ABSTRACT
To increase our understanding of the events that lead to HIV-1 genome packaging, we examined the dynamics of viral RNA and Gag-RNA interactions near the plasma membrane by using total internal reflection fluorescence microscopy. We labeled HIV-1 RNA with a photoconvertible Eos protein via an RNA-binding protein that recognizes stem-loop sequences engineered into the viral genome. Near-UV light exposure causes an irreversible structural change in Eos and alters its emitted fluorescence from green to red. We studied the dynamics of HIV-1 RNA by photoconverting Eos near the plasma membrane, and we monitored the population of photoconverted red-Eos-labeled RNA signals over time. We found that in the absence of Gag, most of the HIV-1 RNAs stayed near the plasma membrane transiently, for a few minutes. The presence of Gag significantly increased the time that RNAs stayed near the plasma membrane: most of the RNAs were still detected after 30 min. We then quantified the proportion of HIV-1 RNAs near the plasma membrane that were packaged into assembling viral complexes. By tagging Gag with blue fluorescent protein, we observed that only a portion, ∼13 to 34%, of the HIV-1 RNAs that reached the membrane were recruited into assembling particles in an hour, and the frequency of HIV-1 RNA packaging varied with the Gag expression level. Our studies reveal the HIV-1 RNA dynamics on the plasma membrane and the efficiency of RNA recruitment and provide insights into the events leading to the generation of infectious HIV-1 virions.
IMPORTANCE Nascent HIV-1 particles assemble on plasma membranes. During the assembly process, HIV-1 RNA genomes must be encapsidated into viral complexes to generate infectious particles. To gain insights into the RNA packaging and virus assembly mechanisms, we labeled and monitored the HIV-1 RNA signals near the plasma membrane. Our results showed that most of the HIV-1 RNAs stayed near the plasma membrane for only a few minutes in the absence of Gag, whereas most HIV-1 RNAs stayed at the plasma membrane for 15 to 60 min in the presence of Gag. Our results also demonstrated that only a small proportion of the HIV-1 RNAs, approximately 1/10 to 1/3 of the RNAs that reached the plasma membrane, was incorporated into viral protein complexes. These studies determined the dynamics of HIV-1 RNA on the plasma membrane and obtained temporal information on RNA-Gag interactions that lead to RNA encapsidation.
INTRODUCTION
The major particle assembly site for HIV-1 is the plasma membrane (1, 2). The HIV-1 Gag protein drives virus assembly; during this process, Gag coordinates multiple activities to generate viral particles, including the packaging of two copies of the viral RNA genome. Most of the HIV-1 particles contain two copies of viral RNA, indicating that RNA packaging is an efficient process (3, 4). Various virological and biochemical studies have analyzed the interactions between Gag and the viral RNA genome (2, 5). These studies showed that the nucleocapsid (NC) domain of Gag plays a critical role in specific recognition of the viral RNA genome and demonstrated that many cis-acting elements in the viral RNA, including the 5′-untranslated region (5′ UTR), are important for Gag-RNA interactions (5–13). The 5′ UTR of the HIV-1 RNA is highly structured; although the proposed structures varied in different studies, it was thought that the sequences from the primer binding site to the beginning of gag formed three stem-loops, termed SL1, SL2, and SL3. The SL3 sequence was the first element identified to be important for RNA packaging, as deletion resulted in a reduction of RNA packaging (14), and it is often referred to as Ψ. SL1 has been shown to play a key role in RNA dimerization. SL2 contains the major splice donor site and was recently shown not to be a stem-loop but to form the base of a three-way junction in the RNA structure (15). Despite the importance of the 5′ UTR, other viral sequences also play a role in RNA packaging, as viral sequences lacking Ψ can still be packaged, albeit at a lower rate than that with wild-type RNA (16, 17).
A series of innovative studies used total internal reflection fluorescence (TIRF) microscopy to examine HIV-1 assembly (16, 18–22). In TIRF microscopy, a shallow volume between the glass and the cell surface is illuminated, making it an ideal technique for studying events near the plasma membrane. In these studies, fluorescent proteins were used to tag viral macromolecules, such as the Gag protein, and the behavior of these molecules was examined in living cells to provide novel dynamic and temporal information on HIV-1 assembly. Several studies used a mixture of untagged Gag and Gag fused with a fluorescent protein to monitor virus assembly, and such a mixture generated morphologically normal viruses (23). Although a single Gag-fluorescent protein fusion molecule is below the detection limit and multiple tagged proteins are needed to detect a signal, the assembly dynamics can be determined by monitoring the increase of fluorescent protein intensity (18, 19, 22). These studies showed that active assembly occurs on the plasma membrane, as HIV-1 Gag signals accumulate with time on the plasma membrane, which often takes ∼5 to ∼30 min to reach the maximal intensity (18, 19, 22). It was also shown that varying the proportion of the tagged Gag molecules did not affect the measured assembly time, indicating that the virus assembly kinetics was not affected by the fluorescent protein tag of the Gag molecules (18). Additionally, interactions between the assembly complex and the host proteins important for the budding of viral particles were also studied to reveal the spatial and/or temporal relationship of the assembly/budding process (20, 21, 24–26).
The interactions between Gag and RNA that lead to virus assembly at the plasma membrane were studied in an elegant report (16). In that study, HIV-1 RNA was modified to contain stem-loop sequences recognized by the bacteriophage MS2 coat protein. The MS2 coat protein tagged with green fluorescent protein (GFP) can bind to HIV-1 RNA, and the RNA's movement can be monitored by the GFP signal. By tracking individual RNA signals, Jouvenet et al. (16) showed that, in the absence of Gag, HIV-1 RNA stayed on the plasma membrane for only a very brief time, ∼3.6 s. In the presence of Gag, HIV-1 RNA signals were first observed near the assembly site, followed by a weak and colocalized Gag signal; the intensity of the Gag signal increased with time, suggesting that Gag was assembled in the presence of viral RNA.
