ABSTRACT
Presently, respiratory syncytial virus (RSV), the main cause of severe respiratory infections in infants, cannot be treated efficiently with antivirals. However, its RNA-dependent polymerase complex offers potential targets for RSV-specific drugs. This includes the recognition of its template, the ribonucleoprotein complex (RNP), consisting of genomic RNA encapsidated by the RSV nucleoprotein, N. This recognition proceeds via interaction between the phosphoprotein P, which is the main polymerase cofactor, and N. The determinant role of the C terminus of P, and more particularly of the last residue, F241, in RNP binding and viral RNA synthesis has been assessed previously. Here, we provide detailed structural insight into this crucial interaction for RSV polymerase activity. We solved the crystallographic structures of complexes between the N-terminal domain of N (N-NTD) and C-terminal peptides of P and characterized binding by biophysical approaches. Our results provide a rationale for the pivotal role of F241, which inserts into a well-defined N-NTD pocket. This primary binding site is completed by transient contacts with upstream P residues outside the pocket. Based on the structural information of the N-NTD:P complex, we identified inhibitors of this interaction, selected by in silico screening of small compounds, that efficiently bind to N and compete with P in vitro. One of the compounds displayed inhibitory activity on RSV replication, thereby strengthening the relevance of N-NTD for structure-based design of RSV-specific antivirals.
IMPORTANCE Respiratory syncytial virus (RSV) is a widespread pathogen that is a leading cause of acute lower respiratory infections in infants worldwide. RSV cannot be treated efficiently with antivirals, and no vaccine is presently available, with the development of pediatric vaccines being particularly challenging. Therefore, there is a need for new therapeutic strategies that specifically target RSV. The interaction between the RSV phosphoprotein P and the ribonucleoprotein complex is critical for viral replication. In this study, we identified the main structural determinants of this interaction, and we used them to screen potential inhibitors in silico. We found a family of molecules that were efficient competitors of P in vitro and showed inhibitory activity on RSV replication in cellular assays. These compounds provide a basis for a pharmacophore model that must be improved but that holds promises for the design of new RSV-specific antivirals.
INTRODUCTION
Human respiratory syncytial virus (HRSV) is the main cause of acute lower respiratory infections in infants worldwide (1). No RSV vaccine is presently available, and the development of pediatric vaccines is particularly challenging. Currently, antiviral therapy is limited to palivizumab, a humanized mouse monoclonal antibody that targets the RSV fusion protein and is licensed for prophylactic use, and ribavirin, which has been used to treat severe infections despite its toxicity, its teratogenicity, and the limited evidence of its antiviral effect (2). Thus, there is an obvious and urgent need to develop new therapeutic strategies and effective antiviral drugs against RSV.
RSV is an enveloped, nonsegmented, negative-strand RNA virus that belongs to the subfamily Pneumovirinae, family Paramyxoviridae, order Mononegavirales. Its genomic RNA is tightly bound to the viral nucleoprotein (N) and maintained as a left-handed helical N-RNA ribonucleoprotein complex (RNP) (3). This RNP is the template for transcription and replication by the RNA-dependent RNA polymerase (RdRp) complex, which consists of the polymerase (L; large protein) and its main cofactor, the phosphoprotein (P). Proteins N, P, and L are sufficient for replication of the viral RNA (4, 5). Transcription requires an additional processivity cofactor, M2-1 (4). Efficient and specific recognition of the RNP template by the RdRp is critical for viral RNA synthesis. It is mediated by P, a multifunctional protein capable of interacting with multiple partners: L (6), N (7), and M2-1 (8), but also cellular proteins (9). P positions the RdRp complex on the RNP and is likely involved in translocation along the RNP (10).
P is a modular protein consisting of a central tetramerization domain (11–13) flanked by two long regions predicted to be intrinsically disordered, termed the N-terminal domain and C-terminal domain of P (P-NTD and P-CTD). We have shown previously that the nine C-terminal residues of P, and particularly the C-terminal F241, are critical for RNP binding and viral RNA synthesis (14, 15). There is currently no structural information at the atomic level for P, but the X-ray structure of ring-shaped RNP particles is known (16). The N protein has a modular organization consisting of two globular domains, the N-NTD (amino acids [aa] 31 to 252) and N-CTD (aa 253 to 360), and two flexible arms at the N and C termini (aa 1 to 30 and 362 to 391). Recently, we showed by a rational approach using N deletions and site-directed mutagenesis that a hydrophobic pocket within the N-NTD corresponds to the P-CTD binding domain. We identified several basic and hydrophobic residues, either inside this pocket or surface exposed, that are critical for the P/N interaction and RNA synthesis (15).
By solving the X-ray structures of the N-NTD in complex with C-terminal peptides of P here, we gained more detailed insight into the N-NTD:P-CTD interaction and the specific roles of the last C-terminal residues of P. These structures underline the pivotal role of the C-terminal residue, P-F241, which is deeply buried in the previously identified N-NTD pocket (15) and acts as an anchor for P. The interaction network between the N-NTD and P-CTD is, furthermore, completed by well-defined electrostatic interactions with P-D240 and less defined interactions outside the pocket. Next, we were able to obtain X-ray structures of the N-NTD in complex with small compounds selected by in silico screening. These compounds, bearing a common 1-benzyl-1H-pyrazole-3,5-dicarboxylate (BPdC) scaffold, displayed affinities similar to that of the P-CTD and efficiently competed with the P-CTD for N-NTD binding in vitro by displacing P-F241 from the hydrophobic pocket. Moreover, the X-ray structures show that the entrance of the N-NTD pocket can be remodeled by the halogen atoms of BPdC compounds. Finally, a molecule derived from the BPdC compound with the highest affinity for the N-NTD was able to specifically inhibit RSV replication in cell assays.
MATERIALS AND METHODS
Protein expression and purification.
Glutathione S-transferase (GST)-PCT and poly-His fused N-NTD proteins were engineered and purified as described previously (11, 14, 15). Protein fractions recovered from the affinity purification step were pooled, concentrated, and further purified by gel filtration using a Superdex75 HiLoad 16/60 column equilibrated in 50 mM Tris, pH 8, 50 mM NaCl. The C-terminal His tag was not removed for N-NTD crystallization trials.
N-NTD samples with stable isotope labeling for nuclear magnetic resonance (NMR) measurements were produced by adapting protocols from rich to minimum culture media. 15N-labeled N-NTD was produced in M9 medium supplemented with 1 g/liter 15NH4Cl (Eurisotop). 13C, 15N, 70% 2H-labeled N-NTD was prepared by starting from a 100-ml preculture in LB medium, used to inoculate 1 liter of unlabeled M9. Cells were harvested at an optical density at 600 nm (OD600) of 0.6 and transferred into 100 ml of 2H13C15N-M9 medium prepared with 95% 2H2O (Eurisotop), 1 g/liter 15NH4Cl, and 3 g/liter [13C]glucose (Cortecnet). After 1 h at 37°C, cells were harvested again by centrifugation and transferred into 900 ml 2H13C15N-M9 medium. After 1 h, induction was started at 28°C overnight. Purification was carried out as for unlabeled N-NTD and followed by a final dialysis against NMR buffer [20 mM MES (morpholineethanesulfonic acid), pH 6.5, 250 mM NaCl, 1.5 mM tris(2-carboxyethyl)phosphine (TCEP)].
P12 peptide synthesis.
Peptide P12 (SEDNDSDNDLSLEDF-COOH) containing the last 12 residues of P, NDSDNDLSLEDF, with three additional N-terminal residues, SED, was synthesized by solid-phase peptide chemistry on an Fmoc (9-fluorenylmethoxy carbonyl)-Phe-SASRIN resin with an Apex 396 Automated Multiple Peptide Synthesizer (Aapptec), using Fmoc chemistry; deprotected by 25% piperidine/NMP; and cleaved from the resin with trifluoroacetic acid (TFA)-triisopropylsilane (TIS)-H2O (95:2.5:2.5) for 1 h 30 min. The product was analyzed by ultraperformance liquid chromatography (UPLC)-mass spectrometry (MS) (Waters Acquity Ultra Performance LC/Micromass Quattro micro API) on an Acquity UPLC BEH C18 column (1.7 μl; 1.0 by 50 mm), and a molecular mass of 1,714.63 g · mol−1 was determined.
Synthesis of M76-diAM.
