Abstract
Cryptococcus neoformans is an opportunistic human pathogen that causes life-threatening meningitis. In this fungus, the cell wall is exceptionally not the outermost structure due to the presence of a surrounding polysaccharide capsule, which has been highly studied. Considering that there is little information about C. neoformans cell wall composition, we aimed at describing proteins and lipids extractable from this organelle, using as model the acapsular mutant C. neoformans cap 67. Purified cell wall preparations were extracted with either chloroform/methanol or hot SDS. Total lipids fractionated in silica gel 60 were analyzed by electrospray ionization-tandem mass spectrometry (ESI-MS/MS), while trypsin digested proteins were analyzed by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). We detected 25 phospholipid species among phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, phosphatidylinositol, and phosphatidic acid. Two glycolipid species were identified as monohexosyl ceramides. We identified 192 non-covalently linked proteins belonging to different metabolic processes. Most proteins were classified as secretory, mainly via nonclassical mechanisms, suggesting a role for extracellular vesicles in transwall transportation. In concert with that, orthologs from 86% of these proteins have previously been reported both in fungal cell wall and/or in extracellular vesicles. The possible role of the presently described structures in fungal-host relationship is discussed.
Keywords: glycolipid, monohexosyl ceramide, pathogenic fungus, phospholipid, proteome
CRYPTOCOCCUS neoformans is a ubiquitous basidiomycete that can be found in diverse types of environments, such as the soil, trees, and bird feces. It is an important opportunistic human pathogen that causes life-threatening meningitis and meningoencephalitis in immunocompromised hosts (Srikanta et al. 2014). Infection occurs when desiccated yeasts or spores are inhaled. Once they reach the pulmonary alveoli, they can rapidly be eliminated by the immune system or remain quiescent until an immunological misbalance leads to central nervous system tropism and disease development. In the last decades, the number of cryptococcosis cases has risen dramatically due to the association with AIDS and patients under immunosuppressive treatment (Srikanta et al. 2014).
A number of factors have been related to C. neoformans virulence, such as melanin production, the ability to grow at 37 °C, phospholipase and urease extracellular activities, induction of the Rim101 transcription factor and the virulence-associated protein VAD1. However, the best studied and most important fungal virulence factor is a polysaccharide capsule that surrounds the fungal cell wall (Vecchiarelli et al. 2013). Strains with larger capsule show higher virulence, primarily attributed to prevention of phagocytosis by host macrophages (Feldmesser et al. 2000). Therefore, investigators have concentrated their studies in capsule components, especially in the major glucuronoxylomannan (GXM). Despite the importance of the fungal cell wall in the relationship with the environment and the host, there is little information about it in Cryptococcus (Perfect and Casadevall 2002).
Fungal cell wall is mainly composed of structural glucans and chitin, with an outer layer of mannan and glycoproteins (Latge 2007). A proportionally minor lipid and protein total content has also been described (San-Blas and San-Blas 1984). In Candida albicans and Saccharomyces cerevisiae, cell wall structural proteins are covalently linked either to β-1,6-glucan via a remnant glycosylphosphatidylinositol (GPI) anchor or directly to the long β-1,3-glucan chain (e.g., proteins with internal repeats – Pir) (De Groot et al. 2005). Additionally, non-covalently linked proteins, associated to the cell wall by disulfide bonds or other unclear ways, have been described (Ruiz-Herrera et al. 2006, Guimaraes et al. 2011, Puccia et al. 2011).
The C. neoformans cell wall consists predominantly of chitin and a mixture of branched and linear glucopyrans (James et al. 1990). As opposed to S. cerevisiae, C. neoformans cell wall lacks structural mannan (James et al. 1990) and mannoproteins (Vartivarian et al. 1989). The literature on cell wall lipids focuses on a glucosylceramide (GlcCer) species localized preferentially to the C. neoformans cell wall, with a role in budding, growth, and pathogenicity (Rodrigues et al. 2000, Rittershaus et al. 2006). There are few studies on cell wall molecules in C. neoformans (Foster et al. 2007).
Considering the scarce literature about cell wall composition, the present work aimed to describe lipid and protein cell wall components using as model an acapsular C. neoformans mutant (cap 67) in order to avoid contamination with capsule components. The role of the presently described structures in fungal-host relationship is speculated.
MATERIALS AND METHODS
Reagents and solvents
Otherwise stated, all reagents and solvents used in this study were HPLC-grade or higher, and mostly from Sigma-Aldrich Co. (St. Louis, MO).
C. neoformans cap 67 growth conditions
Acapsular mutant C. neoformans cap 67 was maintained in slants of modified YPD medium (0.5% yeast extract, 0.5% casein peptone, 1.5% glucose, pH 6.5) at 36 °C. Yeast cells were then seeded to grow at 36 °C for 48 h in 100 ml pre-inoculum flasks containing a defined medium (15 mM glucose, 10 mM MgSO4, 29.5 mM KH2PO4, 13 mM glycine, and 3 µM thiamine, pH 5.5), transferred to fresh medium (200 ml) and cultivated for three more days before cell wall purification.
Cell wall purification
Cell wall purification was performed as previously described for Paracoccidioides brasiliensis (Longo et al. 2013b). Briefly, yeast cell pellets were washed three times in phosphate-buffered saline solution (PBS) and mechanically broken in a cell disruptor (B. Braun Biotech International GmBH, Melsungen, Germany) in the presence of glass beads (425–600 µm, Sigma Aldrich). Mechanical disruption was achieved by six turns of 10-min agitation alternated with a 10-min rest on ice. For proteomic analysis, self-proteolysis was prevented with the use of protease inhibitors (100 mM ethylenediamine tetraacetic acid, EDTA; 10 mM 1,10-phenanthroline; 1 mM phenylmethylsulfonyl fluoride, PMSF; 1 µM pepstatin A; and 15 µM trans-epoxysuccinyl-L-leucylamido(4-guanidino)butane, E-64). The cell wall-containing fraction was pelleted by centrifugation (5,000 × g for 10 min at 4 °C) and washed with deionized water. Cytoplasmic and membranous contents were eliminated by three sequential centrifugations (8,000 × g for 45 min at 4 °C) in 85% sucrose (Kanetsuna et al. 1969). The cell wall enriched fraction was then washed differently for proteomic and lipidomic analysis. Before lipid extraction (detailed below), cell wall fractions were washed 50 times in ultrapure water (Previato JO 1979). In preparations destined for protein extraction (detailed below), cell pellets were first washed five times with ultrapure water. We then sequentially carried out five washes with each NaCl solution (5% NaCl, 2% NaCl and 1% NaCl), to remove non-specifically bound components that could eventually remain in the cell wall preparation, followed by five washes with 1 mM PMSF (Pitarch et al. 2002).
Lipid extraction and fractionation
A lyophilized cell wall preparation (100 mg) from C. neoformans cap 67 was sequentially extracted three times, under shaking with glass beads, with 1 ml chloroform:methanol (2:1, v:v) and chloroform:methanol:water (2:1:0.8, v:v:v), as described (Yichoy et al. 2009). Both extracts were combined, dried under a N2 stream and dissolved in 2 ml chloroform. Lipid fractionation was performed in 500 mg of silica gel 60 spheres (60Å, 200–400 mm, Sigma-Aldrich, St. Louis, MO) packed in a Pasteur pipette. The silica was sequentially washed in 5 ml HPLC-grade methanol, acetone, and chloroform before loading with extracted total lipids. Neutral glycolipids and phospholipids were sequentially eluted with 5 ml of acetone and methanol, and dried under N2 stream.
Glycolipid permethylation
Glycolipids were permethylated as previously described (Ciucanu and Kerek 1984). Glycolipids were stored overnight at −20 °C in desiccant silica and redissolved in 150 µl anhydrous dimethyl sulfoxide (DMSO). A few milligrams of powdered, anhydrous NaOH were added and the mixture was vortexed. After the addition of 80 µl iodomethane, the sample was incubated under shaking for 1 h at room temperature. Two ml of both deionized water and dichloromethane were added, the mixture was vortexed and centrifuged, and the aqueous upper layer was removed. The remaining organic phase was washed three times in 2 ml deionized water and dried under N2 stream.