Although the aforementioned study was informative, many questions remain unanswered. For example, what is the window of opportunity for the RNA near the plasma membrane to interact with Gag? Furthermore, how often do HIV-1 RNAs that have reached the plasma membrane become part of the assembly complexes? We sought to address these unanswered questions regarding Gag-RNA interaction during virus assembly. Previously, we showed that cytoplasmic HIV-1 RNA has dynamic movement and is transported mainly by diffusion (27). We reasoned that because TIRF observations are limited to a very shallow volume, an RNA molecule may move in and out of the observation volume during the observation time due to its dynamic movement. It is possible that once the viral RNA leaves the TIRF observation volume, it rarely or never reenters this region; in this scenario, the length of time an RNA molecule can be tracked, i.e., ∼3 s, is the amount of time the RNA stays near this region. However, we observed that most HIV-1 RNAs move in a nondirectional, random-walking manner in the cytoplasm (27). Thus, it is highly likely that a large portion of the RNAs may leave and reenter the region near the plasma membrane; in this scenario, the time that the RNAs stay near the plasma membrane may be longer than indicated by the track lengths of the viral RNAs.
For this report, we used two approaches different from those from the previous study of Jouvenet et al. to examine the time that the viral RNA stays near the plasma membrane by TIRF microscopy. First, we used a photoconvertible fluorescent protein, Eos (28), to tag viral RNA; this approach allowed us to distinguish between RNAs that were near the plasma membrane at a given time and those arriving later. Eos generally emits signals detectable in the green wavelength; when exposed to near-UV light, Eos undergoes an irreversible structural change and emits signals detectable in the red wavelength (28). We exposed cells at a TIRF angle to photoconvert Eos proteins near the plasma membrane to selectively detect RNAs by the red signals. In contrast, the RNA signals that reached the plasma membrane at a later time carried green signals. This method allowed us to distinguish the existing versus newly arriving viral RNAs. Second, we used a population-based approach to study the photoconverted Eos signals. Rather than following each individual track, we monitored the total number of red signals at various time points to determine how long they stayed near the plasma membrane. Using this system, we addressed the following three experimental questions. How long do HIV-1 RNAs stay near the plasma membrane? How do Gag proteins affect the length of time that HIV-1 RNA stays near the plasma membrane? And what proportion of HIV-1 RNA that reaches the plasma membrane becomes part of the viral assembly complexes? The answers to these questions provide insights into the dynamics and mechanisms of HIV-1 RNA packaging and virus assembly.
MATERIALS AND METHODS
Plasmid construction and cell culture.
The HIV-1 construct 1-Gagnull-BSL was derived from the previously described construct 1-AAG (27) by deleting 8 bases from the CA sequence to generate a frameshift mutation, thereby leading to a premature stop codon 693 nucleotides (nt) downstream of the theoretical translation start of gag. Construct 1-GagBFP-BSL was derived from 1-GagCeFP-BSL (3) by replacing the cerulean fluorescent protein (CeFP) gene with the blue fluorescent protein (BFP) gene (29). Constructs 1-GagΔCTDBFP-BSL and 1-GagΔCTD-BSL were derived from 1-GagBFP-BSL and 1-Gag-BSL (27), respectively, by deleting the region encoding the last 83 amino acids of the capsid. Plasmid 1-GagLZCeFP-BSL was constructed by replacing a portion of the gag sequence of 1-GagCeFP-BSL (3) with the counterpart from 1-GagLZmCherry-BSL (27), which encodes a leucine zipper motif in place of the NC domain. Plasmid Bgl-Eos was derived from Bgl-YFP (27) by replacing the yellow fluorescent protein (YFP) gene with the d2EosFP (Eos) gene, which encodes a dimeric Eos fluorescent protein (28). HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, penicillin (50 U/ml), and streptomycin (50 U/ml). All cultured cells were maintained at 37°C with 5% CO2. For imaging experiments, 2 × 105 HeLa cells were seeded onto a 35-mm glass-bottom dish (MatTek) 24 h before transfection. Transfection was performed with FuGENE HD (Roche) according to the manufacturer's recommendations. Live-cell imaging was performed starting 9 h after transfection. In some experiments, transfected cells were fixed for 10 min in 4% paraformaldehyde in Dulbecco's phosphate-buffered saline for 10 min, followed by two washes in Dulbecco's phosphate-buffered saline.
Microscopy and image acquisition, processing, and analysis.
TIRF microscopy was performed using an inverted Olympus TIRF 3 system microscope and a 60×, 1.49-numerical-aperture Olympus Apo N oil-immersion objective. The microscope was enclosed in a custom-made chamber (Precision Plastic, Inc.), and imaging was performed under controlled conditions at 37°C. For the illumination of BFP, green-Eos, and red-Eos, 405-nm, 488-nm, and 561-nm laser sources (Coherent, Inc.) and BrightLine single-band laser excitation and emission filter sets LF405-A, LF488-A, and LF561-A (Semrock, Inc.), respectively, were used. CeFP signals were imaged under the same conditions as those for BFP. Digital images were acquired by use of an EM-CCD camera (Hamamatsu ImagEM Enhanced model C9100-13). Photoconversion of the Eos protein was performed in the TIRF mode, using a 5-s exposure with the 405-nm laser. Continuous imaging was performed by image streaming with a 100-ms exposure time for 10 s. ImageJ 1.47v was used to process images and to generate time-lapse image stacks. Single-molecule tracking was performed using Localize (30) and a customized MatLab program (Mathworks). Time-lapse TIRF experiments were automated by use of a customized journal (MetaMorph Premier software, 64-bit, version 7.8.3.0). The BFP, green-Eos, and red-Eos channels were aligned with an accuracy of <1 pixel apart prior to imaging, using custom triple-color fiduciary markers. The BFP, green-Eos, and red-Eos channels were sequentially excited, and images were captured for 100 ms, 100 to 300 ms, and 100 to 300 ms, respectively. The positions of the diffraction-limited spots of BFP, green-Eos, and red-Eos in each frame were determined using Localize software (30). BFP-Eos colocalization was defined as the presence of the two signals at a distance of <3 pixels; signal colocalization analysis was performed using a custom MatLab program (Mathworks).
RESULTS
System to study the dynamics of HIV-1 RNA near the plasma membrane.
To characterize the dynamics of HIV-1 RNA on the plasma membrane, we used a modified NL4-3-based HIV-1 construct, 1-Gagnull-BSL. The general structure of this construct is shown in Fig. 1A; 1-Gagnull-BSL contains cis-acting elements required for HIV-1 replication, including 5′ and 3′ UTRs, and expresses functional Tat and Rev proteins. Additionally, 1-Gagnull-BSL contains inactivated gag, pol, and env genes. The gag gene contains two inactivating mutations; the translation start codon AUG was changed to AAG, and a frameshift mutation was introduced in the middle of the capsid region, leading to a premature stop codon. Both the pol and env genes contain large inactivating deletions. The construct also contains 18 copies of stem-loop sequences (BSL) inserted into the pol gene; these sequences are specifically recognized by the Escherichia coli RNA-binding protein BglG.
