ABSTRACT
Cocktails of monoclonal antibodies (MAbs) that target the surface glycoprotein (GP) of Ebola virus (EBOV) are effective in nonhuman primate models and have been used under emergency compassionate-treatment protocols in human patients. However, the amino acids that form the detailed binding epitopes for the MAbs in the ZMapp, ZMAb, and the related MB-003 cocktails have yet to be identified. Other binding properties that define how each MAb functionally interacts with GP—such as affinity, epitope conservation, and epitope accessibility—also remain largely unknown. To help define how each MAb interacts with GP, here we used comprehensive alanine-scanning mutagenesis (shotgun mutagenesis), neutralization escape, and whole virion binding to define each MAb's specific epitope, epitope accessibility, epitope conservation, and apparent affinity. Each of the six therapeutic MAbs binds nonidentical epitopes in the GP base, glycan cap, or mucin-like domain. Their apparent affinity, epitope complementarity, and epitope accessibility helps explain why MAbs 4G7 and 13C6 are more protective than 2G4 and 1H3. The mucin-like domain MAbs 6D8 and 13F6 bind with the strongest apparent affinity, helping to explain their effectiveness in vivo despite their inability to neutralize virus.
IMPORTANCE Ebola virus disease (EVD) can be caused by four different filovirus family members, including Ebola virus (EBOV), which infected 10 times more people in western Africa over the last year than all previous EVD outbreaks combined, with a number of cases distributed across the globe by travelers. Cocktails of inhibitory monoclonal antibodies (MAbs), such as ZMAb, MB-003, and in particular ZMapp, have demonstrated in animal models some of the most significant therapeutic potential for treating EVD, and in 2014, 15 patients were treated with ZMapp or ZMAb under compassionate-use protocols. Here, we have defined the epitope features for the most important therapeutic MAbs against EBOV developed to date. Defining the epitopes and binding characteristics for these MAbs, as well as the commonly used reference MAb KZ52, helps explain their breadth of reactivity against different ebolavirus species, predict viral evasion against these MAbs, and design new cocktails of MAbs with improved complementarity.
INTRODUCTION
The 2014 outbreak of Ebola virus (EBOV, the prototype virus of the Zaire ebolavirus species) centered in Guinea, Liberia, and Sierra Leone has resulted in over 27,000 confirmed cases of Ebola virus disease (EVD) and 11,246 deaths (1), with a number of cases distributed across the globe by travelers. In contrast, from their discovery in 1976 until 2013, the five distinct filoviruses within the genus Ebolavirus (Ebola virus [EBOV], Bundibugyo virus [BDBV], Reston virus [RESTV], Sudan virus [SUDV], and Taï Forest virus [TAFV], each representing an Ebolavirus species [2]) were responsible for a cumulative total of less than 2,300 cases, almost all within local regions in Africa (3). Despite active research into potential vaccines (4, 5) and therapeutics (6, 7), no prophylactic or postinfection therapeutics are yet approved for use against ebolaviruses.
One of the most promising treatments for the often fatal consequences of EBOV infection is the passive administration of antibodies targeting the EBOV surface glycoprotein (GP) (reviewed in references 8 and 9). This was first demonstrated in nonhuman primates in which immunoglobulin from an EBOV-surviving macaque conferred protection in rhesus macaques when administered 2 days after infection with EBOV (10). Numerous studies have shown that treatment with monoclonal antibodies (MAbs) can confer postexposure protection and that their effectiveness is enhanced by their application in combination as a “cocktail” (10–16).
Two of the most studied anti-EBOV cocktails are ZMAb (MAbs 2G4, 4G7, and 1H3) (14, 17) and MB-003 (MAbs 13C6, 6D8, and 13F6) (12, 13). All six MAbs in these cocktails were isolated following immunization of mice, and both cocktails provided some level of protection against EBOV in mice, guinea pigs, and nonhuman primates (12, 13, 17). These MAbs have also been tested individually in animal models, with MAbs 4G7 and 13C6 usually offering relatively better protection, and 2G4, 1H3, 6D8, and 13F6 offering variable protection depending on the animal model and conditions (14, 17, 18). After optimization of the different MAb combinations, two antibodies from ZMAb (2G4 and 4G7) were combined with one MAb from MB-003 (13C6) to create the more potent cocktail ZMapp that reversed clinical signs in six out of six rhesus macaques when given as late as 5 days after EBOV exposure (18).
Based on their success in nonhuman primates, ZMapp and ZMAb have been used under emergency compassionate protocols in humans to treat EBOV infections originating from the 2014 EVD outbreak in western Africa (19). At least seven patients have now been treated with ZMapp, with five surviving (20–22), and six patients have been treated with ZMAb, with all surviving (Gary Kobinger, personal communication). All administrations were reported as well tolerated. It is not clear if the survival of these patients can be directly attributed to treatment with the MAb cocktails, but due to their promise for treating humans, the production and clinical testing of anti-EBOV cocktails are now being rapidly accelerated (23).
Despite the importance of these cocktail MAbs, the mechanisms by which they inhibit GP are not yet well characterized, including exactly how and where they bind GP. On the EBOV surface, GP forms a chalice-shaped trimer, with each constituent monomer comprising GP1 and GP2 derived from proteolytic cleavage of GP (24). The general regions of binding for four cocktail antibodies have been localized on GP by single-particle electron microscopy (EM) at low resolution (21 to 24 Å) (25). Viral escape mutations have also identified three residues (I274, W275, and Q508) that cause resistance to MAb neutralization in replicating EBOV (14, 25, 26). However, the amino acids that form the detailed binding epitopes for all six cocktail MAbs have yet to be identified. Other binding properties that define how each MAb is functionally interacting with GP—such as affinity, epitope conservation, and epitope accessibility—also remain largely unknown. More broadly, conformationally complex epitopes on GP remain highly undercharacterized, with only two cocrystal structures and three individual amino acids determined by virus escape mutation (http://www.iedb.org/).
In this study, we determined the epitope binding properties of all six MAbs in the ZMapp, ZMAb, and MB-003 cocktails, as well as the reference MAb KZ52. Comprehensive alanine scanning across 641 residues of EBOV GP was used to determine the specific epitope residues for each MAb, and results were validated by neutralization escape. Results were combined with previously published single-particle EM data to define the complete footprint of each MAb and the energetically critical “hot-spot” residues within the epitope. Our epitope mapping results indicate that each of the six therapeutic MAbs binds nonidentical epitopes in the GP base, glycan cap, or mucin-like domain. Whole virion binding assays were used to define the apparent affinity of each MAb, with the mucin-like domain MAbs binding the strongest, possibly helping to explain their effectiveness in vivo despite their inability to neutralize virus. Our data help explain why MAbs 4G7 and 13C6 are the most protective in animals, based on their apparent affinity, epitope complementarity, and epitope accessibility. Our results also predict how to improve the effectiveness of new EBOV MAb cocktails.
MATERIALS AND METHODS
Antibodies.