ESI-MS/MS analysis of phospholipids and glycolipids
In order to analyze phospholipids (negative- and positive-ion modes) and glycolipids, we used electrospray ionization tandem mass spectrometry (ESI-MS/MS) on a linear ion-trap mass spectrometer (LTQ XL, ThermoFisher Scientific Inc., San Jose, CA) coupled with an automated nanospray source (TriVersa NanoMate System, Advion, Ithaca, NY), as described previously (Vallejo et al. 2012b). For the negative-ion mode analysis, phospholipid samples were redissolved in methanol containing 0.05% formic acid (FA) and 0.05% NH4OH; 2.5 µM phosphatidylglycerol (C12:0/C12:0-PG) (Avanti Polar Lipids, Alabaster, AL) was used as internal standard. For the positive-ion mode analysis, phospholipid samples were redissolved in 10 mM LiOH/methanol and 2.5 µM phosphatidylcholine (C11:0/C11:0-PC) (Avanti Polar Lipids) was used as internal standard. Full-scan spectra were collected at the 500–1000 m/z range, and samples were subjected to total-ion mapping (TIM) (2 a.m.u. isolation width; pulsed-Q dissociation (PQD) to 29% normalized collision energy; activation Q of 0.7; and activation time of 0.1 ms). We redissolved permethylated glycolipids in methanol and analyzed them as described for phospholipids, using the following parameters: full-scan spectra acquisition at the 500–2000 m/z range, TIM settings at the 700–900 m/z range, 2 a.m.u. isolation width; PQD to 32% normalized collision energy; activation Q of 0.7; and activation time of 0.1 ms. All MS/MS spectra were analyzed manually, as described (Pulfer and Murphy 2003).
SDS extraction of cell wall-associated proteins
Proteins were extracted according to Casanova and coworkers (Casanova et al. 1989), with modifications. Briefly, a lyophilized cell wall preparation (100 mg) was incubated twice for 5 min at 90 °C in a solution containing 100 mM Na-EDTA, 50 mM Tris-HCl, pH 7.8 and 2% SDS. The SDS extracts were collected by centrifugation and filtrated through a sterile 0.22-micron filter. The filtered extract was incubated in ice-cold acetone (1 h at −20 °C), centrifuged for 30 min (16,000 × g at 4 °C) and the protein pellet was removed, washed in acetone and dried.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis
Protein digestion was carried out as described by Russell and coworkers (Russell et al. 2001). After denaturation at 95 °C for 5 min in buffered methanol/50 mM NH4HCO3 (60:40, v/v), proteins were incubated in 5 mM dithiotreitol (DTT) for 30 min at 37 °C for disulfide bond reduction and cysteine residues were alkylated with 10 mM iodoacetamide (90 min at room temperature in the dark). Proteolytic digestion was achieved by overnight incubation at 37 °C with 1 µg sequencing-grade trypsin (Promega), and 0.05% of trifluoroacetic acid (TFA) was added to stop the reaction. In-house POROS R2 microcolumns (Nakayasu et al. 2012) were used for desalting the ensuing peptides, which were then dried in a vacuum concentrator until the LC-MS/MS analysis.
LC-MS/MS analysis and peptide validation
We dissolved tryptic peptides in 20 µL 0.1% formic acid (FA), and loaded 5 µL of the mixture onto a reversed-phase trap column (1 cm × 75 µm, Luna C18, 5 µm, Phenomenex). LC was performed in a capillary reversed-phase column (20 cm × 75 µm, Luna C18, 5 µm, Phenomenex) coupled to a nanoHPLC (1D Plus, Eksigent), exactly as previously described (Vallejo et al. 2012a). Elution of bound peptides was carried out in a linear gradient (5% to 40%) of solvent B [solvent A: 5% acetonitrile (ACN)/0.1% FA; solvent B: 80% ACN/0.1% FA] over 200 min and directly analyzed in a linear ion trap-mass spectrometer equipped at the front end with a TriVersa NanoMate nanospray source. The nanospray was set at 1.45 kV and 0.25 psi N2 pressure using a chip A (Advion). MS spectra were collected in centroid mode at the 400–1700 m/z range and the ten most intense ions were subjected twice to collision-induced dissociation (CID) with 35% normalized collision energy, before being dynamically excluded for 60 s.
Using Bioworks v.3.3.1 (Thermo Fisher), we initially converted into DTA files all MS/MS spectra from peptides bearing 800 to 3,500 Da (over 10 counts) and at least 15 fragments. DTA files were subsequently submitted to database search using TurboSequest (Bioworks 3.3.1, Thermo Fisher Scientific). Sequence information from the C. neoformans var. grubii H99 genome (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html), human genome (International Protein Index), besides porcine trypsin sequences (GenBank), were used in the correct and reverse orientations. The database search parameters included: i) trypsin cleavage in both peptide termini with one missed cleavage site allowed; ii) carbamidomethylation of cysteine residues as a fixed modification; iii) oxidation of methionine residues as a variable modification; and iv) 2.0 Da and 1.0 Da for peptide and fragment mass tolerance, respectively. TurboSequest outputs were filtered with DCn ≥ 0.05, peptide probability ≤ 0.05, and Xcorr ≥ 1.5, 2.0, and 2.5 for singly-, doubly-, and triply-charged peptides, respectively. Files were then exported into XML formats and peptide sequences were assembled into proteins using an in house written script (Nakayasu et al. 2012). Redundant protein hits were assembled into protein groups and protein hits were re-filtered with the sum of peptide Xcorr ≥ 3.0. The false-discovery rate (FDR) was estimated as described previously (Rodrigues et al. 2008, Albuquerque et al. 2008). Proteins with shared peptides were assembled into groups to assess the redundancy issue. Only proteins detected by five or more spectral counts were considered for further analysis.
Bioinformatic analysis
Protein sequences were submitted to Blast2GO algorithm http://www.blast2go.de/ (Conesa et al. 2005) and classified by Gene Ontology (GO) according to their biological functions. Putative secretory proteins were classified according to the Fungal Secretome Database (FSD; http://fsd.snu.ac.kr/)(Choi et al. 2010), and were labeled as not classified, SP (containing signal peptide identified by SignaIP 3.0), SP3 (bearing signal peptide predicted by SigPred, SigCleave or RPSP), SL (subcellular localization predicted by PSort II and/or Target 1.1b), and NS (nonclassical secretion, predicted by SecretomeP 1.0f). Identification of orthologous proteins was performed manually or using the OrthoMCL software v2.0 (http://www.orthomcl.org/cgi-bin/OrthoMclWeb.cgi), with percentage match cutoff = 50 and E- value exponent cutoff = −5.
RESULTS
We carried out lipidomic and proteomic analysis of cell wall components from the acapsular mutant C. neoformans cap 67 by ESI-MS/MS and LC-MS/MS, respectively. Before specific extraction procedures, cell wall pellets were enriched and separated from membrane contaminants through sucrose density centrifugation (Kanetsuna et al. 1969), followed by exhaustive washes in water (for lipid extraction) (Previato JO 1979) or decreasing salt concentration (for protein extraction) (Pitarch et al. 2002), as detailed in Materials and Methods.
ESI-MS/MS analysis of cell wall phospholipids
Phospholipids from the acapsular mutant C. neoformans cap 67 cell wall preparations were analyzed by ESI-MS/MS in the positive- and negative-ion modes (Table 1). The full-scan spectra profile comprised a broad range of mass to charge ratio (m/z), suggesting the presence of phospholipid species from different classes (not shown). The identification of phospholipid species was performed exactly as in previous works from our group focusing on P. brasiliensis extracellular vesicles and cell wall lipids (Vallejo et al. 2012b, Longo et al. 2013a). Briefly, we first searched positive- and negative-ion modes for diagnostic fragment ions and neutral losses exclusive of phospholipids. Then, we looked for specific peaks and losses to identify phospholipid classes. In the negative-ion mode, phospholipid species were identified with chlorine (Cl−) ([M + Cl]−) or formate (HCOO−) ([M + HCOO]−) adducts. In the positive-ion mode, phosphatidylcholine (PC) species were identified with adduct of Li+ ([M + Li]+) or singly protonated ([M + H]+). Fatty acid chains were also identified by neutral fragment losses (positive ion-mode, Fig. S1) or by the negatively charged carboxylate ions at m/z 255 (C16:0), 279 (C18:2), and 281 (C18:1) (Fig. S2–S6).
Table 1.
