ABSTRACT
SSV-type integrases, encoded by fuselloviruses which infect the hyperthermophilic archaea of the Sulfolobales, are archaeal members of the tyrosine recombinase family. These integrases catalyze viral integration into and excision from a specific site on the host genome. In the present study, we have established an in vitro integration/excision assay for SSV2 integrase (IntSSV2). IntSSV2 alone was able to catalyze both integration and excision reactions in vitro. A 27-bp specific DNA sequence is minimally required for the activity of the enzyme, and its flanking sequences influence the efficiency of integration by the enzyme in a sequence-nonspecific manner. The enzyme forms a tetramer through interactions in the N-terminal part (residues 1 to 80), interacts nonspecifically with DNA and performs chemical catalysis in the C-terminal part (residues 165 to 328), and appears to recognize and bind the specific site of recombination in the middle portion (residues 81 to 164). It is worth noting that an N-terminally truncated mutant of IntSSV2 (residues 81 to 328), which corresponded to the putative product of the 3′-end sequence of the IntSSV2 gene of the integrated SSV2 genome, was unable to form tetramers but possessed all the catalytic properties of full-length IntSSV2 except for the slightly reduced recombination activity. Our results suggest that, unlike λ integrase, SSV-type integrases alone are capable of catalyzing viral DNA recombination with the host genome in a simple and reversible fashion.
IMPORTANCE Archaea are host to a variety of viruses. A number of archaeal viruses are able to integrate their genome into the host genome. Many known archaeal viral integrases belong to a unique type, or the SSV type, of tyrosine recombinases. SSV-type integrases catalyze viral integration into and excision from a specific site on the host genome. However, the molecular details of the recombination process have yet to be fully understood because of the lack of an established in vitro recombination assay system. Here we report an in vitro assay for integration and excision by SSV2 integrase, a member of the SSV-type integrases. We show that SSV2 integrase alone is able to catalyze both integration and excision and reveal how different parts of the target DNA and the enzyme serve their roles in these processes. Therefore, our results provide mechanistic insights into a simple recombination process catalyzed by an archaeal integrase.
INTRODUCTION
Tyrosine recombinases (i.e., members of the λ integrase family), named after the nucleophile tyrosine residue in the enzymes that forms a transient 3′-phosphotyrosine covalent linkage to the DNA substrate in the reaction intermediate, are widespread in all three domains of life (1, 2). They catalyze recombination reactions in many important biological processes in both prokaryotes and eukaryotes, such as integration and excision of a phage genome into and out of the host genome (λ Int and HP1 Int), maintenance of plasmid copy number (Flp), resolution of replicon dimers into monomers (Cre and XerC/D), transposition (Tn916/Tn1545 Int), activation or suppression of gene expression (FimB, FimE, XisA, and XisC), generation of genetic diversity (Rci), as well as mobilization of gene cassettes of integrons (IntI) (1–3).
Genetic, biochemical, and structural studies have provided a wealth of molecular details on tyrosine recombinases of both bacterial and eukaryal origins. Site-specific recombination catalyzed by a tyrosine recombinase generally proceeds as follows (1, 4–6). The enzyme binds to DNA at the specific site of recombination, forming a synaptic complex comprising tetrameric recombinase bound to two recombining DNA molecules. The active-site Tyr of the enzyme initiates a nucleophilic attack on the phosphodiester bond of the DNA backbone, catalyzing a variety of sequence-specific DNA rearrangements through two rounds of pairwise cleavage, exchange and rejoining of single strands in the context of the synaptic complex, and producing a Holliday junction intermediate. The synaptic complex is then dissolved, releasing recombination products from the recombinase. The simplest sites of recombination for tyrosine recombinases, such as Cre-loxP, Flp-frt, and XerC/D-dif sites, are core-type recombination sites of ∼30 bp, which contain a central region flanked with a pair of inverted repeats. The central region, where strand cleavage and exchange occur, is 6 to 8 bp long and known as the overlap region. The repeating sequences flanking the central region, known as core-binding sequences, are sites of binding by a pair of recombinase dimers or monomers. More complex sites of recombination contain, in addition to the above-mentioned sequences, hundreds of base pairs of accessory sequences, in which additional binding sites for recombinase as well as accessory proteins reside, as found in the λ Int and XerC/D systems (1, 4). The majority of tyrosine recombinases possess two structural domains, while λ Int has three domains (1, 3). The C-terminal catalytic domain of recombinases, which interacts with the core-binding sequences (1, 3, 4), resembles that of topoisomerase IB in structure and active-site residues, suggesting that these enzymes use similar catalytic mechanisms and are evolutionarily related (7). On the other hand, the N-terminal domain of tyrosine recombinases, conserved neither in structure nor in amino acid sequence, also interacts with the core-binding sequences of the recombination site (1, 3, 4).
Much less is known about tyrosine recombinases in Archaea. An integrase encoded by the fusellovirus SSV1, isolated from the hyperthermophilic crenarchaeon Sulfolobus shibatae, is the prototype of SSV-type integrases and the first archaeal member of the tyrosine recombinase family that has been biochemically characterized (2, 8–10). Integrases encoded by many known archaeal viruses and some archaeal plasmids are of the SSV type (11). SSV-type integrases catalyze site-specific integration of the viral DNA into a tRNA gene in the host chromosome by using viral and chromosomal attachment sites, i.e., attP and attA (also denoted attB), respectively, and the excision of the integrated provirus by using the prophage attachment sites attL and attR. The attP site is located in the integrase gene proximal to the 5′ end (9, 10, 12). Therefore, the SSV-type integrase gene is split upon viral integration into two fragments, termed int(N) and int(C), found at the boundaries of the integrated viral genome (9). This partition feature is unique to archaeal SSV-type integrases (10, 13). For SSV1 integrase (IntSSV1), all the attachment sites share a 44-bp invariant sequence. This 44-bp sequence comprises the downstream half of the tRNAArg gene, the site of integration, and flanks the provirus as direct repeats (14). Consequently, the integration of the SSV1 DNA into the host genome does not result in the disruption of the target tRNAArg gene. SSV1 lacking the IntSSV1 gene is capable of completing the infection cycle but is unable to integrate into the host genome (15). It was reported previously that IntSSV1 catalyzed both integration and excision reactions in vitro, and no additional accessory proteins were apparently required for the excision reaction compared to those for the integration reaction (8, 16). Furthermore, the 44-bp invariant sequence was found to be sufficient to support recombination, although the efficiency of recombination was affected by the flanking sequences (8). An 18-bp sequence within the att site was identified as the binding site for IntSSV1 (16). For unknown reasons, however, the in vitro recombination assays for IntSSV1 have not been successfully established in other laboratories (17, 18). Therefore, subsequent biochemical studies have been mostly targeting steps during the cycle of IntSSV1-catalyzed recombination, such as substrate binding and strand cleavage. In their studies on strand cleavage by IntSSV1, Serre et al. showed that IntSSV1 was able to cleave a 19-bp duplex DNA containing the above-mentioned 18-bp sequence, generating a nick on each of the two DNA strands in vitro. The two points of cleavage were offset by a stretch of 7 bp, referred to as the overlap region, which corresponds perfectly to the anticodon loop of the target tRNA gene (18). The overlap region is flanked by a pair of 10-bp imperfect inverted repeating sequences, and the DNA stretch including all these sequences is proposed to be the minimum substrate for recombination by IntSSV1 (18). Those researchers found that IntSSV1 employed a cleavage mechanism utilizing the active-site residue Tyr314 and formed a 3′-phosphoprotein intermediate during the cleavage reaction, as found with bacterial and eukaryal tyrosine recombinases (18). Like λ Int, IntSSV1 also exhibited type IB topoisomerase activity (17). Recently, the crystal structure of the C-terminal catalytic domain of IntSSV1 was resolved (19, 20). The structure reveals a core fold similar to those of tyrosine recombinases of both bacterial and eukaryal origins, pointing to the conservation of these enzymes among the three domains of life.
