Abstract
Antibiotic administration disrupts the intestinal microbiota, increasing susceptibility to pathogens such as Clostridium difficile. Metronidazole or oral vancomycin can cure C. difficile infection, and administration of these agents to prevent C. difficile infection in high-risk patients, although not sanctioned by Infectious Disease Society of America guidelines, has been considered. The relative impacts of metronidazole and vancomycin on the intestinal microbiota and colonization resistance are unknown. We investigated the effect of brief treatment with metronidazole and/or oral vancomycin on susceptibility to C. difficile, vancomycin-resistant Enterococcus, carbapenem-resistant Klebsiella pneumoniae, and Escherichia coli infection in mice. Although metronidazole resulted in transient loss of colonization resistance, oral vancomycin markedly disrupted the microbiota, leading to prolonged loss of colonization resistance to C. difficile infection and dense colonization by vancomycin-resistant Enterococcus, K. pneumoniae, and E. coli. Our results demonstrate that vancomycin, and to a lesser extent metronidazole, are associated with marked intestinal microbiota destruction and greater risk of colonization by nosocomial pathogens.
Keywords: Clostridium difficile, metronidazole, vancomycin, microbiota, colonization resistance
Clostridium difficile is an intestinal pathogen that causes a wide spectrum of disease in hospitalized patients, ranging from diarrhea to pseudomembranous colitis to potentially lethal toxic megacolon [reviewed in 1]. The past 15 years have seen a dramatic increase in the number of cases, with strains emerging that produce increased amounts of toxin and are resistant to fluoroquinolone antibiotics [2, 3]. The Centers for Disease Control and Prevention (CDC) classifies C. difficile as “threat level urgent,” a rating intended to call for “urgent and aggressive action” against this bacterium [4]. Recent CDC estimates indicate that C. difficile infections kill 14 000 individuals and lead to >$1 billion in medical costs each year in the United States. These infections are also becoming more common outside hospitals, particularly among elderly individuals living in assisted care facilities but also in the general community [5, 6].
Antibiotic treatment is key to both the initiation and resolution of C. difficile infections. One European study found that 92% of patients with a diagnosis of C. difficile infection reported antibiotic use during the preceding 3 months, with third-generation cephalosporins and clindamycin most strongly correlated with the risk of subsequent C. difficile infection [7–9]. Once C. difficile infection has been diagnosed, standard protocols call for administration of more antibiotics: metronidazole for mild and moderate cases and vancomycin for severe infection [10]. Vancomycin yields a slightly higher cure rate in some patients, but concerns over cost and the emergence of vancomycin-resistant pathogens limit its use to patients with severe disease [11–13].
Growing concern over the rise in C. difficile infections has prompted some physicians to prescribe metronidazole prophylactically to high-risk patients [14]. The Infectious Disease Society of America recommends prescribing vancomycin to patients with suspected severe or severe complicated C. difficile illness before infection has been confirmed; however, it is estimated that many clinicians start empiric therapy based on suspicion of mild or moderate disease, with questionable therapeutic benefit [15]. These practices are concerning, because little is known about how metronidazole and vancomycin impact commensal bacteria and how they alter the host's susceptibility to other enteric pathogens commonly encountered in hospital settings [16]. To address this problem, we treated mice with brief courses of metronidazole, vancomycin, or both in combination, and assessed the impact on native bacterial populations as well as colonization resistance to C. difficile spores. We then extended our investigation to other hospital-acquired infections and asked whether the same antibiotic regimens affected the microbiota's ability to suppress infections in mice challenged with vancomycin-resistant Enterococcus (VRE), carbapenem-resistant Klebsiella pneumoniae (KPC), and Escherichia coli.
METHODS
Mouse Husbandry
All experiments were performed with wild-type female C57BL/6 mice, aged 6–8 weeks and purchased from Jackson Laboratories. The mice were housed in the specific pathogen-free facility at Memorial Sloan Kettering's Animal Resource Center, fed irradiated feed, and provided with acidified water. Three of us (B. B. L., C. G. B., and R. A. C.) performed all mouse experiments and changed cages at least once per week. The experiments were performed in compliance with Memorial Sloan Kettering's institutional guidelines and were approved by its Institutional Animal Care and Use Committee.
C. difficile Susceptibility Time Course Experiments
C. difficile susceptibility was assessed as described by Buffie et al [17], with different antibiotic treatments: we used metronidazole (1.0 g L−1; Sigma-Aldrich), vancomycin (1.0 g L−1; NOVAPLUS), or 1.0 g L−1 of both antibiotics. Mice were housed in groups of 5 during antibiotic treatment. At days 1, 3, 7–8, 14–15, and 21–22 after cessation of antibiotics, 1 mouse was removed from each group cage, transferred to an individual cage, and inoculated with 1000 spores of C. difficile strain VPI 10463 (American Type Culture Collection 43255). Each experiment tested 3 groups of mice per antibiotic treatment, and experiments were repeated twice (n = 9 per time point per treatment group tested).