Composition of phospholipids extracted from C. neoformans cap67 cell wall preparations as identified by ESI-MS total-ion mapping
| Phospholipid/Ionmode | Ionspecies | m/z | Proposed composition |
Predicted mass (Da) |
|---|---|---|---|---|
| Phosphatidylethanolamine (PE) | ||||
| (−) | [M−H]− | 478.4 | lyso C18:1 | 479.3 |
| (−) | [M−H]− | 476.4 | lyso C18:2 | 477.3 |
| (−) | [M + Cl]− | 534.4 | ||
| (−) | [M−H]− | 716.6 | C16:0/C18:1 | 717.5 |
| (−) | [M + Cl]− | 774.7 | ||
| (−) | [M−H]− | 714.7 | C16:0/C18:2 | 715.5 |
| (−) | [M−H]− | 740.7 | C18:1/C18:2 | 741.5 |
| (−) | [M + Cl]− | 798.6 | ||
| (−) | [M + HCOO]− | 808.6 | ||
| (−) | [M−H]− | 742.6 | C18:1/C18:1 | 743.5 |
| (−) | [M + Cl]− | 800.6 | ||
| (−) | [M + HCOO]− | 810.6 | ||
| Phosphatidylcholine (PC) | ||||
| (−) | [M + HCOO]− | 802.6 | C16:0/C18:2 | 758.6 |
| (+) | [M + Li]+ | 764.8 | ||
| (−) | [M + Cl]− | 821.7 | C18:1/C18:1 | 786.6 |
| (+) | [M + Li]+ | 793.7 | ||
| (+) | [M + Li]+ | 766.6 | C16:0/C18:1 | 760.6 |
| (+) | [M + Li]+ | 768.7 | C16:0/C18:0 | 762.6 |
| (+) | [M + Li]+ | 790.3 | C18:1/C18:2 | 784.6 |
| (+) | [M + Li]+ | 794.7 | C18:0/C18:1 | 788.6 |
| PhosphatidicAcid (PA) | ||||
| (−) | [M−H]− | 671.5 | C16:0/C18:2 | 672.5 |
| (−) | [M−H]− | 673.5 | C16:0/C18:1 | 674.5 |
| (−) | [M + Cl]− | 731.6 | ||
| (−) | [M−H]− | 695.5 | C18:2/C18:2 | 696.5 |
| (−) | [M−H]− | 697.6 | C18:1/C18:2 | 698.5 |
| (−) | [M + Cl]− | 755.6 | ||
| (−) | [M−H]− | 699.5 | C18:1/C18:1 | 701.0 |
| (−) | [M + HCOO]− | 767.6 | ||
| Phosphatidylserine (PS) | ||||
| (−) | [M−H]− | 758.6 | C16:0/C18:2 | 759.5 |
| (−) | [M−H]− | 760.6 | C16:0/C18:1 | 761.5 |
| (−) | [M−H]− | 784.5 | C18:1/C18:2 | 785.5 |
| (−) | [M−H]− | 786.8 | C18:1/C18:1 | 787.5 |
| Phosphatidylinositol (PI) | ||||
| (−) | [M−H]− | 835.7 | C16:0/C18:1 | 836.5 |
| (−) | [M−H]− | 833.8 | C16:0/C18:2 | 834.5 |
Phospholipid species of the PE (six), PC (two), PA (five), PS (four), and PI (two) classes were detected at m/z ranges between 477.3 and 836.5 in the negative-ion mode, and six PC species were identified between m/z 758.6 and 788.6 in the positive-ion mode (Table 1). Among the identified PE species, two of them showed only one fatty acid, lyso-C18:1-PE (m/z 478.4) and lyso-C18:2-PE (m/z 476.4 and m/z 534.4).
ESI-MS/MS analysis of cell wall glycolipids
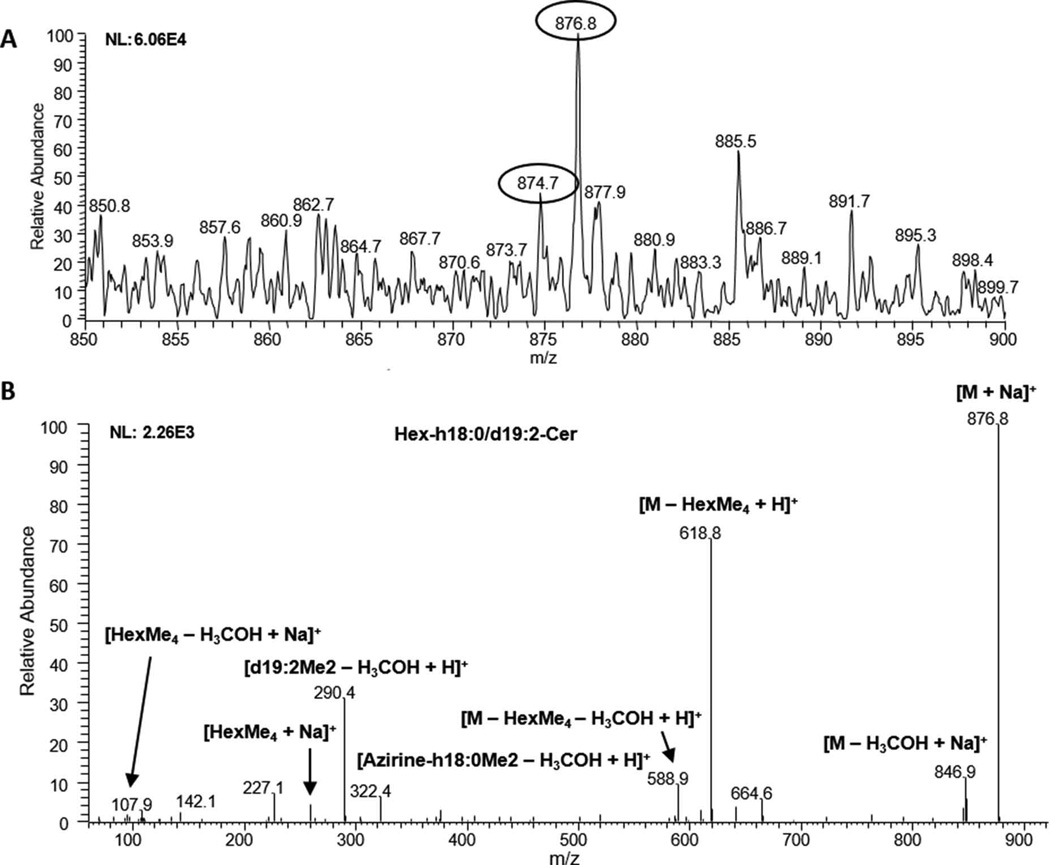
Neutral glycolipids were extracted from cell wall preparations of the acapsular mutant C. neoformans cap 67, isolated in a silica-60 column, permethylated, and subjected to a positive-ion mode ESI-MS/MS analysis (Fig. 1). Fig. 1A shows peaks of two major glycolipid species detected in full-scan spectra at m/z 876.8 and 874.7, which were further characterized by the MS/MS analysis. Glycolipid species were identified as previously reported (Vallejo et al. 2012b, Longo et al. 2013a) by diagnostic fragment ions and specific neutral losses. A C18:0 [azirine-h18:0Me2 – H3COH + H]+ or C18:1 [azirine-h18:1Me2 – H3COH + H]+ fatty acid attached to a remnant of the sphingoid base could be identified at m/z 322.4 and 320.4 fragment ions, respectively in Hex-C18:0-OH/d19:2-Cer (at m/z 876.8) (Fig. 2B) and Hex-C18:1-OH/d19:2-Cer (at m/z 874.7) (Fig. 1B and S7). Although the MS/MS analysis is not able to indicate the unsaturation or the methyl and hydroxyl group positions in the sphingoid base, we believe that the main neutral glycolipid here identified is N-2’-hydroxyoctadecanoate (4E, 8E)-9-methyl-4,8-sphingadienine (m/z 876.8), previously described as the major cryptococcal glycosphingolipid (GSL) and abundantly localized at the fungal cell wall (Rodrigues et al. 2000).
Fig. 1.
ESI-MS analysis of neutral glycolipids from C. neoformans cap 67 mutant cell wall. A. Full-scan spectrum in the positive-ion mode. The m/z of identified glycolipids Hex-C18:1-OH/d19:2-Cer (m/z 874.7) and Hex-C18:0-OH/d19:2-Cer (m/z 876.8) are circled. B. Tandem-MS spectrum of the major glycolipid species identified (m/z 876.8). m/z, mass to charge ratio.
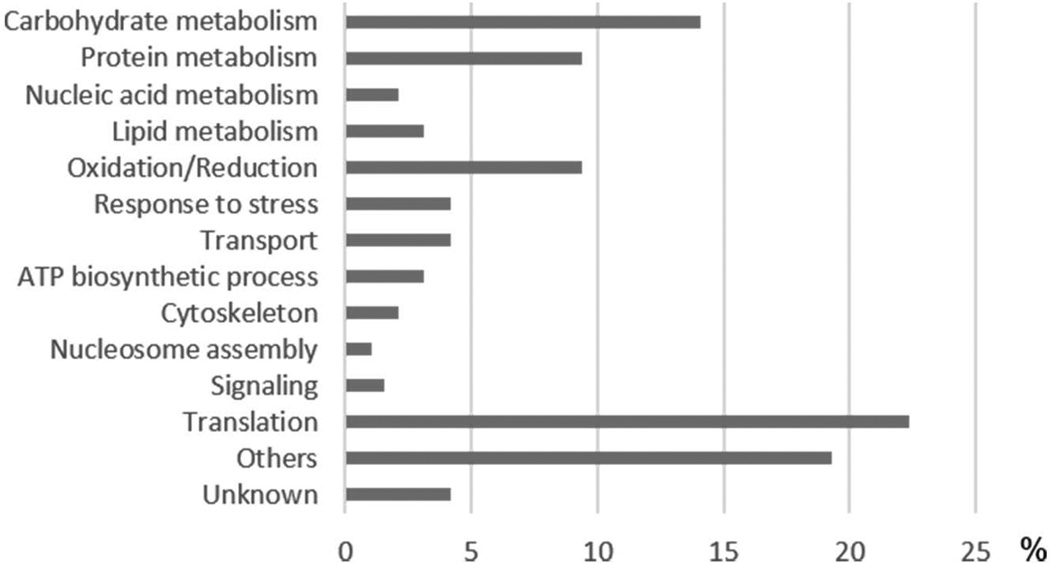
Fig. 2.