FIG 1.
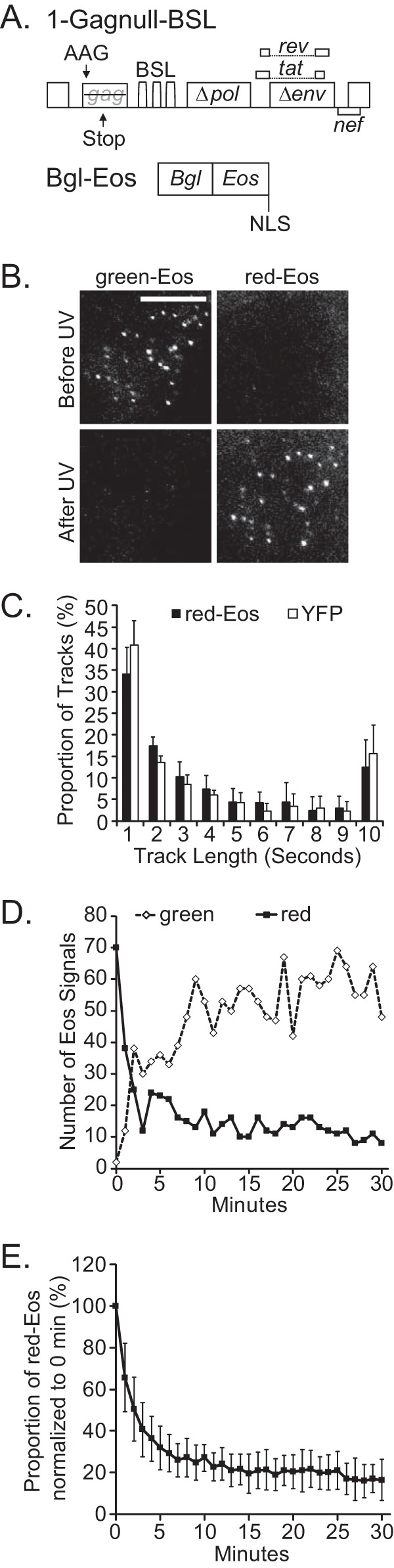
Strategy to study the dynamics of HIV-1 RNA near the plasma membrane. (A) General structures of the modified HIV-1 genome for RNA detection and a construct that expresses the RNA-binding Bgl-Eos fusion protein. BSL, stem-loop sequences recognized by the BglG RNA-binding protein; NLS, nuclear localization signal. (B) Representative TIRF images of HIV-1 RNA labeled with Bgl-Eos near the plasma membrane of HeLa cells before and after photoconversion by near-UV light exposed at a TIRF angle. Bar, 10 μm. An example of a fixed cell is shown; the photoconversion efficiency was 94%. (C) Times that individual HIV-1 RNA tracks stayed near the plasma membrane. RNAs derived from 1-Gagnull-BSL were labeled with Bgl-YFP (open bars) or photoconverted Eos (red-Eos; solid bars). (D) HIV-1 RNA levels in a representative cell, detected via Eos proteins. After near-UV light exposure at a TIRF angle, Eos signals near the plasma membrane were detected at various time points. 0 min, images were captured immediately after photoconversion. Red-Eos signals are shown as solid black squares, and green-Eos signals are shown as open diamonds. (E) Dynamics of HIV-1 RNA near the plasma membrane, detected via photoconverted red-Eos. Results for 18 cells are summarized. Data are averages with standard deviations (error bars). The average photoconversion efficiency for these experiments was 91%.
We generated a plasmid that expresses a truncated BglG protein fused to Eos, a photoconvertible fluorescent protein (Fig. 1A). Bgl-Eos contains a nuclear localization signal (NLS) and is preferentially located in the nucleus. However, when 1-Gagnull-BSL and Bgl-Eos are coexpressed, Bgl-Eos can specifically bind to BSL-containing HIV-1 RNA and traffic back into the cytoplasm. Because the BSL sequence is located in the pol gene and is removed from singly or fully spliced HIV-1 RNAs, only the full-length, unspliced 1-Gagnull-BSL RNA is labeled with Bgl-Eos.
We selected the Eos protein to label viral RNA because of its ability to alter the emission wavelength. Eos emits light at wavelengths in the range of green fluorescence; however, this protein undergoes an irreversible structural change when exposed to near-UV light, i.e., it photoconverts and emits red fluorescence (28). To test the ability of the Bgl-Eos fusion protein to photoconvert when bound to HIV-1 RNA, we transfected HeLa cells with 1-Gagnull-BSL and Bgl-Eos and examined the RNA signals near the plasma membrane by using TIRF microscopy. Our results showed abundant green signals but no red signal (Fig. 1B, upper panels). After exposing cells to near-UV light at a TIRF angle, most of the green signals were replaced by the red signals (94% photoconversion efficiency) (Fig. 1B, lower panels). These results demonstrated that Eos signals were efficiently converted upon near-UV light treatment; additionally, HIV-1 RNA could be labeled by Bgl-Eos and observed when either green or red fluorescence was emitted.
To study HIV-1 RNA movement near the plasma membrane by using TIRF microscopy, we coexpressed 1-Gagnull-BSL with Bgl-Eos, photoconverted the Eos near the plasma membrane by exposing cells to near-UV light at a TIRF angle, and then captured the RNA signals in consecutive frames, with a 100-ms exposure time per frame, for 10 s. We then measured the length of time that individual RNA signals could be followed on the plasma membrane. Our results for a total of 7,579 RNA tracks obtained from 12 cells are summarized in Fig. 1C (black bars). Most of the RNA signals were observed for less than a few seconds, whereas a portion of the viral RNA (∼15%) was observed for the entire imaging time of 10 s. Because all the tracks that could be followed for 10 s or more are shown for the 10-s data point, the RNA proportion for that point is higher than some of the others, such as that at the 9-s data point. For comparison, we also performed experiments using a Bgl-YFP protein to tag 1-Gagnull-BSL RNA, and we observed similar results (Fig. 1C, white bars). These results are consistent with previous observations (16) and indicate that, in the absence of Gag protein, HIV-1 RNA moves dynamically near the plasma membrane. Additionally, these results show that near-UV light treatment does not significantly alter the observed RNA residence time, because similar distributions were obtained when HIV-1 RNA was labeled with Bgl-YFP or photoconverted Bgl-Eos.
