ABSTRACT
The incidence of human cowpox virus (CPXV) infections has increased significantly in recent years. Serological surveys have suggested wild rodents as the main CPXV reservoir. We characterized a CPXV isolated during a large-scale screening from a feral common vole. A comparison of the full-length DNA sequence of this CPXV strain with a highly virulent pet rat CPXV isolate showed a sequence identity of 96%, including a large additional open reading frame (ORF) of about 6,000 nucleotides which is absent in the reference CPXV strain Brighton Red. Electron microscopy analysis demonstrated that the vole isolate, in contrast to the rat strain, forms A-type inclusion (ATI) bodies with incorporated virions, consistent with the presence of complete ati and p4c genes. Experimental infections showed that the vole CPXV strain caused only mild clinical symptoms in its natural host, while all rats developed severe respiratory symptoms followed by a systemic rash. In contrast, common voles infected with a high dose of the rat CPXV showed severe signs of respiratory disease but no skin lesions, whereas infection with a low dose led to virus excretion with only mild clinical signs. We concluded that the common vole is susceptible to infection with different CPXV strains. The spectrum ranges from well-adapted viruses causing limited clinical symptoms to highly virulent strains causing severe respiratory symptoms. In addition, the low pathogenicity of the vole isolate in its eponymous host suggests a role of common voles as a major CPXV reservoir, and future research will focus on the correlation between viral genotype and phenotype/pathotype in accidental and reservoir species.
IMPORTANCE We report on the first detection and isolation of CPXV from a putative reservoir host, which enables comparative analyses to understand the infection cycle of these zoonotic orthopox viruses and the relevant genes involved. In vitro studies, including whole-genome sequencing as well as in vivo experiments using the Wistar rat model and the vole reservoir host allowed us to establish links between genomic sequences and the in vivo properties (virulence) of the novel vole isolate in comparison to those of a recent zoonotic CPXV isolated from pet rats in 2009. Furthermore, the role of genes present only in a reservoir isolate can now be further analyzed. These studies therefore allow unique insights and conclusions about the role of the rodent reservoir in CPXV epidemiology and transmission and about the zoonotic threat that these viruses represent.
INTRODUCTION
Cowpox virus (CPXV), a member of the genus Orthopoxvirus (OPV) in the Poxviridae family, is suspected to be widespread in Western Eurasian rodents, particularly vole species (1, 2). From the presumed reservoir hosts, spill-over infections to accidental hosts are regularly observed (3). The accidental hosts include domestic cats and also exotic animals in zoos, such as large felids and elephants, which regularly develop severe disease (3). As CPXV is a zoonotic virus, humans in direct contact with infected accidental hosts are at risk of infection, while direct infection of humans from presumed reservoir hosts has never been reported. Pet rat-associated human CPXV infections resulted in an epidemiologically linked infection cluster in France and Germany in 2009 (4–6). Interestingly, these human cases resulted from a single CPXV strain that was spread by the infected pet rats and most likely was also responsible for an infection of a woman in France 2 years after the original episode (7). While disease symptoms and virological data are well documented for human infections and infections of domestic and standard laboratory animals, data on the reservoir host consist primarily of serological surveys, with the exception of experimental infection studies in bank voles (Myodes glareolus), field voles (Microtus agrestis), and wood mice (Apodemus sylvaticus) (8, 9). Using a CPXV strain isolated from a cat, only very mild clinical signs were recorded in the experimentally infected rodent hosts, and CPXV could be reisolated from the inoculation sites only (8). Nevertheless, epidemiological surveys based on the serology of different vole populations in Europe demonstrated repeated CPXV infection cycles (10–13). However, CPXV was never recovered from the presumed reservoir hosts in Central, Northern, or Western Europe although the presence of viral genomes was verified (14). Indeed, the only reported vole reservoir sample from which CPXV was isolated originated from a Russian root vole (15), but that virus was never characterized in any detail. In addition, whereas laboratory breeds of selected mouse strains are used for CPXV inhibitor research (16) and whereas rats are used for assessment of viral pathogenicity (17, 18), the nature of CPXV infection in common voles has never been documented.
Here, we describe, to our knowledge, for the first time the in vitro and in vivo characterization of a CPXV strain isolated from a reservoir host, the common vole (Microtus arvalis). Whole-genome sequence analysis of the reservoir-derived CPXV allowed sequence comparisons to a rat isolate and the reference strain Brighton Red (BR). Infection experiments using Wistar rats as a surrogate animal model and common voles as the original reservoir host were performed to evaluate differences in pathogenicity levels of virus strains as well as similarities or differences of clinical symptoms. These experiments revealed various pathogenic potentials among the CPXV strains and suggest mild disease but efficient spread of the reservoir isolate and an increase in virulence after a host switch.
MATERIALS AND METHODS
Viruses.
CPXV strain FM2292 was isolated from the liver of a female common vole, Microtus arvalis (KS11/2292), collected during a large rodent screening project of the network Rodent-borne Pathogens on a grassland site in the federal state of Baden-Wuerttemberg, Germany, in October 2011 (19). The trapping of rodents was coordinated by the Julius Kühn-Institut, Münster, Germany, and approved by the responsible authority, the Regierungspräsidium Stuttgart (Landwirtschaft, ländlicher Raum, Veterinär- und Lebensmittelwesen, permit number BW 35-9185.82/0261). The screening of the rodent samples was performed according to the following protocol. DNA was extracted using a BioSprint 96 instrument and a MagAttract Virus Mini M48 kit (both, Qiagen, Hilden, Germany) from liver samples of individual rodents, and an orthopoxvirus (OPV)-specific quantitative PCR (qPCR) assay was applied (20). The CPXV rat isolate (RatPox09) was obtained from a diseased pet rat, which had infected two humans in southern Germany in 2009, and had already been characterized by experimental infection of Wistar rats (17). CPXV strain BR was used as the reference strain. All CPXV strains were propagated on Vero76 cells (Collection of Cell Lines in Veterinary Medicine [CCLV], Friedrich-Loeffler-Institut, Greifswald-Insel Riems, Germany) and amplified to stock titers of approximately 107 50% tissue culture infective doses (TCID50) ml−1.
NGS.
For next-generation sequencing (NGS), viral DNA was extracted using a High Pure PCR template preparation kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. Subsequently, 0.5 to 1 μg of DNA was fragmented to approximately 300 bp using a Covaris M220 ultrasonicator (Covaris, Brighton, United Kingdom). For library preparation, the fragmented DNA was prepared using NEXTflex DNA bar codes compatible with Illumina systems (Bioo Scientific, Austin, TX, USA) and SPRIworks Fragment Library Cartridge II (Beckman Coulter, Fullerton, CA, USA) on a SPRI-TE library system (Beckman Coulter). As the size selection step was skipped during automated library preparation, upper- and lower-size exclusion of the library was done manually with Ampure XP magnetic beads (Beckman Coulter). The quality of the library was checked on a Bioanalyzer 2100 (Agilent Technologies, Böblingen, Germany) using a high-sensitivity DNA chip and corresponding reagents. Quantity was determined via qPCR with a Kapa Library Quantification kit (Kapa Biosystems, Wilmington, MA, USA). Sequencing was performed on an Illumina MiSeq using a MiSeq reagent kit, version 2 (Illumina, San Diego, CA, USA).
Data analysis.
For de novo assembly and reference mapping, the Genome Sequencer software suite (version 2.8; Roche, Mannheim, Germany) was used. Assembled contigs were connected according to information in the 454ContigGraph.txt file. For reference mapping against the assembled genome, the -rst 0 parameter (repeat score threshold) was used; therefore, reads were mapped to both inverted terminal repeats rather than to just one.
Genome annotation and comparison.
Generated full-genomic sequences were annotated using the nomenclature of CPXV reference strain BR deposited in the NCBI RefSeq database (www.ncbi.nlm.nih.gov/refseq/; complete genome, GenBank accession number AF482758). Open reading frames (ORFs) not annotated in BR were numbered as follows: gCPXV0XXX for forward and reverse genes and pCPXV0XXX for proteins. Nomenclature was chosen in relation to BR nomenclature with additional prefixes to distinguish both. Genomic sequences were aligned in Geneious (version 8.0.5) (21) using the MAFFT algorithm (22). A similarity plot of the aligned sequences was generated using the plotcon tool (EMBOSS, version 6.3.1) (23). Parameters were winsize 180, scorefile EDNAFULL, and graph data. Data were normalized to a range of 0 to 1 and plotted in R (version 3.1.1) (24).
Phylogenetic analysis.
For determination of the CPXV clade, available full-length genomes of OPVs in GenBank were aligned in Geneious (version 8.1.3) using the MAFFT algorithm (22), and the coding region of vaccinia virus (VACV) strain Copenhagen (VACV-Cop C23L-B29R) (GenBank accession number M35027) as described by Carroll et al. (25) was selected. Phylogenetic analysis was done using IQ-Tree (version 1.2.2) (26) with the best-fitting model and 5,000 ultrafast bootstraps.
