ABSTRACT
The mumps virus (MuV) genome encodes a phosphoprotein (P) that is important for viral RNA synthesis. P forms the viral RNA-dependent RNA polymerase with the large protein (L). P also interacts with the viral nucleoprotein (NP) and self-associates to form a homotetramer. The P protein consists of three domains, the N-terminal domain (PN), the oligomerization domain (PO), and the C-terminal domain (PC). While PN is known to relax the NP-bound RNA genome, the roles of PO and PC are not clear. In this study, we investigated the roles of PO and PC in viral RNA synthesis using mutational analysis and a minigenome system. We found that PN and PC functions can be trans-complemented. However, this complementation requires PO, indicating that PO is essential for P function. Using this trans-complementation system, we found that P forms parallel dimers (PN to PN and PC to PC). Furthermore, we found that residues R231, K238, K253, and K260 in PO are critical for P's functions. We identified PC to be the domain that interacts with L. These results provide structure-function insights into the role of MuV P.
IMPORTANCE MuV, a paramyxovirus, is an important human pathogen. The P protein of MuV is critical for viral RNA synthesis. In this work, we established a novel minigenome system that allows the domains of P to be complemented in trans. Using this system, we confirmed that MuV P forms parallel dimers. An understanding of viral RNA synthesis will allow the design of better vaccines and the development of antivirals.
INTRODUCTION
Mumps virus (MuV) is a human pathogen of the Rubulavirus genus of the Paramyxoviridae family that causes an acute infection with symptoms ranging from parotitis to mild meningitis and severe encephalitis (1). The nonsegmented, negative-stranded RNA genome of MuV contains 15,384 nucleotides and encodes nine viral proteins (1). The viral RNA is encapsidated by the nucleoprotein (NP), and this helical nucleocapsid (RNP) functions as the template for viral RNA synthesis. Together, the large protein (L) and the phosphoprotein (P) make up the viral RNA-dependent RNA polymerase (vRdRp) (2). The enzymatic activities of the L protein involve the initiation, elongation, and termination of RNA synthesis, as well as mRNA capping (3). While P is not known to have intrinsic enzymatic activity, P is an essential cofactor of the polymerase. P oligomerizes by itself and forms complexes with L, NP, and RNP. It is thought that P docks the vRdRP to RNP (4).
The P proteins of paramyxoviruses are modular and consist of N-terminal (PN), oligomerization (PO), and C-terminal (PC) domains with flexible linkers between adjoining domains. The self-association of P is observed throughout negative-stranded RNA viruses (NSVs). The oligomerization domain of Sendai virus (SeV) P was the first to be crystallized, and those studies revealed a parallel coiled-coil tetramer (5, 6). The self-association of P is required for transcriptional activity, and the binding site for SeV L was found to neighbor the oligomerization region (5, 7–9). Tetrameric P structures have also been observed for other paramyxoviruses, with crystallization of the P oligomerization domains being found for measles virus, human metapneumovirus, and mumps virus (10–16).
The P protein of vesicular stomatitis virus (VSV), a member of the Rhabdoviridae family, forms a dimer consisting of two parallel α helices that are held together through hydrophobic interactions (17). Oligomerization of VSV P is also needed for P activity, as observed by a VSV minigenome assay (18, 19). The crystal structure of the rabies virus P dimerization domain reveals that each monomer consists of a helical hairpin between two α helices that permits interactions between the N-terminal helix of one monomer and the C-terminal helix of the other monomer (20). The N-terminal domain of rabies virus P interacts with the nascent NP and L, whereas the C-terminal domain binds to RNP (21–23). This structural difference may be important for rabies virus P function since the N-terminal and C-terminal domains of rabies virus are positioned on the same side, whereas these domains are at opposite ends of the oligomerization domains for VSV and SeV.
A C-terminal nucleocapsid-binding domain is found in numerous paramyxovirus P proteins (24–26). The last 49 amino acids (aa) of MuV P (aa 343 to 391) were found to directly mediate binding to the nucleocapsid through their interaction with the assembly domain of NP (27). This nucleocapsid-binding domain is conserved, but MuV P is unique in that the N-terminal domain also binds to the nucleocapsid (16). Electron microscopy revealed uncoiling of the helical nucleocapsid by the N-terminal domain of P, which resulted in enhanced viral RNA synthesis in a minigenome system (28). The crystal structure of the oligomerization domain (defined as residues 213 to 277) revealed two pairs of parallel α helices that are antiparallel to each other, which positions two N-terminal and two C-terminal domains on each end of the oligomerization domain (16). This antiparallel configuration is unique among P proteins of all nonsegmented negative-stranded RNA viruses.
In this study, we developed a novel minigenome system in which the functional domains of P can be studied using trans-complementation. Using this system, we have determined the configuration of the MuV P dimer and tetramer.
MATERIALS AND METHODS
Molecular cloning.
The plasmids used in this work were constructed using standard molecular cloning techniques. The construction details and sequence files of the plasmids are available upon request. The NP, P, and L genes of the MuVIowa/US/06 strain were cloned into the pCAGGS expression vector (29). An MuV minigenome plasmid containing Renilla luciferase (R-Luc) (BH526/pMG-RLuc) and a plasmid containing firefly luciferase (FF-Luc) (pFF-Luc) were described previously (30). P truncations with Flag epitope tags are defined as PN (aa 1 to 194-Flag), PO (aa 213 to 277-Flag), PC (Flag-aa 286 to 391), P lacking the C-terminal domain (PNO; aa 1 to 277-Flag), and P lacking the N-terminal domain (POC; Flag-aa 213 to 391). P chimeric truncations with Flag epitope tags are defined as NMuV-OPIV5 (aa 1 to 212 of MuV-aa 213 to 278 of parainfluenzavirus 5 [PIV5]-Flag), OPIV5-CMuV (Flag-aa 213 to 278 of PIV5-aa 278 to 391 of MuV), NPIV5-OMuV (aa 1 to 212 of PIV5-aa 213 to 277 of MuV-Flag), and OMuV-CPIV5 (Flag-aa 213 to 277 of MuV-aa 279 to 391 of PIV5). The MuV L deletion mutant consisting of domains I through III (LdI-III) with a hemagglutinin (HA) tag is defined as LdI-III aa 1 to 914-HA.