The lack of an efficient in vitro assay for integration and excision by SSV-type integrases has hindered the biochemical analysis of these complex biological processes. In this study, we have successfully set up an in vitro assay for the recombination reactions catalyzed by SSV2 integrase (IntSSV2), a well-studied member of the SSV-type integrase family. Fusellovirus SSV2, isolated from Sulfolobus islandicus REY15/4, bears significant resemblance to SSV1 in both virion morphology and genome organization (21, 22). IntSSV2 (328 residues) shares 34% identity at the amino acid sequence level with IntSSV1 (335 residues) (21). Integration of SSV2 occurs site specifically at a tRNAGly gene in the host genome (23). Taking advantage of the availability of the in vitro assay, we have investigated integration and excision reactions catalyzed by IntSSV2, the substrate requirement, and modular organization and functions of the enzyme. Our results show that a simple recombination system is employed by fuselloviruses for site-specific recombination.
MATERIALS AND METHODS
Recombinant proteins.
To overproduce the full-length and truncated SSV2 integrase proteins, denoted wild-type IntSSV2 (GenBank accession number NP_944456.1), N80 (amino acid residues 1 to 80), C81 (residues 81 to 328), N164 (residues 1 to 164), and C165 (residues 165 to 328), PCRs were performed with specific primer pairs (Table 1), using the SSV2 DNA as the template. PCR fragments were digested and inserted into pET-30a(+) (Novagen) between the NdeI and XhoI sites. The sequences of all inserts were verified by DNA sequencing. Recombinant proteins were overproduced in Escherichia coli BL21(DE3) (Novagen) and purified as described previously (20), except for the purification of N80. For N80, following the (NH4)2SO4 precipitation step, the pellet was resuspended in and dialyzed against a solution containing 20 mM Tris-HCl (pH 6.8), 1 mM dithiothreitol (DTT), 0.1 mM EDTA, and 10% (wt/vol) glycerol. The dialyzed sample was applied onto an equilibrated HiTrap SP column (5 ml; GE) and eluted with a 0 to 1 M NaCl linear gradient in the same buffer. All the recombinant proteins contained no tags. Purified proteins were dialyzed against a solution containing 20 mM Tris-HCl (pH 7.5), 1 mM DTT, 0.1 mM EDTA, 0.3 M NaCl, and 10% (wt/vol) glycerol; aliquoted; and stored at −70°C. Protein concentrations were determined by the Lowry method with bovine serum albumin (BSA) as the standard (26).
TABLE 1.
Oligonucleotides used in this study
| Use | Designation(s)a | Sequence (5′–3′)b |
|---|---|---|
| Construction of expression vectors | SSV2(f), N80(f), N164(f) | CGCCCATATGCCTAACTTTTACGTGGG |
| SSV2(r), C81(r), C165(r) | CCGCCTCGAGTTATCAAAATGTATACAACCATTCAG | |
| N80(r) | CCGCCTCGAGTTAATCATTAGCATCGGGGGT | |
| N164(r) | CCGCCTCGAGTTATCTTTTTATTTTCAAACTTTCG | |
| C81(f) | CGCCCATATGGATGAATTGAAAGGAGTTAGAATA | |
| C165(f) | CGCCCATATGACGAAACCCGATTTACGA | |
| EMSA | A58 top | GATTAGGACGCCGGCCTCCCAAGCCGGTGATCCCGGGTTCAAATCCCGGCGGCCGCAT |
| A58 bottom | ATGCGGCCGCCGGGATTTGAACCCGGGATCACCGGCTTGGGAGGCCGGCGTCCTAATC | |
| Analysis of DNA fragments containing an attachment sitec | A968 (f), L755 (f) | TGGGTACGTCATTTATTGATCTT |
| A968 (r), R943 (r) | GTGTTCTACCTTTTCCACAGTC | |
| P730(f), R943 (f) | GCGGGTTGTAGTCGTTCA | |
| P730(r), L755 (r) | TTTTTGCCGTCTTCTTTCA | |
| A223(f) | GATGATTATCGCTGGAA | |
| A223(r) | AAATTATAGAAATAATGAAATATA | |
| P324(f) | TGTAATTTTTTTCTGCCTAAGTGTA | |
| P324(r) | GTAAAAGAGATTAAAGGTAAGTATTATGTT | |
| P209(f) | TTAGAAGACGTTAGATTAGAG | |
| P209(r) | GATGATGGTAAGCAAAGG |
Forward and reverse primers are indicated by f and r, respectively, in parentheses.
Restriction sites are underlined. The 7-bp overlap region of an att site is shown in boldface type.
The sequences of short attA and attP fragments of various lengths are shown in Fig. 3.
Gel filtration.
Protein samples (100 μl at 33 μM) were loaded onto a Superdex 200 column (10/300 GL; GE) equilibrated in a solution containing 20 mM Tris-HCl (pH 7.5), 1 mM DTT, 0.1 mM EDTA, 0.3 M NaCl, and 10% (wt/vol) glycerol and run at 0.5 ml/min at 10°C on an Äkta fast protein liquid chromatography (FPLC) system (GE). The column was calibrated with gel filtration calibration kits (low molecular weight [LMW] and high molecular weight [HMW]; GE) and alcohol dehydrogenase (150 kDa).
DNA fragments containing an attachment site.
Long DNA fragments (hundreds of base pairs in size) containing an attachment site were prepared by PCR. An attA-containing fragment was made by PCR with a specific primer pair (Fig. 1A and Table 1), using the genomic DNA from Sulfolobus solfataricus P2 as the template (23). A fragment containing attP, attL, or attR was made similarly by using total DNA isolated from SSV2-infected S. solfataricus P2 cells as the template (Fig. 1A; see also Table 1 for primer sequences). The PCR fragments were purified by using the QIAquick gel extraction kit (Qiagen) and inserted into plasmid pGEM-T (Promega). The sequences of the inserts were verified by DNA sequencing. These plasmids were used as the templates for the preparation of att site-containing fragments in subsequent PCRs (see Table 1 for primer sequences). To radiolabel these att fragments, one of the primers for a PCR was first labeled at the 5′ end with [γ-32P]ATP by using T4 polynucleotide kinase. PCRs were then carried out on an att site-containing pGEM-T plasmid by using the radiolabeled primer and a corresponding unlabeled primer with Ex Taq HotStart DNA polymerase (TaKaRa). The PCR fragments were purified by using the QIAquick PCR purification kit (Qiagen).
FIG 1.
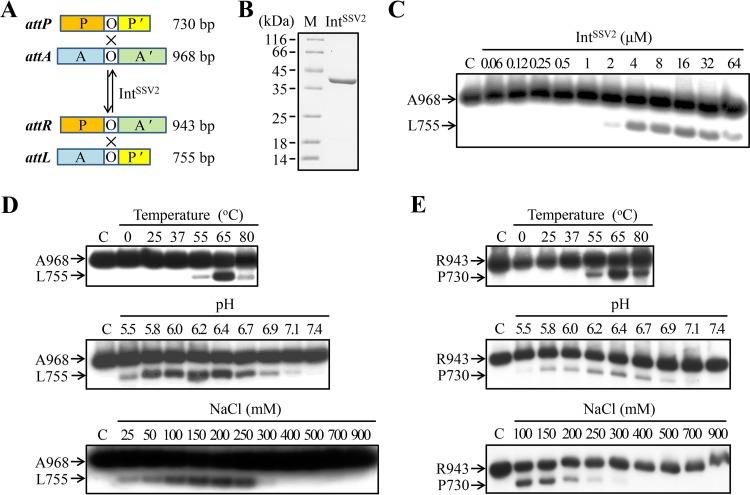
Site-specific recombination by IntSSV2. (A) Schematic representation of DNA fragments containing an attachment site for IntSSV2. O, 7-bp overlap region; P and P′, the two arms of an attP site; A and A′, the two arms of an attA site. Lengths of the att fragments are indicated. (B) SDS-PAGE analysis of recombinant IntSSV2. Recombinant IntSSV2 was purified and subjected to 15% SDS-PAGE. The gel was stained with Coomassie brilliant blue R-250. Lane M shows markers with molecular masses in kilodaltons. (C) Integration by IntSSV2. IntSSV2 was incubated for 4 h at 65°C with 32P-labeled A968 (11 nM) and unlabeled P730 (28 nM). The samples were treated with proteinase K and SDS and subjected to electrophoresis in a 1.5% agarose gel. The gel was exposed to X-ray film. The labeled substrate A968 and product L755 are indicated. For lane C, IntSSV2 was omitted. (D) Effects of temperature, pH, and salt concentrations on the integration activity of IntSSV2. (E) Effects of temperature, pH, and salt concentrations on the excision activity of IntSSV2.