Functional categorization of cell wall-associated proteins from C. neoformans cap 67 mutant. Values represent the percentages relative to all identified proteins.
LC-MS/MS analysis of C. neoformans cap 67 cell wall-associated proteins
Before protein extraction, cell wall preparations were extensively washed in solutions with decreasing concentrations of NaCl in order to remove proteins that might be non-specifically bound to the cell wall preparation during cell lysis. We used two sequential extractions with boiling SDS, which is supposed to release proteins loosely bound to the fungal surface (Casanova et al. 1989). In order to make our analysis more reliable, we used an arbitrary and stringent cut-off corresponding to proteins detected by five or more spectral counts. Using this criterion, a total of 192 proteins belonging to different metabolic processes were identified (Table S1). Proteins belonging to the most represented biological processes are listed in Table 2.
Table 2.
Cell wall-associated proteins identified in C. neoformans cap67 cell wall preparations. The proteins areorganized according to GO functions. Minor and unknown functions are omitted, but can be seen in a complete Supplemental Table 1.
| Codenumber* | Proteinname | CW** | EV** |
|---|---|---|---|
| Transport | |||
| CNAG_02817 | GTP-binding protein ypt2 | - | CnHcPbSc |
| CNAG_02974 | voltage-dependent anion channel protein 2 | Af | Cn |
| CNAG_04851 | transitionalendoplasmicreticulumATPase | Ca Pb | HcSc |
| CNAG_04904 | clathrin heavy chain | - | - |
| CNAG_06101 | ADP, ATP carrier protein | Ca | CnHcPbSc |
| CNAG_06377 | solute carrier family 25, member 3 | - | Cn |
| CNAG_00499 | solute carrier family, member 20/29 | - | Hc |
| CNAG_06096 | tricarboxylatecarrier | - | - |
| Response to stress | |||
| CNAG_00334 | hsp75-like protein | Af Ca Sc | CnHcPbSc |
| CNAG_01727 | hsp71-like protein | Af Ca Sc | CnHcPbSc |
| CNAG_01750 | hsp72-like protein | Ca Sc | CnHcPbSc |
| CNAG_03891 | hsp60-like protein | Ca Pb | CnHcPbSc |
| CNAG_05199 | chaperoneDnaK | Af Ca | HcPbSc |
| CNAG_06150 | hsp90-like protein | Af Ca PbSc | CnHcPbSc |
| CNAG_06208 | heat shock 70kDa protein 4 | Af Ca PbSc | HcPbSc |
| CNAG_06443 | glucose-regulatedprotein | Ca Sc | CnHcPbSc |
| ATP biosyntheticprocess | |||
| CNAG_01004 | ATP synthase F1, delta subunit | - | CnHcPb |
| CNAG_04439 | V-type proton ATPase subunit B | - | CnSc |
| CNAG_05750 | ATP synthase subunit alpha, mitochondrial | Ca Pb | CnHcPbSc |
| CNAG_05918 | ATP synthasesubunit beta, mitochondrial | Ca Pb | CnHcPbSc |
| CNAG_06400 | proton-efflux P-type ATPase | Ca Sc | CnHcPbSc |
| CNAG_02326 | V-type proton ATPase catalytic subunit A | - | HcSc |
| Translation | |||
| CNAG_00034 | largesubunitribosomalprotein L9e | - | CnPbSc |
| CNAG_00096 | small subunit ribosomal protein S3 | - | HcPbSc |
| CNAG_00640 | small subunit ribosomal protein S4-A | Ca | CnHcSc |
| CNAG_00656 | largesubunitribosomalprotein L7e | Ca Sc | CnPbSc |
| CNAG_00672 | small subunit ribosomal protein S9 | Sc | CnHc |
| CNAG_01628 | small subunit ribosomal protein S20 | Ca Sc | CnHc |
| CNAG_01024 | large subunit ribosomal protein L18-A | - | Cn |
| CNAG_02928 | largesubunitribosomalprotein L5e | Ca Sc | CnHcPbSc |
| CNAG_00779 | largesubunitribosomalprotein L27e | Sc | CnHc |
| CNAG_00952 | smallsubunitribosomalprotein S6e | Ca | HcSc |
| CNAG_00953 | smallsubunitribosomalprotein S13e | Ca | CnPbSc |
| CNAG_01884 | large subunit ribosomal protein L3 | Ca Sc | CnHcPb |
| CNAG_01486 | large subunit ribosomal protein L15-A | Ca PbSc | CnSc |
| CNAG_01951 | small subunit ribosomal protein S22-A | - | CnHcSc |
| CNAG_02144 | large subunit ribosomal protein L1-A | - | CnHcPb |
| CNAG_02234 | largesubunitribosomalprotein L6e | Ca Pb | Cn |
| CNAG_02331 | small subunit ribosomal protein S9 | Ca | Cn |
| CNAG_03000 | smallsubunitribosomalprotein S19e | Ca | Cn |
| CNAG_03053 | large subunit ribosomal protein L23 | - | CnSc |
| CNAG_03198 | smallsubunitribosomalprotein S8e | Ca | CnSc |
| CNAG_03510 | largesubunitribosomalprotein L36e | - | HcPb |
| CNAG_03577 | large subunit ribosomal protein LP0 | AfPbSc | HcSc |
| CNAG_03739 | large subunit ribosomal protein L10-like | Ca Sc | CnHcSc |
| CNAG_03780 | small subunit ribosomal protein S16 | Ca | CnSc |
| CNAG_04004 | small subunit ribosomal protein S1 | Ca Sc | CnHcPb |
| CNAG_00409 | large subunit ribosomal protein L37a | - | Cn |
| CNAG_04021 | large subunit ribosomal protein L24 | - | Cn |
| CNAG_00494 | small subunit ribosomal protein S0 | Af Ca Sc | CnSc |
| CNAG_04445 | smallsubunitribosomalprotein S7e | Ca Sc | CnHcPbSc |
| CNAG_04726 | large subunit ribosomal protein L18Ae | Ca | Cn |
| CNAG_04762 | largesubunitribosomalprotein L4e | Ca | CnHcPbSc |
| CNAG_04799 | largesubunitribosomalprotein L14e | - | CnPbSc |
| CNAG_04883 | small subunit ribosomal protein S18 | - | CnHcPb |
| CNAG_00417 | elongationfactor 1-gamma | Pb | CnPbSc |
| CNAG_00785 | ATP-dependent RNA helicase eIF4A | Af Ca Sc | CnHcPbSc |
| CNAG_04609 | argonaute | - | CnHc |
| CNAG_05555 | large subunit ribosomal protein L7Ae | PbSc | CnSc |
| CNAG_06105 | elongationfactor 1-alpha | Ca PbSc | CnHcPbSc |
| CNAG_06605 | small subunit ribosomal protein S2 | AfSc | CnHcPbSc |
| CNAG_00689 | largesubunitribosomalprotein L22e | Pb | Pb |
| CNAG_05232 | large subunit ribosomal protein L8 | Ca PbSc | CnHcSc |
| CNAG_06231 | large subunit ribosomal protein L13 | - | - |
| CNAG_06447 | large subunit ribosomal protein L22 | Ca Sc | CnSc |
| Proteinmetabolism | |||
| CNAG_04009 | aminopeptidase 2 | - | HcPbSc |
| CNAG_00150 | peptidase | - | Cn |
| CNAG_00919 | carboxypeptidase D | Pb | CnSc |
| CNAG_01890 | 5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase | Ca PbSc | CnHcPbSc |
| CNAG_02500 | calnexin | Pb | Cn |
| CNAG_02966 | carboxypeptidase | - | Sc |
| CNAG_03507 | mitochondrial peptidase subunit beta | - | CnHc |
| CNAG_04604 | tryptophan-tRNAligase | - | - |
| CNAG_04625 | cerevisin | Ca Pb | CnHcPbSc |
| CNAG_05725 | ketol-acidreductoisomerase, mitochondrial | Ca | CnHcSc |
| CNAG_05932 | peptidyl-prolylcis-transisomerase D | Pb | HcPb |
| CNAG_06026 | aspartateaminotransferase | Pb | HcPbSc |