Diisopropylethyamine (i-Pr2NEt) (0.5 ml; 2.8 mmol) and bromomethyl acetate (171 mg; 110 μl; 1.12 mmol) were sequentially added to a solution of 1-(2,4-dichloro-benzyl)-1H-pyrazole-3,5-dicarboxylic acid (90 mg; 0.28 mmol) in dimethylformamide (DMF) (2 ml). The reaction mixture was stirred at room temperature for 16 h and concentrated under reduced pressure. The oily residue was taken up into ethyl acetate (AcOEt) (20 ml) and washed with aqueous sodium bicarbonate (2 ml) and brine (2 ml). The organic phase was dried over MgSO4 and concentrated under reduced pressure to leave a yellow oil, which was purified by chromatography over silica gel and eluting with AcOEt-cyclohexane (2:1) to give the protected compound M76-diAM [1-(2,4-dichloro-benzyl)-1H-pyrazole-3,5-dicarboxylic acid diacetoxymethyl ester] as a colorless oil (102 mg; 78%). Infrared (IR) (neat) wave numbers (ν): 3,146, 3,000, 1,769, 1,765, 1,736, 1,591, 1,565, 1,530, 1,476, 1,443, 1,368, 1,259, 1,230, 1,213, 1,187, 1,171, 1,156, 1,087, 1,048, 1,024, 1,101, 985, 956, 861, 836, 811, 760, 739, 685 cm−1. 1H NMR (C6D6, 300 MHz) δ: 7.46 (s, 1H), 7.01 (d, J = 2.1 Hz, 1H), 6.62 (dd, J = 8.4 Hz, J = 2.1 Hz, 1H), 6.28 (d, J = 8.4 Hz, 1H), 5.77 (s, 2H), 5.50 (s, 2H), 5.48 (s, 2H), 1.48 (s, 6H). 13C NMR (C6D6, 300 MHz) δ: 168.9 (CO), 168.6 (CO), 159.8 (CO), 157.3 (CO), 142 (C), 134.4 (C), 133.6 (C), 133.2 (C), 132.8 (C), 129.6 (CH), 129.0 (CH), 127.5 (CH), 115.8 (CH), 79.5 (CH2), 79.4 (CH2), 53.3 (CH2), 20.0 (CH3), 19.9 (CH3).
Crystallization and diffraction data collection.
The N-NTD was concentrated to 8 mg · ml−1 in 20 mM Tris-HCl, pH 7.5, 50 mM NaCl. A nanoscale robot crystallization screen was performed in 96-well sitting-drop plates at 18°C. After optimization, diffraction quality crystals were grown by hanging drop in 25% polyethylene glycol 4000 (PEG 4000) or 28% PEG 5000MME with 100 mM HEPES, pH 7.5, buffer and 200 mM ammonium sulfate; transferred in a solution containing 30% PEG 4000 or PEG 5000MME supplemented with 5% glycerol and 5% PEG 400; and flash-frozen under liquid nitrogen.
P3, P5, P7, and P13 peptides, corresponding to the last 3, 5, 7, and 13 C-terminal residues of the P sequence, respectively, were purchased from Covalab and phenylalanine (P1) and Asp-Phe (P2) from Sigma-Aldrich; they were solubilized at 10 mg/ml in crystallization buffer. M61 was purchased from Aldrich, and M72, M68, M81, and M76 were purchased from Chembridge. Each compound was solubilized at 10 mg/ml in crystallization buffer with 60% ethanol. The N-NTD and peptide or inhibitor solutions were mixed at a ratio of 3:1 and crystallized as the isolated N-NTD, with slight adjustment of precipitant concentrations for each complex.
Structure determination.
X-ray diffraction data were collected at the beamline PX06-SA at the Swiss Light Source (SLS), Proxima-1 at Soleil, and ID14-4 at the European Synchrotron Radiation Facility (ESRF). Data were processed using the XDS package (17) and scaled with SCALA (18). Structures were solved by molecular replacement with PHASER (19) using Protein Data Bank (PDB) entry 2WJ8. Subsequently, careful model building was carried out with COOT (20) alternated with crystallographic refinement with the program BUSTER/TNT (21), which included noncrystallographic symmetry (NCS) restraints and translation, libration, screw rotation (TLS) refinement. Stereochemical restraint dictionaries for BPdC derivatives were generated with the Grade Web server (http://grade.globalphasing.org). In addition, target restraints using the M76 molecular model were used for refinement of the P3, P7, M81, and M72 models. Data collection and refinement statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
| Statistic | Value |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Nat1 | Nat2 | P1 | P2 | P3 | P7 | M76 | M81 | M72 | |
| PDB ID | 4UC6 | 4UC7 | 4UC8 | 4UC9 | 4UCA | 4UCB | 4UCC | 4UCD | 4UCE |
| Data collection | Soleil PX1 | Soleil PX1 | Soleil PX1 | Soleil PX1 | SLS PX06SA | SLS PX06SA | SLS PX06SA | SLS PX06SA | ESRF ID14-4 |
| Space group | P 21 21 2 | P 21 21 21 | P 21 21 2 | P 43 21 2 | P 21 21 21 | P 21 21 21 | P 21 21 21 | P 21 21 21 | P 21 21 21 |
| Unit cell (Å) | |||||||||
| a | 71.71 | 34.31 | 71.81 | 72.51 | 33.84 | 34.52 | 34.82 | 34.68 | 34.0 |
| b | 134.11 | 72.13 | 134.72 | 72.51 | 71.58 | 71.68 | 71.96 | 71.94 | 71.3 |
| c | 50.07 | 181.67 | 50.36 | 383.38 | 177.20 | 178.27 | 178.17 | 178.55 | 178.4 |
| Resolution (Å)a | 40–2.1 (2.21–2.10) | 40–2.4 (2.58–2.45) | 40–2.0 (2.11–2.00) | 40–2.40 (2.53–2.40) | 40–3.22 (3.40–3.22) | 40–2.78 (2.93–2.78) | 40–2.05 (2.16–2.05) | 40–2.66 (2.81–2.66) | 40–2.94 (3.10–2.94) |
| Observations (no.) | 79,621 (9,853) | 87,163 (2,127) | 103,732 (6,816) | 357,987(13,030) | 31,674 (1,938) | 45,949 (1,999) | 170,722(13,444) | 79,506 (4,703) | 39,319 (1,614) |
| Unique reflections (no.) | 28,255 (3,903) | 15,360 (1,157) | 29,895 (2,983) | 39,937 (4,335) | 7,219 (929) | 11,079 (1,096) | 28,687 (3,708) | 13,129 (1,507) | 8,454 (834) |
| Completeness (%) | 98.0 (94.9) | 88.3 (47.7) | 88.4 (61.4) | 96.0 (74.6) | 96.9 (86.0) | 94.7 (71.1) | 98.5 (90.0) | 97.2 (81.1) | 86.8 (61.8) |
| I/σI | 9.9 (1.7) | 13.3 (1.5) | 12.7 (2.0) | 15.8 (1.9) | 7.7 (1.7) | 8.4 (1.7) | 18.1 (2.1) | 14.2 (1.8) | 9.0 (1.9) |
| Rmerge (%) | 6.8 (50.9) | 8.4 (35.0) | 6.9 (40.5) | 10.3 (46.7) | 18.1 (50.7) | 11.6 (42.4) | 4.6 (53.1) | 7.5 (51.9) | 16.7 (45.5) |
| Refinement statistics | 20–2.10 | 15–2.45 | 15–2.00 | 15–2.42 | 20–3.22 | 20–2.78 | 15–2.05 | 20–2.66 | 20–2.94 |
| Work/test reference | 28,202/1,428 | 15,217/1,507 | 29,743/1,507 | 39,586/1,992 | 7,170/707 | 11,000/1,108 | 28,525/2,895 | 13,043/1,286 | 8,353/821 |
| Rcryst./Rfree (%) | 0.187/0.227 | 0.218/0.268 | 0.178/0.212 | 0.194/0.251 | 0.192/0.2,286 | 0.195/0.227 | 0.191/0.236 | 0.174/0.215 | 0.203/0.240 |
| No. of protein atoms | 3,499 | 3,338 | 3,543 | 6,938 | 3,462 | 3,447 | 3,541 | 3,454 | 3,408 |
| No. of heteroatoms | 315 | 122 | 596 | 1,117 | 25 | 76 | 414 | 97 | 70 |
| Root mean square (RMS) deviation from ideal | |||||||||
| Bond length (Å) | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 |
| Bond angle (°) | 1.00 | 1.14 | 1.00 | 1.08 | 1.08 | 1.10 | 1.08 | 1.04 | 1.08 |
| Ramachandran plotb | |||||||||
| Favored (%) | 100 | 97.85 | 98.64 | 98.48 | 99.07 | 98.84 | 98.43 | 98.39 | 99.06 |
| Allowed (%) | 0 | 2.15 | 1.36 | 1.52 | 0.93 | 1.16 | 1.57 | 1.61 | 0.94 |
| Outliers (%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
The highest-resolution shell is shown in parentheses.
Ramachandran statistics were calculated with MolProbity (46).
Drug design.