Short DNA fragments (dozens of base pairs in size) were prepared by annealing a 32P-labeled or unlabeled att site-containing oligonucleotide with its complementary strand (see Fig. 3 for oligonucleotide sequences).
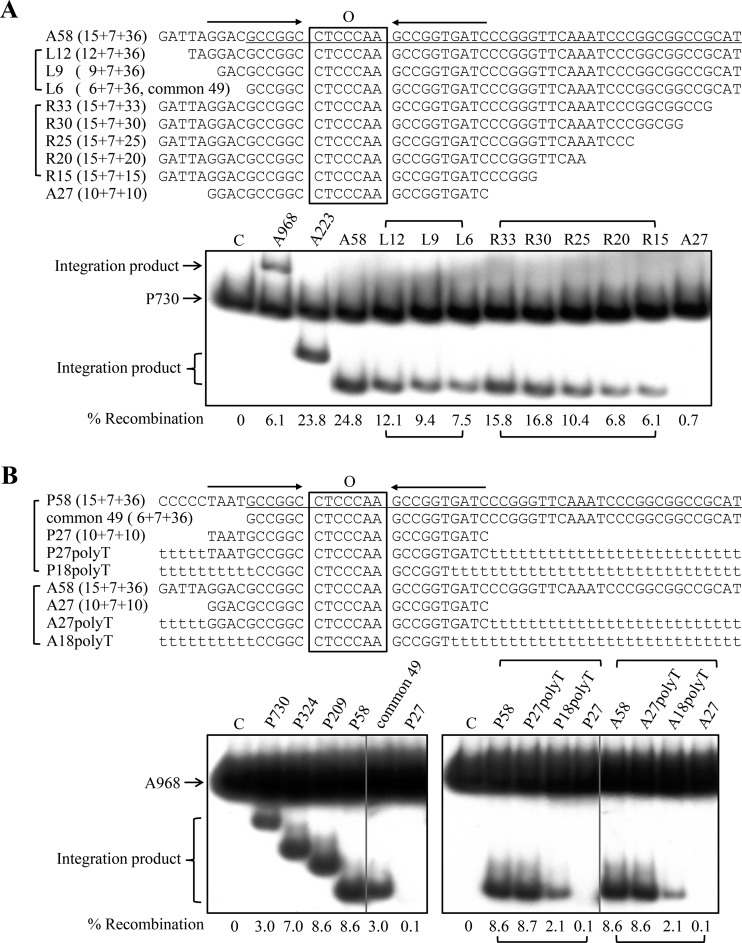
FIG 3.
Substrate requirement for integration by IntSSV2. (A) attA substrates. (Top) Sequences of the attA DNA fragments (top strand) used in the assay. The 10-bp imperfect inverted repeats flanking the 7-bp overlap region (O) are indicated by horizontal arrows. The sequence in A58 identical to the 49-bp DNA (common 49) found at both the attA and attP sites of IntSSV2 is underlined. (Bottom) IntSSV2 (16 μM) was incubated for 4 h at 65°C with 32P-labeled P730 (6 nM) and unlabeled attA DNA fragments of various sizes (30 nM). The samples were treated with proteinase K and SDS and subjected to electrophoresis in a 3.5% nondenaturing polyacrylamide gel. The gel was exposed to X-ray film, and the bands were quantified by phosphorimaging. Recombination efficiencies are indicated. For lane C, IntSSV2 was omitted. (B) attP substrates. (Top) Sequences of the attP DNA fragments (top strand) used in the assay. The 10-bp imperfect inverted repeats flanking the 7-bp overlap region are indicated by horizontal arrows. The sequence in P58 identical to the 49-bp DNA at both the attA and attP sites of IntSSV2 is underlined. Stretches of T's are shown by t's in lowercase type. (Bottom) IntSSV2 (16 μM) was incubated for 4 h at 65°C with 32P-labeled A968 (6 nM) and unlabeled attP DNA fragments of various sizes (30 nM). The samples were processed as described above for panel A.
Integration/excision assays.
Integration/excision assays were performed as described in the legend of Fig. 1A. The standard integration/excision reaction mixture (20 μl) contained a 32P-labeled att fragment (6 nM), an unlabeled att fragment (i.e., the recombination partner of the labeled att fragment) (12 nM), and integrase (16 μM) in a solution containing 50 mM Bis-Tris-HCl (pH 6.7), 150 mM NaCl, 50 μg/ml BSA, and 3% (wt/vol) glycerol. Following incubation for 4 h at 65°C, the sample was treated with SDS (1%) and proteinase K (1 mg/ml) for 1 h at 50°C and extracted with phenol-chloroform-isoamyl alcohol (25:24:1). An aliquot (17 μl) of the sample was mixed with 80% glycerol (3 μl) containing 0.04% bromophenol blue and 0.04% xylene cyanol FF and subjected to electrophoresis in either a 1.5% agarose gel at 5 V/cm in 1× Tris-acetate-EDTA (TAE) or a 3.5% nondenaturing polyacrylamide gel, which had been prerun for 1 h, at 11 V/cm in 0.5× Tris-borate-EDTA (TBE). The gel was dried, exposed to X-ray film, and analyzed by using an ImageQuant Storm PhosphorImager (Amersham Biosciences).
Electrophoretic mobility shift assays.
The standard electrophoretic mobility shift assay (EMSA) reaction mixtures (20 μl) contained 2 nM A58 (an attA fragment) (Table 1) labeled at the 5′ end of the top strand and the indicated amount of wild-type or truncated IntSSV2 in a solution containing 20 mM Bis-Tris-HCl (pH 6.7), 50 μg/ml BSA, 0 to 150 mM NaCl, 1 mM EDTA, and 6% (wt/vol) glycerol. For competitive EMSAs, unlabeled A58 or poly(dI-dC)2 was added as indicated. After incubation for 10 min at 23°C, the reaction mixtures were loaded onto a 5% nondenaturing polyacrylamide gel, which had been prerun to a constant current, and electrophoresis was performed in 0.1× TBE buffer for 3 h at 10 V/cm. The gel was dried, exposed to X-ray film, and analyzed by using a PhosphorImager (Amersham Biosciences).
DNA topoisomerase activity assays.
DNA of negatively supercoiled plasmid pBR322 (0.6 μg; Thermo Fisher Scientific) was incubated for 4 h at 65°C with the indicated amount of wild-type or truncated IntSSV2 in a solution containing 50 mM Bis-Tris-HCl (pH 6.7), 50 μg/ml BSA, 5 mM EDTA, and 150 mM NaCl in a total volume of 20 μl. After incubation, samples were treated with SDS (1%) and proteinase K (1 mg/ml) for 1 h at 50°C and mixed with a 1/10 volume of loading buffer (1% SDS, 50% glycerol, 0.05% bromphenol blue, and 0.05% xylene cyanol FF). An aliquot of each sample (containing 0.3 μg pBR322) was loaded onto a 1.2% agarose gel, and electrophoresis was performed in 1× TAE buffer for 3.5 h at 3 V/cm. DNA was visualized by staining with ethidium bromide. As a positive control, plasmid pBR322 was incubated for 30 min at 37°C with calf thymus topoisomerase I (10 U; TaKaRa), and the sample was treated as described above.