Virus growth kinetics.
Overnight cultures of Vero, HeLa, rat lung, and bank vole kidney cells (27) were infected with CPXV FM2292, CPXV RatPox09, or BR using multiplicities of infection (MOIs) of 0.01 or 3. After an incubation period of 60 min at 37°C, the cells were washed three times with phosphate-buffered saline (PBS). Afterwards, 1 ml of fresh culture medium (Dulbecco's modified Eagle's medium [DMEM]) was added to each well. In total, samples were obtained at five different time points after infection (0 h, 6 h, 12 h, 24 h, and 48 h). Two technical replicates were done, and the whole experiment was performed in duplicate (n = 4 per time point). Virus titers were determined by endpoint dilution and calculated as TCID50 per milliliter.
Electron microscopy.
For morphological examinations of ultrathin sections, Hep-2 cell cultures (CCLV, Friedrich-Loeffler-Institut) were infected with strain BR, RatPox09, or FM2292 at an MOI of 0.1. Cells were fixed at 36 h postinfection (p.i.) for 60 min with 2.5% glutaraldehyde buffered in 0.1 M Na-cacodylate, pH 7.2 (300 mosmol; Merck, Darmstadt, Germany). Cells were then scraped off the plate and pelleted by low-speed centrifugation and embedded in low-melting-point (LMP) agarose (Biozym, Oldendorf, Germany). Small pieces were postfixed in 1.0% OsO4(aq) (Polysciences Europe, Eppelheim, Germany) and stained with uranyl acetate. After stepwise dehydration in ethanol, cells were cleared in propylene oxide, embedded in Glycid ether 100 (Serva, Heidelberg, Germany), and polymerized at 59°C for 4 days. Ultrathin sections of epoxy resin-embedded material, counterstained with uranyl acetate and lead salts, were examined with a 120-kV transmission electron microscope (FEI Tecnai Spirit G2; Eindhoven, The Netherlands).
Animal experiments.
Mixed-sex Wistar rats at 5 to 6 weeks of age (outbred; Charles River, Sulzfeld, Germany) and 3- to 4-month-old mixed-sex common voles (outbred; Federal Environment Agency, Berlin, Germany) were housed in groups of 2 to 4 animals (rats) or separately (common voles) in standard laboratory rodent cages. All animal experiments were approved by the Ministry of Agriculture of Mecklenburg-Vorpommern, Germany (reference numbers LALLF M-V/TSD/7221.3-2.1.-005/09 and LALLF MV 7221.3-1.1-020/13).
The experimental design complied with the experiments published previously (17, 18). Eleven rats and 9 common voles were infected oronasally with CPXV strain FM2292 in two groups using titers of 104 and 106 TCID50/animal, respectively. Additionally, 10 (5 per dose group) common voles were inoculated oronasally with CPXV strain RatPox09 (104 or 106 TCID50/animal). Body temperature, weight, and general health status of all animals were checked daily over a period of 30 days, and every other day oropharyngeal swabs (Bakteriette; EM-TE Vertrieb, Hamburg, Germany) were taken. The animals were either humanely killed for autopsy on days 5 or 30 p.i. or when severe signs of disease were apparent. All animals were dissected for collection of organ specimens. Blood was drawn from freshly deceased animals, and peritoneal lavage samples were taken from common voles at 30 days p.i. In addition, inoculation of BR using titers of 104 or 106 TCID50/animal was performed using Wistar rats (12 animals). Swab samples were taken every other day, and the experiment was finalized at 24 days p.i.
Oropharyngeal swab samples were resuspended in 2 ml of cell culture medium supplemented with antibiotics (enrofloxacin, 1 mg/ml; gentamicin, 0.05 mg/ml; lincomycin, 1 mg/ml). Samples of organ tissue were transferred to reaction tubes containing 1 ml of DMEM supplemented with 10% fetal bovine serum (FBS), antibiotics (1% penicillin-streptomycin [PenStrep]), and stainless steel beads (diameter, 5 mm). The samples were subsequently homogenized (TissueLyser II; Qiagen, Hilden, Germany). DNA from all swab and organ samples was extracted using a BioSprint 96 instrument and a MagAttract Virus Mini M48 kit (both, Qiagen) and then quantified by real-time PCR using an OPV DNA-specific protocol (20). In addition, endpoint dilution assays using the Spearman-Kärber algorithm were done for each swab and organ sample. To detect virus-specific antibodies in sera of infected animals, serum samples were incubated for 30 min at 56°C. Subsequently, CPXV virus-infected Hep2 cells (fixed with methanol-acetone at 1:1 and incubated with Tris-buffered saline plus Tween [TBS-T] for 30 min) were incubated for 1 h at room temperature with a 1:200 or 1:500 dilution of serum. After three washing steps with PBS, commercial anti-mouse and anti-rat conjugates as secondary antibodies (both, Life Technologies) were used. Evans Blue (Sigma-Aldrich, Deisenhofen, Germany) was added to stain the cytoplasm of the infected cells.
Nucleotide sequence accession numbers.
The complete coding sequences of CPXV FM2292 and RatPox09, with the exception of the terminal loop regions, were deposited in the DDBJ/ENA/GenBank under accession numbers LN864566 and LN864565, respectively.
RESULTS
Isolation of a CPXV from a common vole.
During the large-scale screening of rodent tissues, one liver sample of a feral common vole scored positive for the CPXV genome by our diagnostic qPCR. The female common vole was collected during a screening program of rodents in the federal state of Baden-Wuerttemberg in Germany in 2011 (19). Virus isolation using Vero76 cells finally resulted, in the fourth passage, in a high-titer stock (107 TCID50 ml−1) of the vole isolate named FM2292.
Whole-genome analysis of CPXV from a common vole.
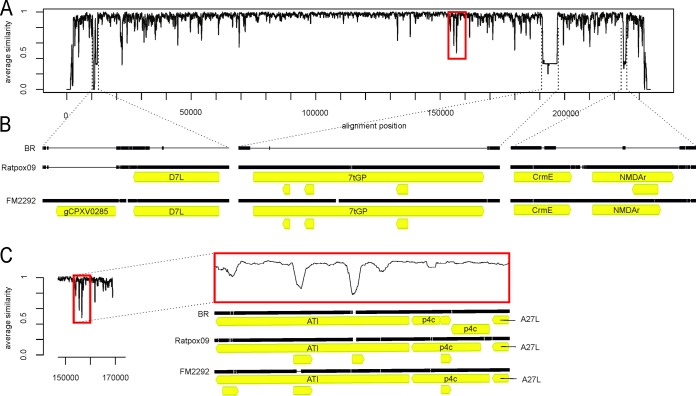
The complete coding sequences of CPXV FM2292 (DDBJ/ENA/GenBank accession number LN864566) and RatPox09 (DDBJ/ENA/GenBank accession number LN864565), with the exception of the terminal loop regions, were generated by NGS. The obtained sequences comprise 227,639 bp for FM2292 and 228,162 bp for RatPox09 (Table 1). The inverted terminal repeat regions (ITRs) are approximately 7.2 kbp and 7.6 kbp in length, respectively, which result in core genome regions of 213,300 bp (FM2292) and 212,800 bp (RatPox09) (Table 1). The nucleotide sequence of FM2292 had the highest identity (98%) to a human isolate from 1998 in Germany (GenBank accession number HQ420897), while CPXV isolates FM2292 and RatPox09 have an overall identity of 96%. Both isolates have a nucleotide sequence identity of approximately 92% to the reference CPXV strain BR. Figure 1 shows a similarity plot of all three genomes over a window size of 180 nucleotides (nt). Clearly, and as expected, the core genome parts tend to be more conserved than the 5′ and 3′ ends of the genome (Fig. 1).
TABLE 1.
Comparison of the genomes of CPXV RatPox09 and FM2292 with the genome of the reference strain Brighton Reda
| Gene group and name | Strain profileb |
Description | |
|---|---|---|---|
| RatPox09 | FM2292 | ||
| Genes absent in Brighton Red | |||
| gCPXV0002 | + | + | NMDA receptor-like proteinc |
| gCPXV0003 | + | + | CrmE homologues proteind |
| gCPXV0030 | + | + | Putative 7-transmembrane G protein-coupled receptor-like protein 7tGP |
| gCPXV0284 | + | + | Kelch-like protein similar to VACV D7L |
| gCPXV0285 | − | + | Unknown function |
| Genes present in Brighton Red | |||
| CPXV001/CPXV229 | − | − | Unknown function |
| CPXV002/CPXV228 | − | − | Unknown function |
| CPXV007/CPXV224 | − | − | Ankyrin repeat-containing protein |
| CPXV192 | − | − | Unknown function |
| CPXV004 | − | − | Unknown function |
| CPXV216 | − | − | Similar to VACV Western Reserve 204.5 |
| CPXV051A | − | + | Unknown function |
| CPXV152A | − | + | Unknown function |
For strains RatPox09 and FM2292, the genomes are characterized as follows (in respective order): genome sizes of 228,162 and 227,639 bp, ITRs of 7.6 and 7.2 kb, and core genomes of 212,800 and 213,300 bp. RatPox09 has 289 ORFs, and FM2292 has 294 ORFs; of these, 280 ORFs are shared.