The X-ray structure of N-NTD complexed with P2 was used as a starting configuration. The missing three-dimensional (3D) coordinates were added using the SWISS-MODEL package. Using the relative positions of the phenylalanine of P2 and the sulfate ion found in the N-NTD:P2 complex, a docking volume was defined using AutoDock Vina (22). By virtual screening of the ZINC database (23), compounds were selected on the basis of the following requirements: (i) fit in the defined volume; (ii) possess an aromatic moiety, like F241; (iii) target R150 of the N-NTD by mimicking the carboxylate of F241; (iv) target R132 of the N-NTD by replacing the interactions of the residue with the sulfate ion. The initial screening identified 1,500 compounds, further reduced to 300 by elimination of those with molecular masses higher than 350 Da and those with demonstrated toxicity. The compounds were then ordered based on favorable van der Waals interaction energies between the ligand and N-NTD. The best 100 compounds were energy minimized and ordered with a free-energy scoring function. Atoms of the N-NTD located within 8 Å from the ligand were left to adopt a relaxed conformation, while the atoms of the rest of the protein were harmonically restrained in their initial positions.
NMR spectroscopy.
Backbone assignment of the N-NTD was based on the acquisition at 293 K of standard triple-resonance NMR experiments [HNCO, HNCA, HN(CO)CA, HN(CO)CBCA, and HNCACB] on a 600-MHz Bruker Avance III spectrometer equipped with a cryogenic TCI probe, using 80 μM 13C,15N, 70%-2H-labeled N-NTD in 20 mM MES, pH 6.5, 250 mM NaCl, 1.5 mM TCEP. Samples contained 7% 2H2O to lock the spectrometer frequency. Spectra were processed with Topspin 3.1 and NmrPipe (24). Analysis of NMR experiments was performed in CCPNMR (25). Ninety-one percent nonproline 1H and 15N amide, 91% 13C′, 96% 13Cα, and 94% 13Cβ chemical shifts were assigned. Assignments for the innermost αN3 helix (residues I160 to V167) are missing due to inefficient back-protonation.
Titration experiments were carried out by recording 1H-15N heteronuclear single-quantum correlation (HSQC) spectra of 50 μM 15N-labeled N-NTD on 700- or 800-MHz Bruker Avance III spectrometers, equipped with TXI and TCI cryoprobes, respectively. P12 (SEDNDSDNDLSLEDF) and P2 peptides were aliquoted, lyophilized, and dissolved step by step into N-NTD solution with molar ratios up to 16:1 and 500:1, respectively. M61, M68, M72, M76, and M81 were added from concentrated stock solutions (5, 10, or 25 mM in ethanol) with molar ratios up to 20. Dissociation constants (Kds) were determined for each perturbed residue by assuming a two-site fast-exchange model with a 1:1 stoichiometry and by fitting 1H and 15N chemical shift differences with Origin 7 (OriginLab), according to equation 1.
| 1 |
where δbound and δfree are the chemical shifts of ligand-bound and free N-NNTD, respectively; α is a factor of proportionality; [N-NTD]tot is the total concentration of N-NTD; r is the molar ratio (ligand)/[N-NTD]tot; B = 1/2 (δbound − δfree), and A is defined by Kd = (A − 1) × [N-NTD]tot.
Correction for the contribution of ethanol during titration with BPdCs was carried out by using a spectrum of N-NTD with 5 μl ethanol and by assuming a linear relationship between these perturbations and the added volumes of ligand solutions.
Combined 1H and 15N amide chemical shift perturbations (CSPs) were calculated using both 1H and 15N chemical differences according to equation 2:
| 2 |
SPR.
Assays were carried out at 25°C in 20 mM Tris-HCl, pH 8, 150 mM NaCl. A goat anti-GST antibody (Biacore GST Capture kit) was covalently coupled to a CM5 sensor chip, using a Biacore 2000 instrument and the Amine Coupling kit (GE Healthcare), reaching an immobilization density of ∼10,000 resonance units (RU) (1 RU ≈ 1 pg · mm−2). This surface was used to capture GST-PΔ161N (P residues 161 to 241) to a density of 1,200 to 1,300 RU or GST (800 RU) as a control. N-NTD (20 μM), alone or mixed and equilibrated for over 2 h with the five BPdC compounds (concentration range, 5 to 750 μM), was then injected over the GST-PΔ161N and GST surfaces for 1 min at a flow rate of 50 ml · min−1. After each injection, the interaction buffer was allowed to flow on the surface until all the N-NTD molecules dissociated, taking advantage of the very transient N-NTD:P-CTD interaction. At the end of the series, the surfaces were regenerated with a 2-min 10 mM glycine-HCl (pH 1.5) wash and two 1-min washes with 0.05% SDS and 20 mM NaOH. The real-time interaction profiles were double referenced using Scrubber 2.0 software (BioLogic Software), that is, the signals both from the reference surface (with GST captured on the anti-GST antibody) and from blank experiments using the compounds alone were subtracted. The steady-state surface plasmon resonance (SPR) responses (Req) were plotted against the compound concentration C and fitted using the BIAevaluation 4.1 software (GE Healthcare), according to equation 3, to determine the inhibition constants (Ki) for each compound:
| 3 |
where the concentration of N-NTD is 20 μM and R0 is the steady-state SPR response for N-NTD alone, allowing us to determine the inhibition constant, Ki, for each compound.
Isothermal titration calorimetry (ITC).
Experiments were performed using the high-precision VP-ITC system (MicroCal; GE Healthcare). All molecules were dissolved in 20 mM Tris-HCl, pH 8, 150 mM NaCl, 5% ethanol. Binding enthalpies were measured by injecting M76 or M61 solution at 1.5 mM into a calorimetric cell containing 30 μM N-NTD. Titrations were performed at 18°C with 7-μl injections of M76 or M61 every 350 s. Heat signals were corrected for the dilution heats and normalized to the amount of compound injected. Binding stoichiometries, enthalpy values, and equilibrium dissociation constants were determined by fitting the corrected data to a standard 1:1 interaction model, allowing for a set of independent and equivalent binding sites, using the Origin7 software (OriginLab) provided by the manufacturer.
rHRSV-mCherry inhibition assay.
The inhibition of RSV replication was measured in cell cultures using an assay based on a human RSV reverse-genetics system in which the red fluorescent protein mCherry was inserted into the RSV genome, recombinant HRSV-Cherry (rHRSV-mCherry), as described previously (26). HEp-2 cells (ATCC number CCL-23) or BHK-21 cells maintained in Eagle's minimum essential medium (EMEM) or Dulbecco modified essential medium (DMEM), respectively, were seeded at 5 × 104 cells per well in 96-well plates the day before and infected for 2 h with 500 PFU of rHRSV-mCherry or 1 h with 50 PFU of vesicular stomatitis virus (VSV)-green fluorescent protein (GFP) (27). The medium was then replaced by medium without phenol red containing dilutions of M76-diAM (stock solution at 70 mM in dimethyl sulfoxide [DMSO]) from 333 μM to 1.37 μM. The experiment was repeated thrice, and each dilution point was made in triplicate. DMSO dilutions in the same range were used as controls. The plates were incubated for 48 h at 37°C, and mCherry fluorescence measurement was performed using a Tecan infinite M200Pro spectrofluorometer with excitation and emission wavelengths of 580 and 620 nm, respectively. Cell survival was quantified using the CellTiter-Glo Luminescent cell viability assay (Promega). Noninfected and nontreated infected HEp-2 cells were used for fluorescence and toxicity standardization and normalization, respectively. The 50% inhibitory concentration (IC50) and the 50% cytotoxic concentration (CC50) were determined by fitting the data to the dose-response curve implemented in Origin version 8.5 software.
Illustrations.
The figures were prepared using Pymol (http://pymol.sourceforge.net) with APBS (28) and PDB2PQR tools (29).
Accession numbers.
Structure factors and coordinates have been deposited in the Protein Data Bank under accession codes 4UC6 and 4UC7 for the Nat1 and Nat2 structures of the N-NTD; 4UC8, 4UC9, 4UCA, and 4UCB for the N-NTD in complex with P1, P2, P3, and P7 P-CTD peptides; and 4UCC, 4UCD, and 4UCE for the N-NTD in complex with M76, M81, and M72 compounds, respectively. Backbone chemical shift assignments for the N-NTD have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under accession number 25705.
RESULTS
A reduced RSV N model for interaction studies.
We recently showed that the N-NTD, the minimal domain of interaction with the P-CTD, is monomeric and mainly alpha-helical (15), consistent with the crystal structure of the N-RNA rings determined previously (16). Two N-NTD crystal forms diffracted to 2.1- and 2.4-Å X-ray resolution, respectively, with two molecules in the asymmetric unit of each of them (Table 1). The structure of the isolated globular domain is essentially identical to that observed within the RNP complex (3, 16) (Fig. 1). Variations are restricted to surface loops, notably the αN7-αN8 loop containing R234, involved in key interactions for lateral N-N contacts in the RNP (3), and the αN3-αN4 loop that closes the RNA groove (16). Other structural variations are observed in protruding regions, the β-hairpin, and the αI2-η1 loop (Fig. 1), which also display high flexibility in the RNP model, as revealed by the high temperature factors of the atoms in this region. Overall, these data show that N polymerization and RNA binding required for RNP assembly do not involve important rearrangements of the N-NTD, other than adaptation of specific loops, indicating that the isolated N-NTD is a good and sufficient model for studying the interactions between the P-CTD and RNP.