| CNAG_06088 | aspartateaminotransferase, mitchondrial | - | - |
| CNAG_00136 | ubiquitin-activatingenzyme E1 | - | Sc |
| CNAG_05722 | alanine-tRNAligase | Sc | Sc |
| CNAG_04906 | 26S protease regulatory subunit 10B | - | - |
| CNAG_05886 | ubiquitin-conjugatingenzyme E2 | Pb | - |
| CNAG_06755 | threonine-tRNAligase | - | Sc |
| Carbohydratemetabolism | |||
| CNAG_00061 | citratesynthase, mitochondrial | Ca | CnHcPbSc |
| CNAG_00937 | aconitatehydratase, mitochondrial | Ca Pb | HcPbSc |
| CNAG_00964 | glutamine-fructose-6-P transaminase | - | - |
| CNAG_01820 | pyruvatekinase | Ca Sc | CnHcSc |
| CNAG_01896 | alcoholdehydrogenase | Ca Pb | CnPbSc |
| CNAG_01984 | transaldolase | Ca Sc | CnHcPbSc |
| CNAG_02035 | triose-phosphateisomerase | Ca Sc | HcPbSc |
| CNAG_02818 | glycine cleavage system T protein | - | CnHc |
| CNAG_03072 | enolase | Af Ca PbSc | CnPbSc |
| CNAG_03225 | malatedehydrogenase, NAD-dependent | Ca | CnHcPbSc |
| CNAG_03358 | phosphoglyceratekinase | Af Ca Sc | CnHcPbSc |
| CNAG_03525 | alpha, alpha-trehalase | - | - |
| CNAG_03674 | oxoglutaratedehydrogenase, E1 | - | Hc |
| CNAG_03725 | dTDP-4-dehydrorhamnose reductase | Pb | Cn |
| CNAG_04640 | ATP-citrate synthase subunit 1 | - | CnHcSc |
| CNAG_04659 | pyruvatedecarboxylase | Ca PbSc | CnSc |
| CNAG_04969 | UDP-glucose 6-dehydrogenase | - | Cn |
| CNAG_05653 | malatesynthase A | - | CnHcSc |
| CNAG_05907 | pyruvatecarboxylase | - | CnHcSc |
| CNAG_06313 | phosphoglucomutase | Ca Pb | HcPbSc |
| CNAG_06666 | starchphosphorylase | - | Cn |
| CNAG_06699 | glyceraldehyde-3-phosphate dehydrogenase | Af Ca PbSc | CnHcPbSc |
| CNAG_06770 | fructose-bisphosphatealdolase 1 | Ca PbSc | HcPbSc |
| CNAG_05351 | phosphomannomutase | Sc | HcSc |
| CNAG_02748 | UTP-glucose-1-Puridylyltransferase | Ca | CnSc |
| CNAG_04217 | phosphoenolpyruvatecarboxykinase | Pb | HcSc |
| CNAG_06501 | 1,3-beta-glucanosyltransferase | Ca PbSc | CnHcSc |
| Lipidmetabolism | |||
| CNAG_02099 | fatty acid synthase subunit beta, fungi type | - | HcSc |
| CNAG_02100 | fatty acid synthase subunit alpha, fungi type | Sc | CnSc |
| CNAG_02562 | acyl-CoAdehydrogenase | - | - |
| CNAG_04308 | 3-hydroxyacyl-CoA dehydrogenase | - | Hc |
| CNAG_04869 | carboxylesterase | - | Cn |
| CNAG_07004 | dihydrolipoyldehydrogenase | - | CnPbSc |
| Nucleicacidmetabolism | |||
| CNAG_01395 | ribose-5-phosphate isomerase | Ca | Pb |
| CNAG_00165 | methylthioadenosinephosphorylase | Pb | Hc |
| CNAG_02285 | nucleosidediphosphatekinase | - | CnHcPbSc |
| CNAG_04577 | nucleoside-diphosphatekinase | - | CnHcPbSc |
| Oxidation/Reduction | |||
| CNAG_00452 | isovaleryl-CoAdehydrogenase | - | Hc |
| CNAG_00938 | cytochrome c peroxidase, mitochondrial | Af | Hc |
| CNAG_01492 | hypotheticalprotein | - | - |
| CNAG_01594 | glycinedehydrogenase | - | Hc |
| CNAG_01981 | sulfide:quinoneoxidoreductase | - | - |
| CNAG_02399 | glutathione-disulfidereductase | PbSc | Cn |
| CNAG_02801 | thioredoxin | Ca Pb | CnSc |
| CNAG_03482 | peroxiredoxin | Ca Sc | CnSc |
| CNAG_03618 | NADPH2:quinone reductase | Ca Sc | HcSc |
| CNAG_04981 | catalase | Pb | CnHcPbSc |
| CNAG_06628 | aldehydedehydrogenase | PbSc | CnHcSc |
| CNAG_06917 | thiol-specific antioxidant protein 3 | Ca Pb | CnHcPb |
| CNAG_00162 | alternative oxidase, mitochondrial | - | - |
| CNAG_03936 | NAD(P)H:quinoneoxidoreductase, type IV | - | PbSc |
| CNAG_05753 | N-acetyl-gamma-glutamyl-Preductase | - | - |
| CNAG_05721 | multifunctional beta-oxidation protein | - | Hc |
| CNAG_05069 | ubiquinol-cytochrome c reductase sub. 10 | - | - |
| CNAG_06431 | acyl-CoA oxidase | - | - |
| Signaling | |||
| CNAG_01523 | CMGC/MAPK/P38 protein kinase | - | Sc |
| CNAG_05235 | protein BMH2 | Ca PbSc | CnHcPbSc |
| CNAG_05465 | guanine nucleotide-binding protein | Af Ca PbSc | HcSc |
| Nucleosomeassembly | |||
| CNAG_06745 | histone H3 | - | - |
| CNAG_06746 | histone H2B | Ca Pb | CnHcPbSc |
| Cytoskeleton | |||
| CNAG_00483 | actin | Ca PbSc | CnHcPbSc |
| CNAG_01840 | tubulin beta chain | - | CnPbSc |
| CNAG_03787 | tubulin alpha-1A chain | Ca | Cn |
| CNAG_04948 | tubulin beta | - | - |
Proteins were classified by the Fungal Secretome Database (FSD) into NS (nonclassical secretion, predicted by SecretomeP 1.0f) (dark gray code number), SP or SP3 (containing signal peptide identified by SignaIP 3.0, SigPred, SigCleave or RPSP) (light gray code number) and not classified as secretory (white code number).
Orthologs previously reported in cell wall (CW) or extracellular vesicles (EV) from H. capsulatum (Hc), P. brasiliensis (Pb), S. cerevisiae (Sc), C. albicans (Ca), and A. fumigatus (Af) are indicated.
We analyzed the biological processes related to the identified proteins using the Blast2GO algorithm (Conesa et al. 2005). Proteins involved in diverse cellular processes were found (Fig. 2), but translation (22%) and carbohydrate metabolism (14%) were the most represented, followed by oxidation/reduction (9%), protein metabolism (9%), transport (4%), and response to stress (4%).
Translation was the most enriched process, in which ribosomal proteins from large and small ribosome subunits prevailed. In addition, two elongation factors (CNAG_00417 and CNAG_06105), a helicase (CNAG_00785), and an argonaute protein (CNAG_04609) were identified. The carbohydrate metabolism group comprised some metabolic enzymes, such as phosphomannomutase (CNAG_05351) and pyruvate kinase (CNAG_01820). We also point out the presence of a 1,3-beta-glucanosyltransferase (CNAG_06501), related to cell wall biosynthesis/remodeling. Some proteins were associated with response to stress and oxidation/reduction, which may potentially account for fungal virulence by playing specific roles in infection. In these groups we identified several heat shock proteins (CNAG_00334, CNAG_01727, CNAG_01750, CNAG_03891, CNAG_06150 and CNAG_06208), a peroxiredoxin (CNAG_03482), and a catalase (CNAG_04981).
Although many proteins identified in this work are typically cytoplasmic, such as ribosomal and carbohydrate metabolic proteins, almost 60% of them have previously been described at the cell surface of other fungal species as well (Table 2; Rodrigues et al. 2013).