Population-based approach to study HIV-1 RNA dynamics near the plasma membrane in the absence of Gag proteins.
Following individual HIV-1 RNA signals by using TIRF microscopy demonstrated that most of the signals could be tracked near the plasma membrane for only seconds (Fig. 1C) (16). It is possible that, without Gag, most HIV-1 RNA can stay near the plasma membrane for only a few seconds. However, because TIRF microscopy has a very shallow observation volume, it is possible that an RNA molecule may stay near the plasma membrane longer than indicated by these studies, but moving into and out of the field of view, thereby generating the impression of a short residence time. With tags such as YFP, RNA signals that reenter the observation volume cannot be distinguished from those that enter this volume for the first time; thus, such a labeling method cannot determine how long an RNA signal stays near the membrane. To distinguish between the two aforementioned possibilities, we utilized the ability of Eos to be photoconverted and to emit light at different wavelengths. Our strategy was to photoconvert RNA signals near the plasma membrane so that they would emit red signals and to monitor the number of red-Eos signals over time.
To monitor the length of time that HIV-1 RNA stays near the plasma membrane, we exposed cells to near-UV light at a TIRF angle to convert the Bgl-Eos near the plasma membrane and then captured images of RNA signals at 1-min intervals for 30 min. In these experiments, instead of tracking the times of individual RNA signals, we examined the number of photoconverted red-Eos RNA signals that appeared on the plasma membrane at each time point. Results for a representative cell are shown in Fig. 1D, and an image was captured immediately after photoconversion (0 min). At 0 min, there were 70 red signals near the plasma membrane, and the number of red signals decreased to 38 and 25 in 1 and 2 min, respectively (Fig. 1D). After the initial sharp decline, the number of red signals decreased slowly; there were 11 and 8 signals at the 25- and 30-min time points, respectively. We also monitored the green signals from RNAs labeled by Eos proteins that were not photoconverted (Fig. 1D). There were two green signals at minute 0, indicating that the photoconversion was efficient. The number of green signals increased with time, and there were 69 and 48 signals at the 25- and 30-min time points, respectively. These results demonstrate that HIV-1 RNAs were moving dynamically during the observation time, entering and leaving the TIRF volume.
Results for 18 cells are summarized in Fig. 1E; for clarity, only the dynamics of red-Eos is shown. The number of red-Eos signals detected at 0 min for each cell was set as 100%, and the numbers of signals at various later time points were expressed as percentages of the signal observed at 0 min. These results showed that ∼50% of the RNAs were detected near the plasma membrane after more than 2 min and ∼20% of the RNAs were detected after 20 to 30 min (Fig. 1E). Therefore, the population-based approach showed that HIV-1 RNAs stay near the plasma membrane at least 30 times longer than estimated using individual tracks.
Determining the effect of Gag binding on HIV-1 RNA dynamics near the plasma membrane.
HIV-1 RNA can interact with Gag, and Gag-RNA complexes may have different dynamics on the plasma membrane. Gag and RNA can also assemble into viral particles at the plasma membrane. To study the effect of Gag binding without the complication of virus assembly, we used a mutant Gag protein that harbors a deletion in the C-terminal domain of capsid (ΔCTD), which maintains the ability to bind HIV-1 RNA and target the plasma membrane but has a multimerization defect and cannot form virus particles (16, 31, 32). Two constructs (Fig. 2A) with general structures similar to that of 1-Gagnull-BSL were used. Each construct expressed a BSL-containing full-length viral RNA that encoded a ΔCTD mutant Gag protein: 1-GagΔCTDBFP-BSL expressed a blue fluorescent protein (BFP)-tagged mutant Gag protein, whereas 1-GagΔCTD-BSL expressed an untagged mutant Gag protein. We cotransfected these two constructs at an equimolar ratio and selected cells with diffuse BFP signals, photoconverted Eos near the plasma membrane, and monitored the red-Eos signals every minute for 30 min. The results for 11 cells are summarized in Fig. 2B. In sharp contrast to HIV-1 RNA in the absence of Gag expression (Fig. 1E), we found that most of the RNA signals (>60%) were detected at 30 min postphotoconversion. We also examined the RNA signals at 5 time points, i.e., 0, 15, 30, 45, and 60 min, and observed similar results (Fig. 2C). Thus, even though the Gag proteins had a defect in multimerization, their presence resulted in a significant increase in the time that HIV-1 RNA stayed near the plasma membrane. These results suggest that most of the RNAs were associated with Gag and that the Gag-RNA complexes stayed near the plasma membrane much longer than HIV-1 RNA not associated with Gag.
FIG 2.