Plus and minus signs indicate presence and absence, respectively, of the gene.
N-methyl-d-aspartate receptor-like protein.
Cytokine response modifier E homologous protein.
FIG 1.
Nucleotide sequence comparison of RatPox09, FM2292, and Brighton Red (BR). (A) Normalized similarity plot of Brighton Red, FM2292, and RatPox09. A value of 1 indicates an identity of 100%, while a value of 0 indicates no conservation at all. Regions of special interest are enlarged. (B) Alignments of genes coding for CPXV0285, D7L-like protein, 7-transmembrane G protein-coupled receptor-like protein (7tGP), CrmE (CPXV0002) protein, and the NMDA receptor-like protein (NMDAr). (C) Alignment of the genes encoding the A-type inclusion protein (ATI), the A26L protein (p4c), and A27L protein.
Phylogenetic analysis showed that RatPox09 clusters within the variola-like clade while FM2292 is part of the CPXV-like clade 2. BR belongs to the CPXV-like clade 3 (Fig. 2), according to Dabrowski et al. (28) and Carroll et al. (25). Detailed analyses of the complete coding sequences were performed for CPXV strains BR, RatPox09, and FM2292 as these viruses were further characterized in the experimental rat model.
FIG 2.
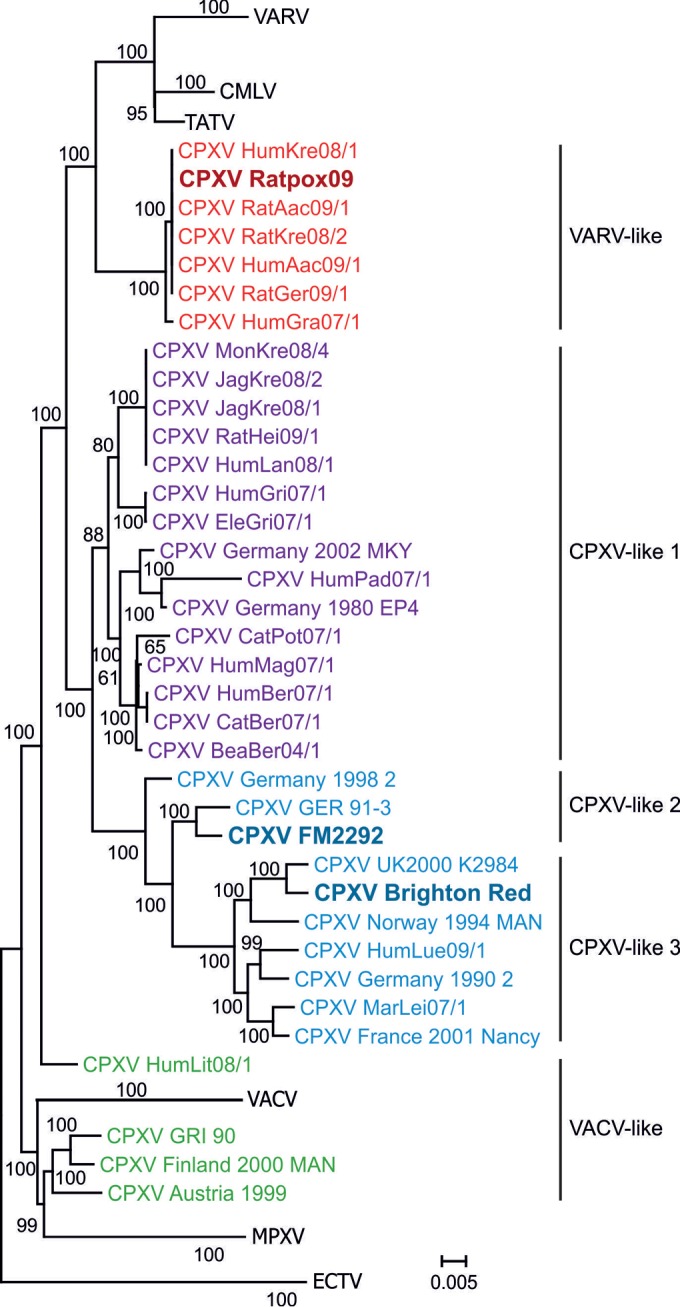
Phylogenetic analysis of whole-genome sequences of orthopoxviruses. CPXV clades (25, 28) are displayed in different colors. RatPox09, FM2292, and BR are highlighted in bold. The variola virus (VARV), camelpox virus (CMLV), taterapox virus (TATV), vaccinia virus (VACV), ectromelia virus (ECTV), and monkeypox virus (MPXV) clusters are presented as collapsed clades and include available whole-genome sequences in GenBank.
RatPox09 and FM2292 share 280 open reading frames (ORFs), out of a total of 289 ORFs for RatPox09 and 294 ORFs for FM2292 (Table 1). The major differences between the three CPXV isolates are (i) a deletion of approximately 660 bp in BR and RatPox09 from position 10600 to position 11200 relative to FM2292 and (ii) three deletions of approximately 770 bp, 5,800 bp, and 1,000 bp in BR relative to the other two strains at positions 11500 to 12300, 191200 to 197000, and 223500 to 224600, respectively. Hence, one complete ORF (gCPXV0285) in BR and RatPox09 and four additional ORFs (gCPXV0284, gCPXV0030, gCPXV0002, and gCPXV0003) in BR are absent when compared to the FM2292 genome (Fig. 1B and Table 1). Within the similarity plot (Fig. 1A), these events are clearly indicated by peaks with values below 0.3 at the respective positions.
ORF gCPXV0030 encodes a putative 7-transmembrane G protein-coupled receptor-like protein (7tGP), a receptor-like protein of unknown function with one predicted C-terminal transmembrane domain, which is encoded only in two other CPXV virus strains, namely, Ger91-3 (GenBank accession number DQ437593; CPXV-like 2 clade) and FRA2001 (GenBank accession number HQ420894; CPXV-like 3 clade).
Only FM2292 and Ger91-3 (both CPXV-like 2) harbor the gCPXV0285 gene, but there is no known or predicted function for the encoded protein. pCPXV0284 is a kelch-like protein that shares homologies with VACV D7L, which has an unknown function in CPXV. Putative proteins pCPXV0002 and pCPXV0003 are homologues of the OPV cytokine response modifier CrmE and the N-methyl-d-aspartate (NMDA) receptor-like proteins, respectively.
The genome of BR harbors 233 annotated ORFs. We did not resolve the sequence of the repeats in the terminal loops and were therefore unable to confirm the presence of CPXV001/CPXV229 (identical ORFs within the ITR) in both rodent CPXV genomes and of CPXV002/CPXV228 in RatPox09 (Table 1). These unresolved tandem repeat regions represent approximately 1,500 bp that are missing in the 3′ and 5′ terminal loop ends of RatPox09 and FM2292 genomes compared to the BR sequence. The most striking feature of the RatPox09 and FM2292 sequences is the absence of the CPXV004, CPXV007/CPXV224, CPXV192, and CPXV216 ORFs that are present in the genome of BR (Table 1). In addition, the CPXV051A and CPXV152A ORFs, which are present in the BR and FM2292 genomes, are absent in the RatPox09 genome (Table 1). These ORFs were identified as incomplete in the two rodent-derived isolates due to missing start codons within the sequences. These findings are, therefore, not reflected by any peaks as shown in Fig. 1. While the CPXV216-encoded protein is similar to the uncharacterized protein of VACV strain Western Reserve 204.5, the CPXV007/CPXV224 ORFs are predicted to encode ankyrin repeat-containing proteins. No putative functions have been assigned to ORFs CPXV004, CPXV051A, CPXV192, and CPXV152A. Additionally, various deletions/insertions and substitutions were found in noncoding regions or led to extended or truncated ORFs, for example, in the p4c-coding region, which will be presented in detail below. Major differences are additionally summarized in Table 1.
In vitro characterization of RatPox09 and FM2292.
Presence or absence of A-type inclusion (ATI) bodies was one of the first properties used to distinguish CPXV from VACV (29). ATI bodies are proteinaceous intracellular aggregates that appear late in infection with some OPVs (30). Three strain-specific phenotypes of ATIs are described: inclusions that have virions embedded within the ATI matrix (V+), those that lack virions within or on the surface of the ATI matrix (V−), and those in which virions are attached to the ATI periphery but are absent from the ATI matrix (V+/) (31, 32). The ATI phenotypes of CPXV FM2292, RatPox09, and BR were evaluated by electron microscopy. Like CPXV BR, RatPox09 produces the ATI V− phenotype, where the inclusion bodies lack virus particles incorporated within or located on the surface of the structure (Fig. 3A and B). In contrast, the common vole isolate FM2292 was clearly characterized by having an ATI V+ phenotype, with numerous virions embedded within the inclusion bodies (Fig. 3C).