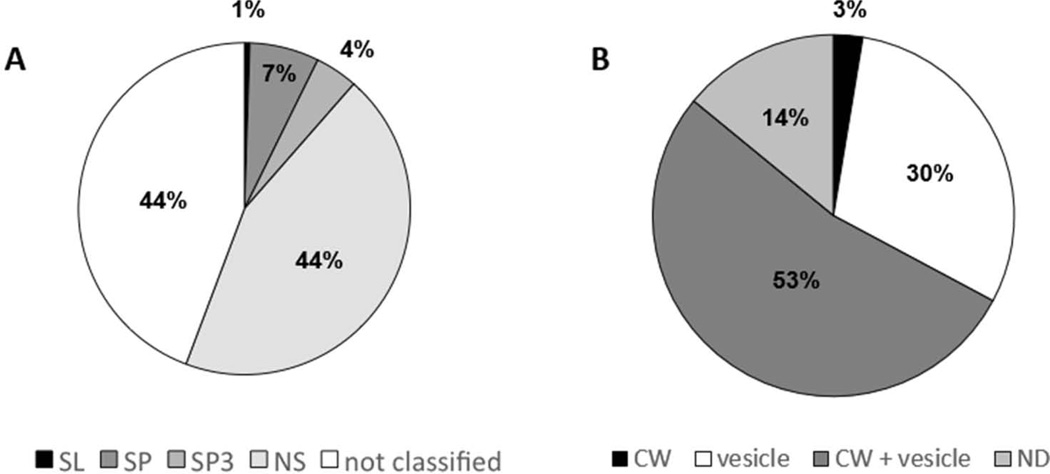
Considering the importance of protein export to the extracellular environment in modulating the host-pathogen relationship, we used the Fungal Secretome Database (FSD) to classify proteins identified in this work according to their secretory features. FSD gathers information from nine secretion prediction programs for a number of fungal genomes, comprising classical and unconventional secretory mechanisms (Choi et al. 2010). The majority of identified proteins were classified as secretory (56%), being 44% via nonclassical (NS) mechanisms, therefore lacking secretory known signals (Fig. 3A).
Fig. 3.
Characterization of cell wall-associated proteins from C. neoformans cap 67. A. Secretory potential classified by the Fungal Secretome Database (FSD): SP (presence of signal peptide as identified by SignaIP 3.0); SP3 (signal peptide predicted by SigPred, SigCleave or RPSP); SL (subcellular localization predicted by PSort II and/or Target 1.1b); NS (nonclassical secretion, predicted by SecretomeP 1.0f), and not classified (described as not secretory). B. Percentage of fungal orthologs previously reported in the cell wall (CW), extracellular vesicles (vesicle), both locations (CW+vesicle), or in neither of these compartments (ND).
We then questioned whether orthologs of the proteins identified in this work have been previously reported in fungal extracellular vesicles or cell wall. Fungal extracellular vesicles constitute an unconventional pathway for cytoplasmic components to reach the cell wall and the extracellular milieu, as already described for C. neoformans (Rodrigues et al. 2008, Wolf et al. 2014), Histoplasma capsulatum (Albuquerque et al. 2008), S. cerevisiae (Oliveira et al. 2010), P. brasiliensis (Vallejo et al. 2012a), and Malassezia sympodialis (Gehrmann et al. 2011). Using the OrthoMCL software, we found that 86% of the identified proteins in this work have been previously reported in fungal extracellular vesicles and/or cell wall (Fig. 3B). Proteins such as hsp90-like (CNAG_06150), ATP-dependent RNA helicase eIF4A (CNAG_00785), and enolase (CNAG_03072) have previously been described in extracellular vesicles and cell wall from several fungal species (Table 2).
DISCUSSION
In the present work we described the structure of phospholipids, neutral glycolipids, and the nature of non-covalently linked proteins extracted from purified cell wall preparations obtained from the acapsular C. neoformans mutant cap 67.
In our cell wall sample, PE and PC were the most represented C. neoformans cap 67 phospholipid classes, each one composed by six species, followed by PA (five species), PS (four species), and PI (two species). Recently, cell wall lipidome of the dimorphic pathogen P. brasiliensis also showed that PC and PE are the phospholipid classes comprising more species in the yeast form (Longo et al. 2013a). Overall, phospholipids from both pathogen cell walls were similar, except for phosphatidylglycerol (PG) that was identified only in P. brasiliensis. Previously, PC has been described as a major cryptococcal membrane phospholipid (Rawat et al. 1984) that has also been identified in the fungal extracellular vesicles (Oliveira et al. 2009). On the other hand, whole phospholipid analysis of extracellular vesicles identified PE, PS, and PC species in H. capsulatum (Albuquerque et al. 2008) and PC, PE, PS, PI, and PG in P. brasiliensis (Vallejo et al. 2012b).
We were able to identify two species of neutral GSLs, specifically Hex-C18:0-OH/d19:2-Cer (m/z 876.8), the most abundant, and Hex-C18:1-OH/d19:2-Cer (m/z 874.7). Considering that our strategy allows extraction of only neutral glycolipids, the presence of acidic GSLs at the cell wall cannot be discarded. Hex-C18:0-OH/d19:2-Cer (m/z 876.8) has previously been identified as a major ceramide monohexoside (CMH) in whole C. neoformans lipid extracts (Rodrigues et al. 2000). Since glucose is the hexose identified in this CMH, we believe that the neutral glycolipids presently characterized are also β-glucosylceramide (GlcCer) species. GlcCer has already been detected in cell wall extracts from P. brasiliensis (Longo et al. 2013a) and C. albicans (Thevissen et al. 2012). It has also been localized by a monoclonal antibody at the surface of P. brasiliensis, H. capsulatum, Sporothrix schenckii yeasts, and mycelia from P. brasiliensis, and Aspergillus fumigatus (Toledo et al. 2001, Longo et al. 2013a). In C. neoformans, transmission electron microscopy images showed GlcCer abundantly distributed along the cell wall, and confocal microscopy revealed GlcCer accumulation in budding sites, correlating with thickened regions of the cell wall (Rodrigues et al. 2000). Identification of this CMH in the lipidome of our cell wall preparation was used as a marker for cell wall enrichment.
In C. neoformans, GlcCer is involved in fungal budding, growth, and pathogenicity (Rodrigues et al. 2000, Rittershaus et al. 2006). It also plays a role in A. fumigatus growth (Noble et al. 2010) and C. albicans virulence (Chattaway et al. 1968). In addition to cell wall localization, GlcCer species have been identified in extracellular vesicles from P. brasiliensis (Vallejo et al. 2012b) and C. neoformans (Rodrigues et al. 2007).
Comparison of the cell wall phospholipids, GSLs, and non-covalently bound proteins (discussed below) here identified in C. neoformans with those described in fungal extracellular vesicles (EVs) strongly reinforces the presumption that, while traversing the cell wall towards the extracellular environment, EVs may be entrapped and release their components, thus participating in the fungal cell wall composition either as transient or as permanent constituents (Longo et al. 2014). The role of EVs in the transportation of virulence factors through the cell wall to the external milieu has already been recognized (Casadevall et al. 2009) and in C. neoformans EVs also traverse the capsule and participate in its synthesis by releasing GXM that can then be incorporated to form capsule fibers (Rodrigues et al. 2007). Most cell wall-associated proteins identified here are typically cytoplasmic; however, it was interesting to find that only 14% of them have not been previously described in the surface or EVs from other fungal species. Additionally, 56% show secretory features, mostly of non-classical (NS) nature. A similar scenario has recently been reported for DTT-extractable cell wall-associated proteins in P. brasiliensis (Longo et al. 2014). Non-classical (NS) secretion includes exportation mechanisms using extracellular vesicles originated from plasma membrane blebbing, exosome release (Rodrigues et al. 2011, Nickel 2010), and the recently proposed biogenesis through cell membrane invagination (Rodrigues et al. 2013). These mechanisms result in membrane and cytoplasmic subtractions, thus helping to justify the finding of some components at the cell wall.
We identified 192 non-covalently bound cell wall proteins related to translation, carbohydrate and protein metabolism, and oxidation/reduction. Although C. neoformans cell wall is not the outermost fungal structure because it is surrounded by a polysaccharide capsule, its components probably play important roles in the host-pathogen relationship, as already reported in other species (Gow and Hube 2012). Proteins isolated from C. neoformans cell wall and membrane stimulated lymphocyte proliferation from both adults and fetal cord blood, and that may account for cell-mediated immunity to this fungus (Mody et al. 1996). Some presently identified proteins may potentially contribute to fungal virulence by playing specific roles in infection, specially those related to response to stress and oxidation/reduction, as exemplified by heat shock proteins, a thioredoxin (CNAG_02801), and a catalase (CNAG_04981).
Tefsen and coworkers (Tefsen et al. 2014) showed that the C. neoformans inactivation of the CAP10 genes, which impairs the polysaccharide capsule formation, leads to enlarged cells prone to aggregation and upregulation of various genes, mainly related to oxidative stress. Some resulting proteins are cell wall-associated according to our present analysis, for e.g., thiol-specific antioxidant protein 3 (CNAG_06917) and peroxiredoxin (alkyl hydroperoxide reductase subunit C) (CNAG_03482), also known as thiol-specific antioxidant protein 1 (Tsa1). Tsa1 has already been detected by confocal microscopy on C. albicans surface of the pathogenic hyphal phase and was shown to be related to oxidative stress resistance and correct cell wall composition (Urban et al. 2003). Tsa1 has been identified among proteins from C. albicans and P. brasiliensis cell wall preparations (Table 2) and we have localized it by confocal microscopy at the P. brasiliensis yeast surface (Longo et al. unpubl. data).