FIG 1.
X-ray structures of N-NTD and P-CTD binding sites. (A) Superposition of isolated N-NTDs in a free form (pale and bright orange, chains A and B for both crystal forms) or in complex with a P C-terminal peptide, P2 (light gray), and N-NTD in the RSV RNP context (black; PDB 2WJ8) in cartoon representation. Secondary structures are labeled according to reference 16. The P C-terminal peptide P2 is shown in stick representation colored by atom type, with carbons in white. (B to E) Final 2Fo-Fc electron density map for P1 (B), P2 (C), M76 (D), and M72 (E) at 2.0-, 2.4-, 2.1-, and 2.9-Å X-ray resolutions, respectively. The maps are drawn in blue mesh contoured at 1.0 sigma, with the ligand displayed as sticks.
Structure of N-NTD:P-CTD complexes.
We also found previously that the nine C-terminal residues of P were necessary and sufficient for the interaction with the N-NTD (14) and that the C-terminal F241 was essential for viral RNA synthesis in vitro (15). Here, we used peptides containing up to 13 residues, starting from the C-terminal P-F241, for cocrystallization. We obtained crystals with P7, P3, P2 (Asp-Phe dipeptide), and P1 (phenylalanine) (Table 1). P2 and P1 crystals diffracted to 2.2- and 1.9-Å X-ray resolution, respectively. Both P2 and P1 had clear electron density in the corresponding crystal (Fig. 1B and C). P7 and P3 crystals diffracted to 2.8- and 3.2-Å X-ray resolution, respectively, but the electron density was clear only for F241, with all other residues being disordered (data not shown). Binding of P peptides did not involve structural rearrangement of the N-NTD (Fig. 1).
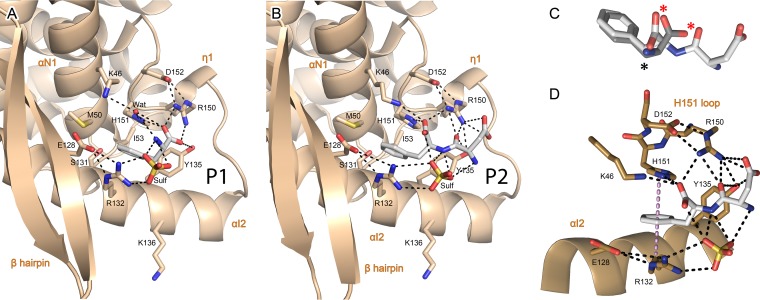
In all complexes, P-F241 is deeply buried in a hydrophobic pocket of the N-NTD located between helices αI2 (residues S131, R132, and Y135) and αN1 (residues M50 and I53) and the H151 loop (Fig. 2). These three regions perfectly correlate with the residues previously found to be critical for RSV polymerase activity in vitro (15). P-F241 is engaged in a dense interaction network, suggesting that it is key for recognition of the N-NTD by the P-CTD. The aromatic moiety of P-F241 fits tightly into the pocket (Fig. 2A and B), and its position is the same in both P1 and P2 complexes (Fig. 2C). One side of the aromatic ring makes a π-π stacking interaction with the imidazole ring of H151. The H151 loop itself is locked by a salt bridge between R150 and D152. The other side of the ring stacks against the aliphatic part of the R132 side chain, while the R132 guanidinium forms a salt bridge with E128, one turn upstream on αI2. This double stacking results in clamping together helix αI2 and the H151 loop (Fig. 2D). In the P2 complex, a network of salt bridges and hydrogen bonds involving K46, R150, H151, and Y135 stabilizes the negative charge of the C-terminal carboxylate, the charged side chain, and the backbone carbonyl of P-D240 (Fig. 2B), even though P-D240 displays some conformational variability (data not shown). In the P1 complex, the negative charge of the C-terminal carboxylate is stabilized by R150, H151, and Y135 (Fig. 2A). Finally, a sulfate molecule from the crystallization solution makes a bridge between the N terminus of P2, the P-F241 amide group, and the R132 side chain.
FIG 2.
Details of the P-CTD binding pocket and interaction network with the N-NTD. (A and B) Enlarged views of the binding of phenylalanine (P1) (A) and Asp-Phe (P2) (B), corresponding to the P C terminus. The N-NTD is in tan and shown in cartoon representation, with secondary structures labeled. Residues involved in binding through electrostatic (dashed lines) or van der Waals interactions are shown in stick representation and colored by atom type, with carbons in tan. P1 and P2 are in stick representation, with the same color scheme as in Fig. 1. (C) Superposition of P2 and P1 conformations. Atoms with conserved polar contacts are indicated by asterisks, notably, two oxygen atoms (red asterisks). (D) Details of the double stacking interaction between the aromatic ring of P2 and the side chains of H151 and R132, with electrostatic interactions between P2 and the N-NTD indicated by black dashed lines. The normal axis to the P-F24 ring plane is indicated by the violet dotted line.
Analysis of P peptide binding to the N-NTD by NMR.
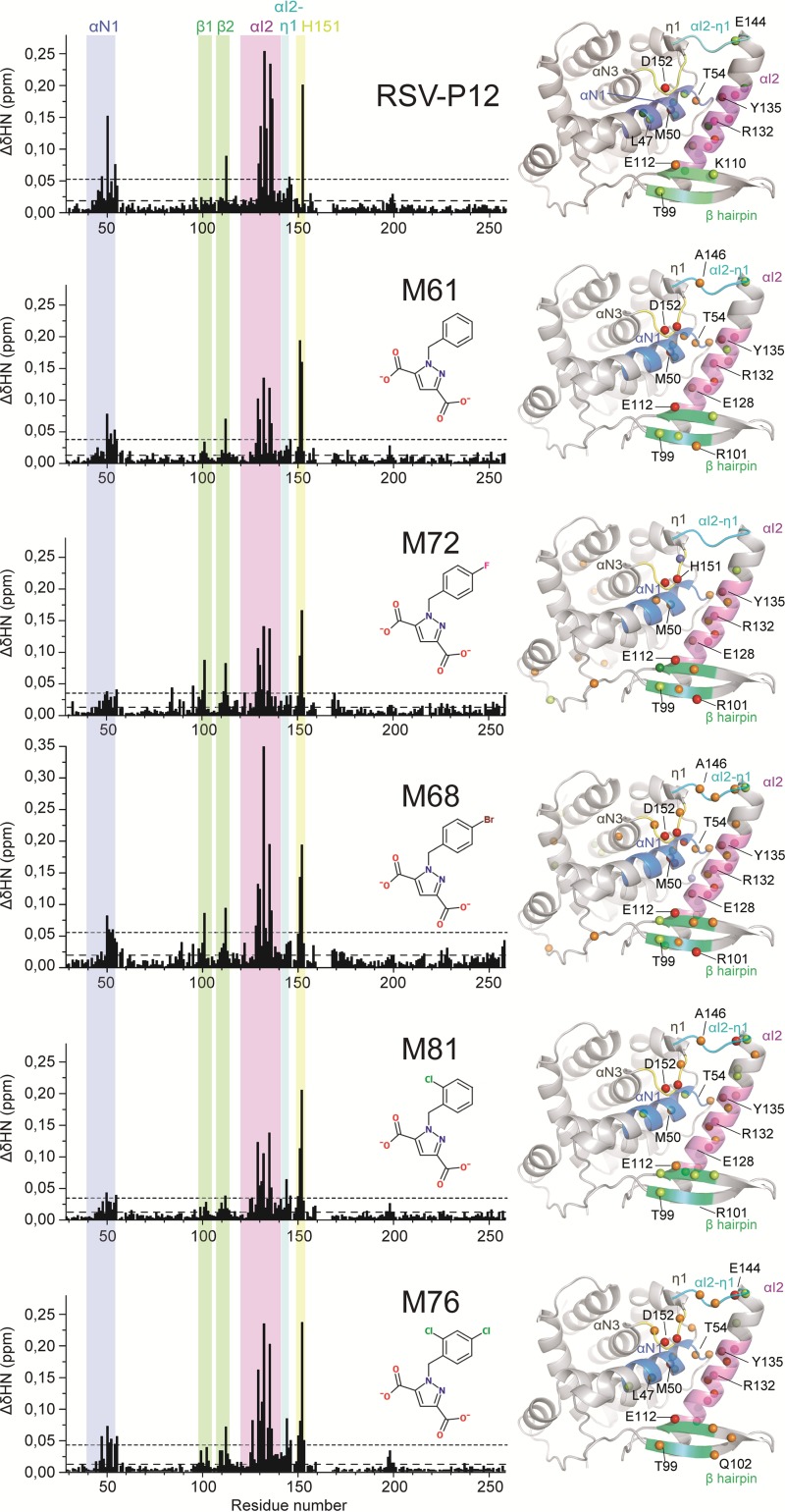
P-CTD binding to the N-NTD in solution was assessed by NMR by using P12, a P-CTD peptide containing the last 12 residues of P. CSPs in 1H-15N HSQC spectra were observed when titrating 15N-labeled N-NTD with P12 (Fig. 3A and B). The C terminus of helix αN1, the center of αI2, and the H151 loop display the largest CSPs, which delineate a common contiguous region (Fig. 4) that matches well with the P-CTD binding site described above (Fig. 2). The αI2-η1 loop and the β-hairpin, two regions proximal to the binding pocket, display smaller CSPs. Since chemical shifts are sensitive both to direct binding and to local geometry changes, these CSPs could be induced by small structural rearrangements mediated via intramolecular contacts with the P-F241 binding site. However, the two regions could also belong to an extended contact surface scanned by the peptide at the entry of the pocket and involving P residues upstream of D240. Assuming an extended conformation, the distance between the hydrophobic pocket and these regions suggests that at least D239 and L238 define this secondary binding site, in agreement with previous data showing that mutation of the 2 residues had an impact on the level of N binding and RNA synthesis (14, 15).