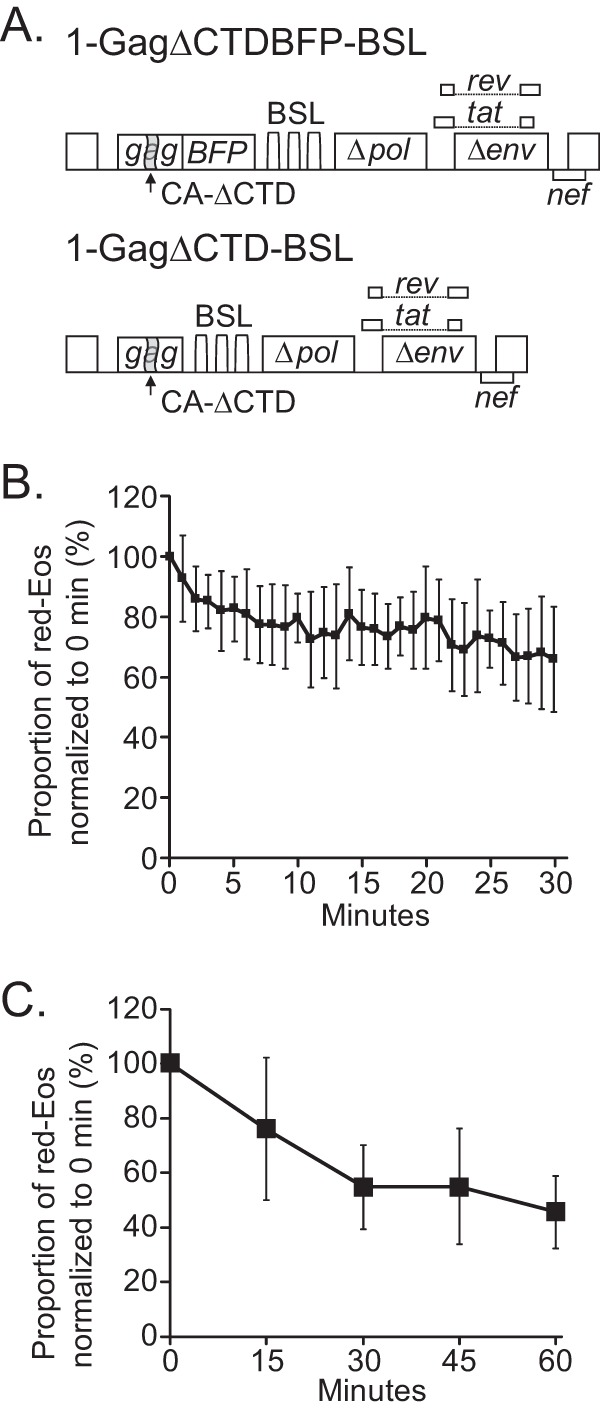
Effects of a mutant Gag protein with multimerization defects on the dynamics of HIV-1 RNA near the plasma membrane. (A) General structures of the HIV-1 constructs used in this study. The GagΔCTD mutant contained a deletion of the region encoding the C-terminal portion of the capsid domain. The proportions of red-Eos signals normalized to those at 0 min are shown in panels B and C. The average photoconversion efficiencies for these experiments were 90% and 91% for panels B and C, respectively.
To confirm that the extended stay of HIV-1 RNA near the plasma membrane was indeed caused by Gag association, we examined the dynamics of HIV-1 RNA signals on the plasma membrane when the RNA was coexpressed with Gag proteins that cannot specifically bind to viral RNA. For this purpose, we used two constructs (Fig. 3A), each encoding a mutant Gag protein (GagLZ) in which the nucleocapsid domain is replaced by a leucine zipper motif (33); 1-GagLZCeFP-BSL expresses a mutant Gag protein tagged with CeFP, whereas 1-GagLZ-BSL expresses an untagged mutant Gag protein. It is well established that these mutant GagLZ proteins can assemble into virus-like particles; however, viral RNA genomes are not packaged in these particles (34, 35). We cotransfected 1-GagLZCeFP-BSL and 1-GagLZ-BSL at an equimolar ratio into cells, ensured that Gag was expressed by observing CeFP particles on the plasma membrane, and used the aforementioned protocol to examine the red-Eos signals near the plasma membrane. The results for 5 cells (summarized in Fig. 3B) show that the red-Eos signals had dynamics similar to those of HIV-1 RNA in the absence of Gag (Fig. 1E). These results confirmed that the ability of Gag to bind HIV-1 RNA is required for HIV-1 RNA to extend its residence time near the plasma membrane.
FIG 3.
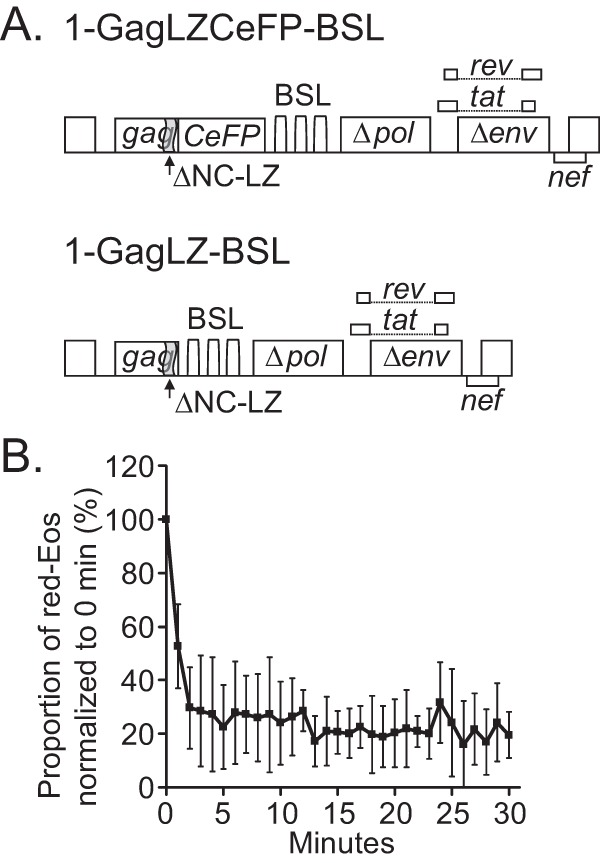
Effects of a mutant Gag protein with an RNA-binding defect on the dynamics of HIV-1 RNA near the plasma membrane. (A) General structures of the HIV-1 constructs used in this study. In the GagLZ protein, the nucleocapsid domain (NC) was replaced with a leucine zipper motif (LZ). (B) Proportions of red-Eos signals detected at various time points. The average photoconversion efficiency for these experiments was 92%.
Understanding HIV-1 RNA dynamics near the plasma membrane during virus assembly.
We used two constructs to study the effects of HIV-1 Gag on the viral genome dynamics. One construct expresses HIV-1 Gag tagged with BFP (1-GagBFP-BSL), and the other expresses untagged Gag (1-Gag-BSL) (Fig. 4A); the full-length RNAs derived from both constructs contain BSL that can be tagged with Bgl-Eos. We used the aforementioned protocol to monitor the Eos signals and also captured BFP signals, once before photoconversion and another time immediately after imaging the last Eos signals at the 60-min time point. In general, we selected and followed cells with few BFP signals at the 0-min time point.
FIG 4.