Cell culture and transfections.
293T cells were maintained in Dulbecco's modified Eagle medium (DMEM) with 5% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S) (Mediatech Inc., Manassas, VA). BSRT7 cells were maintained in DMEM supplemented with 10% FBS, 1% P/S, 10% tryptose phosphate broth (TPB), and 400 μg/ml G418 sulfate antibiotic (Mediatech Inc.). All cell lines were incubated at 37°C with 5% CO2 and passed at an appropriate dilution 1 day prior to use to achieve 80% to 90% confluence upon transfection. Cells were transfected using the JetPRIME transfection reagent (Polyplus Transfection Inc., New York, NY) following the manufacturer's protocols.
MuV minigenome system and dual-luciferase assay.
The MuV minigenome system used in this study was described previously (30). Increasing amounts (0, 10, 20, 40, 80, 160, or 320 ng) of total P or P-domain truncations were transfected along with NP (25 ng), L (500 ng), pMG-RLuc (100 ng), and pFF-Luc (1 ng) into BSRT7 cells. The empty pCAGGS vector was used to normalize the amount of transfected DNA per sample. After 48 h, 2/5 of the lysate from each well was used to carry out the dual-luciferase assay according to the manufacturer's protocol (Promega, Madison, WI), and light intensity was detected using a GloMax 96 microplate luminometer (Promega). Relative luciferase activity was defined as the ratio of R-Luc activity to FF-Luc activity. Six replicates of each sample were used to compare the peak activity of each P mutant to that of wild-type P. Aliquots of cell lysates from the minigenome system were used for Western blot analysis. Mouse monoclonal anti-MuV NP and anti-MuV P antibodies and anti-Flag (M2 clone) antibody (Sigma-Aldrich, St. Louis, MO) were used together to detect the expression of NP, P, and P truncations, respectively.
Cysteine residue replacement and oligomer orientation determination.
293T cells were transfected with 1 μg of PNO or POC containing cysteine mutations. After 24 h, the cells were lysed with whole-cell extraction buffer (WCEB; 50 mM Tris-HCl [pH 8.0], 280 mM NaCl, 0.5% NP-40, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol) with a mixture of protease inhibitors as previously described (31). Prior to lysis, iodoacetamide (25 mM) was added to WCEB where specified. Cleared lysates were divided into equal fractions, mixed with a one-half volume of 3× SDS loading buffer containing dithiothreitol (DTT; reducing conditions) or not containing DTT (nonreducing conditions), and heated at 95°C for 5 min. Samples were resolved in 12.5% SDS-polyacrylamide gels and transferred to a polyvinylidene difluoride membrane (GE Healthcare, Piscataway, NJ).
Immunoblotting was performed as previously described (19). The membrane was blocked with enhanced chemiluminescence prime blocking agent (GE Healthcare Life Sciences, Piscataway, NJ), incubated with mouse anti-Flag (1:1,000 dilution) followed by incubation with goat Cy3-labeled anti-mouse IgG secondary antibody (1:2,500 dilution; KPL, Gaithersburg, MD), and scanned using a Typhoon 9700 imager (GE Healthcare Life Sciences).
Coimmunoprecipitation.
To determine the P domains that interact with L, 6-well plates of 293T cells were transfected with 2 μg of P truncations with 3 μg of LdI-III. At 18 h posttransfection (hpt), the cells were starved with DMEM lacking cysteine-methionine and then labeled with 36.4 μCi/ml 35S-EasyTag Express35S protein labeling mix (PerkinElmer, Waltham, MA) for 4 h. The cells were then lysed with WCEB, and the lysate was precleared with recombinant protein G-Sepharose 4B conjugate (Invitrogen) for 1 h at 4°C and was then subjected to immunoprecipitation (IP) using recombinant protein G-Sepharose 4B conjugate and mouse anti-Flag or mouse anti-HA (Sigma-Aldrich). The IP products were washed with WCEB and resolved by 12.5% SDS-polyacrylamide gels. The gels were fixed (20% methanol, 7% acetic acid) and dried, and the proteins were visualized using a Typhoon 9700 imager (GE Healthcare).
Statistical analysis.
Statistical analysis was performed using GraphPad Prism (version 5.00) software for Windows (GraphPad Software, San Diego, CA). Student's t test was used to calculate P values.
RESULTS
Functional activity of P domains.
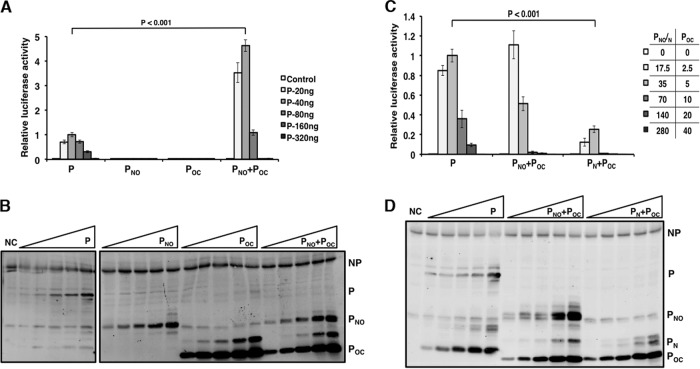
The MuV P protein contains an N-terminal domain (PN), an oligomerization domain (PO), and a C-terminal domain (PC) which are connected with flexible linker regions. To determine the roles of these domains, a series of truncations was generated (Fig. 1). To examine the role of the individual P domains in viral RNA synthesis in the absence of viral infection, these deletion mutants were tested in the MuV minigenome system that has been previously described (30). A range of concentrations of the P plasmids was used to obtain the maximal minigenome activity since expression levels varied between the P domains. Previously, we showed that the individual domains do not maintain the P function in the MuV minigenome system (28). P truncations lacking the N-terminal domain or the C-terminal domain also had no activity (Fig. 2A). When PNO and POC were transfected together in equal amounts, the luciferase activity was detected and was significantly greater than that of full-length P (Fig. 2A). The expression levels of P and NP were determined using immunoblotting (Fig. 2B). These results suggest that all P domains are required for P function. Most interestingly and importantly, PNO and POC can trans-complement each other to restore the biological activity of P.