FIG 3.
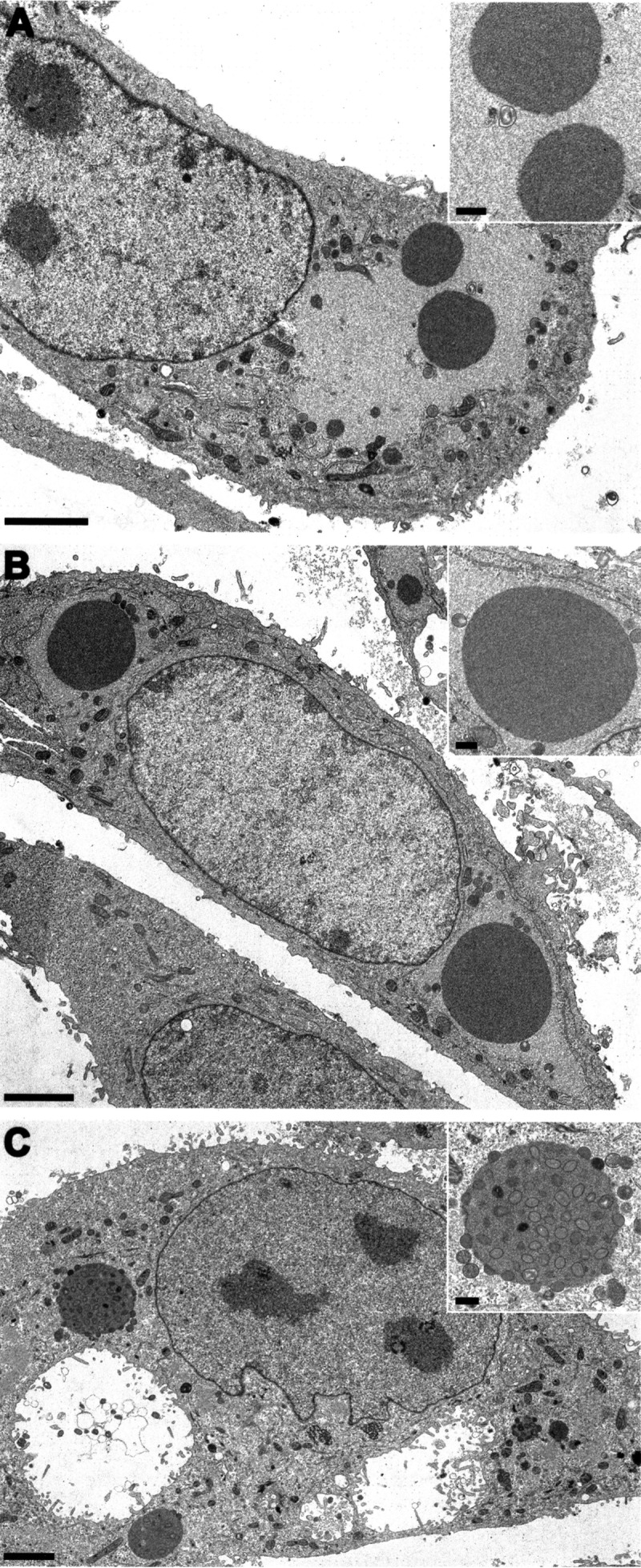
A-type inclusions (ATIs) produced by the three CPXVs. Electron microscopy images of thin sections of Vero76 cells infected with Brighton Red (A), RatPox09 (B), and FM2292 (C) at 36 h postinfection are shown. ATIs were seen in all experiments; however, the V+ phenotype could be observed only for FM2292 (C). Scale bars: 2.5 μm and 500 nm (inset).
It was shown that three proteins, the homologues of the VACV A25L, A26L, and A27L, are necessary for formation of V+ ATIs. The ATI matrix is formed by multiple copies of the A25L polypeptide, which corresponds to the CPXV158 protein (30). A26L, also named p4c or CPXV159 protein, is required to direct intracellular mature virions (IMVs) into ATIs (33); the protein has a bridging function between the A25L matrix protein and IMVs containing the membrane-associated A27L (CPXV162 protein) (34). In addition, the capability to embed mature virions in the ATIs was recently shown to influence the virulence of CPXV (35). We showed that the RatPox09 p4c amino acid sequence (A26; CPXV159 protein) results in a 59-amino-acid (aa) truncation at the N terminus, which likely is responsible for the observed V− phenotype; in contrast, FM2292 harbors a full-length p4c gene (Fig. 1C). CPXV162 corresponding to VACV A27L (Fig. 1C) and CPXV159 corresponding to VACV A25L (96 to 99% identity) show differences in length and sequence, especially within the repeat regions, but the translational initiation and stop sequences of the two ORFs are identical to each other.
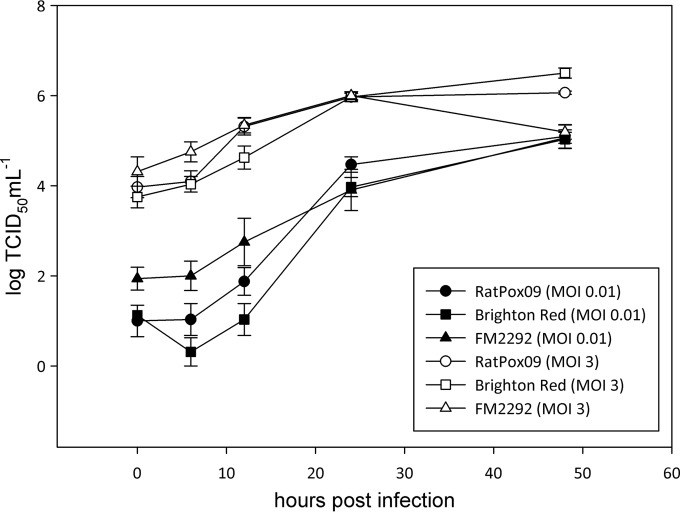
CPXV FM2292 was further characterized in terms of its in vitro growth characteristics and compared to RatPox09 and the BR reference strain. In two independent experiments using technical duplicates as internal controls, growth kinetics of the CPXV strains were determined in Vero76 cells at different MOIs. Generally speaking, the three CPXV isolates replicated similarly in cultured cells, irrespective of whether we used an MOI of 0.01 or 3 (Fig. 4). At 48 h p.i. titers of ∼104 TCID50 ml−1 (MOI of 0.01) or ∼105 (MOI of 3) were determined. Using an MOI of 3, only the FM2292 virus reached a lower titer at 48 h p.i. (∼104.1 TCID50 ml−1) than the 24-h value (105 TCID50 ml−1). In addition, viral growth kinetics were evaluated using HeLa, bank vole kidney, and rat lung cells; however, the shapes of the growth curves for the different viruses were virtually identical in all of the cell types investigated (data not shown).
FIG 4.
Comparison of replication characteristics of FM2292, RatPox09, and Brighton Red. Vero76 cells were infected with different CPXV strains at an MOI of 0.01 or 3. Infected cells were harvested at several time points postinfection. Virus titers were determined by endpoint dilution assays. The means and standard deviations of two independent experiments are shown, including two technical replicates (n = 4).
In vivo characterization.
Inoculation of BR into Wistar rats was performed to compare the in vivo data of CPXV FM2292 and RatPox09 with those of an experimentally well-defined virus isolate (see, for example, reference 36). For the determination of the in vivo characteristics of CPXV FM2292, we infected common voles and Wistar rats. In addition, we also infected common voles with RatPox09 in order to test this zoonotic isolate in a reservoir host species.
After oronasal infection with FM2292 or RatPox09, the first clinical signs were respiratory in nature and included dyspnea, sneezing, and nasal discharge at 4 to 6 days p.i. in all animal groups, i.e., regardless of whether voles or Wistar rats were infected. An exception was the common vole group that had been infected with the low-titer preparation of FM2292, where obvious clinical signs were not observed. In addition, Wistar rats inoculated with strain BR exhibited no or only very mild clinical symptoms. In contrast, common voles inoculated with the high-titer preparation of RatPox09 showed the earliest onset of respiratory symptoms at 4 days p.i. and the most severe course of disease in general. Respiratory signs vanished quickly in common voles infected with high-titer FM2292 and low-titer RatPox09, whereas the average duration of clinical disease in Wistar rats was 16 days in the low-titer FM2292 group and 16 days in the high-titer FM2292 group. In addition to the respiratory symptoms, Wistar rats also developed pox-like rashes on noses, ears, paws, and tails (Fig. 5) at approximately 14 days p.i., which began to heal around 20 days p.i. Interestingly, CPXV infection in common voles was not associated with any obvious skin lesions.
FIG 5.