MAbs 2G4, 4G7 and 1H3 (ZMAb, obtained as individual murine IgGs, was a kind gift of Gary Kobinger and Xiangguo Qiu) were previously obtained following immunization of mice with packaged Venezuelan equine encephalitis virus replicons encoding the EBOV Mayinga isolate GP or with vesicular stomatitis virus (VSV) with the VSV glycoprotein gene replaced with one encoding EBOV Mayinga isolate GP (27, 28). MB-003 cocktail MAbs 13C6, 6D8, and 13F6 (IgGs obtained as mouse variable regions chimerized with human constant regions and produced from tobacco [Nicotiana benthamiana]; purchased from IBT Bioservices) were originally derived from mice injected subcutaneously with Venezuelan equine encephalitis virus replicons encoding EBOV Mayinga isolate GP (27). All MAbs are referred to in the text by their original names, irrespective of chimerization.
Construction of EBOV GP mutation library.
As previously described for other viral envelope proteins (29–31), comprehensive high-throughput alanine scanning (shotgun mutagenesis) was carried out on an expression construct for EBOV GP (Ebola virus H.sapiens-tc/COD/1976/Yambuku-Mayinga [2]; UniProt accession number Q05320) with a C-terminal V5-His tag. Residues 33 to 676 of full-length EBOV GP were mutagenized to create a library of clones, each representing an individual point mutant. Residues were changed to alanine (with alanine residues changed to serine), which provides a controlled method of defining the side chain contributions of each residue. GP residues 1 to 32, which constitute the GP signal peptide, were not mutagenized. The resulting EBOV GP alanine scan library covered 99.5% of target residues (641 of 644). Each individual mutation was confirmed by DNA sequencing, and clones were arrayed into 384-well plates, one mutant per well.
Immunofluorescence assay.
The EBOV GP mutation library, arrayed in 384-well microplates, was transfected into HEK-293T cells and allowed to express for 22 h. The immunoreactivity of each MAb was first optimized by determining reactivity with fixed or unfixed cells over a range of MAb concentrations to identify optimal signal-to-background ratios (>5:1) and to ensure that signals were within the linear range of detection. Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) plus calcium and magnesium, or were left unfixed, and were then incubated with an anti-EBOV MAb diluted in 10% normal goat serum (NGS; Sigma-Aldrich, St. Louis, MO). The cells were incubated with anti-EBOV antibody for 1 h at room temperature, followed by a 30-min incubation with Alexa Fluor 488-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) in 10% NGS. Cells were washed twice with PBS without calcium or magnesium (PBS−/−) and resuspended in Cellstripper (Cellgro, Manassas, VA) plus 0.1% bovine serum albumin (BSA; Sigma-Aldrich, St. Louis, MO). Cellular fluorescence was detected using an Intellicyt high-throughput flow cytometer (Intellicyt, Albuquerque, NM). Background fluorescence was determined by fluorescence measurement of vector-transfected control cells. MAb reactivities against each mutant EBOV GP clone were calculated relative to wild-type EBOV GP reactivity by subtracting the signal from mock-transfected controls and normalization to the signal from wild-type GP-transfected controls.
Epitope identification.
Mutated residues within critical clones were identified as critical to the MAb epitope if they did not support reactivity of the test MAb but did support reactivity of other control EBOV MAbs. This counterscreen strategy facilitates the exclusion of GP mutants that are locally misfolded or that have an expression defect (30, 32). The detailed algorithms used to interpret shotgun mutagenesis data are described elsewhere (Kahle KM, Doranz BJ, patent application 61/938,894 and reference 33). All epitopes solved here have been deposited with the Immune Epitope Database (submission ID 1000673; http://www.iedb.org/) (34).
Infectivity and neutralization assays.
Lentiviral reporter pseudotypes with EBOV GP (Yambuku-Mayinga isolate) were produced essentially as described previously (35, 36) by cotransfecting EBOV GP plasmid (or mutants thereof) with plasmids encoding HIV-1 core (gag-pol, based on reference 37) and luciferase (pNL-luc, based on pNL4-3-R−E− [38]). Cells were incubated at 37°C in 5% CO2 to allow for transfection and pseudotype production. Forty-eight to 72 h posttransfection, supernatants were harvested and stored at −80°C. Target HEK-293T cells were plated at 0.4 × 105 cells/well in Dulbecco's modified Eagle medium (DMEM; ThermoScientific, Waltham, MA) containing additives and incubated at 37°C in 5% CO2 overnight. The following day, serial dilutions of MAb and pseudotypes preincubated for 45 min were added to the HEK-293T cells. Cells were then incubated at 37°C. At 24 h postransduction, 100 μl of fresh medium was added to each well. Transduced target cells were lysed 72 h postinfection, and lysates were assayed for luciferase activity (Promega, Madison, WI).
ELISA binding assays.
Ninety-six-well white, flat-bottom microtiter plates were coated with murine leukemia virus Gag-based virus-like particles (“lipoparticles”) pseudotyped with EBOV GP or EBOV Δmucin GP (lacking residues 313 to 462) at 0.5 μg (2 U)/well and incubated overnight at 4°C. The plates were blocked with 3% BSA (Sigma) for 15 min at room temperature. Primary MAb was added to the plates and allowed to incubate for 1 h at room temperature. The plates were washed three times with PBS-/-, and then horseradish peroxidase (HRP)-conjugated rabbit anti-human secondary antibody diluted at 1:5,000 was added for 30 min at room temperature. The plates were washed three times with PBS-/-, and reactivity was detected using SuperSignal West Pico chemiluminescent substrate (ThermoScientific, Waltham, MA).
RESULTS
Identification of critical residues on GP required for EBOV MAb binding.
The most protective combination of MAbs in an antiviral cocktail involves balancing epitope exposure and accessibility, epitope complementarity, epitope conservation, the functional importance of the targeted residues, and binding affinity. To help define these epitope features for the therapeutic EBOV MAbs currently in use, the residues required for binding of each MAb were first determined. We accomplished this by using comprehensive alanine scanning, in which MAb binding was assessed against a shotgun mutagenesis mutation library of EBOV GP with 641 of 644 target residues mutated. The entire mutation library was transfected into human HEK-293T cells in a 384-well array format (one clone per well) and assessed for immunoreactivity using high-throughput flow cytometry (Fig. 1A). Five hundred eighty-six of the 641 alanine scan mutants (91%) were expressed at >70% of the wild type at the cell surface (by MAb KZ52 surface staining). The expression of full-length GP1/GP2 in human cells and detection of the complex at the cell surface help ensure that only correctly folded GP is used for epitope mapping.
FIG 1.