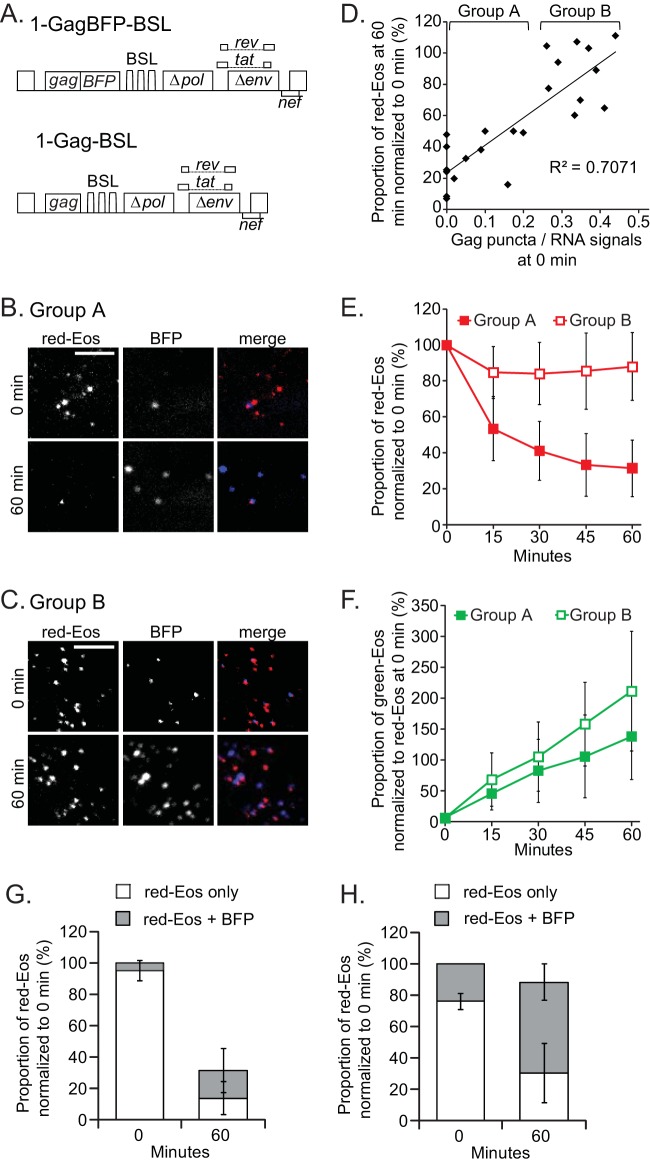
Effects of Gag on HIV-1 RNA dynamics near the plasma membrane. (A) General structures of the HIV-1 constructs used in this study. Images of representative cells that had lost more RNA signals (B) or fewer RNA signals (C) at 60 min are shown. Only a region of each cell is shown. Bars, 5 μm. (D) Correlation between RNA retention and the ratio of Gag to RNA at the 0-min time point. Each solid diamond represents a datum point for one cell. The data for 23 cells are summarized. For analysis and discussion, these cells were separated into two groups, A and B, as indicated. (E) Dynamics of red-Eos-labeled HIV-1 RNAs of groups A and B. The results for group A, summarized for 13 cells, are shown as solid squares, whereas the results for group B, summarized for 10 cells, are shown as open squares. (F) Dynamics of green-Eos-labeled HIV-1 RNAs of groups A and B. The average photoconversion efficiency for these experiments was 94%. The proportions of red-Eos (RNA) signals associated with BFP (Gag) signals were summarized for cells in group A (G) and group B (H). Open bar, red-Eos signal without an observable BFP signal; gray bar, red-Eos signal colocalized with a BFP signal. Error bars show standard deviations.
In these experiments, we observed large cell-to-cell variations in the time that Eos signals stayed near the plasma membrane. In some cells, the number of red-Eos signals decreased drastically after 60 min, whereas in other cells, most of the Eos signals remained in the observation volume after 60 min. Examples of each type of cell are shown in Fig. 4B and C, respectively. For the cell shown in panel B, 45 and 17 red-Eos signals and 4 and 34 BFP puncta were detected at the 0- and 60-min time points, respectively. For the cell shown in panel C, 127 and 119 red-Eos signals and 37 and 205 BFP puncta were detected at the 0- and 60-min time points, respectively. Further comparisons suggested that the level of Gag expression affects the dynamics of the RNA signals. In a comparison of these two groups of cells, those that lost a large portion of the Eos signals after 60 min were characterized by very few or no Gag puncta at the beginning of the imaging time (Fig. 4B). On the other hand, the cells that retained most of the Eos signals after 60 min were characterized by more Gag puncta (Fig. 4C). We then analyzed the relationship between RNA retention time and the ratio of Gag to RNA signals at the 0-min time point. These results are shown in Fig. 4D: the y axis shows the proportion of the red-Eos signals detected at the 60-min time point, whereas the x axis shows the ratio of Gag and RNA signals (BFP puncta/red-Eos signals) at the 0-min time point. Some of the cells did not have BFP puncta at time zero, which generated a 0 value on the x axis. There was a positive correlation (R = 0.7) (Fig. 4D) between the Gag expression level and the ratios of Eos signals retained after 60 min. The variation of the Gag expression level was continuous; however, we defined two groups of cells to better illustrate the results. Cells in group A had lower BFP/Eos ratios at 0 min, and cells in group B had higher ratios (Fig. 4D). The results for the two groups are summarized in Fig. 4E. The cells in group A had lost a significant portion of the Eos signals at the end of 60 min, and the loss was gradual over the monitored time points. In contrast, cells in group B retained most of the Eos signals observed at 0 min. The BFP signals measured Gag expression; hence, these results indicate that with sufficient Gag expression, most of the HIV-1 RNAs stayed near the plasma membrane for 60 min; however, with less Gag expression, a significant amount of RNA could not be detected after 60 min.
RNA-Gag association and efficiency of RNA packaged into assembly complexes.
The positive correlation between Gag expression and the proportion of HIV-1 RNA signals that remained on the plasma membrane suggests that Gag proteins interact with viral RNA and retain the complex at the plasma membrane. To examine the association of Gag signals with RNA signals, we analyzed the locations of the BFP and Eos signals on the plasma membrane. We first analyzed group A, which consisted of cells with lower Gag expression levels (Fig. 4G). At the 0-min time point, most of the red-Eos signals (95%) were not associated with BFP signals. Only 5% of the RNA signals were associated with Gag signals. At the 60-min time point, the majority of the red signals were not detected; only 32% of the red-Eos signals remained in the observation volume, and approximately half of the red-Eos signals were associated with Gag signals. The colocalized Eos-BFP signals detected at the 0-min time point were likely either at the membrane or trapped between the membrane and the glass, thereby remaining detectable in the observation volume at the 60-min time point. Thus, of the 95% of red-Eos signals not associated with Gag signals at the 0-min time point, only a small portion, 13%, stayed near the plasma membrane and became associated with Gag signals by 60 min. In contrast, a much larger proportion of the viral RNA became associated with Gag when there was more Gag expression (Fig. 4H). As shown in Fig. 4E, most of the red-Eos RNA signals (88%) remained near the plasma membrane at the 60-min time point, and 58% of the RNA signals were associated with Gag signals. Considering that 24% of the Eos signals were associated with BFP signals at time zero, under the assumption that these signals remained in the observation volume at the 60-min time point, 34% of the red-Eos RNA signals became associated with Gag during the 1-h observation time.