FIG 1.
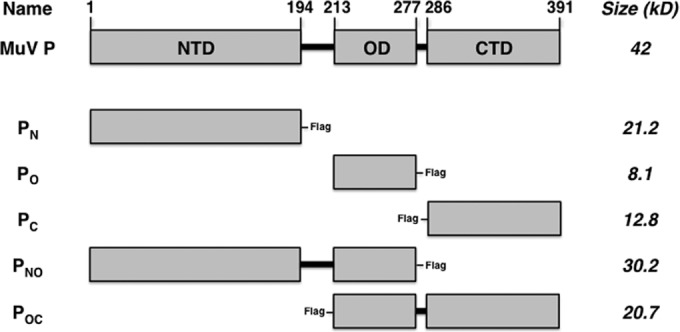
Schematic representation showing deletion mutants of MuV P. The amino acid residues for PN, PO, and PC are provided. Mutant names correspond to the domains included in the P deletion mutants, and approximate sizes are provided. The terminal Flag tag locations are included for each mutant. NTD, N-terminal domain; OD, oligomerization domain; CTD, C-terminal domain.
FIG 2.
trans-Complementation of P in minigenome system. (A) Minigenome activity of P, PNO, and POC. Increasing amounts of P or P deletion mutants were transfected together with other plasmids as described in Materials and Methods. For PNO-POC, the amount of each deletion mutant transfected was one-half of the total amount of P transfected. Renilla luciferase was the reporter gene in the minigenome, and firefly luciferase expression was used as a transfection control. The minigenome activity was measured and normalized as the ratio of Renilla luciferase activity to firefly luciferase activity (relative luciferase activity). (C) Minigenome activity of P, PN, PNO, and POC. Increasing amounts of P or P deletion mutants were used. P values were calculated using Student's t test. Error bars represent the SEMs of data from six replicates. (B and D) Immunoblotting was performed to detect the expression levels of NP, P, and the P deletion mutants. Lanes NC, control.
We previously found that addition of PN with full-length P resulted in enhanced minigenome activity due to the role of the N-terminal domain in inducing uncoiling of the nucleocapsid (28). To determine whether trans-complementation requires the oligomerization domain, the N-terminal domain alone was transfected with POC and tested in the minigenome system. Transfection of PN with POC resulted in some activity; however, the luciferase activity was significantly less than that of full-length P (Fig. 2C). The expression levels of P and NP were determined using immunoblotting (Fig. 2D). A lower ratio of POC was used due to the higher level of expression of the plasmid carrying this construct. These results suggest that self-oligomerization via PO is required for functional trans-complementation of P in viral transcription and replication. The fact that PN and PC can trans-complement each other indicates that they indeed likely form functional domains.
P oligomerizes to form parallel dimers.
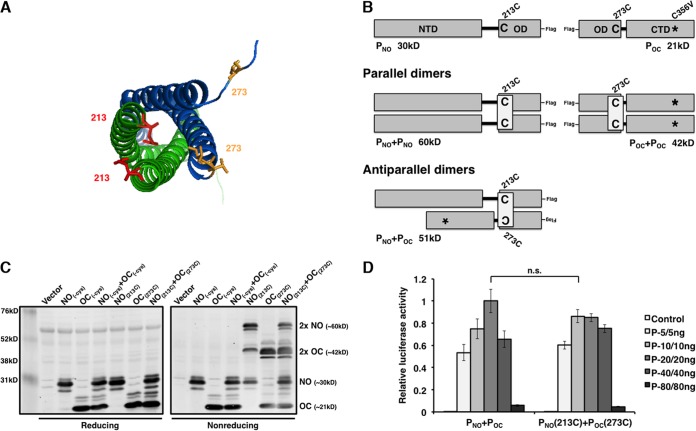
Crystal structure analysis of PO shows the formation of a tetramer consisting of two pairs of parallel α helices that are antiparallel to each other (16). This is unique among known paramyxovirus P proteins. To determine the preferential α-helical orientation during P dimerization, cysteine residues were strategically incorporated into the P protein for disulfide bond cross-linking. However, wild-type P contains one natural cysteine residue at amino acid 356 in the C-terminal domain. To avoid potential interference from this wild-type cysteine residue, a series of C356 mutants with mutations in full-length P and POC was generated and tested in the minigenome system in order to identify a mutation that maintains P function (Fig. 3A and B). P-C356V maintained the highest activity among the cysteine mutants; therefore, this modification was incorporated for all remaining P alterations.
FIG 3.
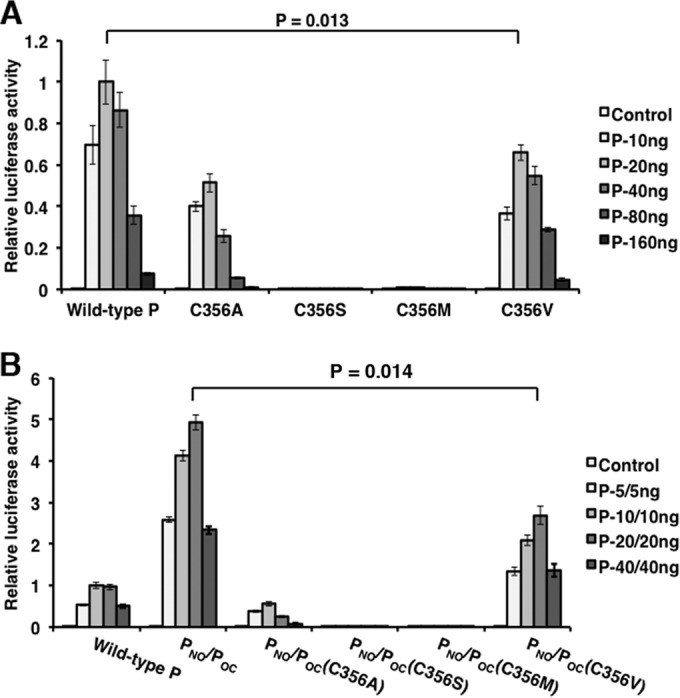
Mutation analysis of cysteine residue 356 of P. Residue C356 in full-length P and POC was mutated to alanine, serine, methionine, and valine and tested in the minigenome system. Increasing amounts of P or P deletion mutants were transfected together with other plasmids as described in Materials and Methods. The amounts transfected are provided in each graph. P values were calculated using Student's t test. Error bars represent the SEMs of data from six replicates.