FIG 3.
NMR interaction experiments using 1H-15N HSQC spectra of the N-NTD. (A) 1H-15N HSQC spectrum at 800 MHz 1H frequency of 50 μM 15N-labeled N-terminal domain of RSV nucleoprotein at 293 K. Peak assignments are indicated according to the numbering in the native sequence. Unassigned amide or side chain peaks are indicated by asterisks. The inset (top left) shows detailed assignment of the central region of the spectrum (boxed area). Asn and Gln NH2 side chain resonances are connected by horizontal lines. (B) Enlarged views of superimposed 1H-15N HSQC spectra obtained by titrating either RSV-P12 peptide (top) or M76 (bottom) into 15N N-NTD. Chemical shift perturbations of large amplitude are indicated by arrows. Molar ligand/protein ratios and color codes are indicated on the right of each spectrum.
FIG 4.
(Left) Mapping of chemical shift perturbations induced by P peptide and BPdC ligands. CSP profiles were extracted from 1H-15N HSQC spectra of 15N-labeled N-NTD, with the P12 peptide and BPdC ligands at the titration midpoint, where CSPs were half of those at saturation. The bar graphs represent combined CSPs containing both 1H and 15N contributions (Δδ1H15N). Mean values (long dashes) and means plus 1 standard deviation (short dashes) are plotted for each ligand. (Right) CSPs were mapped onto the 3D structure of the N-NTD. Amide nitrogen atoms of residues with large CSPs are represented as spheres, with a color code reflecting the contributions of 1H and 15N chemical shifts: gray, Δδ1H > 0.06 ppm; dark green, Δδ15N > 0.3 ppm; light green, Δδ15N > 0.2 ppm; red; Δδ1H15N > 0.09 ppm; and orange, Δδ1H15N > 0.06 ppm. The regions where most CSPs are observed are labeled on the diagrams and on the structure: the C terminus of helix αN1 in blue, the center of helix αI2 in magenta, the αI2-η1 loop in cyan, the H151-loop in yellow, and the β-hairpin in green.
The linear evolution of N-NTD chemical shifts during titration (Fig. 3B) indicates a fast chemical exchange between the free and bound forms of P12. Accordingly, we determined a mean dissociation constant of 54 ± 9 μM. This weak affinity is consistent with recent work by Shapiro et al. (30). Whether P-F241 and P-D240 are sufficient for binding was tested with the P2 peptide, also used for X-ray crystallography, as described above. It also bound to the pocket, but with an affinity of 4.2 ± 1.0 mM, confirming that residues upstream of F241 and D240 contribute to binding interactions.
In silico screening of small compounds targeting the P-binding pocket of the N-NTD.
Based on the structure of the N-NTD:P2 complex, we screened the ZINC database (23) for compounds with an aromatic ring, which could mimic P-F241 and bind to the hydrophobic pocket, a second functional group mimicking the C-terminal carboxylate of P-F241, and a third one mimicking the bridging sulfate ion to build electrostatic interactions with R150 and R132. Virtual screening with AutoDock generated a set of 300 compounds, restricted to an upper molecular mass of 350 Da. These compounds were ranked based on the interaction energy between the ligand and N. Among the top 50 compounds, based on initial screens by NMR and crystallography, we selected five compounds that form a chemical series with a common BPdC scaffold and a variable pattern of ortho and para halogen substitutions on the benzene ring (Table 2), for further experimental studies.
TABLE 2.
BPdC biochemical profiles

a Kd errors were estimated from standard deviations of Kd values determined for individual residues at 1H or 15N resonance frequencies.
Binding properties of BPdC compounds.
We investigated the binding potentials of the five BPdC compounds selected by in silico screening by NMR. BPdCs induced CSPs and affected the same regions as the P12 peptide, indicating that they target the P-binding site (Fig. 3B and 4). Chemical exchange between free and bound species was also fast, and dissociation constants between 20 and 680 μM were determined. The compounds were ranked by affinity as follows: M72 < M61 < M68 < M81 < M76 (Table 2). Since the five BPdCs differ only by substitutions on the benzyl moiety, the difference of more than 1 order of magnitude between M72 and M76 indicates that halogens on the benzene ring can significantly modulate BPdC binding to the N-NTD. In particular, the ortho Cl atoms in M81 and M76, as well as the para Cl in M76, seem to induce tighter binding.
In parallel, we investigated the N-NTD binding of the most substituted M76 and unsubstituted M61 by ITC. Titration experiments yielded a stoichiometry of 0.99 in both cases and Kds of 48 and 510 μM for M76 and M61, respectively (Table 2), consistent with the NMR data. Thermodynamic parameters derived from the ITC isotherms show that binding of both M76 and M61 to the N-NTD is driven by favorable enthalpy (ΔH = −11.0 ± 3.6 and −6.42 ± 2.42 kcal · mol−1 for M76 and M61, respectively), which could be associated with favorable electrostatic and van der Waals interactions. This is partly counterbalanced by unfavorable entropic contributions (−TΔS = 5.4 ± 3.8 and 2.2 ± 1.8 kcal · mol−1 for M76 and M61, respectively), which may reflect the high flexibility of the compounds, resulting in an entropic penalty.
Structure of N-NTD:BPdC complexes.
We obtained N-NTD crystals in complex with the compounds with the highest and lowest affinities, M76, M81, and M72, which diffracted to 2.0-, 2.7-, and 2.9-Å X-ray resolutions, respectively (Table 1). The compounds were clearly visible in electron density maps (Fig. 1D and E). Similarly to P-CTD peptides, BPdC binding did not induce any large structural rearrangement of the N-NTD. The structures confirm that the benzyl moiety of BPdCs fits into the hydrophobic pocket of the N-NTD (Fig. 5A and B). Overall BPdCs interact with the same residues of the N-NTD as P peptides, but the three X-ray structures reveal subtle side chain rearrangements that stabilize N-NTD complexes under certain conditions.
FIG 5.
X-ray structures of N-NTD:BPdC complexes. (A and B) Enlarged view of the binding of M76 (A) and M72 (B) in the N-NTD pocket. The N-NTD is displayed as in Fig. 1, with M76 and M72 in stick representation colored by atom type, with magenta and yellow carbons, respectively. Stabilizing electrostatic interactions are displayed as black dashed lines. The insets show superposition of M76 and P2, and a black star marks the most deeply buried atom of both ligands in the binding pocket (A) and superposition of M76 and M72 interacting with S131 through a halogen bond (yellow dashed line) and an H bond (black dashed line) (B). (A, C, and D) The green arrow indicates the water molecule in head-on interaction with the para Cl. (C) Details of the double stacking interaction involving the H151 imidazole, the benzene ring of M76, and the R132 side chain. H bonds and halogen bonds are plotted as in panels A and B. The normal axis to the benzene ring plane of M76 running through H151 NE2 and R132 NE is shown as a violet dotted line. (D) Electrostatic interactions of the two halogen substituents in M76, with H bonds and halogen bonds displayed as in panels A and B. The final 2Fo-Fc (blue) and Fo-Fc (red) electron density maps are displayed around M76 at 2σ and 3σ, respectively. Additional intraprotein H bonds stabilizing N-NTD secondary-structure elements and involved in M76 binding or involving direct contacts between solvent molecules and M76 are displayed in light blue.
The benzyl moiety of M76/M81 inside the pocket is slightly shifted compared to P-F241: the para Cl atom takes the place of the most deeply buried benzyl C atom of P-F241 in P2 (Fig. 5A, inset). This results in a double π-π stacking interaction with H151 and R132, respectively (Fig. 5C). It is reminiscent of that in the P complexes, but the positions of the R132 guanidinium and H151 indole groups relative to the normal axis of the benzyl plane (Fig. 2D and 5C) provide a more favorable interaction than in the P complexes, as described previously (31, 32).
