JB.01443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poulter S, Carlton TM, Spring DR, Salmond GP. 2011. The Serratia LuxR family regulator CarR 39006 activates transcription independently of cognate quorum sensing signals. Mol Microbiol 80:1120–1131. doi: 10.1111/j.1365-2958.2011.07634.x. [DOI] [PubMed] [Google Scholar]
- 24.Slater H, Crow M, Everson L, Salmond GP. 2003. Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol Microbiol 47:303–320. doi: 10.1046/j.1365-2958.2003.03295.x. [DOI] [PubMed] [Google Scholar]
- 25.Fineran PC, Slater H, Everson L, Hughes K, Salmond GP. 2005. Biosynthesis of tripyrrole and beta-lactam secondary metabolites in Serratia: integration of quorum sensing with multiple new regulatory components in the control of prodigiosin and carbapenem antibiotic production. Mol Microbiol 56:1495–1517. doi: 10.1111/j.1365-2958.2005.04660.x. [DOI] [PubMed] [Google Scholar]
- 26.Ryan GT, Wei Y, Winans SC. 2013. A LuxR-type repressor of Burkholderia cenocepacia inhibits transcription via antiactivation and is inactivated by its cognate acylhomoserine lactone. Mol Microbiol 87:94–111. doi: 10.1111/mmi.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malott RJ, O'Grady EP, Toller J, Inhulsen S, Eberl L, Sokol PA. 2009. A Burkholderia cenocepacia orphan LuxR homolog is involved in quorum-sensing regulation. J Bacteriol 191:2447–2460. doi: 10.1128/JB.01746-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z, Wang W, Zhu Y, Gong Q, Yu W, Lu X. 2013. Antibiotics at subinhibitory concentrations improve the quorum sensing behavior of Chromobacterium violaceum. FEMS Microbiol Lett 341:37–44. doi: 10.1111/1574-6968.12086. [DOI] [PubMed] [Google Scholar]
- 29.Yeom DH, Im SJ, Kim SK, Lee JH. 2014. Activation of multiple transcriptional regulators by growth restriction in Pseudomonas aeruginosa. Mol Cells 37:480–486. doi: 10.14348/molcells.2014.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandler JR, Heilmann S, Mittler JE, Greenberg EP. 2012. Acyl-homoserine lactone-dependent eavesdropping promotes competition in a laboratory co-culture model. ISME J 6:2219–2228. doi: 10.1038/ismej.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An D, Danhorn T, Fuqua C, Parsek MR. 2006. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc Natl Acad Sci U S A 103:3828–3833. doi: 10.1073/pnas.0511323103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duerkop BA, Varga J, Chandler JR, Peterson SB, Herman JP, Churchill ME, Parsek MR, Nierman WC, Greenberg EP. 2009. Quorum-sensing control of antibiotic synthesis in Burkholderia thailandensis. J Bacteriol 191:3909–3918. doi: 10.1128/JB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bainton NJ, Stead P, Chhabra SR, Bycroft BW, Salmond GP, Stewart GS, Williams P. 1992. N-(3-oxohexanoyl)-L-homoserine lactone regulates carbapenem antibiotic production in Erwinia carotovora. Biochem J 288:997–1004. doi: 10.1042/bj2880997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Limmathurotsakul D, Wongratanacheewin S, Teerawattanasook N, Wongsuvan G, Chaisuksant S, Chetchotisakd P, Chaowagul W, Day NP, Peacock SJ. 2010. Increasing incidence of human melioidosis in Northeast Thailand. Am J Trop Med Hyg 82:1113–1117. doi: 10.4269/ajtmh.2010.10-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dance D. 2014. Treatment and prophylaxis of melioidosis. Int J Antimicrob Agents 43:310–318. doi: 10.1016/j.ijantimicag.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bazzini S, Buroni S, Udine C, Pasca MR, Riccardi G. 2014. Molecular basis for antibiotic resistance in the genus Burkholderia, p 183–196. In Coenye T. (ed), Burkholderia: from genomes to function. Caister Acadmic Press, Norfolk, United Kingdom. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