PO is defined as residues 213 to 277 (16). To determine if P forms parallel dimers, residues Q213 and V273 were changed to cysteine residues on the basis of the crystal structure of the oligomerization domain (Fig. 4A). PNO and POC could be differentiated by their sizes (Fig. 4B). 293T cells were transfected with plasmids expressing PNO and POC that contained no cysteine residues or PNO with a cysteine residue at position 213 (PNO-213C) and POC-273C. Polypeptides were separated by SDS-PAGE analysis under reducing and nonreducing conditions and detected by immunoblotting (Fig. 4C). As expected, only monomers were detected for PNO (30 kDa) and POC (21 kDa) in all denaturing gels. Transfection with PNO-213C and POC-273C resulted in bands corresponding to monomer and dimer (2PNO, 60 kDa; 2POC, 42 kDa) polypeptides. Bands with molecular masses higher than the molecular mass of a monomer were eliminated under reducing conditions, suggesting that oligomerization was due to disulfide bond formation (Fig. 4C). To confirm that the cysteine mutations maintain P function, PNO-213C and POC-273C were tested in the minigenome system (Fig. 4D). Luciferase activity was similar for PNO/POC and PNO-213C/POC-273C, suggesting that addition of 213C and 273C does not disrupt P function. These results suggest that P forms parallel dimers.
FIG 4.
P mutants with engineered cysteine residues dimerize in parallel orientation with biological activity. (A) Pairs of parallel chains are shown in green or blue. Side chains of mutated amino acids 213 (red) and 273 (yellow) are highlighted. The amino acid 213 residue not contained in the file was superimposed from the adjacent parallel chain. The figure was generated using the PyMOL program (43). (B) Schematic representation of incorporated cysteine residues for disulfide bond engineering. The sizes of the dimers in the parallel and antiparallel orientations are provided. (C) Assessment of the parallel oligomerization status of P deletion mutants containing cysteine residues under reducing and nonreducing conditions. (D) Minigenome activity of engineered P deletion mutants. Increasing amounts of P mutants were transfected with other plasmids as described in Materials and Methods. P values were calculated using Student's t test. Error bars represent the SEMs of data from six replicates. n.s., not significant.
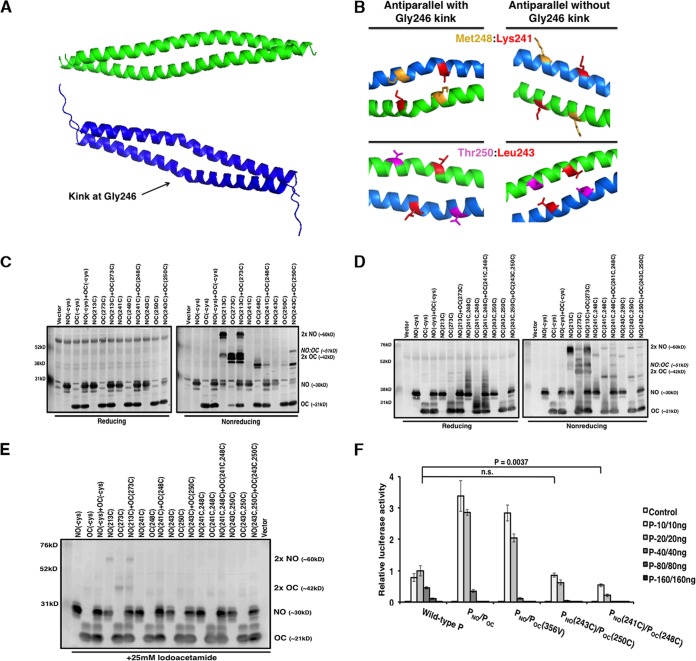
Presence of antiparallel P molecules in a tetramer.
In the P tetramer, the two pairs of parallel dimers are antiparallel to each other. The crystal structure of PO was also utilized to strategically incorporate cysteine residues at possible sites of antiparallel interactions. It is important to note that a kink at Gly246 is found in only one molecule in the pair of parallel α helices, thus distinguishing interacting side chains of each parallel pair (Fig. 5A). The kink at Gly246 is responsible for structural differences between the antiparallel dimer pairs; therefore, in each antiparallel dimer pair, separate amino acid residues were identified as having side chains in close proximity. To determine if there are antiparallel P molecules with a kink or without a kink, cysteine residues were introduced at residues K241 and M248 or L243 and T250, respectively (Fig. 5B).
FIG 5.
P mutants with engineered cysteine residues for cross-linking of antiparallel helices. (A and B) Pairs of antiparallel chains are shown in green or blue. The kink at Gly246 located on only one pair of helices is highlighted. (B) Residues with side chains in close proximity are highlighted. These residues were mutated to cysteine to engineer disulfide bonds between antiparallel α helices in chains containing and lacking the Gly246 kink. The figure was generated using the PyMOL program (43). (C and D) Assessment of the antiparallel oligomerization status of P deletion mutants containing single (C) or double (D) cysteine residues under reducing and nonreducing conditions. (E) Iodoacetamide treatment (25 mM) to achieve the natural dimerization orientation of P mutants with engineered parallel or antiparallel disulfide bonds. (F) Minigenome activity of engineered P mutants. Increasing amounts of P mutants were transfected with other plasmids as described in Materials and Methods. P values were calculated using Student's t test. Error bars represent the SEMs of data from six replicates.