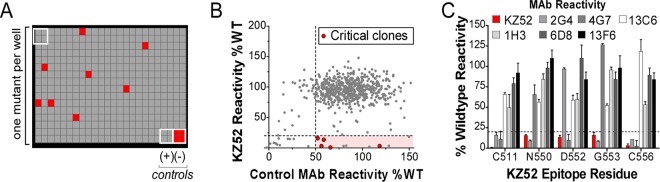
Shotgun mutagenesis epitope mapping of EBOV MAbs. (A) A mutation library for EBOV envelope protein encompassing 641 GP mutations was constructed in which each amino acid was individually mutated. Residues were changed to alanine (with alanine residues changed to serine) to provide a controlled method for defining the side chain contributions of each residue. Each well of each mutation array plate contains cells expressing one mutant with a defined substitution. A representative 384-well plate of reactivity results is shown. Eight positive (wild-type GP) and four negative (mock-transfected) control wells are included on each plate. Wells low in signal (<20% of wild type) are colored red. (B) Human HEK-293T cells expressing the EBOV envelope mutation library were tested for immunoreactivity with MAb KZ52 and a control MAb (13C6 shown) and measured using an Intellicyt high-throughput flow cytometer. The highlighted region identifies clones with high GP expression but low KZ52 binding, and clones that were confirmed as critical are shown in red. Other clones in the highlighted region were not confirmed as critical based on likely misfolding. (C) Mutation of five individual residues reduced KZ52 binding (red bars) but did not affect binding of most other MAbs (gray, black, and white bars). Error bars represent the mean and range (half of the maximum minus minimum values) of at least two replicate data points.
Residues critical for each MAb epitope were initially identified as those where GP mutations resulted in less than 20% reactivity for the MAb of interest (relative to wild-type EBOV GP) yet greater than 50% wild-type binding by a reference MAb (Fig. 1B). Residues were further validated as critical by comparing their reactivities across the panel of MAbs to verify that the mutation did not globally disrupt the binding of diverse MAbs (Fig. 1C). By testing each MAb against the comprehensive alanine scan mutation library, we systematically mapped the detailed epitopes of each of the MAbs in the ZMapp, ZMAb, and MB-003 cocktails (Table 1).
TABLE 1.
Residues critical for EBOV MAb bindinga
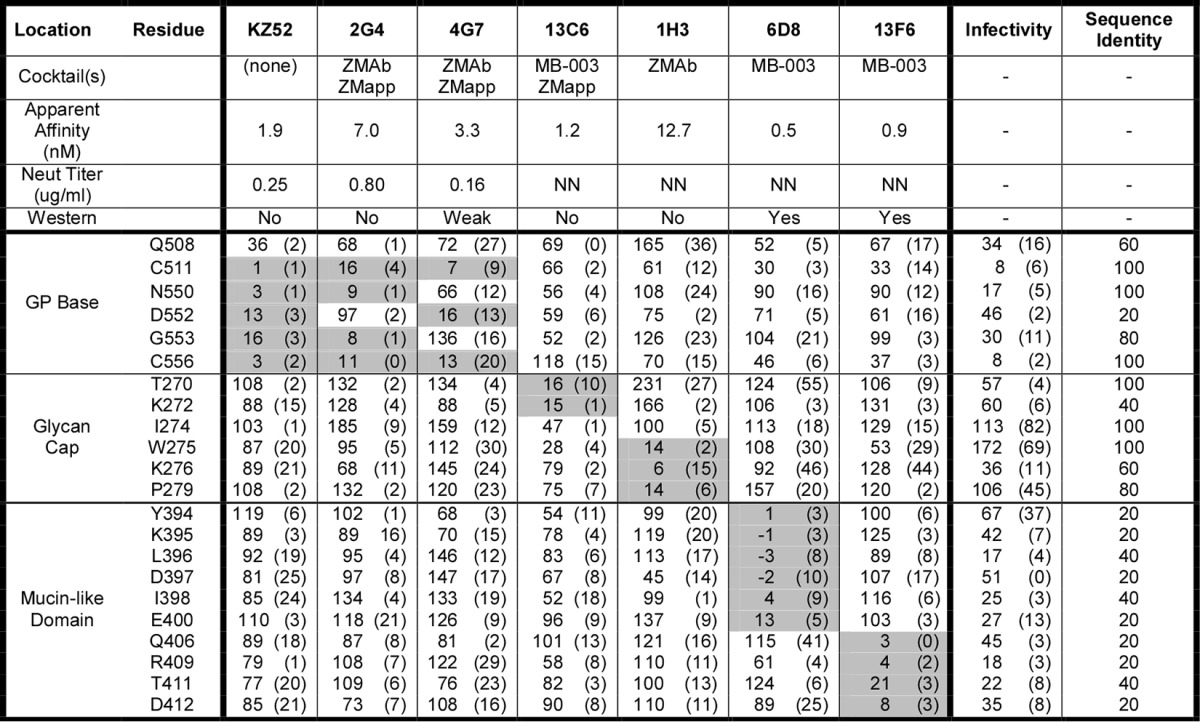
Summary data for EBOV GP MAbs are shown. MAb reactivities for each alanine scan mutant are expressed as a percentage of wild-type reactivity with ranges (half of the maximum minus minimum values) in parentheses. Values for critical residues are shaded in gray. At least two replicate values were obtained for each experiment. The infectivity of each mutant and neutralization (Neut) titer of each MAb were determined using lentiviral reporter pseudotypes. The percent sequence identity at individual residues was determined by comparing sequences representing the five Ebolavirus species (EBOV, BDBV, TAFV, SUDV, and RESTV). The ability of each MAb to detect GP by Western blotting (an indication of the conformational or linear nature of its epitope) is indicated in references 27 and 28. NN, nonneutralizing.
Epitope mapping of protective EBOV antibodies.
Our results indicate that the seven MAbs tested bind to three different regions of GP—the base of GP, the tip of the glycan cap, and the heavily glycosylated mucin-like domain. For MAb 2G4, we identified four critical residues at the GP base whose mutation impaired MAb binding to wild-type GP: C511, N550, G553, and C556 (Fig. 2A). MAb 4G7 bound in the same area but instead used critical residue D552 (Fig. 2B). Binding of all GP base MAbs was affected by mutation of C511 or C556, suggesting that elimination of the C511-C556 disulfide bond disrupts the structure of this entire epitope region. The overlapping footprint of the GP base MAbs explains why they compete with each other for binding (25), but the different residues that they each engage suggests that there are important functional differences between these MAbs in how they bind GP, their breadth of reactivity against ebolavirus species, and how susceptible each MAb may be to viral evasion.
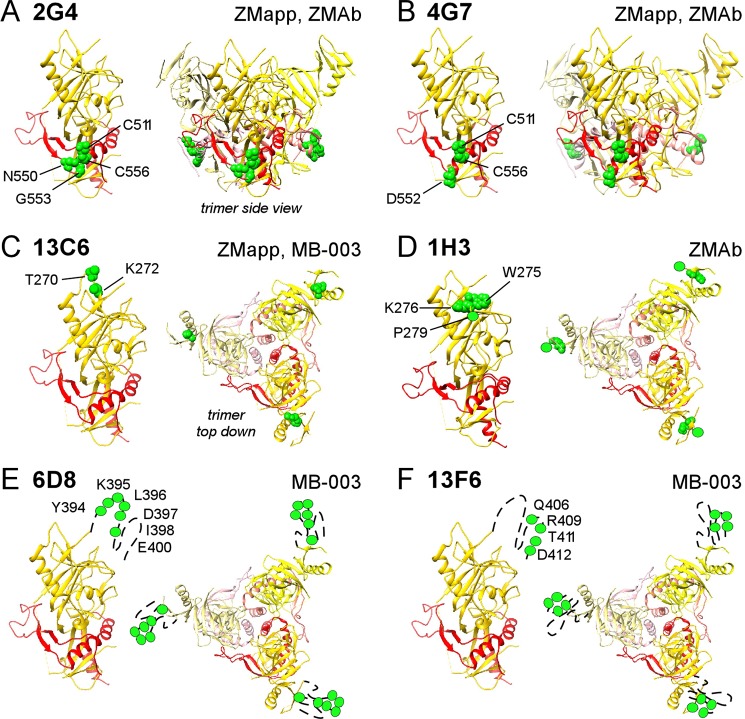
FIG 2.