RESULTS
IntSSV2 catalyzes both integration and excision reactions in vitro.
To test the integration activity of IntSSV2, we designed two att fragments of different sizes, i.e., a 968-bp attA-containing fragment (A968) and a 730-bp attP-containing fragment (P730) (Fig. 1A). Integration would result in the production of a 943-bp attR fragment (R943) and a 755-bp attL fragment (L755). The top strand of A968 was labeled at the 5′ end so that only L755 would be labeled. The excision activity of IntSSV2 was determined in the same fashion, except that R943, labeled at the 5′ end of the top strand, and L755, instead of A968 and P730, were used as the substrates (Fig. 1A). Recombinant IntSSV2 protein was purified to near homogeneity (Fig. 1B). As shown in Fig. 1C, IntSSV2 alone was able to catalyze both integration and excision reactions. In contrast, bacteriophage λ integrase requires different sets of accessory proteins for the integration and excision reactions.
We then examined the effects of temperature, pH, and salt concentrations on the integration activity of IntSSV2. IntSSV2 was significantly active in integration only at temperatures of ≥55°C (Fig. 1D) and optimally active at 65°C. Integration catalyzed by the integrase was highly sensitive to pH, and significant integrase activity was found only in a narrow pH range of 5.8 to 6.7, with pH 6.2 being optimum (Fig. 1D). The integrase activity of IntSSV2 increased with increasing NaCl concentrations at moderate levels but was drastically inhibited at concentrations of ≥300 mM NaCl (Fig. 1D). Similar temperature, pH, and salt dependences were observed with the excision activity of IntSSV2, but the enzyme appeared to be more sensitive to salt in the excision reaction than in the integration reaction (Fig. 1E). We conclude that IntSSV2 catalyzes both types of recombination optimally at slightly acidic pH (i.e., pH 6.2), high temperatures (i.e., ∼65°C), and moderate salt concentrations (i.e., 100 to 150 mM NaCl).
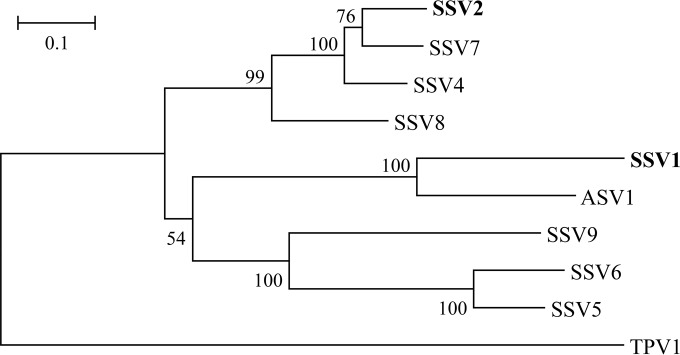
We also prepared att fragments for IntSSV1 and performed the integration and excision assays on the enzyme in a similar manner. However, IntSSV1 was inactive in those assays (our unpublished results). Given the similarity between the two viruses, the discrepancy observed with the two enzymes is somewhat surprising. However, the biochemical differences between IntSSV2 and IntSSV1 appear to be consistent with the observation that the two enzymes are clearly separated phylogenetically (Fig. 2). Clarification of the differences between these two enzymes in biochemical properties awaits further investigation.
FIG 2.
Phylogenetic tree of the SSV-type integrases. A sequence search in GenBank with the amino acid sequence of IntSSV2 as a query was performed by using the BLASTp algorithm. The retrieved sequences were aligned by using ClustalW, a phylogenetic tree was reconstructed by using the neighbor-joining method implemented in the MEGA software package (version 6.06), and the TPV1 integrase was used as the outgroup. Numbers at nodes represent percentages of bootstrap support based on a neighbor-joining analysis of 1,000 resampled data sets (only values of >50% are shown). Evolutionary distances were calculated by using Kimura's two-parameter model. GI numbers of sequences are as follows: 38639805 for IntSSV2, 270281826 for IntSSV7, 160688429 for IntSSV4, 42495064 for IntSSV8, 9625521 for IntSSV1, 270281754 for IntASV1, 42494935 for IntSSV9, 270281793 for IntSSV6, 198449240 for IntSSV5, and 378554453 for IntTPV1.
Substrate requirement of IntSSV2 for integration.
The SSV2 genome is capable of integrating into the S. solfataricus P2 genome at the site of the tRNAGly (CCC) gene (23). The attP and attA attachment sites share a 49-bp common sequence (i.e., common 49), which is comprised of a 7-bp overlap region, where strand exchange occurs, and portions of the left and right arms of the attA or attP site (Fig. 3). A pair of 10-bp imperfect sequence repeats flanks the 7-bp-overlap region. These repeats, which are probably recognized and bound by IntSSV2, and the overlap region form a 27-bp core-type site. As found in the SSV1 genome, the 49-bp common sequence comprises the downstream half of the integrated tRNAGly (CCC) gene, and the 7-bp overlap region corresponds perfectly to the anticodon loop of the target tRNA gene. The minimum sequence requirement for the integration of a fuselloviral genome has not been experimentally determined. Some researchers have suggested that the 44-bp common sequence is essential for viral integration (8), while others believe that the core-type site is minimally required for integration in SSV1 (18). In this study, we tested a range of attachment site-containing substrates in our SSV2 integration assays. First, we determined the dependence of the integration activity of IntSSV2 on the length of the attA-containing substrate by employing radiolabeled P730 and a series of unlabeled attA fragments of different sizes in the integration reactions (Fig. 3A, top). As shown in Fig. 3A, bottom, significant integration was observed when the unlabeled attA fragment was shortened from 968 bp (A968) to 58 bp (A58). The efficiency of recombination was lower on A968 than on A223 or A58, presumably because the longer fragment allowed more sequence-nonspecific binding by the integrase and thus reduced the availability of the enzyme for the site of recombination. Further tests were then carried out with progressively shortened derivatives of A58 (Fig. 3A). As the left arm of A58 was shortened from 15 to 6 bp, the integration activity of the enzyme decreased gradually. However, even with the 6-bp left arm left, or when A58 was converted into a fragment of common 49, approximately one-third of the recombination activity observed with A58 remained. Similarly, when the right arm of A58 was reduced from 36 to 15 bp, the efficiency of recombination between the labeled attP fragment and the A58 derivative decreased successively from 24.8% to 6.1%. Importantly, however, integration with A27, or the core-type site, was barely detectable (0.7%). Based on these results, we conclude that IntSSV2 was able to catalyze significant integration on attA fragments shorter than common 49 but longer than the core-type site, and the efficiency of recombination closely depended on the lengths of both the left and right arms of the attA fragment, at least up to those in A58.
We then determined the influence of the size of an attP-containing fragment on the integration activity of IntSSV2 in a similar fashion by using labeled A968 and a series of unlabeled attP fragments of various lengths in the reactions (Fig. 3B, top). As shown in Fig. 3B, among the tested attP fragments, maximum recombination was observed on P209 and P58. The longer attP fragments (i.e., P730 and P324) were less efficient in the integration reactions, as found with recombination on A968. The efficiency of recombination on the fragment of common 49 was reduced to one-third of that on P58. Little integration was detected on P27, the core-type site.