Cells were transfected with plasmids expressing PNO and POC that contained no cysteine residues, PNO-213C and POC-273C as parallel dimer controls, PNO-241C, POC-248C, PNO-243C, and POC-250C. Polypeptides were separated by SDS-PAGE analysis under reducing and nonreducing conditions and detected by immunoblotting (Fig. 5C). Cotransfection with PNO-243C and POC-250C resulted in the separation of an additional band whose size corresponded to that of a PNO-POC antiparallel dimer (51 kDa). Since each pair of cysteine residues forms only a single disulfide bond, cysteine pairs were incorporated into each set of P truncations and polypeptides were examined to see if a cross-linked tetramer could be detected (Fig. 5D). Cotransfection with PNO-241C,248C and POC-241C,248C also resulted in separation of an additional band whose size corresponded to that of a PNO-POC antiparallel dimer. Extra bands corresponding to oligomer populations likely due to nonspecific interactions between the additional cysteine residues were observed. Even though the higher-molecular-mass bands were eliminated in Fig. 5C and D under reducing conditions, cell lysates were treated with iodoacetamide to confirm disulfide bond formation between dimers in their native conformation. As seen in Fig. 5E, only polypeptides corresponding to parallel dimers were observed for PNO-213C and POC-273C.
To confirm that the putative antiparallel cysteine mutations maintain P function, mutants with these mutations were tested in the minigenome system (Fig. 5F). Luciferase activity was reduced for the cysteine mutants; however, activity was still observed, indicating that incorporation of cysteine residues at these sites may hinder but not eliminate P function. The activity of PNO-241C and POC-248C was comparable to that of PNO and POC-248C (data not shown). These results suggest that even though the incorporated antiparallel cysteine residues can interact to form disulfide bonds, the P oligomerization domain preferentially polymerizes to form functional parallel dimers before formation of a more dynamic tetramer.
Charged PO zipper residues critical for P function.
To validate the amino acid residues critical for tetramer formation, we examined residues likely to disrupt the parallel and antiparallel associations of the P subunits. The crystal structure of PO revealed that the tetramer is primarily formed with hydrophobic interactions and two zippers of charged side chain interactions formed by Asp229, Glu236, and Asp240 (chain A) and by Arg231 and Lys238 (chain B) between the parallel pair of helices and by Lys253 and Lys260 (chain B) and by Asp229, Glu236, and Asp240 (chain B′) between the antiparallel pair of helices (16). To examine the roles of these residues in viral RNA synthesis, Arg231 and Lys238 were mutated to alanine to disrupt the charge interactions between the pair of parallel helices, Lys253 and Lys260 were mutated to alanine to disrupt the charge interactions between the pair of antiparallel helices, and the constructs were tested in the minigenome system. Disruption of the charged zipper interactions resulted in significantly reduced activity and completely abolished the activity for the mutant with Lys253Ala (Fig. 6A). The expression levels of P and NP were determined using immunoblotting (Fig. 6B).
FIG 6.
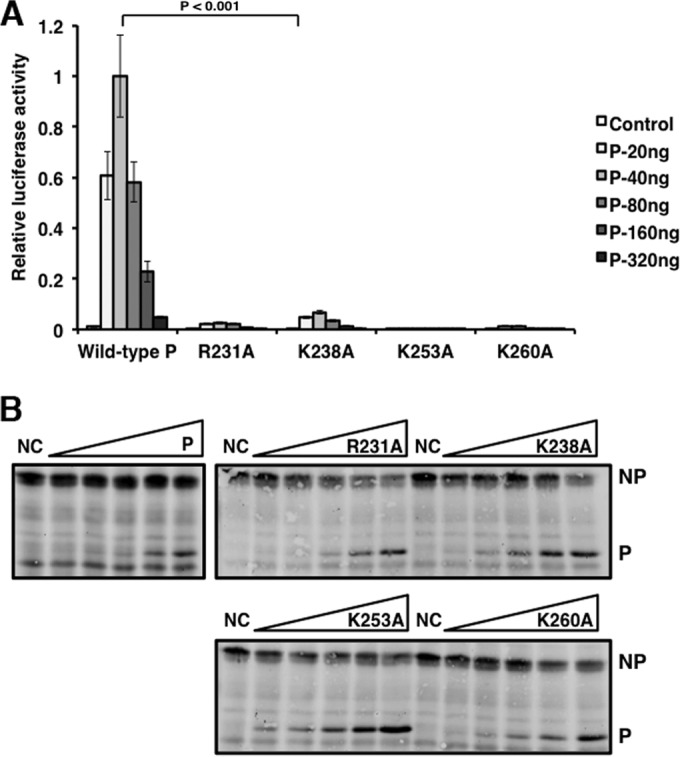
Charged residues in the P oligomerization domain are critical for P activity in the minigenome system. (A) Residues found in charged zippers were mutated to alanine and tested in the minigenome system. Increasing amounts of P mutants were transfected with other plasmids as described in Materials and Methods. P values were calculated using Student's t test. Error bars represent the SEMs of data from six replicates. (B) Immunoblotting was performed to detect the expression levels of NP, P, and the P mutants. NC, negative control (no P).
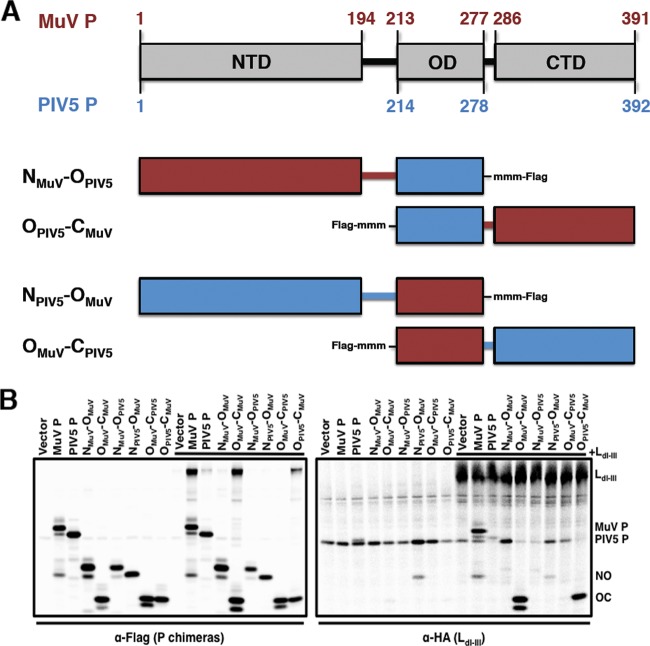
MuV PC contains a putative L-binding site.