Distance and angle requirements for Cl atoms in M76 and M81 to be halogen bond acceptors are fulfilled (33) (Table 3). The carbonyl group of S131 makes a halogen bond with the ortho Cl in M76/M81 at the bottom of the binding pocket (Fig. 5C and D and Table 3). This interaction is completed by two H bonds between the same ortho Cl atom and the S131 hydroxyl and R132 amide groups (Fig. 5C and D and Table 3). The second para Cl atom in M76 makes two halogen bonds, one with the E128 carboxylate and another with a water molecule (Fig. 5C and D and Table 3). The last atoms also establish a complex network of H bonds, ultimately connecting the β-hairpin via E112 main-chain atoms (Fig. 5D). Moreover, synergy between ortho and para Cl atoms takes place for M76, reflected in the higher affinity of M76 than of M81. The final 2Fo-Fc electron density map shows a strong peak at 2σ around the ortho Cl but no density around the para Cl (Fig. 5D), whereas a positive residual density is observed at 3σ in the Fo-Fc map on the para Cl substituent.
TABLE 3.
Polar contacts of halogen atomsa

Relevant polar contacts within 4 Å of halogen atoms in M76, M81, and M72 complexes. The bond geometry was defined following Auffinger et al. (33).
b Values are provided for the two independent N-NTD domains present in each crystal asymmetric unit (chain A/chain B).
c M72 has the lowest affinity for N-NTD, and it was observed only in the chain A binding site.
The F atom in M72 also forms a halogen bond with the S131 carbonyl (Table 3). It induces an alternative conformation of the M72 benzyl with a para F in place of the ortho Cl in M76/M81 and, in turn, a 90° rotation of the pyrazole moiety (Fig. 5B). In the case of M76/M81, the pyrazole makes a π-π stacking interaction with Y135 (Fig. 5A), which is not the case for M72 (Fig. 5B). This strongly impacts the formation of salt bridges by the BPdC carboxylate groups on the pyrazole cycle. One carboxylate of M76/M81 (Fig. 5C) occupies the position of the sulfate present in the P complexes (Fig. 2D), which is left unoccupied by M72 (Fig. 5B). The second M76/M81 carboxylate forms a salt bridge with R150, thereby bridging the ligand over the hydrophobic pocket. In the case of M72, both carboxylates interact with R150, and the interaction with helix αI2 is lost.
In summary, an optimal charge and shape complementarity is achieved for M76 with a para and ortho Cl combination, compared to M72 (Fig. 6), accounting for the higher affinity of M76 for the N-NTD. In addition, examination of the bottom of the binding pocket in apo and complexed forms reveals the presence of an internal cavity in the N-NTD large enough to accommodate several water molecules and flanked by conserved residues (Fig. 6). In all BPdC complexes, this highly organized water network is linked to the ligand binding site via E112, E128, and sometimes R132 and may contribute to ligand stabilization.
FIG 6.

N-NTD surface complementarity for P peptide and BPdC ligands. Shown are enlarged views of the binding of P2 (A), M76 (B), and M72 (C). The N-NTD van der Waals surface is represented according to the electrostatic potential, with positive (blue) and negative (red) potentials shown. Ligands are displayed as van der Waals spheres and colored by atom type, with white carbons.
Inhibition of P-CTD binding to the N-NTD by BPdCs.
Since BPdC compounds bind to the same site as the P-CTD in the N-NTD, we investigated the inhibition of P-CTD binding to the N-NTD by BPdCs using surface plasmon resonance. Since SPR signals are proportional to the molecular weight of the analytes, we resorted to indirect competition due to the small size of BPdCs (Fig. 7). The PΔ161N peptide (residues 161 to 241) was immobilized via a GST tag, and serial dilutions of the N-NTD were injected. The interaction between the two domains was transient, with a high dissociation rate (koff > 1 s−1) and a Kd of 30 μM. Subsequent competition experiments with all five BPdC compounds showed that their inhibitory potentials followed the same trend as affinity: M72 < M61 < M68 < M81 < M76 (Table 1). M76 and M81 best inhibited the N-NTD:PΔ161N interaction, with inhibition constants, Ki, of 155 and 247 μM, respectively, underlining the role of ortho and para Cl substituents in N-NTD binding. Unsubstituted M61 and compounds with an alternative para F (M72) or Br (M68) appeared to be less potent, with Ki values of 893, 1,660, and 610 μM, respectively, illustrating the influence of the nature of the halogen on BPdC binding to the N-NTD.
FIG 7.
BPdC competition with P-CTD for N-NTD binding by SPR. (Top) Real-time association and dissociation profiles corresponding to the injection over immobilized P-CTD of the N-NTD premixed with different concentrations of M61 (left) or M76 (right). (Bottom) BPdC concentration dependence of the steady-state SPR response upon N-NTD injection over a P-CTD surface relative to the response in the absence of BPdC. The lines represent the best fit of the experimental data to a single class of binding site model.
Antiviral activities of BPdCs.
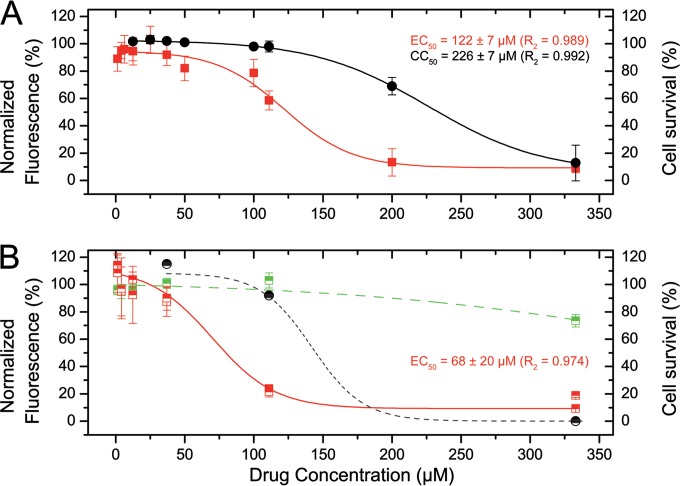
To assess the relevance of the results obtained in vitro with BPdCs, we used a recombinant virus, rHRSV-mCherry (26), to monitor RSV multiplication by following the expression of the red fluorescent mCherry protein in cell cultures. Preliminary experiments with the highest-affinity compound, M76, did not show significant inhibitory activity. Since the two negatively charged carboxylates in M76 might hinder membrane passage, we designed the pH-sensitive prodrug M76-diAM by attaching acylal groups on both carboxylates of M76. This electrically neutral molecule can be internalized by the cell, and the carboxylates are regenerated by hydrolysis in the cytosol (34). The replication rate of rHRSV-mCherry in Hep-2 cells in the presence of M76-diAM clearly decreased in a dose-dependent manner (Fig. 8A), with an IC50 of 122 ± 7 μM. The toxicity was characterized with a rather high CC50 of 226 ± 7 μM, which could be expected, since hydrolysis of acetoxymethyl esters releases formaldehyde. Alternative pH-sensitive protective groups that do not produce toxic metabolites should address this issue.
FIG 8.
Inhibition of rHRSV-mCherry replication by M76-diAM. (A) HEp-2 cells in 96-well plates were infected with 500 PFU of rHRSV-mCherry in the presence of serial dilutions of M76-diAM. The red fluorescence (red symbols) and the luminescence reporting on cell survival (black symbols) were read at 48 h postinfection by automatic counting and normalized based on the fluorescence and luminescence of nontreated infected cells. Each data point represents the mean of the results of three experiments done in triplicate. The standard errors of the mean (SEM) are represented by the error bars. The lines correspond to the data fitted to a dose-response curve. (B) BHK-21 cells in 96-well plates were infected with 500 PFU of rHRSV-mCherry or 50 PFU VSV-GFP in the presence of serial dilutions of M76-diAM. Red (red symbols) and green (green symbols) fluorescence, as well as cell survival (black symbols), were read at 48 h postinfection by automatic counting and normalized based on the fluorescence and luminescence of nontreated infected cells. Each data point represents the mean (±SEM) of two experiments done in triplicate. rHRSV-mCherry fluorescence was fitted to a dose-response curve (solid line). The approximate fit of VSV-GFP fluorescence and cytotoxicity to dose response curves are plotted with green and black dashed lines, respectively.
To test the specificity of M76-diAM for RSV, BHK-21 cells were infected with either rHRSV-mCherry or VSV (a rhabdovirus belonging to the same order, Mononegavirales) expressing GFP and treated as previously with M76-diAM. With 111 μM M76-diAM, RSV replication was reduced by 80%, while no effect was observed for VSV (Fig. 8B).