However, IntSSV2 showed similar integration activities on P58 and P27polyT, a 58-bp DNA fragment which resembles P58 except for the replacement of the sequences flanking the core-type site with two strings of A·T pairs (Fig. 3B). This observation suggests that P27 contains the sequence information necessary for integration by IntSSV2 and that the DNA stretches flanking the core-type site affect the efficiency of integration in a sequence-nonspecific manner. It has been shown that an 18-bp core region-containing fragment, which corresponds to the anticodon stem-loop of the tRNAArg gene, was protected by IntSSV1 in a footprinting experiment (16). Therefore, we tested the recombination efficiency of P18polyT, a 58-bp fragment containing a region equivalent to the 18-bp region in SSV1 and flanking A·T regions, in the integration assays (Fig. 3B, top). As shown in Fig. 3B, bottom, the efficiency of recombination on P18polyT was about one-quarter of that on P58, indicating that further shortening of the core-type site would result in a significant loss of integration by IntSSV2.
We also examined the ability of IntSSV2 to recombine two attA fragments. As shown in Fig. 3B, bottom, the efficiencies of recombination between labeled A968 and unlabeled A58, A27polyT, and A27 were highly similar to those between labeled A968 and unlabeled P58, P27polyT, and P27, respectively. Therefore, IntSSV2 is capable of catalyzing recombination between two att fragments, and the sequence shared by the attA and attP core-type sites is likely involved in sequence-specific interactions with the integrase. Taken together, these data indicate that the core-type sequences at attA and attP are required for efficient integration catalyzed by IntSSV2.
Oligomerization of IntSSV2.
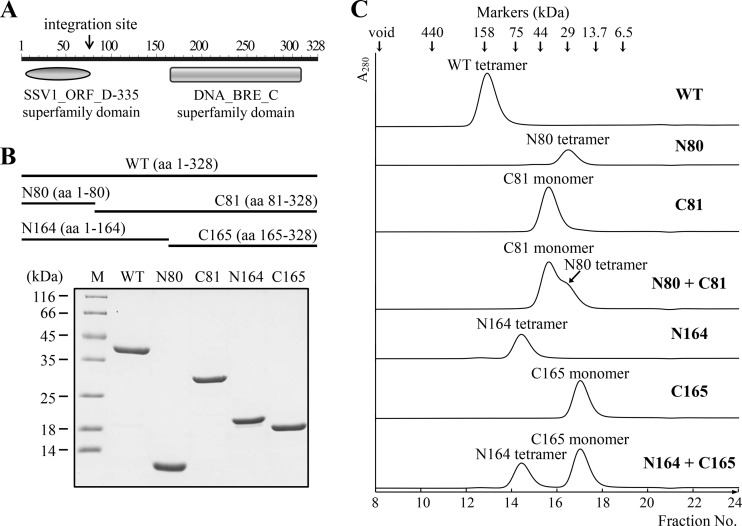
Since the formation of a synaptic complex during recombination catalyzed by a tyrosine recombinase depends on the protein-protein interaction between protomers of the enzyme, we sought to examine the ability of IntSSV2 to form an oligomer. As shown in Fig. 4, purified wild-type IntSSV2 eluted as a single peak with a calculated molecular mass of 143.8 kDa on a Superdex 200 column. Given the theoretical molecular mass of 38.4 kDa for IntSSV2, the protein existed as a tetramer in solution. Domain analysis (Conserved Domain Search Service [http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi]) suggests that IntSSV2 contains two structural domains, i.e., an N-terminal SSV1_ORF_D-335 superfamily domain, which is shared by all SSV-type integrases, and a C-terminal catalytic domain belonging to the DNA breaking-rejoining enzyme superfamily, which includes tyrosine recombinases/type IB topoisomerases (Fig. 4A). The N-terminal domain terminates at the sequence corresponding to the attP site (Fig. 4A), and its function is unclear. The C-terminal domain accounts for about one-half of the size of the integrase and resembles the C-terminal catalytic domain of type IB topoisomerases in structure. Based on the above-described analysis, we constructed two sets of deletion mutants of IntSSV2 (Fig. 4B). One set of mutant proteins includes N80 (amino acid residues 1 to 80), which contains the SSV1_ORF_D-335 superfamily domain, and C81 (residues 81 to 328). The other set consists of C165 (residues 165 to 328), which has the catalytic domain, and N164 (residues 1 to 164), which correspond to the deletion mutant proteins C174 and N173, respectively, of IntSSV1 (20). All of the mutant proteins were purified to homogeneity (Fig. 4B). To determine their oligomeric states, these truncated mutant proteins were subjected to gel filtration. Each of the four mutant proteins eluted as a single peak (Fig. 4C). N80, C81, N164, and C165 displayed apparent molecular masses of 24.6, 38.0, 68.6, and 19.0 kDa, respectively. Based on the theoretical molecular masses of 9.1, 29.3, 19.0, and 19.4 kDa for N80, C81, N164, and C165, respectively, we inferred that N164 existed as a tetramer and that C81 and C165 existed as monomers in solution. N80 was likely present as a nonglobular tetramer since it came off the column slightly later than expected for an N80 tetramer. It appears, therefore, that IntSSV2 protomers form a tetramer in solution through interactions in the N-terminal SSV1_ORF_D-335 superfamily domain. When an equal molar mixture of N80 and C81 was applied to the gel filtration column, two separate peaks typical of the two individual proteins were observed. The same observation was made with a mixture of N164 and C165. Therefore, no stable interaction between N80 and C81 or between N164 and C165 was detected.
FIG 4.
Oligomerization of wild-type and truncated IntSSV2. (A) Domain architecture of the IntSSV2 protein. The N-terminal domain terminates at the sequence corresponding to the integration site attP. (B) SDS-PAGE analysis of wild-type (WT) and truncated IntSSV2 proteins. Recombinant wild-type and truncated IntSSV2 proteins were purified and subjected to 15% SDS-PAGE. The gel was stained with Coomassie brilliant blue R-250. Lane M indicates markers with molecular masses in kilodaltons. aa, amino acids. (C) Analysis of wild-type and truncated IntSSV2 proteins by gel filtration. A sample (100 μl; 33 μM) of the indicated protein was loaded onto a Superdex 200 column. The void volume of the column and protein size markers are shown with arrows at the top. The proposed oligomeric states of the proteins, as deduced based on a calibration curve prepared from the elution profiles of the protein size markers, are indicated. N80 + C81 and N164 + C165 indicate N80 and N164 mixed with C81 and C165, respectively, at a molar ratio of 1.
Substrate binding and catalytic activities of the truncated mutants of IntSSV2.
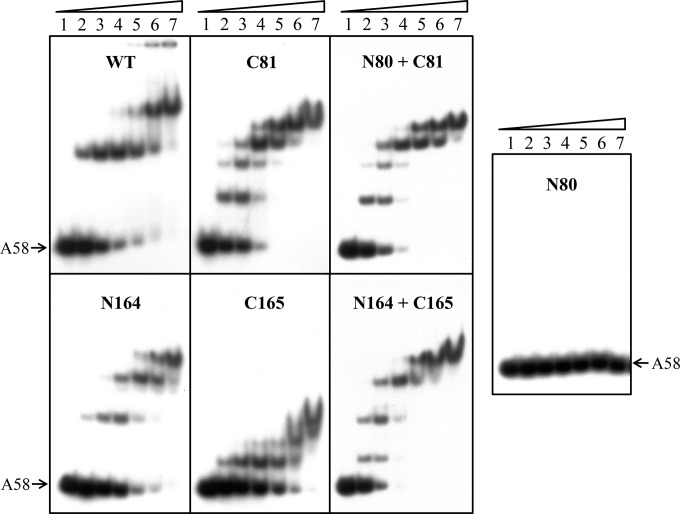
To understand the roles of the structural domains or portions of IntSSV2 in substrate binding by the protein, we conducted electrophoretic mobility shift assays on the full-length and truncated versions of the protein using labeled A58 as the probe. C81, N164, and C165 bound as well as wild-type IntSSV2 to DNA, with similar affinities (KD [equilibrium dissociation constant] of 10 to 50 nΜ), as estimated based on the amount of the protein required to retard one-half of the labeled probe (Fig. 5). It appears that the binding affinity of IntSSV2 is similar to that of IntSSV1 under the same experimental conditions (20). Notably, N80 failed to bind to the DNA. Different binding patterns were generated by the full-length protein and the three truncated proteins (C81, N164, and C165). The full-length protein produced two major retardation bands. C81 generated four shifts, whereas N164 and C165 formed three shifts of distinctly different patterns. From the retardation patterns (including spacing between adjacent shifts), molecular masses, and gel filtration profiles of these proteins, we infer that both the full-length protein and N164 bound the DNA as dimers, whereas C81 and C165 interacted with the DNA as monomers. The observations that the full-length protein and N164 existed as a tetramer in gel filtration assays and bound the DNA as a dimer in gel retardation assays presumably indicate that the proteins were present as dimers at low concentrations (e.g., 0.025 μM in EMSAs) and as tetramers at high concentrations (e.g., ∼3.3 to 6.6 μM in gel filtration assays, considering a 5- to 10-fold dilution effect). The addition of both N80 and C81 to the binding reaction mixture did not restore the binding pattern of the full-length protein and, instead, produced a pattern identical to that generated by C81 alone, in agreement with the failure of N80 to bind the DNA. Similarly, a mixture of N164 and C165 did not produce a gel retardation pattern typical of that of the full-length protein. The two truncated proteins appeared to bind the DNA independently.