Epitope mapping of EBOV cocktail MAbs. GP residues critical for binding each MAb are shown in green on one GP monomer (left diagram of each panel) and the GP trimer (right diagram of each panel) of the EBOV Δmucin GP structure (PDB accession no. 3CSY) (24) for MAbs 2G4 (A), 4G7 (B), 13C6 (C), 1H3 (D), 6D8 (E), and 13F6 (F). The locations of residue P279 (D) and residues in the mucin-like domain (E and F) are approximated, as the structures of these regions (residues 279 to 298 and mucin-like domain residues 313 to 464) are not yet solved. Also indicated (top right of each panel) are the cocktails that contain the MAb.
For MAb 13C6, we identified T270 and K272 as critical binding residues in an exposed area at the tip of GP1 glycan cap distal from the viral membrane (Fig. 2C). MAb 1H3 also bound to the GP1 glycan cap, but using residues W275, K276, and P279 (Fig. 2D). The locations of these epitopes are in agreement with their general location by cryo-EM and help explain binding competition results (25).
Finally, for MAb 6D8, we identified consecutive residues Y394, K395, L396, D397, and I398 and adjacent residue E400 as critical for binding within the GP mucin-like domain (Fig. 2E). MAb 13F6 bound close by, to nonoverlapping residues Q406, R409, T411, and D412 (Fig. 2F). The location of these residues is consistent with low-resolution cryo-EM localization of these epitopes within the mucin-like domain (25, 39), the reactivity of the MAbs to peptides derived from the mucin-like domain (27), the crystal structure of 13F6 with a peptide substrate (40), and the inability of peptides containing Q406A or R409A mutations to compete with 13F6 binding to wild-type GP (40). Glycosylation across the mucin-like domain is expected to shield many otherwise immunogenic sequences of this domain, but our results suggest that these particular residues are accessible within the mucin-like domain.
Hot-spot binding residues for the cocrystallized human MAb KZ52.
To further understand the implications of our results, we also epitope mapped the conformationally sensitive MAb KZ52. KZ52 is currently one of the only MAbs cocrystallized with GP (24) and therefore is an important structural reference. We identified as critical five (C511, N550, D552, G553, and C556) of the 15 GP residues in close proximity to KZ52 in the cocrystal structure (<3.9 Å in PDB structure 3CSY) (Fig. 3). Thus, the epitope for KZ52 is very similar to the 2G4 epitope but also includes D552. KZ52 has been postulated to neutralize EBOV by preventing the rearrangement of GP2 helical regions HR1A and HR1B in later stages of infectivity (24). The localization of all critical epitope residues for KZ52 (as well as for 2G4 and 4G7) at the interface of GP1/GP2 is consistent with this mechanism of action. Because KZ52 was derived directly from a human patient (41), its epitope at the GP base and mechanism of action represent part of the native human B-cell response against EBOV.
FIG 3.
Mapping of MAb KZ52 identifies the energetically critical epitope residues of the interaction. (A) Residues are shown on the crystal structure of EBOV Δmucin GP (PDB accession no. 3CSY) (24). One monomer is shown, with GP1 colored gold and GP2 colored red. KZ52 contact residues identified from crystallographic analysis (left diagram) are shown in blue. The residues identified here by mutagenesis are shown in green (right diagram). (B) The KZ52 epitope (residues identified here in green) forms a conformational epitope that lies at the interface of GP1 and GP2 at the base of the GP trimer (monomers indicated as I, II, III).
Our data suggest that residues N550, D552, and G533 on GP2 and in the approximate center of all the contact residues form the “functional epitope” for KZ52, often called hot-spot residues that provide the primary energetic interactions of an epitope and are usually surrounded by energetically less important contact residues (42, 43). Their energetic importance is supported by the cocrystal structure, where N550, D552, and G553 form hydrogen bonds with KZ52 heavy-chain complementarity-determining regions (CDRs) (24) (Table 2). The 10 other KZ52 contact residues on GP surrounding the hot-spot residues were not identified as critical for binding by KZ52 (or 2G4 or 4G7), although Q508 significantly reduced the binding of KZ52 (but not beyond our 20% threshold; see Discussion). Many of these residues form energetically less significant van der Waals interactions with KZ52, consistent with their inability to disrupt MAb binding when mutated to alanine. Cysteine residues C511 and C556 also form weaker van der Waals interactions with KZ52 in the cocrystal structure but were identified as critical residues by shotgun mutagenesis likely because the disulfide bond they form is required to pin together a loop and α-helix that contain the epitope. It is notable that cocrystallography alone cannot directly discriminate which residues are the most energetically important among the contact residues, and even contact residues in cocrystallography are inferred primarily by distance, so our results are highly complementary to structural analysis.
TABLE 2.
Comparison of KZ52 cocrystal contacts to binding dataa
All 15 predicted contact residues (<3.9 Å) from the KZ52-GP cocrystal structure (23) are shown, as well as the type of bond identified from analysis of the crystal structure and the KZ52 immunoreactivity data (with ranges [the maximum minus minimum values]) derived in the current study. Residues identified as critical for binding are shaded in gray. All residues in the cocrystal structure are predicted to make hydrogen bonds and/or van der Waals interactions, as indicated; no salt bridges are formed in the cocrystal structure.
b Wild-type residue T42 (T42A in our mutation library) was mutated to V42 in the GP crystal structure to eliminate glycosylation at N40 (23).
c Contact of the indicated residues with KZ52 is only through alpha-carbon interactions on GP, so mutation to alanine is less likely to disrupt the MAb-GP interaction.
Validation of antibody epitopes by single-particle EM and neutralization escape.
Previous results using low-resolution EM localized the general footprint and angle of antibody binding for four cocktail MAbs of interest (25). These results were compared to our mutagenesis results to resolve the specific epitope for these antibodies and validate both results using independent approaches. Interestingly, for each of the four EM footprints available, the critical residues identified by shotgun mutagenesis were at the approximate center of the EM footprint (Fig. 4). These results suggest that the residues that we identified by mutagenesis are the energetically critical hot-spot residues for each MAb.
FIG 4.
Visualization of epitopes within the EM footprint. Visualization of epitopes obtained by mutagenesis with the Fab footprints from electron microscopy (25) suggest that the critical residues identified by mutagenesis are the energetically critical hot-spot residues at the center of the MAb epitope. The EM footprint residues (purple) and mutagenesis hot-spot residues (green) are shown in the context of the Fab EM density information (solid structures). Docking of each MAb to GP was obtained by fitting the EBOV GP crystal structure (25) into the structural information for the EM reconstructions of Fabs bound to EBOV GP, identified by the EMDataBank ID emd-6151 (2G4) (A), emd-6152 (4G7) (B), emd-6152 (13C6) (C), or emd-6150 (1H3) (D) (25). Insets show the view of the footprints and hot spots from the angle of the Fab. The location of residue P279 (D) is approximated, as the structure of this region (residues 279 to 298) is not yet solved.