Therefore, our results indicate that only a portion of the full-length HIV-1 RNAs that reach the plasma membrane are incorporated into assembly complexes. Furthermore, the level of Gag expression dictates the proportion of HIV-1 RNAs that become encapsidated, indicating that during the assembly process, Gag, not the viral RNA genome, is the limiting factor.
DISCUSSION
During the assembly of infectious HIV-1 particles, Gag proteins interact with full-length viral RNAs to mediate their encapsidation. Many aspects of the HIV-1 genome encapsidation process are poorly understood, including the dynamics of the RNA genome near the assembly site, the location(s) where Gag-RNA molecules initially interact, and what proportion of the RNA that reaches the plasma membrane becomes incorporated into assembly complexes.
We used two methods to examine the dynamics of HIV-1 RNA near the plasma membrane. By tracking individual RNA signals, we observed that HIV-1 RNA stayed near the plasma membrane for only a few seconds (Fig. 1C), consistent with previously published results (16). Using a population-based approach to monitor the number of photoconverted red-Eos signals, we observed that the RNA stayed near the observation volume at least 30 times longer than estimated by tracking individual signals. We postulate that HIV-1 RNA moves dynamically and can leave and reenter the shallow TIRF volume, causing short track lengths for individual signals and resulting in the different estimates of RNA residence time near the plasma membrane obtained using these two methods.
Biochemical studies indicated that Gag can interact with full-length HIV-1 RNA and various cellular RNAs in the cytoplasm. However, it is unclear what proportion of the full-length HIV-1 RNAs in the cytoplasm interacts with Gag and what proportion of the HIV-1 RNAs that reach the plasma membrane is associated with Gag. Our results concur with a previous TIRF microscopy study showing that HIV-1 RNA signals are detected on the plasma membrane before detection of Gag signals (16). Multiple copies of Gag proteins are required for their detection on the plasma membrane. It is unclear whether HIV-1 RNAs that have reached the membrane without Gag signals are unassociated with Gag or are associated with a few Gag proteins but below the limit of detection. In this report, we compared the dynamics of the HIV-1 RNA populations in multiple cells in the absence of Gag (Fig. 1E) and in the presence of a mutant Gag protein that is unable to form virus particles (Fig. 2B and C). Our results demonstrate an increased overall time that RNA stays near the membrane, which suggests that most of the RNAs are associated with Gag when they are near the plasma membrane.
During HIV-1 expression, full-length RNA is exported from the nucleus to the cytoplasm, where translation occurs to generate Gag and Gag-Pol proteins. The cellular level of Gag is lower at the early phase of gene expression than at the later phase. We observed that the duration of HIV-1 RNA retention near the plasma membrane varied with the level of functional Gag expression (Fig. 4D and E). When fewer Gag puncta were observed, a large portion of red-Eos RNA signals left the observation volume during the monitored time, and only ∼31% of the red-Eos signals were detected at the 60-min time point. In contrast, when more Gag puncta were observed at 0 min, most (∼90%) of the red-Eos signals were detected at the 60-min time point. The precise mechanism that causes the difference in the duration that RNA stays near the membrane is unclear. One possibility is that with a lower Gag expression level, some of the RNAs that travel to the plasma membrane are either not associated with Gag or associated with only a few Gag proteins, and these RNAs stay near the plasma membrane for a shorter time. It is also possible that more Gag proteins must be added to the initial Gag-RNA complex to extend its retention time and that this process is less efficient when the Gag expression level is low, leading to the shorter time that RNA signals stay near the plasma membrane. Future experiments are needed to distinguish between these possibilities.
We examined the association of viral RNA with Gag near the plasma membrane within 60 min. In our experimental system, Gag is tagged with BFP that is excited at a 405-nm wavelength; however, exposing cells to this wavelength can also inadvertently convert green-Eos to red-Eos. Thus, we captured the Gag image once before the photoconversion and once at the end of the 60-min time span. Using this protocol, we determined the proportion of RNAs that became colocalized with Gag or were unassociated with Gag puncta after 60 min. We found that 13% and 34% of the RNAs were associated with Gag signals at the end of the observation time for the two studied groups (group A, with fewer Gag puncta at the 0-min time point, and group B, with more Gag puncta at the 0-min time point). Thus, only a limited number of the full-length HIV-1 RNAs at the plasma membrane become viral genomes in the particle, and this frequency is affected by the level of Gag expression. Interestingly, even when there was sufficient Gag to retain most of the RNAs near the plasma membrane at the 60-min time point, a significant portion of the red-Eos signals were not associated with sufficient Gag to be visualized as BFP puncta at the end of the observation time. These results indicate that not all of the RNA reaching the membrane becomes part of the assembly complexes, especially considering the large number of green-Eos signals that appeared near the plasma membrane during the same time frame (Fig. 4F), some of which were associated with Gag-BFP puncta. What determines the fate of the HIV-1 RNA near the plasma membrane remains unclear. We hope that future experiments can address whether it is pure chance that some RNAs become associated with Gag puncta and others do not or if there are differences in the regions where these RNAs are located near the plasma membrane and in the concentrations of Gag proteins in those regions.
The interactions between Gag and full-length viral RNA are complex during genome encapsidation and virus assembly. HIV-1 Gag can bind to RNA in the cytoplasm but does not multimerize to form particles in the cytoplasm. At the plasma membrane, Gag must interact with RNA to ensure genome encapsidation but limit genome packaging to one RNA dimer. A further understanding of the interactions among viral molecules, i.e., RNA-RNA, Gag-RNA, and Gag-Gag interactions, and their dynamics is important to advance our knowledge of how infectious HIV particles are assembled.