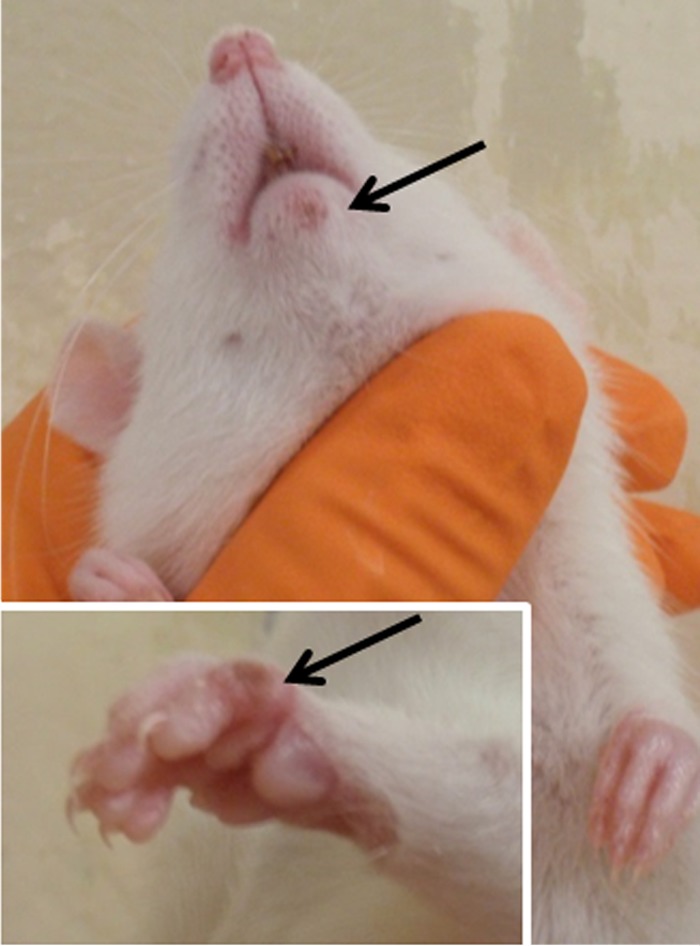
Typical pox-like lesions of a Wistar rat. Wistar rats infected with FM2292 at both doses developed pox-like lesions on the nose, ears, paws, and tail. Images were taken at 9 days p.i. (mouth) and 11 days p.i. (paws).
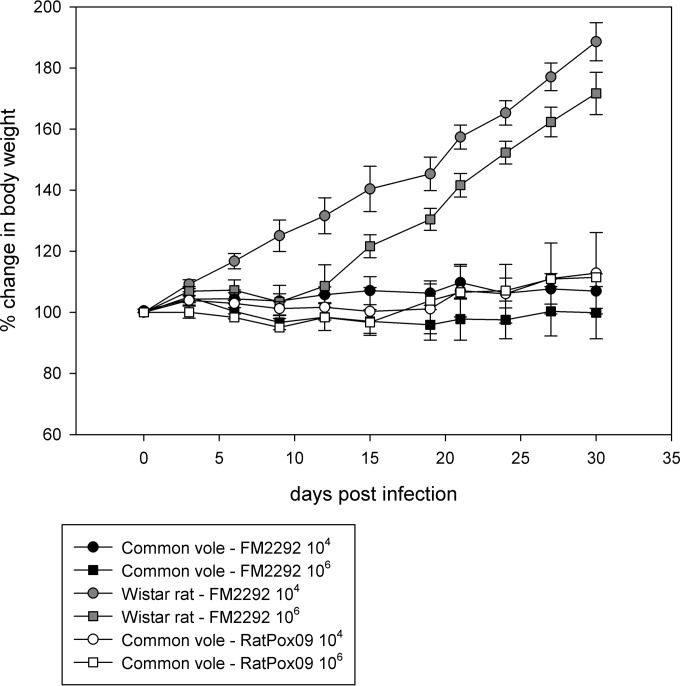
Overall, Wistar rats showed more prominent symptoms after FM2292 infection than the common voles in which subclinical disease was recorded, with the notable exception of one animal in the FM2292 high-dose group. Nevertheless, all Wistar rats survived the infection and recovered. In contrast, 4 of the 5 common voles infected with high-titer RatPox09 virus had to be euthanized between 4 and 8 days p.i. as their health deteriorated dramatically. Two additional common voles, one in the high-titer CPXV FM2292 group and one in the low-titer CPXV RatPox09 group, succumbed to infection. Body temperatures of the individual animals correlated with subnormal temperatures of terminally diseased voles, while such correlation was not detected with the milder symptoms (data not shown). Body weight data demonstrated a clear increase for Wistar rats, while common voles had relatively steady weights throughout the experiment (Fig. 6). Interestingly, body weight increases in rats inoculated with high doses of FM2292 were delayed compared to increases in rats inoculated with the low dose.
FIG 6.
Body weight evolution of common voles and Wistar rats inoculated with FM2292 and RatPox09, respectively. The starting weight was set as 100%, and values are the means with standard deviations.
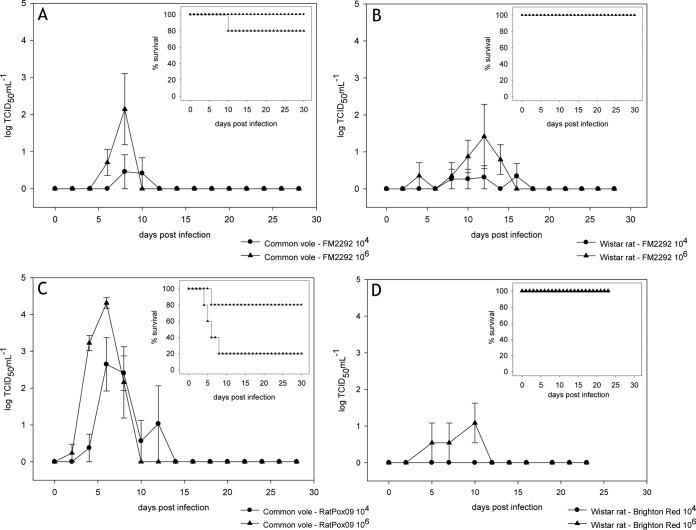
Oropharyngeal shedding was detected in all animal groups (Fig. 7). Common voles infected with FM2292 showed nearly the same virus shedding pattern as the Wistar rats. Generally, the first excretion of virus was recorded between 4 and 8 days p.i. The largest amount of virus was shed on day 8 p.i. in common voles (101.375 TCID50 ml−1) (Fig. 7A) and on day 10 p.i. in Wistar rats (103 TCID50 ml−1) (Fig. 7B). Between 10 and 16 days p.i., small amounts of virus were excreted from individual animals (<101.5 TCID50 ml−1) (Fig. 7A and B). Only one common vole, inoculated with a high dose of FM2292, had to be euthanized on day 8 p.i. as a consequence of its deteriorating health (Fig. 7A, inset). Viral shedding in this particular animal was 104 TCID50 ml−1, while all other animals shed less than 101.5 TCID50 ml−1 of virus.
FIG 7.
Oropharyngeal viral shedding patterns of common voles and Wistar rats. Shedding patterns are shown for common voles inoculated with FM2292 (A) or RatPox09 (C) and for Wistar rats infected with FM2292 (B) or BR (D). Values are the means with standard deviations.
Viral shedding following RatPox09 infection of the common voles was detected between 2 and 4 days p.i. (Fig. 7C). Generally, animals in the high-dose group shed more virus than those in the low-dose group (Fig. 7C). On day 6 p.i., single voles shed up to 104.5 TCID50 ml−1 (high-dose group) or 103.6 TCID50 ml−1 (low-dose group) of virus. Due to progressive clinical signs, these animals had to be euthanized between 4 and 8 days p.i. (Fig. 7C, inset). The remaining common voles excreted virus at maximal titers from day 4 p.i. until day 8 p.i., and virus titers reached up to 104.56 TCID50 ml−1. After 12 days p.i., no virus shedding was detected in any of the animals (Fig. 7C). Only Wistar rats inoculated with the high-dose BR preparation shed virus from 5 to 10 days p.i., when small amounts of excreted virus were detected (Fig. 7D).
Virus distribution in various organs was determined by real-time PCR and also by endpoint dilution assay (Tables 2 and 3). All voles that succumbed to CPXV infection scored positive for virus in turbinates (6/6 animals) (Table 2) and rhinarium (5/5 animals) (Table 2). Turbinate samples taken from Wistar rats euthanized at 5 days p.i. were also positive (Table 3). The turbinates showed the highest viral load of all tissues analyzed. Common voles infected with RatPox09 had viral loads of up to 106 TCID50 ml−1 in this organ, while virus titers in the turbinates of Wistar rats infected with FM2292 reached 104 TCID50 ml−1, and BR-infected Wistar rats shed virus at mean titers of 103.875 TCID50 ml−1. Furthermore, virus was detected in several organ samples of voles that succumbed to the infection (Table 2).