To further verify the epitopes identified from binding assays, key critical residues were tested for escape from neutralization. All critical mutants were first tested for infectivity (Table 1). Interestingly, many of the mutants identified in our studies demonstrate no or low infectivity with just a single alanine substitution, indicating that these epitope residues are important for GP functionality and suggesting that binding by MAbs to these functionally important locations could inhibit infectivity. The low infectivity of these critical residues also implies that these residues might be difficult to identify using traditional viral escape strategies using live virus that may not be viable with these substitutions.
All MAbs were tested for the ability to neutralize the infectivity of lentiviral reporter pseudotypes, but only the three GP base MAbs (KZ52, 2G4, and 4G7) demonstrate any significant neutralizing activity against wild-type EBOV GP in vitro (Fig. 5), consistent with prior reports. Based on their ability to support infectivity and their importance for binding by neutralizing GP base MAbs, D552A and G553A critical mutants were chosen to test their resistance to the three neutralizing MAbs. Neutralization results mirrored binding results. Mutation D552A eliminated EBOV neutralization by MAbs KZ52 and 4G7, while mutation G553A eliminated neutralization by MAbs KZ52 and 2G4, confirming that these residues are indeed a critical part of their respective epitopes, as binding results suggest. Thus, results from neutralization and single-particle EM corroborate mutagenesis localization of the critical residues for each MAb.
FIG 5.
Neutralization escape studies validate epitope residues. MAbs were tested for their ability to neutralize the infectivity of lentiviral reporter pseudotypes with full-length EBOV GP. Reporter pseudotypes were preincubated with MAbs, and infection of HEK-293T target cells was detected by the expression of Renilla luciferase. (A) MAbs 13C6, 1H3, 6D8, and 13F6 demonstrated no neutralization at the concentrations tested. (B to D) Known neutralizing MAbs KZ52 (B), 2G4 (C), and 4G7 (D) were tested for neutralization of reporter pseudotypes with wild-type (WT) EBOV GP and with GPs containing mutants critical for binding by KZ52 (D552A, G553A), 2G4 (G553A), and 4G7 (D552A). Data points represent the mean of three replicates (± standard deviation), and data are representative of two independent experiments.
Epitope conservation among ebolaviruses.
With the critical residues for each MAb identified, we next determined how these specific epitope residues differ between species of ebolavirus and currently circulating isolates of EBOV. The reactivity of each MAb was first tested using cells expressing GP from EBOV, BDBV, TAFV, and SUDV ebolaviruses, as well as EBOV Δmucin GP. Of the seven MAbs tested, only 2G4 and 13C6 showed cross-reactivity with any ebolaviruses other than EBOV, with some reactivity against TAFV (>3:1 signal-to-background ratio) (Fig. 6). Interestingly, MAb 2G4 has an epitope nearly identical to that of KZ52, except that 2G4 does not require residue D552. In contrast, D552 was the primary critical residue for MAb 4G7, and this MAb was also determined to bind specifically to EBOV. Therefore, we propose that residue D552, which is present only in EBOV GP, contributes to the ebolavirus reactivity restriction of GP base MAbs KZ52 and 4G7.
FIG 6.
MAb cross-reactivity with Ebolavirus species GP. MAbs were tested by flow cytometry for immunoreactivity with HEK-293T cells transfected with constructs expressing GP from EBOV, BDBV, TAFV, or SUDV, an EBOV Δmucin GP construct, or empty vector. All ebolavirus GPs were fully reactive with a control MAb against the V5 epitope tag included on the C terminus of each protein as well as with other anti-GP MAbs (not shown). Data shown represent the mean and standard deviation of at least four data points. RFU, relative fluorescence units.
We next compared MAb epitope residues to the sequences of circulating isolates of EBOV. The GP of the 1976 EBOV Yambuku-Mayinga isolate used in our studies differs at 19 residues from the GP of the EBOV Makona isolates from the 2013-to-present day EVD outbreak (44–46) (Fig. 7A). However, only one of the GP sequence changes is at a critical residue for the epitopes we identified, i.e., mutation T411A in the mucin-like domain (Fig. 7B). Since this residue is critical only for MAb 13F6 in the MB-003 cocktail, this suggests that the most important cocktails (ZMapp and ZMAb) should be as effective against the currently circulating Makona variants of EBOV as they have been against the Kikwit EBOV isolate tested in preclinical studies.
FIG 7.
Conservation of epitope residues. (A) Diagram of EBOV GP1 and GP2 showing the location of 19 residue changes (red circles) that have occurred between the EBOV Makona isolate from the 2013-to-present day outbreak and the 1976 EBOV Yambuku-Mayinga isolate (45). The locations of critical epitope residues identified in our study are shown in green. SP, signal peptide; TM, transmembrane region. (B) Alignment of the GP domains representing the five species of ebolaviruses (EBOV, BDBV, TAFV, SUDV, and RESTV), showing the critical epitope residues for ZMapp, ZMAb, and MB-003 MAbs. The epitope residues identified for the indicated MAbs are highlighted green, and the locations of residues altered by changes in EBOV are marked with red circles. Conservation among the five ebolaviruses is shown below the alignment: asterisks indicate complete conservation, colons indicate conservative mutations, and periods indicate semiconservative mutations.
Reactivity and relative affinity of EBOV MAbs.
To further define their binding mechanism, each MAb was tested for relative affinity by enzyme-linked immunosorbent assay (ELISA) using retroviral virus-like particles (VLPs) pseudotyped with wild-type EBOV GP. Wild-type GP pseudotyped on retroviral VLPs is functionally active (similar particles were used for infectivity assays [Table 1]), so these data represent the apparent affinity (Kd,app) of the MAbs and maximum number of GP molecules recognized (Bmax) on these particles. Interestingly, our results suggest that the mucin-like domain MAbs 6D8 and 13F6 bind GP with the highest affinity (0.5 to 0.9 nM Kd,app), as well as recognizing the most numbers of GP molecules on the virion surface (Bmax, 3.2 × 106 to 3.5 × 106), compared to the other MAbs (Fig. 8A and Table 3). The high affinity of the mucin-like domain MAbs and their ability to recognize the most numbers of GP molecules on the virion surface help explain their contribution to protection in vivo despite their lack of neutralization in vitro.
FIG 8.
Relative binding affinities of EBOV cocktail MAbs. ELISAs using VLPs pseudotyped with EBOV wild-type GP (A) or EBOV Δmucin GP (B) were used to test 2-fold dilutions of each MAb to compare apparent binding affinities against GP on virions. VLPs were also tested for reactivity with a V5 antibody against a V5 epitope tag incorporated onto the C terminus of EBOV WT GP and EBOV Δmucin GP (dotted line).
TABLE 3.