DISCUSSION
Specificity of RNP:P recognition in RSV.
Transcription and replication in the Mononegavirales is carried out by a unique RNA-dependent RNA polymerase complex. Although a common scheme for RNA synthesis is recognized, in which the catalytic unit L is delivered to the RNP by the viral phosphoprotein, it appears that the precise molecular mechanisms have largely diverged. Our data add a new piece of evidence to the diversity of recognition mechanisms of the RNP by P among the Mononegavirales. Since N proteins display an overall conserved fold consisting of two domains closing similar RNA binding grooves and flexible arms that lock lateral N-N interactions in the RNP (35–39), the variety found in RNP:P complexes essentially stems from the different structures of phosphoproteins recognizing alternative regions of their specific N partners. In the case of VSV, representative of the family Rhabdoviridae, the RNP:P complex is formed between a well-defined α-helical P-CTD and N-CTD (40). In the Paramyxovirinae, a 20-residue region of the large disordered C-terminal tail of N folds into a helix upon binding to a three-helix bundle at the C terminus of P, denoted P-XD, as exemplified by measles (41) and Hendra (42) viruses.
RSV, belonging to the subfamily Pneumovirinae and to the same family, Paramyxoviridae, as the Paramyxovininae, has developed an alternative strategy for RNP:P recognition. We have shown previously that the RSV P-CTD, but not the P-NTD, binds to nucleocapsid-like particles (NLPs) (14) and that the P-CTD binding region is located in the N-NTD (15), indicating that the RSV N-NTD:P-CTD interaction is relevant for the RSV RNP:P complex. Here, we show how the very last C-terminal residue of RSV P, F241, participates in recognition of N, and more particularly of the RNP, by inserting into a hydrophobic binding pocket with a predefined fold in the N-NTD. Only this residue of P is fully immobilized in N-NTD complexes with P peptides, and mutation of this residue to alanine completely abrogates RSV minigenome activity (15). Replacement by another aromatic amino acid could possibly maintain the function of P, as suggested by the ability of the P-F241W mutant to bind to N (14). In contrast, the penultimate P-D240 is less ordered than P-F241 in the N-NTD complex (data not shown), which can be correlated with the previous observation that the P-D240A mutation did not significantly affect minigenome activity (15).
Roles of residues upstream of F241 in the N-NTD:P-CTD interaction.
While the N/P interaction relies heavily on P-F241, critical for viral RNA synthesis (15), the residue is not sufficient for binding the P-CTD to the N-NTD. The affinity of the P2 Asp-Phe peptide is lower than that of P12 or the P-CTD by 2 orders of magnitude at moderate ionic strength. Similar observations were made by Shapiro et al. at low ionic strength (30). Residues upstream of P-FD240 thus play a role in the P-CTD:N-NTD interaction. The P-CTD is predicted to be intrinsically disordered, and it was shown that such regions may remain disordered upon complex formation (43). This seems to be the case for the N-NTD:P-CTD complex, as shown by the N-NTD:P7 structure, where only P7-F241 is well defined. Multiple transient contacts between the P-CTD and N-NTD would result in an extended interaction surface and could explain why long-range chemical shift perturbations of the N-NTD can be observed on opposite sides of the P-F241 binding pocket (Fig. 4). The driving force for formation of a fuzzy complex outside this pocket is likely provided by a network of electrostatic interactions, which were evidenced in the X-ray structures of P1 and P2 complexes and which could be reinforced by phosphorylation of this region (30). It extends at least to P-L238, as suggested by the observation that the polymerase activity of P-E239A/D240A and P-L238A mutants is reduced by nearly 50% compared to wild-type (WT) P (15).
Still, the N-NTD:P-CTD interaction is rather weak. This is nevertheless compatible with the function of P, which must open the RNP structure for the L protein to access the viral RNA (44). A loose association between P and the RNP allows the polymerase to progress along the RNP. However, based on the present work, the possibility that neighboring N protomers in the RNP context may participate in the binding cannot be excluded (Fig. 9), as reported for VSV (40). This contribution would not be seen with our reduced model using only the isolated N-NTD. The possibility that the tetrameric form of P may induce synergy for binding to the RNP also cannot be excluded, but due to the length and flexibility of the P-CTD, it is tempting to assume that the binding events on two different N protomers are independent and that multiple P anchors merely prevent dissociation of P from the RNP.
FIG 9.
Evolutionary conservation of the N-NTD binding pocket in the RNP context. (Top) Surface representation of N-NTD colored according to the evolutionary conservation of amino acids, calculated using the ConSurf server (45), with turquoise-to-magenta indicating variable-to-conserved. The result is displayed as the crystallographic model of the RSV RNP (PDB 2WJ8 [16]) restricted to the N-NTD, without terminal arms or the N-CTD. The two neighboring N-NTD protomers in the RNP are displayed in cartoon (tan). The RNA molecule and M76 are displayed in stick form and colored by atom type, with white and magenta carbons, respectively. (Bottom) Location of the P-CTD binding site on the model of the authentic left-handed helical RNP (PDB 4BKK [3]). The N-NTD (tan), N-CTD (yellow), and terminal arms (gray) are shown. The RNA molecule and M76 are shown as white sticks and ball and stick colored as in panel A, respectively. In the N-NTD, residues corresponding to N mutants with a minigenome replication activity of <33%, <66%, or >133% that of the wild type are colored red, orange, and green, respectively (15). On the right is an enlarged view of the N-NTD.
The P-CTD binding site in the N-NTD is a druggable pocket.
In the context of the RNP, conserved areas correspond to N-N lateral contacts, the RNA binding site, and the P-CTD binding site, respectively. The P-binding site is located on the ridge formed by the bulky N-NTD domain projecting away from the RNP helix (Fig. 9). It is easily accessible between two N protomers and two consecutive turns of the RNP. This region of N was rated as the most divergent in the three-dimensional structures of N in the Mononegavirales but is conserved within the Pneumovirinae (16) and thus provides an ideal target for antiviral drugs.
Here, we present a chemical series with a common BPdC scaffold, identified on the basis of structural determinants derived from the N-NTD:P2 complex. We show that the P-binding pocket on the N-NTD can indeed be targeted by small compounds. BPdCs are able to replace P-F241 in the hydrophobic pocket and to establish similar electrostatic interactions at the entrance of the pocket. Furthermore, they display specific interactions driven by halogen atoms on the benzyl moiety, with no equivalents in P, which allow them to improve their N-NTD binding properties and inhibitory potential for the P-CTD. While the cavity in the N-NTD appears to be rather rigid, its entrance and more remote regions, like the αI2-η1 loop and the β-hairpin, can be partly remodeled by the BPdC ligands (Fig. 7). This is an advantage for drug development and reinforces the hypothesis of a binding model with a dual mode.
Negatively charged M76 does not spontaneously cross the cell membrane, and to gain an antiviral response with M76, a prodrug strategy was necessary. M76-diAM displayed anti-RSV activity (Fig. 8A), and the control with VSV (Fig. 8B) showed that it specifically targets the RSV RNP:P recognition mechanism. However, it afforded only a very narrow therapeutic window, showing that M76, together with the prodrug strategy, needs to be improved. Taken together, the data presented here provide a preliminary assessment for the design of potent RSV antivirals based on the N-NTD:P-CTD interaction.
ACKNOWLEDGMENTS
This work was supported by the Agence Nationale de la Recherche (grant ANR 11 BSV8 024 02 to S.D., F.A.R., J.-F.E., and C.S.) and the Grand Equipement National de Calcul Intensif (grant 2012-076378 to A.S.-S. and B.T. for access to IDRIS HPC resources). Doctoral fellowships from Université Pierre et Marie Curie (Ecole Doctorale Iviv) to M.O.-O., Institut de Chimie des Substances Naturelles to N.P., and Région Ile-de-France DIM Malinf to S.L. are acknowledged. We thank the staffs at beam lines PROXIMA-1 at the Soleil synchrotron (St Aubin, France), ID14-4 at the European Synchrotron Radiation Facility (Grenoble, France), and PX06SA at the SLS synchrotron (Villigen, Switzerland); Ahmed Haouz and Patrick Weber (Protein Crystallization Platform, Institut Pasteur Paris); Origène Nyanguile and Vanessa Gaillard (HES-SO Valais-Wallis, Sion, Switzerland) for providing the P12 peptide; and Jacques Perrault (San Diego, CA) and Christophe Chevalier (Jouy-en-Josas, France) for providing and amplifying VSV-GFP, respectively.
C.S., J.-F.E., M.G., P.E., and S.D. designed experiments. A.S.-S. and B.T. carried out in silico screening. J.F., M.G., M.O.-O., N.P., and S.L. purified proteins. M.O.-O. and S.D. collected X-ray data and calculated X-ray structures. B.B., M.O.-O., P.E., and S.H. performed and analyzed ITC and SPR experiments. C.S., N.P., and S.L. performed and analyzed NMR experiments. J.-F.E. and M.G. performed and analyzed viral replication inhibition assays. D.D. and P.C. synthesized M76-diAM. F.B, F.A.R., and M.A.T. participated in early stages of the project and provided critical readings of the manuscript. A.S.-S., C.S., and S.D. wrote the manuscript.