In one group of experiments, mice were administered antibiotics (metronidazole or metronidazole in combination with vancomycin) by oral gavage instead of the drinking water; 3.5 mg of antibiotics was dissolved in 200 µL of water and administered every day for 3 days. At 24 hours after the final dose, corresponding to day 1 of the previous experiments, mice were then challenged with C. difficile spores, as described elsewhere [19].
Quantitative C. difficile Culture
The C. difficile burden in mouse ceca 24 hours after infection was assessed as described elsewhere [17].
Sample Collection, DNA Extraction, and Quantification of 16S Copy Number Density
Intestinal content samples were collected from mice on days −3, 1, 3, 7–8, 14–15, and 21–22 (feces) and 24 hours after infection on days 2, 4, 8–9, 15–16, and 22–23 (colon). The samples were immediately flash-frozen and DNA was extracted as described elsewhere [18]. Briefly, the frozen samples (approximately 100 mg) were suspended in 500 µL extraction buffer (200 mmol/L Tris, pH 8.0/200 mmol/L sodium chloride/20 mol/L ethylenediaminetetraacetic acid), 200 µL of 20% sodium dodecyl sulfate, 500 µL of phenol–chloroform–isoamyl alcohol (24:24:1), and 500 µL of 0.1-mm-diameter zirconia/silica beads (BioSpec Products). Bacterial cells were lysed with bead beating (BioSpec Products) for 2 minutes, and DNA was isolated with 2 rounds of phenol–chloroform–isoamyl alcohol extraction. After extraction, the DNA was precipitated in ethanol and resuspended in 200 µL of Tris-EDTA buffer with 100 µg/mL RNAse, and further purified with QIAmp Mini Spin Columns (Qiagen).
The DNA extracted from fecal samples was then subjected to reverse-transcription polymerase chain reaction (PCR) of 16S RNA gene sequences. We used the broad-range bacterial 16S primers 517F (5′- GCCAGCAGCCGCGCTAA -3′) and 798R (5′- AGGGTATCTAATCCT -3′) at 0.2 mmol/L concentrations with the DyNAmo SYBR Green quantitative PCR kit (Finnzymes). Sample amplification was compared with standard curves to quantify 16S copy number. Cycling conditions were as follows: 95°C for 15 minutes and then 40 cycles of 94°C for 15 seconds, 52°C for 30 seconds, and 72°C for 30 seconds. The program finished with 95°C for 15 minutes, 60°C for 1 minute, and 95°C for 15 minutes.
16S Ribosomal RNA Gene Amplification, Multiparallel Sequencing, and Sequence Analysis
Amplicons of the V4-V5 region of 16S ribosomal RNA (rRNA) were amplified and sequenced with the Illumina MiSeq platform, as described elsewhere [19]. Sequences were analyzed using the mothur pipeline, version 1.33.3 [20], as described [19]. Operational taxonomic units (OTUs) were classified with a modified Greengenes reference database [21]. OTU-based microbial diversity was estimated by calculating the inverse Simpson index and Bray–Curtis dissimilarity index. Phylogenetic trees were inferred by using Clearcut [22] on the alignment created by mothur, and then unweighted UniFrac [23] was run on the resulting tree, followed by a principal coordinate analysis of the distance matrix.
VRE, KPC, and E. coli Susceptibility Time Course Experiments
Mice were treated with metronidazole (1.0 g L−1), vancomycin (1.0 g L−1), or no antibiotic for 3 days, then switched to untreated water. At day 1, 7, or 14 after stopping antibiotic treatment, mice were orally gavaged with 50 000 colony-forming units (CFUs) of vancomycin-resistant Enterococcus faecium (American Type Culture Collection 700221), KPC, or E. coli (KPC and E. coli were both isolated from blood cultures collected from patients at Memorial Sloan Kettering Cancer Center). All mice were singly housed after infection with the indicated pathogen. At 24 hours after infection, fecal samples were collected from the mice, and the corresponding pathogen burden was enumerated by plating serial dilutions on selective agar plates (VRE: EnterococcoseI agar plates [Difco] with vancomycin [8 µg/mL]; KPC: Luria Broth with agar [Difco], carbenacillin [100 µg/mL; LabScientific], and neomycin [50 µg/mL; Sigma-Aldrich]; E. coli: Luria Broth with agar and neomycin [50 µg/mL]).