Apparent affinity and maximal binding of MAbs to EBOV WT GP and EBOV Δmucin GPa
| MAb | EBOV WT GP |
EBOV Δmucin GP |
||
|---|---|---|---|---|
| Kd,app (nM) | Bmax signal (105) | Kd,app (nM) | Bmax signal (105) | |
| KZ52 | 1.9 ± 0.5 | 4.7 ± 0.6 | 1.9 ± 0.8 | 26 ± 0.4 |
| 2G4 | 7.0 ± 3.8 | 13 ± 2.7 | 2.8 ± 0.4 | 39 ± 9.0 |
| 4G7 | 3.3 ± 0.5 | 6.6 ± 2.6 | 1.3 ± 0.3 | 26 ± 5.0 |
| 13C6 | 1.2 ± 0.2 | 17 ± 1.7 | 0.4 ± 0.1 | 39 ± 3.9 |
| 1H3 | 12.7 ± 4.0 | 6.6 ± 1.0 | 4.2 ± 0.3 | 26 ± 5.7 |
| 6D8 | 0.5 ± 0.1 | 32 ± 0.0 | ND | ND |
| 13F6 | 0.9 ± 0.4 | 35 ± 3.7 | ND | ND |
| V5 | NA | 20 ± 0.4 | NA | 39 ± 1.49 |
The values for apparent affinity (Kd,app) and maximal binding (Bmax, shown as relative fluorescence units) were derived from the ELISAs using VLPs pseudotyped with EBOV WT GP or EBOV Δmucin GP, as shown in Fig. 8. Values are averages (and ranges) of two experiments. NA, not applicable; ND, not detectable.
Because the mucin-like domain is removed by cellular cathepsin cleavage during virus endocytosis (and often also experimentally to improve GP expression), each MAb was also tested for relative affinity using retroviral VLPs pseudotyped with EBOV Δmucin GP. EBOV Δmucin GP was expressed approximately 2-fold better than wild-type GP, as measured using an identical V5 epitope tag on both constructs (Fig. 8B). Independent of the increased expression of EBOV Δmucin GP, MAbs 13C6 and 1H3 against the glycan cap demonstrated a 3-fold-better Kd,app against EBOV Δmucin GP (Table 3). This suggests that exposure of glycan cap epitopes is enhanced when the mucin-like domain is removed. As EBOV Δmucin GP is commonly used by laboratories due to its high expression levels, our results suggest that its use could result in systematic biases (e.g., artificially increased glycan cap reactivity), for example, in serum reactivity experiments.
DISCUSSION
The mechanisms of inhibition for the MAbs in the ZMapp, ZMAb, and MB-003 cocktails are not well understood, despite their compassionate use as therapeutics. Here we identified the critical binding properties for all six of the MAbs being used in therapeutic antibody cocktails, as well as the human reference MAb KZ52. Five of the seven epitopes mapped here are conformational in nature so could not have been fully mapped with peptides or denatured protein. With only limited epitopes for any ebolavirus MAbs characterized previously, our results significantly enhance our understanding of the humoral immune response against ebolaviruses.
Our shotgun mutagenesis approach identifies epitopes irrespective of MAb neutralization status or viral fitness, so it is not limited to inhibitory MAbs or to mutations that are compatible with virus replication. Nonetheless, this strategy cannot detect the contribution of alpha carbons to an interaction, mutations other than those to alanine could result in somewhat different results, and the ability to differentiate direct from indirect or allosteric effects on MAb interactions is dependent on the number and diversity of other available control MAbs. However, the concordance of mutagenesis data, single-particle EM data, and neutralization escape data provides high confidence that the specific epitope residues identified here are accurate. Other residues may also be involved in each epitope, but their mutation to alanine did not disrupt the energetics of binding significantly enough to be identified as critical.
Previously, viral escape studies identified the mutations I274M, W275L, Q508R, and Q508H, which confer resistance to MAb neutralization in replicating EBOV (14, 25, 26). Consistent with these studies, we identified W275, as well as nearby K276 and P279, as energetically important for 1H3 binding (and significantly affecting 13C6 binding, although not below our chosen 20% threshold) (Table 1). However, our I274A mutation had no apparent effect on 1H3 binding and only a modest effect on 13C6 binding, possibly due to the different type of residue substitution. Similarly, our Q508A mutation significantly reduced the binding of KZ52 but not beyond our 20% threshold. However, Q508 is known to contact KZ52 only through alpha carbon interactions (Table 2), which our methodology may not be able to detect, and we did not test the more structurally disruptive Q508R or Q508H substitutions.
Our data predict that MAbs KZ52, 4G7, and 13C6 may be particularly susceptible to viral evasion, since critical residues within each of their epitopes (D552 and K272) are relatively variable among ebolaviruses (Table 1). Conversely, MAbs 2G4 and 1H3 may be more resistant to viral evasion, since critical residues within each of their epitopes are relatively conserved among ebolaviruses. However, none of the MAbs tested are highly cross-reactive between ebolaviruses or demonstrate absolutely conserved epitopes, suggesting that improved MAbs with more conserved epitopes and higher cross-reactivity would be better for inclusion in protective MAb cocktails.
The ZMapp cocktail combines the two best MAbs from ZMAb (2G4 and 4G7) with the best MAb from MB-003 (13C6) (18) (Fig. 9). Thus, ZMapp and ZMAb are identical except for the replacement of 1H3 with 13C6 in the more protective ZMapp cocktail; both MAbs bind at structurally adjacent residues at the tip of the glycan cap. Our results suggest that MAb 13C6 provides better protection in vivo than MAb 1H3 primarily due to its stronger apparent affinity (10-fold better for both wild-type GP and EBOV Δmucin GP).
FIG 9.
Inhibitory epitopes on EBOV GP. Residues identified as critical for the MAbs assayed in this study are represented on the crystal structure of EBOV Δmucin GP (PDB accession no. 3CSY) (24). (A) All epitopes. (B) MB-003 cocktail epitopes. (C) ZMAb cocktail epitopes. (D) ZMapp cocktail epitopes. For reference, the epitope for KZ52 is highlighted in panel A, overlapping the 2G4 (red outline) and 4G7 (green) epitopes. For each cocktail (B to D), a value for the percent survival is indicated, obtained from a direct comparison of the results of the cocktails in guinea pigs (5-mg doses given 3 days postinfection) (18).
Epitopes at the base of GP have inherent advantages in neutralization that may be informative for both vaccine design and therapeutic development. The glycan cap and mucin-like domain epitopes are removed from the trimer by cathepsin cleavage in the host endosome prior to GP interaction with receptors and membrane fusion (47, 48). Thus, MAbs that bind the glycan cap and mucin-like domain likely neutralize EBOV by using complement, antibody-dependent cell-mediated cytotoxicity (ADCC), or other Fc-mediated mechanisms. The identification of epitopes for KZ52, 2G4, and 4G7 in the base of GP, as well as the localization of another neutralizing MAb (16F6) to the equivalent region in SUDV (49, 50), suggests that the base of GP contains highly protective epitopes. It is also possible that the specific epitope residues identified here at the base of GP could serve as structural targets for small-molecule inhibition.