TABLE 2.
qPCR and virus titration results for the infection experiments with CPXV isolates FM2292 and RatPox09 in common voles
| Virus and tissue typea | Virus distribution by time point and method |
|||||
|---|---|---|---|---|---|---|
| 5 dpib |
30 dpi |
Deathe |
||||
| PCRc | Endpoint dilutiond | PCR | Endpoint dilution | PCR | Endpoint dilution | |
| FM2292 | ||||||
| Turbinates | 2/3 | 2/3 | 3/5 | 0/5 | 1/1 | 1/1 |
| Trachea | 0/3 | 0/3 | 0/5 | 0/5 | 0/1 | 0/1 |
| Lung | 0/3 | 0/3 | 0/5 | 0/5 | 1/1 | 0/1 |
| Esophagus | 0/3 | 0/3 | 0/5 | 0/5 | 1/1 | 0/1 |
| Stomach | 1/3 | 0/3 | 0/5 | 0/5 | 1/1 | 0/1 |
| Liver | 1/3 | 0/3 | 0/5 | 0/5 | 1/1 | 1/1 |
| Kidney | 0/3 | 0/3 | 0/5 | 0/5 | 1/0 | 0/1 |
| Bladder | 0/3 | 0/3 | 1/1 | 1/1 | ||
| Gonads | 0/3 | 0/3 | 1/1 | 0/1 | ||
| Spleen | 2/3 | 0/3 | 0/5 | 0/5 | 1/1 | 0/1 |
| Thymus | 0/3 | 0/3 | 1/1 | 0/1 | ||
| Skin | 0/3 | 0/3 | 0/5 | 0/5 | 0/1 | 0/1 |
| Myocardium | 0/3 | 0/3 | 0/1 | 0/1 | ||
| Brain | 0/3 | 0/3 | 0/1 | 0/1 | ||
| Rhinarium | 2/3 | 2/3 | 0/5 | 0/5 | 1/1 | 1/1 |
| RatPox09 | ||||||
| Turbinates | 2/2 | 2/2 | 1/3 | 1/3 | 5/5 | 5/5 |
| Trachea | 0/2 | 1/2 | 0/3 | 0/3 | 5/5 | 4/5 |
| Lung | 0/2 | 0/2 | 0/3 | 0/3 | 2/5 | 2/5 |
| Esophagus | 0/2 | 0/2 | 0/3 | 0/3 | 3/5 | 5/5 |
| Stomach | 0/2 | 0/2 | 0/3 | 0/3 | 4/5 | 3/5 |
| Liver | 0/2 | 0/2 | 0/3 | 0/3 | 5/5 | 1/5 |
| Kidney | 0/2 | 0/2 | 0/3 | 0/3 | 1/5 | 0/5 |
| Bladder | 0/2 | 0/2 | 1/5 | 2/5 | ||
| Gonads | 0/2 | 0/2 | 1/5 | 0/5 | ||
| Spleen | 0/2 | 0/2 | 0/3 | 0/3 | 4/5 | 1/5 |
| Thymus | 0/2 | 0/2 | 2/5 | 1/5 | ||
| Skin | 0/2 | 0/2 | 0/3 | 0/3 | 4/5 | 3/5 |
| Myocardium | 0/2 | 0/2 | 0/5 | 2/5 | ||
| Brain | 0/2 | 0/2 | 2/5 | 2/5 | ||
| Rhinarium | 0/2 | 0/2 | 0/3 | 0/3 | 4/4 | 4/4 |
CPXV infection in common voles was not associated with any obvious skin lesions.
dpi, days postinfection.
Determined by qPCR as the number of positive samples (quantification cycle value of <38)/total number of samples tested.
Number of samples testing positive for CPXV by endpoint dilution assay/total number of samples tested.
Succumbed to infection or had to be euthanized.
TABLE 3.
qPCR and virus titration results for the infection experiments with CPXV isolates FM2292 and BR in Wistar rats
| Tissue type | Virus distribution by time point and method for the indicated virus |
|||||||
|---|---|---|---|---|---|---|---|---|
| FM2292 |
BR |
|||||||
| 5 dpia |
30 dpi |
5 dpi |
24 dpi |
|||||
| PCRb | Endpoint dilutionc | PCR | Endpoint dilution | PCRc | Endpoint dilution | PCR | Endpoint dilution | |
| Turbinates | 4/4 | 4/4 | 0/7 | 0/7 | 3/3 | 3/3 | 1/6 | 1/6 |
| Trachea | 0/4 | 0/4 | 0/7 | 0/7 | 0/3 | 0/3 | 0/6 | 0/6 |
| Lung | 0/4 | 0/4 | 0/7 | 0/7 | 0/3 | 0/3 | 0/6 | 0/6 |
| Esophagus | 0/3 | 0/3 | 0/6 | 0/6 | ||||
| Stomach | 0/3 | 0/3 | 0/6 | 0/6 | ||||
| Liver | 1/4 | 0/4 | 0/7 | 0/7 | 0/3 | 0/3 | 0/6 | 0/6 |
| Kidney | 0/3 | 0/3 | 0/6 | 0/6 | ||||
| Bladder | 0/3 | 0/3 | 0/6 | 0/6 | ||||
| Gonads | 0/6 | 0/6 | ||||||
| Spleen | 2/4 | 0/4 | 0/7 | 0/7 | 0/3 | 0/3 | 0/6 | 0/6 |
| Thymus | 0/3 | 0/3 | 0/6 | 0/6 | ||||
| Skin | 0/4 | 0/4 | 0/7 | 0/7 | 0/3 | 0/3 | 0/6 | 0/6 |
| Myocardium | 0/3 | 0/3 | 0/6 | 0/6 | ||||
| Brain | 0/3 | 0/3 | 0/6 | 0/6 | ||||
| Rhinarium | 4/4 | 4/4 | 2/3 | 2/3 | 0/6 | 0/6 | ||
| Skin Lesion | 6/7 | 5/7 | ||||||
dpi, days postinfection.
Determined by qPCR as the number of positive samples (quantification cycle value of <38)/total number of samples tested.
Number of samples testing positive for CPXV by endpoint dilution assay/total number of samples tested.
In single FM2292-infected Wistar rats necropsied on day 30 p.i., the skin lesions still contained virus with loads up to 103.875 TCID50 ml−1, while BR-infected Wistar rats did not display skin lesions, and only the turbinate sample of one animal scored positive (102.375 TCID50 ml−1) at 24 days p.i. Likewise, one turbinate sample of a common vole infected with RatPox09 also still contained infectious virus (101.375 TCID50 ml−1) at day 30 p.i. Other organs of voles and rats scored positive only when animals succumbed or were examined at 5 days p.i. (Tables 2 and 3). Blood samples from common voles that had succumbed to infection or animals dissected on day 5 p.i. were negative for viral DNA. Antibodies against CPXV were detected in all but one of the individual serum samples obtained, which originated from a common vole in the low-titer CPXV FM2292-infected group (data not shown).
DISCUSSION
The number of human CPXV infections is growing, possibly due to the cessation of smallpox vaccination since its official eradication was made public in 1978 (37). Above all, animal caretakers and veterinarians having contact with infected accidental hosts are at risk of infection with CPXV. Accidental hosts, except humans, become initially infected after direct or indirect contact with infected reservoir host species (3). Despite the presumed continued presence of CPXV in the reservoir hosts, successful virus isolation from wild rodents is extremely rare. In the present study, we characterized CPXV FM2292, which we isolated recently from a rodent reservoir, specifically, a female common vole (Microtus arvalis). Using NGS, we generated a whole-genome sequence of FM2292 and compared it to that of RatPox09, a pet rat strain from 2009, and reference strain BR. The FM2292 and RatPox09 strains have a sequence identity of 96%. Both RatPox09 and FM2292 have sequence identities of approximately 92% to the reference strain BR. A first analysis of whole-genome sequences identified four genes that are present in RatPox09 and FM2292 but absent in strain BR, namely, gCPXV0284, gCPXV0002, gCPXV0030, and gCPXV0003. As we detected differences in the virulence of RatPox09 and FM2292 compared to that of BR, these four gene products might represent bona fide virulence genes. All genes have homologues in other OPVs and encode the kelch-like protein D7L, the cytokine response modifier CrmE, the putative 7-transmembrane G protein-coupled receptor-like protein 7tGP, and the NMDA receptor-like protein. For example, a contribution to virulence was demonstrated for BTB/kelch proteins (where BTB is broad-complex, tramtrack, and bric a brac) other than D7L (38–43) and for CrmE in the case of VACV (44, 45).
The annotation of the genome of FM2292 revealed that this strain is equipped with additional ORFs compared to the standard BR genome and even to that of the RatPox09 isolate. Clearly, the annotation used here accounts for the increased number of ORFs relative to the number in BR. However, it is clear that FM2292 absolutely and relatively specifies more ORFs (294 in FM2292 versus 289 in RatPox09). The fact that shorter genomes are isolated from accidental host species supports the hypothesis of gene loss as a result of adaptation to a new host and may even confirm the hypothesis that CPXV-like genomes represent the phylogenetically oldest representatives of this group of viruses (46, 47). Certainly, the functions of the newly annotated ORFs have to be analyzed in more detail.