FIG 5.
Substrate DNA binding by wild-type and truncated IntSSV2 proteins. Wild-type or truncated IntSSV2 was incubated for 10 min at 23°C with 32P-labeled A58 DNA (2 nM). The samples were subjected to electrophoresis in a 5% polyacrylamide gel. The gel was exposed to X-ray film. Lanes 1 to 7 indicate proteins at concentrations of 0, 12.5, 25, 50, 100, 200, and 400 nM, respectively. N80 + C81 and N164 + C165 indicate N80 and N164 mixed with C81 and C165, respectively, at a molar ratio of 1.
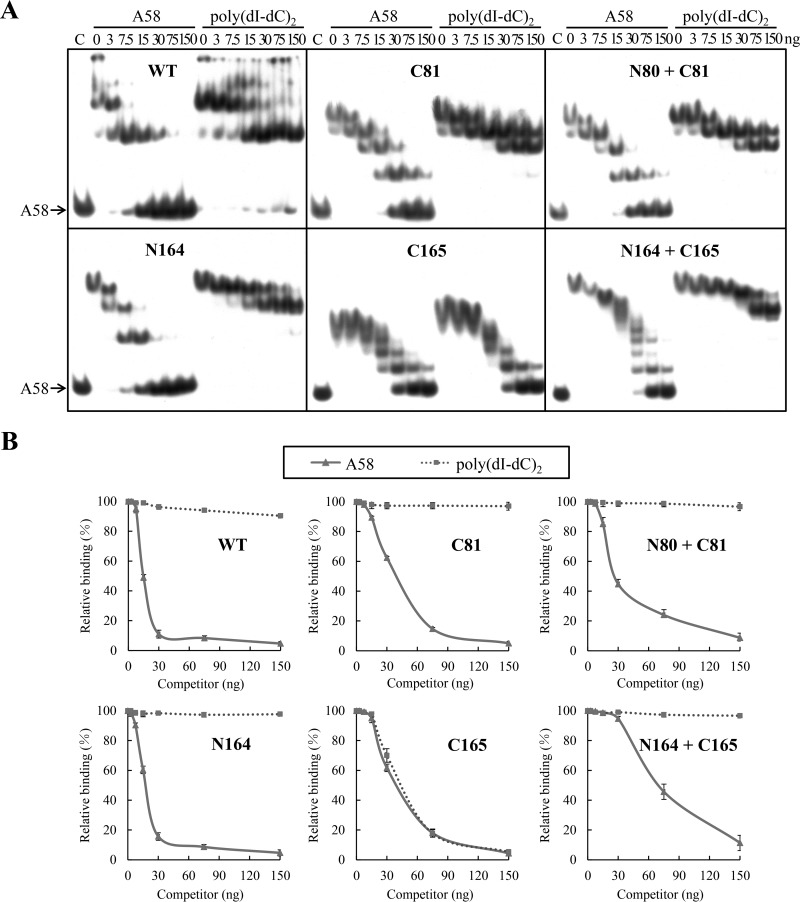
To examine the contribution of various portions of IntSSV2 to the sequence specificity of binding of the protein to DNA, we performed competitive binding assays. Binding of the full-length and truncated proteins to labeled A58 in the presence of increasing amounts of unlabeled A58 or poly(dI-dC)2 was analyzed (Fig. 6). IntSSV2, C81, and N164 showed significant binding specificity, but C165 did not. In addition, IntSSV2 and N164 had similar sequence-specific binding affinities, and both proteins showed stronger sequence-specific binding than did C81. The addition of N80 did not affect the sequence-specific binding of C81 to the DNA. Intriguingly, comparison of the gel retardation patterns obtained in the presence and in the absence of the unlabeled competitors reveals that, as in the case of the full-length protein, binding of C81 and N164 to labeled A58 formed two stable complexes, since only two shifts were resistant to competition from an excess amount of unlabeled poly(dI-dC)2 (Fig. 5 and 6). This is consistent with the prediction that each of the two 10-bp imperfect repeats flanking the 7-bp overlap region in the labeled probe is capable of being bound by a single IntSSV2 molecule. Based on these results, we conclude that the sequence specificity of substrate binding by IntSSV2 is determined by the N-terminal half, most likely residues 81 to 164 (i.e., a region shared by C81 and N164), of the protein.
FIG 6.
Sequence specificity of DNA binding by wild-type and truncated IntSSV2 proteins. (A) Competitive EMSA. IntSSV2 (300 nM) was incubated for 10 min at 23°C with 32P-labeled A58 (2 nM) and increasing amounts (0, 3, 7.5, 15, 30, 75, and 150 ng) of unlabeled A58 or poly(dI-dC)2 in the presence of 150 mM NaCl. The samples were subjected to electrophoresis in a 5% polyacrylamide gel. The gel was exposed to X-ray film. N80 + C81 and N164 + C165 indicate N80 and N164 mixed with C81 and C165, respectively, at a molar ratio of 1. For lane C, IntSSV2 was omitted. (B) Quantitative analysis. The gels shown in panel A were quantified by phosphorimaging. Data represent averages of results from three independent measurements.
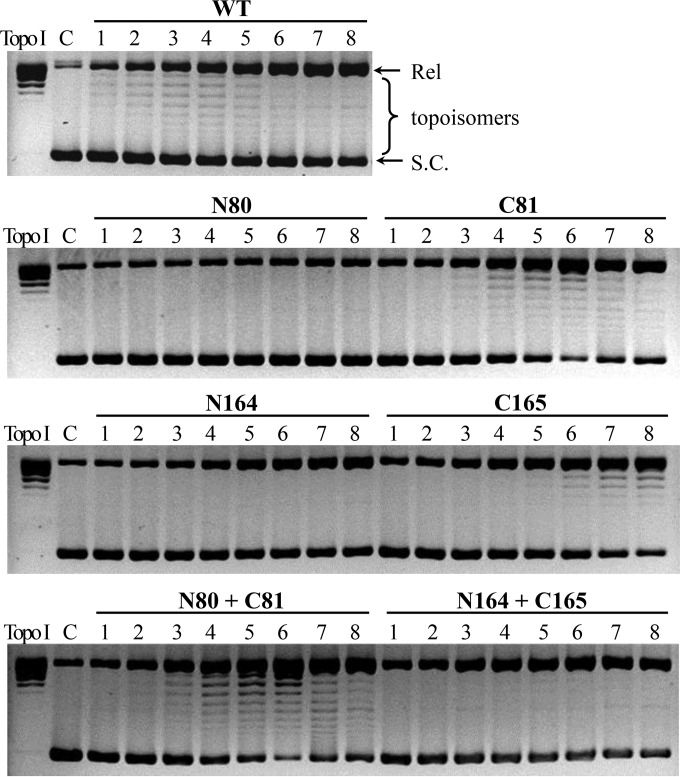
We also examined the catalytic activities of the truncated mutants of IntSSV2. First, we tested the cleavage/religation or topoisomerase activity of the full-length and deletion mutant proteins by using negatively supercoiled plasmid pBR322. As shown in Fig. 7, C81 was nearly as active as the full-length protein in relaxing the negatively supercoiled plasmid. The activity of C81 was not enhanced in the presence of N80. C165 showed lower topoisomerase activity than that of either the full-length enzyme or C81. Unexpectedly, the topoisomerase activity of C165 was further reduced in the presence of N164, probably as a result of the competition of N164 for binding to the plasmid DNA. Neither N80 nor N164 was active in the topoisomerase activity assays. These observations were consistent with the presence of all six conserved residues (R203, R234, K271, R274, R297, and Y307) required for cleavage and religation in the structural domain contained in C165.