The “best” combination of MAbs in a cocktail will involve balancing affinity, epitope accessibility, epitope conservation, epitope complementarity, and the functional importance of the targeted residues. Our results help define these epitope features of the most important therapeutic MAbs against EBOV developed to date and can be used to rationally design better cocktails of MAbs with potentially better neutralizing potency, epitope complementarity, and resistance properties.
ACKNOWLEDGMENTS
This work was supported by NIH contract HHSN272201400058C.
We thank David Tucker, Silveria Rodriguez, Soma Banik, and Andrew McNeal for valuable technical assistance. We thank Gary Kobinger and Xiangguo Qiu for helpful advice and for providing MAbs 2G4, 4G7, and 1H3.
REFERENCES
- 1.World Health Organization. 2015. Ebola situation report—8 July 2015. World Health Organization, Geneva, Switzerland: http://apps.who.int/ebola/current-situation/ebola-situation-report-8-july-2015. [Google Scholar]
- 2.Kuhn JH, Andersen KG, Bao Y, Bavari S, Becker S, Bennett RS, Bergman NH, Blinkova O, Bradfute S, Brister JR, Bukreyev A, Chandran K, Chepurnov AA, Davey RA, Dietzgen RG, Doggett NA, Dolnik O, Dye JM, Enterlein S, Fenimore PW, Formenty P, Freiberg AN, Garry RF, Garza NL, Gire SK, Gonzalez JP, Griffiths A, Happi CT, Hensley LE, Herbert AS, Hevey MC, Hoenen T, Honko AN, Ignatyev GM, Jahrling PB, Johnson JC, Johnson KM, Kindrachuk J, Klenk HD, Kobinger G, Kochel TJ, Lackemeyer MG, Lackner DF, Leroy EM, Lever MS, Muhlberger E, Netesov SV, Olinger GG, Omilabu SA, Palacios G, et al. 2014. Filovirus RefSeq entries: evaluation and selection of filovirus type variants, type sequences, and names. Viruses 6:3663–3682. doi: 10.3390/v6093663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leroy EM, Gonzalez JP, Baize S. 2011. Ebola and Marburg haemorrhagic fever viruses: major scientific advances, but a relatively minor public health threat for Africa. Clin Microbiol Infect 17:964–976. doi: 10.1111/j.1469-0691.2011.03535.x. [DOI] [PubMed] [Google Scholar]
- 4.Marzi A, Feldmann H. 2014. Ebola virus vaccines: an overview of current approaches. Expert Rev Vaccines 13:521–531. doi: 10.1586/14760584.2014.885841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohammadi D. 2015. Ebola vaccine trials back on track. Lancet 385:214–215. doi: 10.1016/S0140-6736(15)60035-6. [DOI] [PubMed] [Google Scholar]
- 6.Roddy P, Colebunders R, Jeffs B, Palma PP, Van Herp M, Borchert M. 2011. Filovirus hemorrhagic fever outbreak case management: a review of current and future treatment options. J Infect Dis 204(Suppl 3):S791–S795. doi: 10.1093/infdis/jir297. [DOI] [PubMed] [Google Scholar]
- 7.Clark DV, Jahrling PB, Lawler JV. 2012. Clinical management of filovirus-infected patients. Viruses 4:1668–1686. doi: 10.3390/v4091668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saphire EO. 2013. An update on the use of antibodies against the filoviruses. Immunotherapy 5:1221–1233. doi: 10.2217/imt.13.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong G, Kobinger GP, Qiu X. 2014. Characterization of host immune responses in Ebola virus infections. Expert Rev Clin Immunol 10:781–790. doi: 10.1586/1744666X.2014.908705. [DOI] [PubMed] [Google Scholar]
- 10.Dye JM, Herbert AS, Kuehne AI, Barth JF, Muhammad MA, Zak SE, Ortiz RA, Prugar LI, Pratt WD. 2012. Postexposure antibody prophylaxis protects nonhuman primates from filovirus disease. Proc Natl Acad Sci U S A 109:5034–5039. doi: 10.1073/pnas.1200409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeitlin L, Pettitt J, Scully C, Bohorova N, Kim D, Pauly M, Hiatt A, Ngo L, Steinkellner H, Whaley KJ, Olinger GG. 2011. Enhanced potency of a fucose-free monoclonal antibody being developed as an Ebola virus immunoprotectant. Proc Natl Acad Sci U S A 108:20690–20694. doi: 10.1073/pnas.1108360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pettitt J, Zeitlin L, Kim do H, Working C, Johnson JC, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, Pauly MH, Whaley KJ, Ingram MF, Zovanyi A, Heinrich M, Piper A, Zelko J, Olinger GG. 2013. Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Sci Transl Med 5:199ra113. [DOI] [PubMed] [Google Scholar]
- 13.Olinger GG Jr, Pettitt J, Kim D, Working C, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, Pauly M, Whaley KJ, Lear CM, Biggins JE, Scully C, Hensley L, Zeitlin L. 2012. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc Natl Acad Sci U S A 109:18030–18035. doi: 10.1073/pnas.1213709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu X, Audet J, Wong G, Pillet S, Bello A, Cabral T, Strong JE, Plummer F, Corbett CR, Alimonti JB, Kobinger GP. 2012. Successful treatment of ebola virus-infected cynomolgus macaques with monoclonal antibodies. Sci Transl Med 4:138ra181. doi: 10.1126/scitranslmed.3003876. [DOI] [PubMed] [Google Scholar]
- 15.Marzi A, Yoshida R, Miyamoto H, Ishijima M, Suzuki Y, Higuchi M, Matsuyama Y, Igarashi M, Nakayama E, Kuroda M, Saijo M, Feldmann F, Brining D, Feldmann H, Takada A. 2012. Protective efficacy of neutralizing monoclonal antibodies in a nonhuman primate model of Ebola hemorrhagic fever. PLoS One 7:e36192. doi: 10.1371/journal.pone.0036192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takada A, Feldmann H, Stroeher U, Bray M, Watanabe S, Ito H, McGregor M, Kawaoka Y. 2003. Identification of protective epitopes on ebola virus glycoprotein at the single amino acid level by using recombinant vesicular stomatitis viruses. J Virol 77:1069–1074. doi: 10.1128/JVI.77.2.1069-1074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu X, Audet J, Wong G, Fernando L, Bello A, Pillet S, Alimonti JB, Kobinger GP. 2013. Sustained protection against Ebola virus infection following treatment of infected nonhuman primates with ZMAb. Sci Rep 3:3365. doi: 10.1038/srep03365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP. 2014. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyon GM, Mehta AK, Varkey JB, Brantly K, Plyler L, McElroy AK, Kraft CS, Towner JS, Spiropoulou C, Stroher U, Uyeki TM, Ribner BS, Emory Serious Communicable Diseases Unit. 2014. Clinical care of two patients with Ebola virus disease in the United States. N Engl J Med 371:2402–2409. doi: 10.1056/NEJMoa1409838. [DOI] [PubMed] [Google Scholar]
- 20.Bishop BM. 2015. Potential and emerging treatment options for Ebola virus disease. Ann Pharmacother 49:196–206. doi: 10.1177/1060028014561227. [DOI] [PubMed] [Google Scholar]