ACKNOWLEDGMENTS
We thank Malika Kuzembayeva and Elizabeth Anderson for discussions and critical readings of the manuscript, Dominic Esposito (Frederick National Laboratory for Cancer Research) for sharing the pdsEos-758 plasmid, and Daniel Larson (National Cancer Institute) for sharing the Localize software.
This research was supported by the Intramural Research Program of the NIH, NCI, CCR, by Intramural AIDS Targeted Antiviral Program grant funding to W.-S.H. and V.K.P., and, in part, by federal funds from the National Cancer Institute, National Institutes of Health (contract HHSN261200800001E).
REFERENCES
- 1.Balasubramaniam M, Freed EO. 2011. New insights into HIV assembly and trafficking. Physiology 26:236–251. doi: 10.1152/physiol.00051.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK, Hu WS. 2009. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc Natl Acad Sci U S A 106:13535–13540. doi: 10.1073/pnas.0906822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikolaitchik OA, Dilley KA, Fu W, Gorelick RJ, Tai SH, Soheilian F, Ptak RG, Nagashima K, Pathak VK, Hu WS. 2013. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog 9:e1003249. doi: 10.1371/journal.ppat.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuzembayeva M, Dilley K, Sardo L, Hu WS. 2014. Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 454–455:362–370. doi: 10.1016/j.virol.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkowitz R, Fisher J, Goff SP. 1996. RNA packaging. Curr Top Microbiol Immunol 214:177–218. [DOI] [PubMed] [Google Scholar]
- 7.Lever AM. 2007. HIV-1 RNA packaging. Adv Pharmacol (San Diego, Calif) 55:1–32. doi: 10.1016/S1054-3589(07)55001-5. [DOI] [PubMed] [Google Scholar]
- 8.Johnson SF, Telesnitsky A. 2010. Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog 6:e1001007. doi: 10.1371/journal.ppat.1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell RS, Liang C, Wainberg MA. 2004. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology 1:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rein A. 1994. Retroviral RNA packaging: a review. Arch Virol Suppl 9:513–522. [DOI] [PubMed] [Google Scholar]
- 11.Berkhout B. 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol 54:1–34. doi: 10.1016/S0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 12.Butsch M, Boris-Lawrie K. 2002. Destiny of unspliced retroviral RNA: ribosome and/or virion? J Virol 76:3089–3094. doi: 10.1128/JVI.76.7.3089-3094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Souza V, Summers MF. 2005. How retroviruses select their genomes. Nat Rev Microbiol 3:643–655. [DOI] [PubMed] [Google Scholar]
- 14.Lever A, Gottlinger H, Haseltine W, Sodroski J. 1989. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol 63:4085–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, Barton S, Hosic A, Florwick A, Santos J, Bolden NC, McCowin S, Case DA, Johnson BA, Salemi M, Telesnitsky A, Summers MF. 2015. RNA structure. Structure of the HIV-1 RNA packaging signal. Science 348:917–921. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jouvenet N, Simon SM, Bieniasz PD. 2009. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci U S A 106:19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBride MS, Panganiban AT. 1996. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol 70:2963–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jouvenet N, Bieniasz PD, Simon SM. 2008. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanchenko S, Godinez WJ, Lampe M, Krausslich HG, Eils R, Rohr K, Brauchle C, Muller B, Lamb DC. 2009. Dynamics of HIV-1 assembly and release. PLoS Pathog 5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumgartel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Krausslich HG, Brauchle C, Muller B, Lamb DC. 2011. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol 13:469–474. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- 21.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol 13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ku PI, Miller AK, Ballew J, Sandrin V, Adler FR, Saffarian S. 2013. Identification of pauses during formation of HIV-1 virus like particles. Biophys J 105:2262–2272. doi: 10.1016/j.bpj.2013.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larson DR, Johnson MC, Webb WW, Vogt VM. 2005. Visualization of retrovirus budding with correlated light and electron microscopy. Proc Natl Acad Sci U S A 102:15453–15458. doi: 10.1073/pnas.0504812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bleck M, Itano MS, Johnson DS, Thomas VK, North AJ, Bieniasz PD, Simon SM. 2014. Temporal and spatial organization of ESCRT protein recruitment during HIV-1 budding. Proc Natl Acad Sci U S A 111:12211–12216. doi: 10.1073/pnas.1321655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Engelenburg SB, Shtengel G, Sengupta P, Waki K, Jarnik M, Ablan SD, Freed EO, Hess HF, Lippincott-Schwartz J. 2014. Distribution of ESCRT machinery at HIV assembly sites reveals virus scaffolding of ESCRT subunits. Science 343:653–656. doi: 10.1126/science.1247786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prescher J, Baumgartel V, Ivanchenko S, Torrano AA, Brauchle C, Muller B, Lamb DC. 2015. Super-resolution imaging of ESCRT-proteins at HIV-1 assembly sites. PLoS Pathog 11:e1004677. doi: 10.1371/journal.ppat.1004677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Grunwald D, Sardo L, Galli A, Plisov S, Nikolaitchik OA, Chen D, Lockett S, Larson DR, Pathak VK, Hu WS. 2014. Cytoplasmic HIV-1 RNA is mainly transported by diffusion in the presence or absence of Gag protein. Proc Natl Acad Sci U S A 111:E5205–E5213. doi: 10.1073/pnas.1413169111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Rocker C, Salih A, Spindler KD, Nienhaus GU. 2004. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci U S A 101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Gruenwald D, Souslova EA, Chudakov DM, Verkhusha VV. 2008. Conversion of red fluorescent protein into a bright blue probe. Chem Biol 15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zenklusen D, Larson DR, Singer RH. 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kutluay SB, Bieniasz PD. 2010. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog 6:e1001200. doi: 10.1371/journal.ppat.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutluay SB, Zang T, Blanco-Melo D, Powell C, Jannain D, Errando M, Bieniasz PD. 2014. Global changes in the RNA binding specificity of HIV-1 Gag regulate virion genesis. Cell 159:1096–1109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Barklis E. 1995. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol 69:5716–5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Barklis E. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J Virol 71:6765–6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crist RM, Datta SA, Stephen AG, Soheilian F, Mirro J, Fisher RJ, Nagashima K, Rein A. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J Virol 83:2216–2225. doi: 10.1128/JVI.02031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]