FIG 7.
Topoisomerase activity of wild-type and truncated IntSSV2 proteins. Wild-type or truncated IntSSV2 was incubated for 4 h at 65°C with negatively supercoiled pBR322 DNA (0.6 μg). The samples were treated with proteinase K and SDS and subjected to electrophoresis in a 1.2% agarose gel. The gel was stained with ethidium bromide and photographed under UV light. Lanes 1 to 8 show enzyme concentrations of 0.125, 0.25, 0.5, 1, 2, 4, 8, and 16 μM, respectively. N80 + C81 and N164 + C165 indicate N80 and N164 mixed with C81 and C165, respectively, at a molar ratio of 1. Topo I indicates a positive control in which pBR322 DNA was relaxed with calf thymus topoisomerase I (10 U). For lane C, IntSSV2 was omitted. Positions of negatively supercoiled (S.C.) and relaxed (Rel) plasmids are indicated.
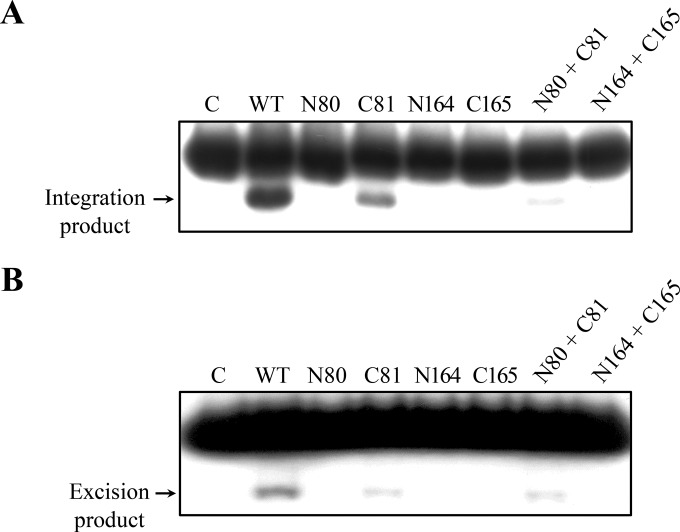
Next, we determined both the integration and excision activities of the truncated mutant proteins of IntSSV2, as described above. Among the four deletion mutant proteins, only C81 was active in integration as well as excision, although both activities were lower than those of the full-length protein (Fig. 8). Like the full-length protein, C81 appeared to be less efficient in excision than in integration. None of the other mutant proteins was active in either integration or excision. The integration, but not the excision, activity of C81 was inhibited by N80. Our results suggest that N-terminally truncated IntSSV2 (amino acid residues 81 to 328) contains all the necessary sequence elements required for the recombination reaction. N80 probably affected the integration activity of C81 through transient or unstable protein-protein interactions, since no stable interaction between the two proteins was detected.
FIG 8.
Substrate recombination by wild-type and truncated IntSSV2 proteins. (A) Integration activity analysis. Wild-type or truncated IntSSV2 (16 μM) was incubated for 4 h at 65°C with 32P-labeled A968 (6 nM) and unlabeled P730 (60 nM). The samples were treated with proteinase K and SDS and subjected to electrophoresis in a 3.5% polyacrylamide gel. The gel was exposed to X-ray film. (B) Excision activity analysis. Wild-type or truncated IntSSV2 (16 μM) was incubated for 4 h at 65°C with 32P-labeled R943 (6 nM) and unlabeled L755 (60 nM). The samples were processed as described above for panel A. N80 + C81 and N164 + C165 indicate N80 and N164 mixed with C81 and C165, respectively, at a molar ratio of 1. For lane C, IntSSV2 was omitted. The labeled products are indicated by arrows.
DISCUSSION
There are both simple and complex systems for site-specific recombination catalyzed by tyrosine recombinases. In simple systems, such as those involving Cre and Flp, recombination occurs at a specific site with a simple composition and limited size (i.e., ∼30 bp) and is catalyzed by recombinase alone (1, 4). On the other hand, in a complex system, exemplified by that of λ integrase, accessory proteins, in addition to recombinase, are required for the recombination reaction, and the site of recombination is long (e.g., ∼250 bp for the λ attP site) and complex to accommodate binding by the accessory proteins (1, 4). Furthermore, accessory proteins involved in integration and excision reactions are not identical. SSV-type integrases are the only archaeal tyrosine recombinases that have been extensively characterized biochemically. IntSSV1, the prototype of SSV-type integrases, was shown to catalyze both integration and excision reactions, and no additional accessory proteins were apparently required for the excision reaction, compared to those for the integration reaction, in early biochemical studies (8, 16). Like other laboratories, we failed to establish in vitro recombination assays for IntSSV1 (17, 18; our unpublished results). However, we were able to set up an in vitro IntSSV2 assay that permitted the biochemical analysis of the recombination activity of SSV-type integrases in this study.
IntSSV2 was capable of efficient integration and excision on its own. Since no additional accessory proteins were required for the reactions, recombination catalyzed by the enzyme was readily reversible. This is consistent with the presence of an apparent dynamic equilibrium between the free and the integrated forms of SSV2 DNA in Sulfolobus host cells infected with SSV2 (23). It was also noticed that integration was slightly favored over excision in vitro. However, the underlying mechanism for this is unknown. Obviously, our results do not exclude the possibility that the balance between the integration and excision activities of IntSSV2 is regulated by host- and/or virus-encoded proteins in the cell. Notably, IntSSV2 was able to catalyze recombination on short attA and attP substrates. A 27-bp core-type sequence, comprising a 7-bp-overlap region flanked with 10-bp imperfect inverted repeats, satisfied the requirement of the integrase for the sequence-specific recognition of the site of recombination. However, the efficiency of integration was drastically influenced by sequences flanking the core-type site in a sequence-nonspecific fashion. The ability of IntSSV2 to catalyze both integration and excision by itself and the characteristics of the site of recombination for the enzyme suggest that SSV-type integrases function in a simple type of site-specific recombination. Recently, a similar observation was made for XerA, a tyrosine recombinase involved in chromosome resolution, from Pyrococcus abyssi (24, 25). The protein was shown to be able to recombine dif sites in vitro in the absence of any protein partners.
Deletion analyses revealed that the N-terminal portion (residues 1 to 80) of IntSSV2 (N80), which corresponds to the N-terminal SSV1_ORF_D-335 superfamily domain of SSV-type integrases, did not bind DNA and was involved in the tetramerization of the integrase. The C-terminal catalytic domain (C165; residues 165 to 328) interacted with DNA in a sequence-nonspecific fashion and was responsible for chemical catalysis during the cleavage and religation steps of the recombination reaction. The remaining middle portion of the protein (residues 81 to 164) appeared to be involved in the recognition and binding of the specific site of recombination.