- 21.Meyers L, Frawley T, Goss S, Kang C. 2015. Ebola virus outbreak 2014: clinical review for emergency physicians. Ann Emerg Med 65:101–108. doi: 10.1016/j.annemergmed.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Roos R. (ed). 2014. Experimental Ebola drug may have helped 2 US patients. CIDRAP News and Perspective Center for Infectious Disease Research and Policy, University of Minnesota, Minneapolis, MN: http://www.cidrap.umn.edu/news-perspective/2014/08/experimental-ebola-drug-may-have-helped-2-us-patients. [Google Scholar]
- 23.McCarthy M. 2014. US signs contract with ZMapp maker to accelerate development of the Ebola drug. BMJ 349:g5488. doi: 10.1136/bmj.g5488. [DOI] [PubMed] [Google Scholar]
- 24.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murin CD, Fusco ML, Bornholdt ZA, Qiu X, Olinger GG, Zeitlin L, Kobinger GP, Ward AB, Saphire EO. 2014. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. Proc Natl Acad Sci U S A 111:17182–17187. doi: 10.1073/pnas.1414164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Audet J, Wong G, Wang H, Lu G, Gao GF, Kobinger G, Qiu X. 2014. Molecular characterization of the monoclonal antibodies composing ZMAb: a protective cocktail against Ebola virus. Sci Rep 4:6881. doi: 10.1038/srep06881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, Hart MK. 2000. Epitopes involved in antibody-mediated protection from Ebola virus. Science 287:1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- 28.Qiu X, Alimonti JB, Melito PL, Fernando L, Stroher U, Jones SM. 2011. Characterization of Zaire ebolavirus glycoprotein-specific monoclonal antibodies. Clin Immunol 141:218–227. doi: 10.1016/j.clim.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Christian EA, Kahle KM, Mattia K, Puffer BA, Pfaff JM, Miller A, Paes C, Davidson E, Doranz BJ. 2013. Atomic-level functional model of dengue virus envelope protein infectivity. Proc Natl Acad Sci U S A 110:18662–18667. doi: 10.1073/pnas.1310962110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fong RH, Banik SS, Mattia K, Barnes T, Tucker D, Liss N, Lu K, Selvarajah S, Srinivasan S, Mabila M, Miller A, Muench MO, Michault A, Rucker JB, Paes C, Simmons G, Kahle KM, Doranz BJ. 2014. Exposure of epitope residues on the outer face of the chikungunya virus envelope trimer determines antibody neutralizing efficacy. J Virol 88:14364–14379. doi: 10.1128/JVI.01943-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castelli M, Clementi N, Sautto GA, Pfaff J, Kahle KM, Barnes T, Doranz BJ, Dal Peraro M, Clementi M, Burioni R, Mancini N. 2014. HCV E2 core structures and mAbs: something is still missing. Drug Discov Today 19:1964–1970. doi: 10.1016/j.drudis.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paes C, Ingalls J, Kampani K, Sulli C, Kakkar E, Murray M, Kotelnikov V, Greene TA, Rucker JB, Doranz BJ. 2009. Atomic-level mapping of antibody epitopes on a GPCR. J Am Chem Soc 131:6952–6954. doi: 10.1021/ja900186n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davidson E, Doranz BJ. 2014. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology 143:13–20. doi: 10.1111/imm.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, Peters B. 2015. The immune epitope database (IEDB) 3.0. Nucleic Acids Res 43:D405-412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salvador B, Zhou Y, Michault A, Muench MO, Simmons G. 2009. Characterization of Chikungunya pseudotyped viruses: identification of refractory cell lines and demonstration of cellular tropism differences mediated by mutations in E1 glycoprotein. Virology 393:33–41. doi: 10.1016/j.virol.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ, Bates P. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci U S A 101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 38.Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 39.Tran EE, Simmons JA, Bartesaghi A, Shoemaker CJ, Nelson E, White JM, Subramaniam S. 2014. Spatial localization of the Ebola virus glycoprotein mucin-like domain determined by cryo-electron tomography. J Virol 88:10958–10962. doi: 10.1128/JVI.00870-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JE, Kuehne A, Abelson DM, Fusco ML, Hart MK, Saphire EO. 2008. Complex of a protective antibody with its Ebola virus GP peptide epitope: unusual features of a V lambda x light chain. J Mol Biol 375:202–216. doi: 10.1016/j.jmb.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maruyama T, Rodriguez LL, Jahrling PB, Sanchez A, Khan AS, Nichol ST, Peters CJ, Parren PW, Burton DR. 1999. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J Virol 73:6024–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bogan AA, Thorn KS. 1998. Anatomy of hot spots in protein interfaces. J Mol Biol 280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 43.Lo Conte L, Chothia C, Janin J. 1999. The atomic structure of protein-protein recognition sites. J Mol Biol 285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 44.Gire SK, Goba A, Andersen KG, Sealfon RS, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, Wohl S, Moses LM, Yozwiak NL, Winnicki S, Matranga CB, Malboeuf CM, Qu J, Gladden AD, Schaffner SF, Yang X, Jiang PP, Nekoui M, Colubri A, Coomber MR, Fonnie M, Moigboi A, Gbakie M, Kamara FK, Tucker V, Konuwa E, Saffa S, Sellu J, Jalloh AA, Kovoma A, Koninga J, Mustapha I, Kargbo K, Foday M, Yillah M, Kanneh F, Robert W, Massally JL, Chapman SB, Bochicchio J, Murphy C, Nusbaum C, Young S, Birren BW, Grant DS, Scheiffelin JS, et al. 2014. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kugelman JR, Sanchez-Lockhart M, Andersen KG, Gire S, Park DJ, Sealfon R, Lin AE, Wohl S, Sabeti PC, Kuhn JH, Palacios GF. 2015. Evaluation of the potential impact of Ebola virus genomic drift on the efficacy of sequence-based candidate therapeutics. mBio 6(1):e02227-14. doi: 10.1128/mBio.02227-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ponomarenko J, Vaughan K, Sette A, Maurer-Stroh S. 3 November 2014. Conservancy of mAb epitopes in ebolavirus glycoproteins of previous and 2014 outbreaks. PLoS Curr 6. doi: 10.1371/currents.outbreaks.f1a7028a13ce1c5f0bdbb4b0cc0b919b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol 80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dias JM, Kuehne AI, Abelson DM, Bale S, Wong AC, Halfmann P, Muhammad MA, Fusco ML, Zak SE, Kang E, Kawaoka Y, Chandran K, Dye JM, Saphire EO. 2011. A shared structural solution for neutralizing ebolaviruses. Nat Struct Mol Biol 18:1424–1427. doi: 10.1038/nsmb.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bale S, Dias JM, Fusco ML, Hashiguchi T, Wong AC, Liu T, Keuhne AI, Li S, Woods VL Jr, Chandran K, Dye JM, Saphire EO. 2012. Structural basis for differential neutralization of ebolaviruses. Viruses 4:447–470. doi: 10.3390/v4040447. [DOI] [PMC free article] [PubMed] [Google Scholar]