The ATI phenotypes of the viruses studied here were explored in more detail, and our analyses revealed that both rodent-derived CPXV strains produced ATI bodies, but only in the case of FM2292 were virions present in the ATI matrix (V+). In contrast, RatPox09 produced ATIs with no virions in either the ATI matrix or on its surface (V−). Most natural CPXV strains produce V+ or V− ATIs as the dominant but stable phenotype. Recently, the first poxvirus isolate, CPXV-No-H2, was described that produced V+/ ATI bodies as a stable phenotype (48). Nevertheless, all other CPXVs isolated from Fennoscandia represent the V+ ATI phenotype and, hence, encode full-length ati and p4c genes (49). Since RatPox09, unlike FM2292, was isolated from rats and not a reservoir host, it is tempting to speculate that it may have lost the ability to form V+ ATIs. Kastenmayer and coworkers (35) constructed and characterized a p4c deletion mutant of the GER1991_3 CPXV strain. Mice inoculated intranasally with the mutant lost slightly more weight than mice infected with the parental wild-type virus, but the difference did not reach statistical significance. Consistent with this finding, RatPox09 seemed more virulent than FM2292 in our study, which may at least in part be caused by the p4c mutation and the inability of its encoded protein to incorporate virions into ATIs. However, final testing of this hypothesis will require the production of isogenic RatPox09 and FM2292 mutants, experiments that are under way.
From analyzing the growth kinetics of FM2292 in comparison to those of BR and RatPox09, we deduced that replication in vitro is not influenced by the differences between the viruses in the number of genes they carry. Whether reservoir hosts, which according to serological data include mainly vole species (the common vole Microtus arvalis, the bank vole Myodes glareolus, and the field vole Microtus agrestis) and wood mice (Apodemus sylvaticus) (12), are truly clinically affected by CPXV infection is unknown, but it was suggested that overt infection is unlikely (8, 9). The comparative experimental infection of captivity-bred common voles and regular Wistar rats described here is the first controlled experimental infection with different CPXV strains in reservoir hosts instead of an accidental host. CPXV FM2292 isolated from the liver of a common vole was used to inoculate common voles and Wistar rats as a surrogate model and compared to an already characterized virus isolated from a pet rat in 2009 (17). Based on data evaluated after experimental infection of Wistar rats using RatPox09 (17), low- and high-dose infections via the oronasal route were assessed.
Unexpectedly, common voles exhibited clear signs of respiratory disease rather than skin lesions, irrespective of the virus strain used. This might suggest that CPXV could also be transmitted via droplet infection within the reservoir host population and also to accidental hosts. In contrast, Wistar rats inoculated with the reservoir host-derived FM2292 isolate developed severe and characteristic pox lesions, primarily on noses, ears, paws, and tails, but recovered after infection. In addition, body weight increase was delayed in a dose-dependent fashion in Wistar rats inoculated with FM2292. Therefore, FM2292 seems to be less virulent than RatPox09 in the surrogate model, which is also reflected by smaller amounts of virus shed. This is evidenced by the observation that rats inoculated with the high dose of RatPox09 shed up to 104.5 TCID50 ml−1 for up to 12 days (17). Likewise, organ samples of animals in this group yielded higher titers, >106 TCID50 ml−1 (17), than those of FM2292-infected rats. Similarly, RatPox09 caused higher mortality and more robust shedding in the common vole. The rat-adapted CPXV strain from 2009 has, however, gained virulence in the new host, probably as a result of consecutive animal passages that have occurred primarily in rat breeding farms. However, one out of five low-dose-infected common vole individuals did not show seroconversion, which might indicate that an infectious dose of 104 TCID50 ml−1 approaches the minimal infectious dose for that species. As common voles inoculated were older than the Wistar rats, body weight increases were not detected, but diseased voles did not show a prominent drop in weight.
Taken together, our data confirmed the variable pathogenic potential of CPXV isolates. Clearly, virulence of CPXV has to be correlated to the affected host species as the manifestation of disease induced by CPXV is host dependent. Future research will focus mainly on the correlation between virus genotype and phenotype/pathotype in accidental and reservoir species. Here, the four additional ORFs found in both isolates will be the first targets to be investigated through the generation of isogenic mutants.
ACKNOWLEDGMENTS
We are indebted to Mareen Lange for excellent technical assistance. We are grateful to Matthias Lenk, Greifswald-Insel Riems, for providing cell lines and to Hermann Meyer, Institut für Mikrobiologie der Bundeswehr, Munich, Germany, for kindly providing the RatPox09 isolate.
This project was funded by the German Research Foundation SPP1596 project awarded to M.B., R.G.U., and N.O. and by the Federal Ministry of Education and Research through the National Research Platform for Zoonoses to R.G.U. (Network Rodent-borne Pathogens; project number 01KI1303). The vole KS11/2292 was kindly provided by Daniela Reil and Christian Imholt from UFOPLAN projects 3709 41 401 and 3713 48 401. Sequencing was supported by the ANIHWA-project EPiSeq (grant number 2811ERA094).
Common voles used for animal experiments were kindly provided by Erik Schmolz (Federal Environment Agency, Section Health Pests and Their Control, Berlin, Germany), and the bank vole cell line by Sandra Essbauer, Munich.
REFERENCES
- 1.Vorou RM, Papavassiliou VG, Pierroutsakos IN. 2008. Cowpox virus infection: an emerging health threat. Curr Opin Infect Dis 21:153–156. doi: 10.1097/QCO.0b013e3282f44c74. [DOI] [PubMed] [Google Scholar]
- 2.Essbauer S, Pfeffer M, Meyer H. 2010. Zoonotic poxviruses. Vet Microbiol 140:229–236. doi: 10.1016/j.vetmic.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chantrey J, Meyer H, Baxby D, Begon M, Bown KJ, Hazel SM, Jones T, Montgomery WI, Bennett M. 1999. Cowpox: reservoir hosts and geographic range. Epidemiol Infect 122:455–460. doi: 10.1017/S0950268899002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker C, Kurth A, Hessler F, Kramp H, Gokel M, Hoffmann R, Kuczka A, Nitsche A. 2009. Cowpox virus infection in pet rat owners: not always immediately recognized. Dtsch Arztebl Int 106:329–334. doi: 10.3238/arztebl.2009.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campe H, Zimmermann P, Glos K, Bayer M, Bergemann H, Dreweck C, Graf P, Weber BK, Meyer H, Büttner M, Busch U, Sing A. 2009. Cowpox virus transmission from pet rats to humans, Germany. Emerg Infect Dis 15:777–780. doi: 10.3201/eid1505.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ninove L, Domart Y, Vervel C, Voinot C, Salez N, Raoult D, Meyer H, Capek I, Zandotti C, Charrel RN. 2009. Cowpox virus transmission from pet rats to humans, France. Emerg Infect Dis 15:781–784. doi: 10.3201/eid1505.090235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elsendoorn A, Agius G, Le Moal G, Aajaji F, Favier AL, Wierzbicka-Hainault E, Béraud G, Flusin O, Crance JM, Roblot F. 2011. Severe ear chondritis due to cowpox virus transmitted by a pet rat. J Infect 63:391–393. doi: 10.1016/j.jinf.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Bennett M, Crouch AJ, Begon M, Duffy B, Feore S, Gaskell RM, Kelly DF, McCracken CM, Vicary L, Baxby D. 1997. Cowpox in British voles and mice. J Comp Pathol 116:35–44. doi: 10.1016/S0021-9975(97)80041-2. [DOI] [PubMed] [Google Scholar]
- 9.Feore SM, Bennett M, Chantrey J, Jones T, Baxby D, Begon M. 1997. The effect of cowpox virus infection on fecundity in bank voles and wood mice. Proc Biol Sci 264:1457–1461. doi: 10.1098/rspb.1997.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Begon M, Telfer S, Burthe S, Lambin X, Smith MJ, Paterson S. 2009. Effects of abundance on infection in natural populations: field voles and cowpox virus. Epidemics 1:35–46. doi: 10.1016/j.epidem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Burthe S, Telfer S, Begon M, Bennett M, Smith A, Lambin X. 2008. Cowpox virus infection in natural field vole Microtus agrestis populations: significant negative impacts on survival. J Anim Ecol 77:110–119. doi: 10.1111/j.1365-2656.2007.01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hazel SM, Bennett M, Chantrey J, Bown K, Cavanagh R, Jones TR, Baxby D, Begon M. 2000. A longitudinal study of an endemic disease in its wildlife reservoir: cowpox and wild rodents. Epidemiol Infect 124:551–562. doi: 10.1017/S0950268899003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Telfer S, Lambin X, Birtles R, Beldomenico P, Burthe S, Paterson S, Begon M. 2010. Species interactions in a parasite community drive infection risk in a wildlife population. Science 330:243–246. doi: 10.1126/science.1190333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinnunen PM, Henttonen H, Hoffmann B, Kallio ER, Korthase C, Laakkonen J, Niemimaa J, Palva A, Schlegel M, Ali HS, Suominen P, Ulrich RG, Vaheri A, Vapalahti O. 2011. Orthopox virus infections in Eurasian wild rodents. Vector Borne Zoonotic Dis 11:1133–1140. doi: 10.1089/vbz.2010.0170. [DOI] [PubMed] [Google Scholar]