IntSSV2 resembles IntSSV1 in sequence and properties (21). However, the two enzymes differ in several aspects. Full-length IntSSV2 exists as a stable tetramer, whereas IntSSV1 is present as a dimer in solution. The former exhibited greater sequence specificity than the latter in DNA binding (20). Furthermore, IntSSV2 recognizes and binds the binding site primarily through the middle portion of the protein (residues 81 to 164), which corresponds to the C-terminal half of the N173 domain of IntSSV1. In comparison, the binding specificity of IntSSV1 was conferred by neither its N-terminal domain (N173) nor its C-terminal catalytic domain (C174) alone and depended strongly on the covalent linkage between the two domains (20). Elucidation of these differences between the two SSV-type integrases will help us understand the mechanisms of site-specific recombination by archaeal tyrosine recombinases.
Interestingly, C81, an N-terminally truncated mutant of IntSSV2 (residues 81 to 328), was unable to form tetramers but possessed all the catalytic properties of full-length IntSSV2 except for the slightly reduced recombination activity. Since attP is located in the IntSSV2 gene at the junction between the sequences encoding N80 and C81, integration of SSV2 DNA into the host genome would lead to a separation of the integrase gene into two parts, i.e., int(N) and int(C) (23). Sequence analysis suggests that int(C) could potentially be expressed, and the resulting protein would be slightly larger than C81. However, we were unable to detect the presence of the int(C) product in S. solfataricus cells infected with SSV2 under our experimental conditions by immunoblotting (data not shown). Therefore, it will be of interest to investigate if int(C) would be induced under certain growth conditions and how the product of int(C) would function in SSV2-infected cells.
ACKNOWLEDGMENTS
We are grateful to Qunxin She for valuable discussions. We thank Ziyi Wang for assisting in part of this study.
This work was supported by grant 31130003 from the National Natural Science Foundation of China.
REFERENCES
- 1.Grindley ND, Whiteson KL, Rice PA. 2006. Mechanisms of site-specific recombination. Annu Rev Biochem 75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 2.Esposito D, Scocca J. 1997. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res 25:3605–3614. doi: 10.1093/nar/25.18.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y, Rice PA. 2003. New insight into site-specific recombination from Flp recombinase-DNA structures. Annu Rev Biophys Biomol Struct 32:135–159. doi: 10.1146/annurev.biophys.32.110601.141732. [DOI] [PubMed] [Google Scholar]
- 4.Rajeev L, Malanowska K, Gardner JF. 2009. Challenging a paradigm: the role of DNA homology in tyrosine recombinase reactions. Microbiol Mol Biol Rev 73:300–309. doi: 10.1128/MMBR.00038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gopaul DN, Duyne GD. 1999. Structure and mechanism in site-specific recombination. Curr Opin Struct Biol 9:14–20. doi: 10.1016/S0959-440X(99)80003-7. [DOI] [PubMed] [Google Scholar]
- 6.Grainge I, Jayaram M. 1999. The integrase family of recombinase: organization and function of the active site. Mol Microbiol 33:449–456. doi: 10.1046/j.1365-2958.1999.01493.x. [DOI] [PubMed] [Google Scholar]
- 7.Cheng C, Kussie P, Pavletich N, Shuman S. 1998. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell 92:841–850. doi: 10.1016/S0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 8.Muskhelishvili G, Palm P, Zillig W. 1993. SSV1-encoded site-specific recombination system in Sulfolobus shibatae. Mol Gen Genet 237:334–342. [DOI] [PubMed] [Google Scholar]
- 9.She Q, Shen B, Chen L. 2004. Archaeal integrases and mechanisms of gene capture. Biochem Soc Trans 32:222–226. doi: 10.1042/bst0320222. [DOI] [PubMed] [Google Scholar]
- 10.She Q, Brugger K, Chen L. 2002. Archaeal integrative genetic elements and their impact on genome evolution. Res Microbiol 153:325–332. doi: 10.1016/S0923-2508(02)01331-1. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Peng N, Shah SA, Huang L, She Q. 2015. Archaeal extrachromosomal genetic elements. Microbiol Mol Biol Rev 79:117–152. doi: 10.1128/MMBR.00042-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palm P, Schleper C, Grampp B, Yeats S, McWilliam P, Reiter W-D, Zillig W. 1991. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185:242–250. doi: 10.1016/0042-6822(91)90771-3. [DOI] [PubMed] [Google Scholar]
- 13.She Q, Peng X, Zillig W, Garrett RA. 2001. Gene capture in archaeal chromosomes. Nature 409:478. doi: 10.1038/35054138. [DOI] [PubMed] [Google Scholar]
- 14.Reiter WD, Palm P, Yeats S. 1989. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res 17:1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clore AJ, Stedman KM. 2007. The SSV1 viral integrase is not essential. Virology 361:103–111. doi: 10.1016/j.virol.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Muskhelishvili G. 1994. The archaeal SSV integrase promotes intermolecular excisive recombination in vitro. Syst Appl Microbiol 16:605–608. [Google Scholar]
- 17.Letzelter C, Duguet M, Serre MC. 2004. Mutational analysis of the archaeal tyrosine recombinase SSV1 integrase suggests a mechanism of DNA cleavage in trans. J Biol Chem 279:28936–28944. doi: 10.1074/jbc.M403971200. [DOI] [PubMed] [Google Scholar]
- 18.Serre MC, Letzelter C, Garel JR, Duguet M. 2002. Cleavage properties of an archaeal site-specific recombinase, the SSV1 integrase. J Biol Chem 277:16758–16767. doi: 10.1074/jbc.M200707200. [DOI] [PubMed] [Google Scholar]
- 19.Eilers BJ, Young MJ, Lawrence CM. 2012. The structure of an archaeal viral integrase reveals an evolutionarily conserved catalytic core yet supports a mechanism of DNA cleavage in trans. J Virol 86:8309–8313. doi: 10.1128/JVI.00547-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhan Z, Ouyang S, Liang W, Zhang Z, Liu Z-J, Huang L. 2012. Structural and functional characterization of the C-terminal catalytic domain of SSV1 integrase. Acta Crystallogr D Biol Crystallogr 68:659–670. doi: 10.1107/S0907444912007202. [DOI] [PubMed] [Google Scholar]
- 21.Stedman K, She Q, Phan H, Arnold H, Holz I, Garrett R, Zillig W. 2003. Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res Microbiol 154:295–302. doi: 10.1016/S0923-2508(03)00074-3. [DOI] [PubMed] [Google Scholar]
- 22.Wiedenheft B, Stedman K, Roberto F, Willits D, Gleske AK, Zoeller L, Snyder J, Douglas T, Young M. 2004. Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae viruses. J Virol 78:1954–1961. doi: 10.1128/JVI.78.4.1954-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Contursi P, Jensen S, Aucelli T, Rosé M, Bartolucci S, She Q. 2006. Characterization of the Sulfolobus host-SSV2 virus interaction. Extremophiles 10:615–627. doi: 10.1007/s00792-006-0017-2. [DOI] [PubMed] [Google Scholar]
- 24.Cortez D, Quevillon-Cheruel S, Gribaldo S, Desnoues N, Sezonov G, Forterre P, Serre MC. 2010. Evidence for a Xer/dif system for chromosome resolution in archaea. PLoS Genet 6:e1001166. doi: 10.1371/journal.pgen.1001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serre MC, El Arnaout T, Brooks MA, Durand D, Lisboa J, Lazar N, Raynal B, van Tilbeurgh H, Quevillon-Cheruel S. 2013. The carboxy-terminal alphaN helix of the archaeal XerA tyrosine recombinase is a molecular switch to control site-specific recombination. PLoS One 8:e63010. doi: 10.1371/journal.pone.0063010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275. [PubMed] [Google Scholar]