Statistical Analysis
Statistical analysis was conducted using the R (version 3.1.1) and GraphPad Prism (version 6.0c) software packages. Analysis of the number of 16S rRNA copies, inverse Simpson indices, and Bray–Curtis dissimilarity was performed using 2-way analysis of variance with Bonferroni correction and Prism software. Analysis of differential VRE, KPC, or E. coli burden was performed using a Kruskal–Wallis test with Dunn correction. Differences were considered significant at P < .05. For the principal coordinate analysis, the analysis of molecular variance method [24] was used to compare samples that supported C. difficile growth (susceptible samples) with samples in which no C. difficile was detected (resistant samples).
RESULTS
Differential Impact of Metronidazole and Vancomycin Treatment on C. difficile Susceptibility
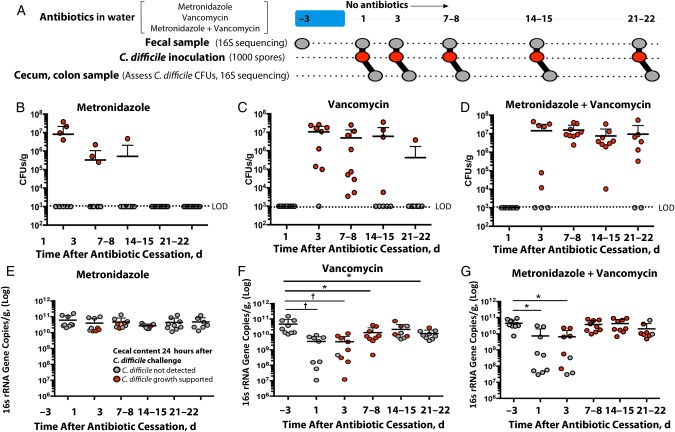
In the absence of pretreatment with antibiotics, administration of C. difficile spores does not lead to infection or colonization of conventionally housed wild-type C57BL/6 mice [25, 17]. To determine the impact of metronidazole and oral vancomycin on susceptibility to C. difficile infection, we treated mice with metronidazole, vancomycin, or metronidazole plus vancomycin in their drinking water for 3 days (1.0 g L−1 for each). Each treatment protocol was tested in 3 replicate cages. After 3 days, antibiotic-containing water was replaced with untreated water for the remainder of the experiment (Figure 1A). At days 1, 3, 7–8, 14–15, and 21–22 after stopping antibiotic treatment, 1 mouse was removed from each cage at random and challenged with 1000 spores of C. difficile strain VPI 10463 by oral gavage. Each infected mouse was housed individually to prevent cross-contamination. At 24 hours after infection, the mice were euthanized, and cecal and colonic contents were collected for assessment of C. difficile burden and sequencing, respectively. Each experiment tested 3 mice per antibiotic treatment and was conducted 3 separate times (n = 9 for each treatment and time point).
Figure 1.
Exposure to metronidazole, vancomycin, or both increases susceptibility to Clostridium difficile infection. A, Summary of experimental protocol. B–D, Burden of C. difficile in ceca of mice 24 hours after infection. E–G, Intestinal bacterial density of animals before and after antibiotic exposure. Horizontal lines represent means; error bars, standard deviations (n = 9 except for metronidazole + vancomycin–treated mice on days 21–22, where n = 8). *P < .05; †P < .01. Abbreviations: CFUs, colony-forming units; LOD, limit of detection.
Of the mice treated with metronidazole alone, approximately half (56%) had no detectable C. difficile in their ceca when challenged 1 day after antibiotic removal (Figure 1B). When mice were challenged 3 and 7–8 days after stopping metronidazole treatment, the proportion with undetectable C. difficile levels increased to 67% and 89% of mice, respectively. All mice had recovered colonization resistance 2 weeks after stopping metronidazole treatment.
In contrast, none of the mice treated with vancomycin had detectable C. difficile in their ceca 1 day after stopping treatment (Figure 1C). Vancomycin has been shown to persist in the lower intestine of mice and humans days after the last dose has stopped, so it is likely that residual vancomycin killed germinating spores at this early time point [16]. Indeed, by days 3 and 7–8, almost all mice become heavily colonized (89% of mice on day 3, 100% on days 7–8). By the last time point studied, only 1 mouse had detectable C. difficile CFUs in its cecum. The disruption in colonization resistance was more pronounced in mice treated with both metronidazole and vancomycin, 78% of which had ceca that supported C. difficile growth 3 weeks after stopping antibiotic treatment (Figure 1D).
To investigate the causes of the loss of colonization resistance to C. difficile infection, we first assessed changes in total bacterial density by quantitative reverse-transcription PCR targeted to bacterial 16S rRNA-encoding genes. We found that the density in metronidazole-treated mice remained relatively stable throughout the course of the experiment (Figure 1E), in contrast to the vancomycin-treated mice (Figure 1F) and metronidazole plus vancomycin-treated mice (Figure 1G). Bacterial density approached baseline levels by day 14–15 of the experiment. However, because many of the mice had not reestablished colonization resistance to C. difficile at this time, the change in bacterial density did not fully explain changes in C. difficile colonization resistance (Figure 1B–D).