- 15.L'Vov SD, Gromashevskiĭ VL, Marennikova SS, Bogoiavlenskiĭ GV, Baĭluk FN. 1988. Isolation of poxvirus (Poxviridae, Orthopoxvirus, the cowpox complex) from the root vole Microtus (M) oeconomus Pal. 1776 in the forest tundra of the Kola Peninsula. Vopr Virusol 33:92–94. (In Russian.) [PubMed] [Google Scholar]
- 16.Smee DF. 2013. Orthopoxvirus inhibitors that are active in animal models: an update from 2008 to 2012. Future Virol 8:891–901. doi: 10.2217/fvl.13.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalthoff D, König P, Meyer H, Beer M, Hoffmann B. 2011. Experimental cowpox virus infection in rats. Vet Microbiol 153:382–386. doi: 10.1016/j.vetmic.2011.05.048. [DOI] [PubMed] [Google Scholar]
- 18.Breithaupt A, Kalthoff D, Deutskens F, König P, Hoffmann B, Beer M, Meyer H, Teifke JP. 2012. Clinical course and pathology in rats (Rattus norvegicus) after experimental cowpox virus infection by percutaneous and intranasal application. Vet Pathol 49:941–949. doi: 10.1177/0300985812439077. [DOI] [PubMed] [Google Scholar]
- 19.Jacob J, Ulrich RG, Freise J, Schmolz E. 2014. Monitoring populations of rodent reservoirs of zoonotic diseases. Projects, aims and results. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 57:511–518. (In German.) doi: 10.1007/s00103-013-1924-x. [DOI] [PubMed] [Google Scholar]
- 20.Scaramozzino N, Ferrier-Rembert A, Favier AL, Rothlisberger C, Richard S, Crance JM, Meyer H, Garin D. 2007. Real-time PCR to identify variola virus or other human pathogenic orthopox viruses. Clin Chem 53:606–613. doi: 10.1373/clinchem.2006.068635. [DOI] [PubMed] [Google Scholar]
- 21.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 24.R Development Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 25.Carroll DS, Emerson GL, Li Y, Sammons S, Olson V, Frace M, Nakazawa Y, Czerny CP, Tryland M, Kolodziejek J, Nowotny N, Olsen-Rasmussen M, Khristova M, Govil D, Karem K, Damon IK, Meyer H. 2011. Chasing Jenner's vaccine: revisiting cowpox virus classification. PLoS One 6:e23086. doi: 10.1371/journal.pone.0023086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Essbauer SS, Krautkrämer E, Herzog S, Pfeffer M. 2011. A new permanent cell line derived from the bank vole (Myodes glareolus) as cell culture model for zoonotic viruses. Virol J 8:339. doi: 10.1186/1743-422X-8-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dabrowski PW, Radonić A, Kurth A, Nitsche A. 2013. Genome-wide comparison of cowpox viruses reveals a new clade related to variola virus. PLoS One 8:e79953. doi: 10.1371/journal.pone.0079953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Downie AW. 1939. A study of the lesions produced experimentally by cowpox virus. J Pathol Bacteriol 48:361–379. doi: 10.1002/path.1700480212. [DOI] [Google Scholar]
- 30.Patel DD, Pickup DJ, Joklik WK. 1986. Isolation of cowpox virus A-type inclusions and characterization of their major protein component. Virology 149:174–189. doi: 10.1016/0042-6822(86)90119-4. [DOI] [PubMed] [Google Scholar]
- 31.Okeke MI, Adekoya OA, Moens U, Tryland M, Traavik T, Nilssen Ø. 2009. Comparative sequence analysis of A-type inclusion (ATI) and P4c proteins of orthopoxviruses that produce typical and atypical ATI phenotypes. Virus Genes 39:200–209. doi: 10.1007/s11262-009-0376-8. [DOI] [PubMed] [Google Scholar]
- 32.Shida H, Tanabe K, Matsumoto S. 1977. Mechanism of virus occlusion into A-type inclusion during poxvirus infection. Virology 76:217–233. doi: 10.1016/0042-6822(77)90298-7. [DOI] [PubMed] [Google Scholar]
- 33.McKelvey TA, Andrews SC, Miller SE, Ray CA, Pickup DJ. 2002. Identification of the orthopoxvirus p4c gene, which encodes a structural protein that directs intracellular mature virus particles into A-type inclusions. J Virol 76:11216–11225. doi: 10.1128/JVI.76.22.11216-11225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howard AR, Weisberg AS, Moss B. 2010. Congregation of orthopoxvirus virions in cytoplasmic A-type inclusions is mediated by interactions of a bridging protein (A26p) with a matrix protein (ATIp) and a virion membrane-associated protein (A27p). J Virol 84:7592–7602. doi: 10.1128/JVI.00704-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kastenmayer RJ, Maruri-Avidal L, Americo JL, Earl PL, Weisberg AS, Moss B. 2014. Elimination of A-type inclusion formation enhances cowpox virus replication in mice: implications for orthopoxvirus evolution. Virology 452–453:59–66. doi: 10.1016/j.virol.2013.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duraffour S, Mertens B, Meyer H, van den Oord JJ, Mitera T, Matthys P, Snoeck R, Andrei G. 2013. Emergence of cowpox: study of the virulence of clinical strains and evaluation of antivirals. PLoS One 8:e55808. doi: 10.1371/journal.pone.0055808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shchelkunov SN. 2013. An increasing danger of zoonotic orthopoxvirus infections. PLoS Pathog 9:e1003756. doi: 10.1371/journal.ppat.1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beard PM, Froggatt GC, Smith GL. 2006. Vaccinia virus kelch protein A55 is a 64 kDa intracellular factor that affects virus-induced cytopathic effect and the outcome of infection in a murine intradermal model. J Gen Virol 87:1521–1529. doi: 10.1099/vir.0.81854-0. [DOI] [PubMed] [Google Scholar]
- 39.Froggatt GC, Smith GL, Beard PM. 2007. Vaccinia virus gene F3L encodes an intracellular protein that affects the innate immune response. J Gen Virol 88:1917–1921. doi: 10.1099/vir.0.82815-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kochneva G, Kolosova I, Maksyutova T, Ryabchikova E, Shchelkunov S. 2005. Effects of deletions of kelch-like genes on cowpox virus biological properties. Arch Virol 150:1857–1870. doi: 10.1007/s00705-005-0530-0. [DOI] [PubMed] [Google Scholar]
- 41.Pires de Miranda M, Reading PC, Tscharke DC, Murphy BJ, Smith GL. 2003. The vaccinia virus kelch-like protein C2L affects calcium-independent adhesion to the extracellular matrix and inflammation in a murine intradermal model. J Gen Virol 84:2459–2471. doi: 10.1099/vir.0.19292-0. [DOI] [PubMed] [Google Scholar]
- 42.Balinsky CA, Delhon G, Afonso CL, Risatti GR, Borca MV, French RA, Tulman ER, Geary SJ, Rock DL. 2007. Sheeppox virus kelch-like gene SPPV-019 affects virus virulence. J Virol 81:11392–11401. doi: 10.1128/JVI.01093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q, Burles K, Couturier B, Randall CM, Shisler J, Barry M. 2014. Ectromelia virus encodes a BTB/kelch protein, EVM150, that inhibits NF-κB signaling. J Virol 88:4853–4865. doi: 10.1128/JVI.02923-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reading PC, Khanna A, Smith GL. 2002. Vaccinia virus CrmE encodes a soluble and cell surface tumor necrosis factor receptor that contributes to virus virulence. Virology 292:285–298. doi: 10.1006/viro.2001.1236. [DOI] [PubMed] [Google Scholar]
- 45.Saraiva M, Alcami A. 2001. CrmE, a novel soluble tumor necrosis factor receptor encoded by poxviruses. J Virol 75:226–233. doi: 10.1128/JVI.75.1.226-233.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hendrickson RC, Wang C, Hatcher EL, Lefkowitz EJ. 2010. Orthopoxvirus genome evolution: the role of gene loss. Viruses 2:1933–1967. doi: 10.3390/v2091933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hatcher EL, Hendrickson RC, Lefkowitz EJ. 2014. Identification of nucleotide-level changes impacting gene content and genome evolution in orthopoxviruses. J Virol 88:13651–13668. doi: 10.1128/JVI.02015-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okeke MI, Hansen H, Traavik T. 2012. A naturally occurring cowpox virus with an ectromelia virus A-type inclusion protein gene displays atypical A-type inclusions. Infect Genet Evol 12:160–168. doi: 10.1016/j.meegid.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 49.Okeke MI, Okoli AS, Nilssen Ø, Moens U, Tryland M, Bøhn T, Traavik T. 2014. Molecular characterization and phylogenetics of Fennoscandian cowpox virus isolates based on the p4c and atip genes. Virol J 11:119. doi: 10.1186/1743-422X-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]