Recognition of cell wall proteins by immune cells or antibodies can interfere in the host immune response (Gow et al. 2012). Many proteins presently identified at the C. neoformans cell wall proteins have also been identified in P. brasiliensis cell wall extracts from live cells (Table 2). Some are antigenic and recognized by paracoccidioidomycosis (PCM) patients’ sera, such as hsp60-like protein (CNAG_03891) (Cunha et al. 2002), heat shock 70 kDa protein 4 (CNAG_06208) (Bisio et al. 2005), triose-phosphate isomerase (TPI) (CNAG_02035), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH)(CNAG_06699) (da Fonseca et al. 2001). Vaccination using Hsp60 induced protective cellular immune response against experimental pulmonary PCM (de Bastos Ascenco Soares et al. 2008), while in H. capsulatum it is involved in fungal attachment to macrophage CD11/CD18 receptors (Long et al. 2003). Importantly, TPI, GAPDH and Hsp60 are predicted adhesins in several fungal species (Chaudhuri et al. 2011) and may prompt fungal invasion. Enolase (CNAG_03072) can bind and activate plasminogen to plasmin, a serine protease involved in P. brasiliensis invasion to host tissues (Nogueira et al. 2010). In C. albicans, cell wall enolase and TPI can bind to human kininogen, a kinin precursor related to human host response to microbial infections (Karkowska-Kuleta et al. 2011).
Overall, the present work improves our knowledge about the poorly studied C. neoformans cell wall, opening doors to understanding its possible roles in fungal biology and pathogenesis. In addition, comparisons of the proteomic and lipidomic data with those from fungal EVs support the idea that these structures are at least partially responsible for adding components to the cell wall (Rodrigues et al. 2011).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Marcio L. Rodrigues, Leonardo Nimrichter, and Luiz R. Travassos for discussion. We thank Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for the grant Proc. No. 2013/25950-1 and for scholarships (RPS, MCV), CAPES for the Graduation Program financial support, and CNPq for RP a productivity fellowship. ICA was partially supported by the grant 2G12MD007592 from the National Institute on Minority Health and Health Disparities (NIMHD), a component of the National Institutes of Health (NIH). FGL was supported by a fellowship from the Research Initiative for Scientific Enhancement (RISE) program (NIH/NIGMS grant # 2R25GM069621-09) at UTEP. ICA is special visiting researcher of the Ciência Sem Fronteiras (Science Without Borders) Program, Brazil. We are grateful to the Biomolecule Analysis Core Facility at the BBRC/Biology/UTEP, funded by the NIH/NIMHD grant # 2G12MD007592, for the access to the ESI-MS and LC-MS/MS instruments.
LITERATURE CITED
- Albuquerque PC, Nakayasu ES, Rodrigues ML, Frases S, Casadevall A, Zancope-Oliveira RM, Almeida IC, Nosanchuk JD. Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell Microbiol. 2008;10:1695–1710. doi: 10.1111/j.1462-5822.2008.01160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisio LC, Silva SP, Pereira IS, Xavier MA, Venancio EJ, Puccia R, Soares CM, Felipe MS. A new Paracoccidioides brasiliensis 70-kDa heat shock protein reacts with sera from paracoccidioidomycosis patients. Med Mycol. 2005;43:495–503. doi: 10.1080/13693780400029478. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Nosanchuk JD, Williamson P, Rodrigues ML. Vesicular transport across the fungal cell wall. Trends Microbiol. 2009;17:158–162. doi: 10.1016/j.tim.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova M, Gil ML, Cardenoso L, Martinez JP, Sentandreu R. Identification of wall-specific antigens synthesized during germ tube formation by Candida albicans. Infect Immun. 1989;57:262–271. doi: 10.1128/iai.57.1.262-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattaway FW, Holmes MR, Barlow AJ. Cell wall composition of the mycelial and blastospore forms of Candida albicans. J Gen Microbiol. 1968;51:367–376. doi: 10.1099/00221287-51-3-367. [DOI] [PubMed] [Google Scholar]
- Chaudhuri R, Ansari FA, Raghunandanan MV, Ramachandran S. FungalRV: adhesin prediction and immunoinformatics portal for human fungal pathogens. BMC Genomics. 2011;12:192. doi: 10.1186/1471-2164-12-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Park J, Kim D, Jung K, Kang S, Lee YH. Fungal secretome database: integrated platform for annotation of fungal secretomes. BMC Genomics. 2010;11:105. doi: 10.1186/1471-2164-11-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucanu I, Kerek F. A Simple and Rapid Method for the Permethylation of Carbohydrates. Carbohydr. Res. 1984;131:209–217. [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cunha DA, Zancope-Oliveira RM, Sueli M, Felipe S, Salem-Izacc SM, Deepe GS, Jr, Soares CM. Heterologous expression, purification, and immunological reactivity of a recombinant HSP60 from Paracoccidioides brasiliensis. Clin Diagn Lab Immunol. 2002;9:374–377. doi: 10.1128/CDLI.9.2.374-377.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca CA, Jesuino RS, Felipe MS, Cunha DA, Brito WA, Soares CM. Two-dimensional electrophoresis and characterization of antigens from Paracoccidioides brasiliensis. Microbes Infect. 2001;3:535–542. doi: 10.1016/s1286-4579(01)01409-5. [DOI] [PubMed] [Google Scholar]
- de Bastos Ascenco Soares R, Gomez FJ, de Almeida Soares CM, Deepe GS., Jr Vaccination with heat shock protein 60 induces a protective immune response against experimental Paracoccidioides brasiliensis pulmonary infection. Infect Immun. 2008;76:4214–4221. doi: 10.1128/IAI.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot PW, Ram AF, Klis FM. Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet Biol. 2005;42:657–675. doi: 10.1016/j.fgb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Feldmesser M, Kress Y, Novikoff P, Casadevall A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster AJ, Bird RA, Smith SN. Biotinylation and characterization of Cryptococcus neoformans cell surface proteins. J Appl Microbiol. 2007;103:390–399. doi: 10.1111/j.1365-2672.2006.03259.x. [DOI] [PubMed] [Google Scholar]
- Gehrmann U, Qazi KR, Johansson C, Hultenby K, Karlsson M, Lundeberg L, Gabrielsson S, Scheynius A. Nanovesicles from Malassezia sympodialis and host exosomes induce cytokine responses--novel mechanisms for host-microbe interactions in atopic eczema. PLoS One. 2011;6:e21480. doi: 10.1371/journal.pone.0021480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NA, Hube B. Importance of the Candida albicans cell wall during commensalism and infection. Curr Opin Microbiol. 2012;15:406–412. doi: 10.1016/j.mib.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Guimaraes AJ, de Cerqueira MD, Nosanchuk JD. Surface architecture of Histoplasma capsulatum. Front Microbiol. 2011;2:225. doi: 10.3389/fmicb.2011.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James PG, Cherniak R, Jones RG, Stortz CA, Reiss E. Cell wall glucans of Cryptococcus neoformans Cap 67. Carbohydr Res. 1990;198:23–38. doi: 10.1016/0008-6215(90)84273-w. [DOI] [PubMed] [Google Scholar]
- Kanetsuna F, Carbonell LM, Moreno RE, Rodriguez J. Cell wall composition of the yeast and mycelial forms of Paracoccidioides brasiliensis. J Bacteriol. 1969;97:1036–1041. doi: 10.1128/jb.97.3.1036-1041.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkowska-Kuleta J, Kedracka-Krok S, Rapala-Kozik M, Kamysz W, Bielinska S, Karafova A, Kozik A. Molecular determinants of the interaction between human high molecular weight kininogen and Candida albicans cell wall: Identification of kininogen-binding proteins on fungal cell wall and mapping the cell wall-binding regions on kininogen molecule. Peptides. 2011;32:2488–2496. doi: 10.1016/j.peptides.2011.10.021. [DOI] [PubMed] [Google Scholar]
- Latge JP. The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol. 2007;66:279–290. doi: 10.1111/j.1365-2958.2007.05872.x. [DOI] [PubMed] [Google Scholar]
- Long KH, Gomez FJ, Morris RE, Newman SL. Identification of heat shock protein 60 as the ligand on Histoplasma capsulatum that mediates binding to CD18 receptors on human macrophages. J Immunol. 2003;170:487–494. doi: 10.4049/jimmunol.170.1.487. [DOI] [PubMed] [Google Scholar]