To determine the putative L-binding site within P, P deletion mutants were utilized. In order to investigate L binding within each individual P domain, P chimeras were generated with the corresponding regions of parainfluenza virus type 5 (PIV5), the virus most closely related to MuV, to enhance the structural stability of the MuV P domains (Fig. 7A). Visualization of full-length MuV L expression was limited; therefore, L deletion mutants were generated. The N-terminal residues of the L proteins of related viruses have been found to be essential for P binding; therefore, a sequence alignment of these L proteins with MuV L was analyzed to generate a MuV L truncation consisting of domains I through III (LdI-III; aa 1 to 914) (data not shown) (32–35). Interactions between the P chimeras and MuV L were examined using coimmunoprecipitation of transfected 293T cells. The radioactively labeled cell lysates were immunoprecipitated with anti-MuV-P (full-length MuV P), anti-PIV5-P (full-length PIV5 P), anti-Flag (chimeric P truncations), or anti-HA (LdI-III). As shown in Fig. 7B, only chimeras containing the C-terminal domain of MuV P interacted with LdI-III. Coimmunoprecipitation was observed when both the P constructs or LdI-III was immunoprecipitated, suggesting that the C-terminal domain of MuV P interacted with LdI-III. It is important to note that the coimmunoprecipitation band corresponding to full-length PIV5 P is not above the background, suggesting that PIV5 P does not interact with MuV LdI-III. These results demonstrate that the C-terminal domain of MuV P interacts with LdI-III.
FIG 7.
The C-terminal domain of P binds to L. (A) A schematic representation of the predicted amino acid residues for PIV5 PN, PO, and PC from sequence analysis with MuV P is provided. Mutant names correspond to the domains included in the P deletion mutants. The terminal Flag tag locations as well as additional methionine residues (mmm), used to aid with visualization by radioactive labeling with 35S, are included for each mutant. (B) Interaction between P domains and LdI-III in 293T cells. Cells were transfected with full-length P, P chimeras, or LdI-III or cotransfected with P and LdI-III and labeled with 35S. Cell lysates were precleared with Sepharose G beads and coimmunoprecipitated with monoclonal anti-MuV P (full-length MuV P), anti-PIV5 P (full-length PIV5 P), anti-Flag (chimeric P truncations), or anti-HA (LdI-III) antibody and resolved on a 12.5% SDS-polyacrylamide gel.
DISCUSSION
The P proteins of paramyxoviruses have a modular structure, and it is thought that the individual P domains can be modified or deleted without affecting the function of the other P domains (5). The MuV P domains have been systematically mapped and characterized (16). In this study, we generated P truncations to examine the function of the P domains in viral RNA synthesis. Previously, we found that transfecting additional PN with full-length P resulted in enhanced activity in the MuV minigenome system due to uncoiling of the nucleocapsid (28). In the present study, we confirmed that all P domains are required for full P function. P multimerization has been found to be essential for viral transcription of rinderpest virus and VSV (10, 19). Interestingly, when MuV P truncations lacking either the N-terminal domain (POC) or the C-terminal domain (PNO) were transfected together, P activity was restored due to trans-complementation through PO. Thus, P multimerization via the central oligomerization domain is likely required for MuV P to function in viral RNA synthesis.
The crystal structure of MuV PO revealed the formation of a tetramer consisting of two pairs of parallel α helices that are antiparallel to each other (16). It is unknown whether MuV P favorably forms parallel or antiparallel dimers during oligomerization. To further characterize the P oligomerization structure during viral RNA synthesis, the preferential α-helical orientation of P dimers was examined using engineered disulfide bond cross-linking. The introduction of cysteine residue pairs for cross-linking analysis is an applicable strategy for examination of protein structures. MuV P residues were strategically mutated to cysteine on the basis of the close proximity of side chains found in either the parallel or the antiparallel orientation of the crystal structure (Fig. 4 and 5). Our studies showed that P dimerization was preferentially parallel and these mutants were biologically active in the MuV minigenome. The selected antiparallel residues did not always cross-link antiparallel molecules, which resulted in reduced minigenome activity, possibly because mutation of these residues disrupted the native P structure. However, when multimerization was investigated with the addition of iodoacetamide, parallel dimers were observed, thus suggesting that disulfide bonds can readily form between the parallel helices. The cross-link between antiparallel molecules was not clearly observed in several cases, suggesting that the tetramer may be a more dynamic complex or the selection of the cysteine mutations was not appropriate. However, tetramer formation is required for P activity, as supported by the findings of mutational studies of the charge zippers (Fig. 6A).
A necessity of phosphorylation for oligomerization differs between P proteins. Phosphorylation of P is required for multimerization of the rhabdoviruses VSV and Chandipura virus and the more closely related paramyxovirus respiratory syncytial virus (11, 18, 36–38). The oligomerization of unphosphorylated P proteins expressed in bacteria suggests that phosphorylation is not required, as has been shown for rinderpest virus (10). Phosphorylation has also been found to be dispensable for P oligomerization of MuV, SeV, and measles virus (5, 9). We previously screened the serine and threonine (S/T) residues of MuV P to determine the phosphorylated residues critical for viral transcription and replication. Unexpectedly, six of the nine identified residues are located in the oligomerization domain (T250, S257, T258, T261, T262, and T265) (30). Analysis of these amino acids within the crystallized MuV PO structure revealed that all six side chains are positioned outward to the tetramer; thus, it is unlikely that these residues are important for oligomerization and instead they are more accessible to interactions with host proteins, such as kinases. Further analysis of the phosphorylation of these S/T residues is ongoing.