REFERENCES
- 1.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olszewska W, Openshaw P. 2009. Emerging drugs for respiratory syncytial virus infection. Expert Opin Emerg Drugs 14:207–217. doi: 10.1517/14728210902946399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakker SE, Duquerroy S, Galloux M, Loney C, Conner E, Eleouet JF, Rey FA, Bhella D. 2013. The respiratory syncytial virus nucleoprotein-RNA complex forms a left-handed helical nucleocapsid. J Gen Virol 94:1734–1738. doi: 10.1099/vir.0.053025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins PL, Hill MG, Camargo E, Grosfeld H, Chanock RM, Murphy BR. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci U S A 92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Q, Hardy RW, Wertz GW. 1995. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J Virol 69:2412–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khattar SK, Yunus AS, Samal SK. 2001. Mapping the domains on the phosphoprotein of bovine respiratory syncytial virus required for N-P and P-L interactions using a minigenome system. J Gen Virol 82:775–779. [DOI] [PubMed] [Google Scholar]
- 7.Garcia J, Garcia-Barreno B, Vivo A, Melero JA. 1993. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195:243–247. doi: 10.1006/viro.1993.1366. [DOI] [PubMed] [Google Scholar]
- 8.Mason SW, Aberg E, Lawetz C, DeLong R, Whitehead P, Liuzzi M. 2003. Interaction between human respiratory syncytial virus (RSV) M2-1 and P proteins is required for reconstitution of M2-1-dependent RSV minigenome activity. J Virol 77:10670–10676. doi: 10.1128/JVI.77.19.10670-10676.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveira AP, Simabuco FM, Tamura RE, Guerrero MC, Ribeiro PG, Libermann TA, Zerbini LF, Ventura AM. 2013. Human respiratory syncytial virus N, P and M protein interactions in HEK-293T cells. Virus Res 177:108–112. doi: 10.1016/j.virusres.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Kolakofsky D, Le Mercier P, Iseni F, Garcin D. 2004. Viral DNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology 318:463–473. doi: 10.1016/j.virol.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 11.Castagne N, Barbier A, Bernard J, Rezaei H, Huet JC, Henry C, Da Costa B, Eleouet JF. 2004. Biochemical characterization of the respiratory syncytial virus P-P and P-N protein complexes and localization of the P protein oligomerization domain. J Gen Virol 85:1643–1653. doi: 10.1099/vir.0.79830-0. [DOI] [PubMed] [Google Scholar]
- 12.Llorente MT, Garcia-Barreno B, Calero M, Camafeita E, Lopez JA, Longhi S, Ferron F, Varela PF, Melero JA. 2006. Structural analysis of the human respiratory syncytial virus phosphoprotein: characterization of an alpha-helical domain involved in oligomerization. J Gen Virol 87:159–169. doi: 10.1099/vir.0.81430-0. [DOI] [PubMed] [Google Scholar]
- 13.Llorente MT, Taylor IA, Lopez-Vinas E, Gomez-Puertas P, Calder LJ, Garcia-Barreno B, Melero JA. 2008. Structural properties of the human respiratory syncytial virus P protein: evidence for an elongated homotetrameric molecule that is the smallest orthologue within the family of paramyxovirus polymerase cofactors. Proteins 72:946–958. doi: 10.1002/prot.21988. [DOI] [PubMed] [Google Scholar]
- 14.Tran TL, Castagne N, Bhella D, Varela PF, Bernard J, Chilmonczyk S, Berkenkamp S, Benhamo V, Grznarova K, Grosclaude J, Nespoulos C, Rey FA, Eleouet JF. 2007. The nine C-terminal amino acids of the respiratory syncytial virus protein P are necessary and sufficient for binding to ribonucleoprotein complexes in which six ribonucleotides are contacted per N protein protomer. J Gen Virol 88:196–206. doi: 10.1099/vir.0.82282-0. [DOI] [PubMed] [Google Scholar]
- 15.Galloux M, Tarus B, Blazevic I, Fix J, Duquerroy S, Eleouet JF. 2012. Characterization of a viral phosphoprotein binding site on the surface of the respiratory syncytial nucleoprotein. J Virol 86:8375–8387. doi: 10.1128/JVI.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagne N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, Eleouet JF, Rey FA. 2009. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326:1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 17.Kabsch W. 2010. Xds. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 19.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. 2004. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr 60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 22.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. 2012. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model 52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. [DOI] [PubMed] [Google Scholar]
- 25.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. 2005. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 26.Rameix-Welti MA, Le Goffic R, Herve PL, Sourimant J, Remot A, Riffault S, Yu Q, Galloux M, Gault E, Eleouet JF. 2014. Visualizing the replication of respiratory syncytial virus in cells and in living mice. Nat Commun 5:5104. doi: 10.1038/ncomms6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruedas JB, Perrault J. 2009. Insertion of enhanced green fluorescent protein in a hinge region of vesicular stomatitis virus L polymerase protein creates a temperature-sensitive virus that displays no virion-associated polymerase activity in vitro. J Virol 83:12241–12252. doi: 10.1128/JVI.01273-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res 32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapiro AB, Gao N, O'Connell N, Hu J, Thresher J, Gu RF, Overman R, Hardern IM, Sproat GG. 2014. Quantitative investigation of the affinity of human respiratory syncytial virus phosphoprotein C-terminus binding to nucleocapsid protein. Virol J 11:191. doi: 10.1186/s12985-014-0191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flocco MM, Mowbray SL. 1994. Planar stacking interactions of arginine and aromatic side-chains in proteins. J Mol Biol 235:709–717. doi: 10.1006/jmbi.1994.1022. [DOI] [PubMed] [Google Scholar]
- 32.Chakrabarti P, Bhattacharyya R. 2007. Geometry of nonbonded interactions involving planar groups in proteins. Prog Biophys Mol Biol 95:83–137. doi: 10.1016/j.pbiomolbio.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Auffinger P, Hays FA, Westhof E, Ho PS. 2004. Halogen bonds in biological molecules. Proc Natl Acad Sci U S A 101:16789–16794. doi: 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slayman CL, Moussatos VV, Webb WW. 1994. Endosomal accumulation of pH indicator dyes delivered as acetoxymethyl esters. J Exp Biol 196:419–438. [DOI] [PubMed] [Google Scholar]
- 35.Ruigrok RW, Crepin T, Kolakofsky D. 2011. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol 14:504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Yabukarski F, Lawrence P, Tarbouriech N, Bourhis JM, Delaforge E, Jensen MR, Ruigrok RW, Blackledge M, Volchkov V, Jamin M. 2014. Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat Struct Mol Biol 21:754–759. doi: 10.1038/nsmb.2868. [DOI] [PubMed] [Google Scholar]
- 37.Gutsche I, Desfosses A, Effantin G, Ling WL, Haupt M, Ruigrok RW, Sachse C, Schoehn G. 2015. Structural virology. Near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science 348:704–707. doi: 10.1126/science.aaa5137. [DOI] [PubMed] [Google Scholar]
- 38.Alayyoubi M, Leser GP, Kors CA, Lamb RA. 2015. Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein-RNA complex. Proc Natl Acad Sci U S A 112:E1792-1799. doi: 10.1073/pnas.1503941112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong S, Yang P, Li G, Liu B, Wang W, Liu X, Xia B, Yang C, Lou Z, Guo Y, Rao Z. 2015. Insight into the Ebola virus nucleocapsid assembly mechanism: crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution. Protein Cell 6:351–362. doi: 10.1007/s13238-015-0163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Green TJ, Luo M. 2009. Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc Natl Acad Sci U S A 106:11713–11718. doi: 10.1073/pnas.0903228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kingston RL, Hamel DJ, Gay LS, Dahlquist FW, Matthews BW. 2004. Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc Natl Acad Sci U S A 101:8301–8306. doi: 10.1073/pnas.0402690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Communie G, Habchi J, Yabukarski F, Blocquel D, Schneider R, Tarbouriech N, Papageorgiou N, Ruigrok RW, Jamin M, Jensen MR, Longhi S, Blackledge M. 2013. Atomic resolution description of the interaction between the nucleoprotein and phosphoprotein of Hendra virus. PLoS Pathog 9:e1003631. doi: 10.1371/journal.ppat.1003631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uversky VN. 2011. Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem Soc Rev 40:1623–1634. doi: 10.1039/C0CS00057D. [DOI] [PubMed] [Google Scholar]
- 44.Collins PL, Melero JA. 2011. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res 162:80–99. doi: 10.1016/j.virusres.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. 2010. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]