- Longo LV, Nakayasu ES, Gazos-Lopes F, Vallejo MC, Matsuo AL, Almeida IC, Puccia R. Characterization of cell wall lipids from the pathogenic phase of Paracoccidioides brasiliensis cultivated in the presence or absence of human plasma. PLoS One. 2013a;8:e63372. doi: 10.1371/journal.pone.0063372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo LV, Nakayasu ES, Matsuo AL, Peres da Silva R, Sobreira TJ, Vallejo MC, Ganiko L, Almeida IC, Puccia R. Identification of human plasma proteins associated with the cell wall of the pathogenic fungus Paracoccidioides brasiliensis. FEMS Microbiol Lett. 2013b;341:87–95. doi: 10.1111/1574-6968.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo LVG, Cunha JPC, Sobreira TJP, Puccia R. Proteome of cell wall-extracts from pathogenic Paracoccidioides brasiliensis: Comparison among morphological phases, isolates, and reported fungal extracellular vesicle proteins. EuPA Open Proteomics. 2014;3:216–228. [Google Scholar]
- Mody CH, Sims KL, Wood CJ, Syme RM, Spurrell JC, Sexton MM. Proteins in the cell wall and membrane of Cryptococcus neoformans stimulate lymphocytes from both adults and fetal cord blood to proliferate. Infect Immun. 1996;64:4811–4819. doi: 10.1128/iai.64.11.4811-4819.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu ES, Sobreira TJ, Torres R, Jr, Ganiko L, Oliveira PS, Marques AF, Almeida IC. Improved proteomic approach for the discovery of potential vaccine targets in Trypanosoma cruzi. J Proteome Res. 2012;11:237–246. doi: 10.1021/pr200806s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel W. Pathways of unconventional protein secretion. Curr Opin Biotechnol. 2010;21:621–626. doi: 10.1016/j.copbio.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira SV, Fonseca FL, Rodrigues ML, Mundodi V, Abi-Chacra EA, Winters MS, Alderete JF, de Almeida Soares CM. Paracoccidioides brasiliensis enolase is a surface protein that binds plasminogen and mediates interaction of yeast forms with host cells. Infect Immun. 2010;78:4040–4050. doi: 10.1128/IAI.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DL, Nakayasu ES, Joffe LS, Guimaraes AJ, Sobreira TJ, Nosanchuk JD, Cordero RJ, Frases S, Casadevall A, Almeida IC, Nimrichter L, Rodrigues ML. Characterization of yeast extracellular vesicles: evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS One. 2010;5:e11113. doi: 10.1371/journal.pone.0011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR, Casadevall A. Cryptococcosis. Infect Dis Clin North Am. 2002;16:837–874. v–vi. doi: 10.1016/s0891-5520(02)00036-3. [DOI] [PubMed] [Google Scholar]
- Pitarch A, Sanchez M, Nombela C, Gil C. Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol Cell Proteomics. 2002;1:967–982. doi: 10.1074/mcp.m200062-mcp200. [DOI] [PubMed] [Google Scholar]
- Previato JO, G P, Haskins RH, Travassos LR. Soluble and Insoluble Glucans from Different Cell-Types of the Human Pathogen Sporothrix schenckii. Experimental Mycology. 1979;3:92–105. [Google Scholar]
- Puccia R, Vallejo MC, Matsuo AL, Longo LV. The Paracoccidioides cell wall: past and present layers toward understanding interaction with the host. Front Microbiol. 2011;2:257. doi: 10.3389/fmicb.2011.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulfer M, Murphy RC. Electrospray mass spectrometry of phospholipids. Mass Spectrom Rev. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- Rawat DS, Upreti HB, Das SK. Lipid composition of Cryptococcus neoformans. Microbiologica. 1984;7:299–307. [PubMed] [Google Scholar]
- Rittershaus PC, Kechichian TB, Allegood JC, Merrill AH, Jr, Hennig M, Luberto C, Del Poeta M. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J Clin Invest. 2006;116:1651–1659. doi: 10.1172/JCI27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Franzen AJ, Nimrichter L, Miranda K. Vesicular mechanisms of traffic of fungal molecules to the extracellular space. Curr Opin Microbiol. 2013;16:414–420. doi: 10.1016/j.mib.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Rodrigues ML, Nosanchuk JD, Schrank A, Vainstein MH, Casadevall A, Nimrichter L. Vesicular transport systems in fungi. Future Microbiol. 2011;6:1371–1381. doi: 10.2217/fmb.11.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, Almeida IC, Casadevall A. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell. 2008;7:58–67. doi: 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Travassos LR, Miranda KR, Franzen AJ, Rozental S, de Souza W, Alviano CS, Barreto-Bergter E. Human antibodies against a purified glucosylceramide from Cryptococcus neoformans inhibit cell budding and fungal growth. Infect Immun. 2000;68:7049–7060. doi: 10.1128/iai.68.12.7049-7060.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, Alvarez M, Nakouzi A, Feldmesser M, Casadevall A. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell. 2007;6:48–59. doi: 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera J, Elorza MV, Valentin E, Sentandreu R. Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res. 2006;6:14–29. doi: 10.1111/j.1567-1364.2005.00017.x. [DOI] [PubMed] [Google Scholar]
- Russell WK, Park ZY, Russell DH. Proteolysis in mixed organic-aqueous solvent systems: applications for peptide mass mapping using mass spectrometry. Anal Chem. 2001;73:2682–2685. doi: 10.1021/ac001332p. [DOI] [PubMed] [Google Scholar]
- San-Blas G, San-Blas F. Molecular aspects of fungal dimorphism. Crit Rev Microbiol. 1984;11:101–127. doi: 10.3109/10408418409105474. [DOI] [PubMed] [Google Scholar]
- Srikanta D, Santiago-Tirado FH, Doering TL. Cryptococcus neoformans: historical curiosity to modern pathogen. Yeast. 2014;31:47–60. doi: 10.1002/yea.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefsen B, Grijpstra J, Ordonez S, Lammers M, van Die I, de Cock H. Deletion of the CAP10 gene of Cryptococcus neoformans results in a pleiotropic phenotype with changes in expression of virulence factors. Res Microbiol. 2014 doi: 10.1016/j.resmic.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Thevissen K, de Mello Tavares P, Xu D, Blankenship J, Vandenbosch D, Idkowiak-Baldys J, Govaert G, Bink A, Rozental S, de Groot PW, Davis TR, Kumamoto CA, Vargas G, Nimrichter L, Coenye T, Mitchell A, Roemer T, Hannun YA, Cammue BP. The plant defensin RsAFP2 induces cell wall stress, septin mislocalization and accumulation of ceramides in Candida albicans. Mol Microbiol. 2012;84:166–180. doi: 10.1111/j.1365-2958.2012.08017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo MS, Suzuki E, Levery SB, Straus AH, Takahashi HK. Characterization of monoclonal antibody MEST-2 specific to glucosylceramide of fungi and plants. Glycobiology. 2001;11:105–112. doi: 10.1093/glycob/11.2.105. [DOI] [PubMed] [Google Scholar]
- Urban C, Sohn K, Lottspeich F, Brunner H, Rupp S. Identification of cell surface determinants in Candida albicans reveals Tsa1p, a protein differentially localized in the cell. FEBS Lett. 2003;544:228–235. doi: 10.1016/s0014-5793(03)00455-1. [DOI] [PubMed] [Google Scholar]
- Vallejo MC, Nakayasu ES, Matsuo AL, Sobreira TJ, Longo LV, Ganiko L, Almeida IC, Puccia R. Vesicle and vesicle-free extracellular proteome of Paracoccidioides brasiliensis: comparative analysis with other pathogenic fungi. J Proteome Res. 2012a;11:1676–1685. doi: 10.1021/pr200872s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo MC, Nakayasu ES, Longo LV, Ganiko L, Lopes FG, Matsuo AL, Almeida IC, Puccia R. Lipidomic analysis of extracellular vesicles from the pathogenic phase of Paracoccidioides brasiliensis. PLoS One. 2012b;7:e39463. doi: 10.1371/journal.pone.0039463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartivarian SE, Reyes GH, Jacobson ES, James PG, Cherniak R, Mumaw VR, Tingler MJ. Localization of mannoprotein in Cryptococcus neoformans. J Bacteriol. 1989;171:6850–6852. doi: 10.1128/jb.171.12.6850-6852.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli A, Pericolini E, Gabrielli E, Kenno S, Perito S, Cenci E, Monari C. Elucidating the immunological function of the Cryptococcus neoformans capsule. Future Microbiol. 2013;8:1107–1116. doi: 10.2217/fmb.13.84. [DOI] [PubMed] [Google Scholar]
- Wolf J, Espadas-Moreno J, Luque-Garcia JL, Casadevall A. Interaction of Cryptococcus neoformans extracellular vesicles with the cell wall. Eukaryot Cell. 2014 doi: 10.1128/EC.00111-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yichoy M, Nakayasu ES, Shpak M, Aguilar C, Aley SB, Almeida IC, Das S. Lipidomic analysis reveals that phosphatidylglycerol and phosphatidylethanolamine are newly generated phospholipids in an early-divergent protozoan, Giardia lamblia. Mol Biochem Parasitol. 2009;165:67–78. doi: 10.1016/j.molbiopara.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.