We found a putative binding site of L to be in the C-terminal domain of P. Alterations of MuV L were required for the visualization of protein expression in our assays. The N-terminal 1,147 residues of the SeV L protein (LdI-IV) were found to bind to SeV P; however, shorter truncations abolished P binding (32). Similarly, for PIV5, human PIV3, and measles virus, P binds to the N-terminal portion of L, with the shortest truncation of L being from measles virus, which included the 408 amino acids through domain I (34, 39, 40). An alignment of the sequences of these L proteins with the MuV L sequence was analyzed to generate MuV L truncations consisting of domains I through III (aa 1 to 914), domains II through VI (aa 506 to 2261), and domains IV through VI (aa 882 to 2261) (35, 41). Visualization of L deletion mutant expression was achieved only for LdI-III. The generation of truncations of L and P to determine binding sites may cause misfolding of the viral proteins. Additional binding sites within P may be unidentified because the failure of P domains to bind to L in our studies may be a result of structural changes rather than the deletion of putative binding sites. It is important to note that the C-terminal domain of MuV P alone was unable to coimmunoprecipitate with LdI-III; therefore, the P sequences of PIV5, the virus most closely related to mumps virus, were utilized to stabilize MuV P-domain structures. Further studies are needed to more precisely locate the L-binding residues in PC.
The oligomerization of MuV P results in two parallel dimers that are in an antiparallel orientation to each other during tetramer formation. This unique tetramer alignment likely has functional significance. Both the N-terminal and C-terminal domains of P bind to the nucleocapsid, and it has been proposed that P oligomerization brings the terminal regions together in close proximity for effective nucleocapsid binding (16). Our studies reveal that the N-terminal and C-terminal domains of P can trans-complement each other to achieve functional activity in a minigenome system, likely through oligomerization of the P truncations. A cartwheel model has been proposed from studies with SeV, where P is thought to simultaneously make and break contacts with the nucleocapsid to ensure polymerase processivity during viral RNA synthesis (42). This model also proposes that binding of P to the nucleocapsid or L may open the RNP structure so that the polymerase can access the viral template. Recent studies with MuV have shown that the nucleocapsid uncoils to expose the RNA access to the polymerase through an interaction between the N-terminal domain of P and the nucleocapsid (28). Our studies suggest that MuV P is able to place the polymerase complex on the NP-RNA template via an L-binding site in the C-terminal domain of P.
ACKNOWLEDGMENTS
We appreciate the helpful discussions and technical assistance from all members of Biao He's laboratory.
This work was supported by grants (R01AI097368 and R01AI106307) from the National Institutes of Health.
REFERENCES
- 1.Carbone KM, Wolinsky JS. 2001. Mumps virus, p 1381–1441. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Emerson S, Yu Y. 1975. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J Virol 15:1348–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamb RA, Kolakofsky D. 2001. Paramyxoviridae: the viruses and their replication, p 1305–1340. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.Banerjee AK, Barik S, De BP. 1991. Gene expression of nonsegmented negative strand RNA viruses. Pharmacol Ther 51:47–70. doi: 10.1016/0163-7258(91)90041-J. [DOI] [PubMed] [Google Scholar]
- 5.Curran J, Boeck R, Lin-Marq N, Lupas A, Kolakofsky D. 1995. Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology 214:139–149. doi: 10.1006/viro.1995.9946. [DOI] [PubMed] [Google Scholar]
- 6.Tarbouriech N, Curran J, Ruigrok RWH, Burmeister WP. 2000. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat Struct Biol 7:777–781. [DOI] [PubMed] [Google Scholar]
- 7.Smallwood S, Ryan KW, Moyer SA. 1994. Deletion analysis defines a carboxyl-proximal region of Sendai virus P protein that binds to the polymerase L protein. Virology 202:154–163. doi: 10.1006/viro.1994.1331. [DOI] [PubMed] [Google Scholar]
- 8.Curran J, Pelet T, Kolakofsky D. 1994. An acidic activation-like domain of the Sendai virus P protein is required for RNA synthesis and encapsidation. Virology 202:875–884. doi: 10.1006/viro.1994.1409. [DOI] [PubMed] [Google Scholar]
- 9.Kolakofsky D, Le Mercier P, Iseni F, Garcin D. 2004. Viral RNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology 318:463. doi: 10.1016/j.virol.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 10.Rahaman A, Srinivasan N, Shamala N, Shaila MS. 2004. Phosphoprotein of the rinderpest virus forms a tetramer through a coiled coil region important for biological function: a structural insight. J Biol Chem 279:23606–23614. doi: 10.1074/jbc.M400673200. [DOI] [PubMed] [Google Scholar]
- 11.Asenjo A, Villanueva N. 2000. Regulated but not constitutive human respiratory syncytial virus (HRSV) P protein phosphorylation is essential for oligomerization. FEBS Lett 467:279–284. doi: 10.1016/S0014-5793(00)01171-6. [DOI] [PubMed] [Google Scholar]
- 12.Castagne N, Barbier A, Bernard J, Rezaei H, Huet J-C, Henry C, Da Costa B, Eleouet J-F. 2004. Biochemical characterization of the respiratory syncytial virus P-P and P-N protein complexes and localization of the P protein oligomerization domain. J Gen Virol 85:1643–1653. doi: 10.1099/vir.0.79830-0. [DOI] [PubMed] [Google Scholar]
- 13.Chattopadhyay S, Banerjee AK. 2009. Phosphoprotein, P of human parainfluenza virus type 3 prevents self-association of RNA-dependent RNA polymerase, L. Virology 383:226–236. doi: 10.1016/j.virol.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leyrat C, Renner M, Harlos K, Grimes JM. 2013. Solution and crystallographic structures of the central region of the phosphoprotein from human metapneumovirus. PLoS One 8:e80371. doi: 10.1371/journal.pone.0080371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Communie G, Crépin T, Maurin D, Jensen MR, Blackledge M, Ruigrok RWH. 2013. Structure of the tetramerization domain of measles virus phosphoprotein. J Virol 87:7166–7169. doi: 10.1128/JVI.00487-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox R, Green TJ, Purushotham S, Deivanayagam C, Bedwell GJ, Prevelige PE, Luo M. 2013. Structural and functional characterization of the mumps virus phosphoprotein. J Virol 87:7558–7568. doi: 10.1128/JVI.00653-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding HT, Green TJ, Lu SY, Luo M. 2006. Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J Virol 80:2808–2814. doi: 10.1128/JVI.80.6.2808-2814.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y, Lenard J. 1995. Cooperative binding of multimeric phosphoprotein (P) of vesicular stomatitis virus to polymerase (L) and template: pathways of assembly. J Virol 69:7718–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen M, Ogino T, Banerjee AK. 2006. Mapping and functional role of the self-association domain of vesicular stomatitis virus phosphoprotein. J Virol 80:9511–9518. doi: 10.1128/JVI.01035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanov I, Crépin T, Jamin M, Ruigrok RWH. 2010. Structure of the dimerization domain of the rabies virus phosphoprotein. J Virol 84:3707–3710. doi: 10.1128/JVI.02557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chenik M, Schnell M, Conzelmann KK, Blondel D. 1998. Mapping the interacting domains between the rabies virus polymerase and phosphoprotein. J Virol 72:1925–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoehn G, Iseni F, Mavrakis M, Blondel D, Ruigrok RWH. 2001. Structure of recombinant rabies virus nucleoprotein-RNA complex and identification of the phosphoprotein binding site. J Virol 75:490–498. doi: 10.1128/JVI.75.1.490-498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mavrakis M, Méhouas S, Réal E, Iseni F, Blondel D, Tordo N, Ruigrok RWH. 2006. Rabies virus chaperone: identification of the phosphoprotein peptide that keeps nucleoprotein soluble and free from non-specific RNA. Virology 349:422–429. doi: 10.1016/j.virol.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 24.Ryan KW, Kingsbury DW. 1988. Carboxyl-terminal region of Sendai virus P protein is required for binding to viral nucleocapsids. Virology 167:106–112. doi: 10.1016/0042-6822(88)90059-1. [DOI] [PubMed] [Google Scholar]
- 25.García-Barreno B, Delgado T, Melero JA. 1996. Identification of protein regions involved in the interaction of human respiratory syncytial virus phosphoprotein and nucleoprotein: significance for nucleocapsid assembly and formation of cytoplasmic inclusions. J Virol 70:801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De BP, Hoffman MA, Choudhary S, Huntley CC, Banerjee AK. 2000. Role of NH2- and COOH-terminal domains of the P protein of human parainfluenza virus type 3 in transcription and replication. J Virol 74:5886–5895. doi: 10.1128/JVI.74.13.5886-5895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kingston RL, Baase WA, Gay LS. 2004. Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins. J Virol 78:8630–8640. doi: 10.1128/JVI.78.16.8630-8640.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox R, Pickar A, Qiu S, Tsao J, Rodenburg C, Dokland T, Elson A, He B, Luo M. 2014. Structural studies on the authentic mumps virus nucleocapsid showing uncoiling by the phosphoprotein. Proc Natl Acad Sci U S A 111:15208–15213. doi: 10.1073/pnas.1413268111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu P, Li Z, Sun D, Lin Y, Wu J, Rota PA, He B. 2011. Rescue of wild-type mumps virus from a strain associated with recent outbreaks helps to define the role of the SH ORF in the pathogenesis of mumps virus. Virology 417:126–136. doi: 10.1016/j.virol.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pickar A, Xu P, Elson A, Li Z, Zengel J, He B. 2014. Roles of serine and threonine residues of mumps virus P protein in viral transcription and replication. J Virol 88:4414–4422. doi: 10.1128/JVI.03673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu P, Luthra P, Li Z, Fuentes S, D'Andrea JA, Wu J, Rubin S, Rota Pa, He B. 2012. The V protein of mumps virus plays a critical role in pathogenesis. J Virol 86:1768–1776. doi: 10.1128/JVI.06019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandrika R, Horikami SM, Smallwood S, Moyer SA. 1995. Mutations in conserved domain I of the Sendai virus L polymerase protein uncouple transcription and replication. Virology 213:352–363. doi: 10.1006/viro.1995.0008. [DOI] [PubMed] [Google Scholar]
- 33.Cevik B, Holmes DE, Vrotsos E, Feller JA, Smallwood S, Moyer SA. 2004. The phosphoprotein (P) and L binding sites reside in the N-terminus of the L subunit of the measles virus RNA polymerase. Virology 327:297–306. doi: 10.1016/j.virol.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Parks GD. 1994. Mapping of a region of the paramyxovirus L protein required for the formation of a stable complex with the viral phosphoprotein P. J Virol 68:4862–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sidhu MS, Menonna JP, Cook SD, Dowling PC, Udem SA. 1993. Canine distemper virus L gene: sequence and comparison with related viruses. Virology 193:50–65. doi: 10.1006/viro.1993.1102. [DOI] [PubMed] [Google Scholar]
- 36.Pattnaik AK, Hwang L, Li T, Englund N, Mathur M, Das T, Banerjee AK. 1997. Phosphorylation within the amino-terminal acidic domain I of the phosphoprotein of vesicular stomatitis virus is required for transcription but not for replication. J Virol 71:8167–8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raha T, Samal E, Majumdar A, Basak S, Chattopadhyay D, Chattopadhyay DJ. 2000. N-terminal region of P protein of Chandipura virus is responsible for phosphorylation-mediated homodimerization. Protein Eng 13:437–444. doi: 10.1093/protein/13.6.437. [DOI] [PubMed] [Google Scholar]
- 38.Barik S, McLean T, Dupuy LC. 1995. Phosphorylation of Ser232 directly regulates the transcriptional activity of the P protein of human respiratory syncytial virus: phosphorylation of Ser237 may play an accessory role. Virology 213:405–412. doi: 10.1006/viro.1995.0013. [DOI] [PubMed] [Google Scholar]
- 39.Malur AG, Choudhary SK, De BP, Banerjee AK. 2002. Role of a highly conserved NH2-terminal domain of the human parainfluenza virus type 3 RNA polymerase. J Virol 76:8101–8109. doi: 10.1128/JVI.76.16.8101-8109.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horikami SM, Smallwood S, Bankamp B, Moyer SA. 1994. An amino-proximal domain of the L protein binds to the P protein in the measles virus RNA polymerase complex. Virology 205:540–545. doi: 10.1006/viro.1994.1676. [DOI] [PubMed] [Google Scholar]
- 41.Poch O, Blumberg BM, Bougueleret L, Tordo N. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J Gen Virol 71:1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- 42.Curran J. 1998. A role for the Sendai virus P protein trimer in RNA synthesis. J Virol 72:4274–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delano W. 2002. The PyMOL user's manual. Delano Scientific, San Carlos, CA. [Google Scholar]