Prolonged Impact of Metronidazole and Vancomycin Treatment on Intestinal Bacterial Populations
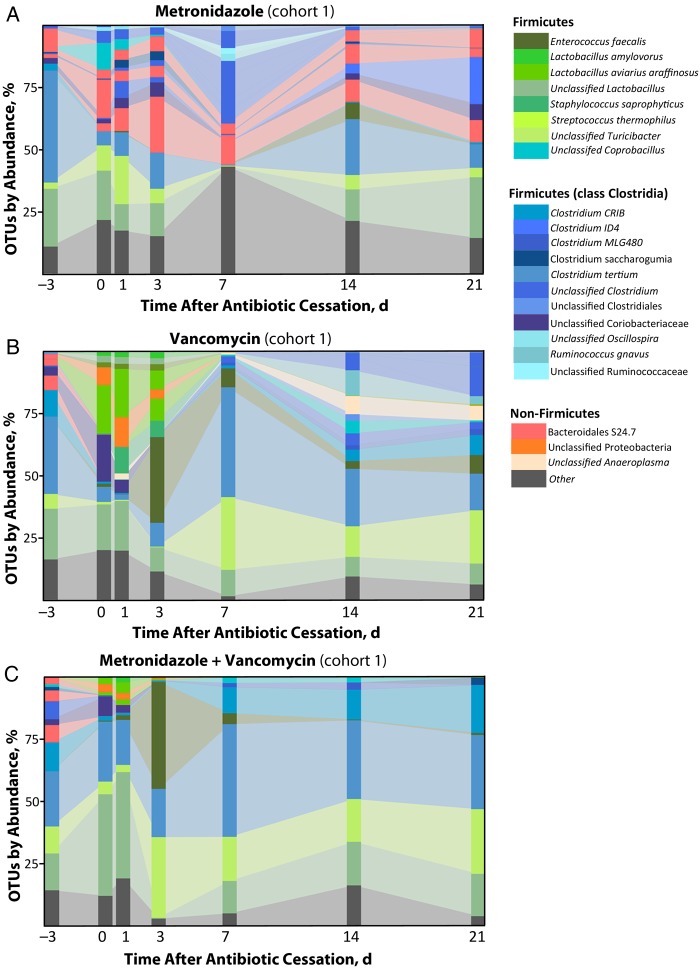
We next expanded our investigation beyond density measures to examine changes in bacterial composition over time. DNA extracted from fecal samples was sequenced on the Illumina MiSeq platform and reads were analyzed using an in-house pipeline mainly derived from the mothur software [20] (see “Methods” section). We found that before antibiotic treatment was initiated, fecal samples were composed of a rich community of bacterial OTUs spanning multiple taxa, including the Bacteroidales class S24.7 as well as many members of the Lactobacillus and Clostridium genera, among others (Figure 2A–C; day −3 time point).
Figure 2.
Exposure to metronidazole, vancomycin, or both disrupts commensal bacterial species found in the lower intestine. Fecal samples were collected from mice before antibiotic treatment (day −3) and at the indicated times after cessation of antibiotics. Samples were assessed for bacterial operational taxonomic units (OTUs), as described in “Methods” section. Each stacked bar represents the mean microbiota composition of 3 independently housed mice.
Animals treated with metronidazole alone experienced relatively transient disruptions in their fecal microbial communities, and returned to a state similar to pretreatment composition by 1–2 weeks after stopping treatment (Figure 2A). In contrast, animals treated with vancomycin or metronidazole plus vancomycin experienced a profound shift in their microbiota composition, with taxa such as Bacteroidales S24–7 falling permanently below the limit of detection (Figure 2B and 2C). In parallel, bacterial OTUs present at low or undetectable levels before antibiotics expanded greatly, including members of the genus Enterococcus, unclassified Proteobacteria, and novel members of Lactobacillus and Clostridium.
Repeating these experiments with new cohorts of mice revealed that the expanding bacterial populations were highly dependent on the initial commensal bacteria present in the mice, and that different mouse cohorts (all wild-type C57BL/6 mice obtained from the Jackson Labs) were colonized with different communities (Supplementary Figure 1). In 3 separate experiments performed with mice obtained ≥1 month apart, vancomycin treatment led to the expansion of primarily Lactobacillus aviarius and Enterococcus faecalis (Figure 2B, cohort 1), Klebsiella oxytoca (Figure 3C, cohort 3), or K. oxytoca and Akkermansia muciniphilia (Figure 3D, cohort 4).
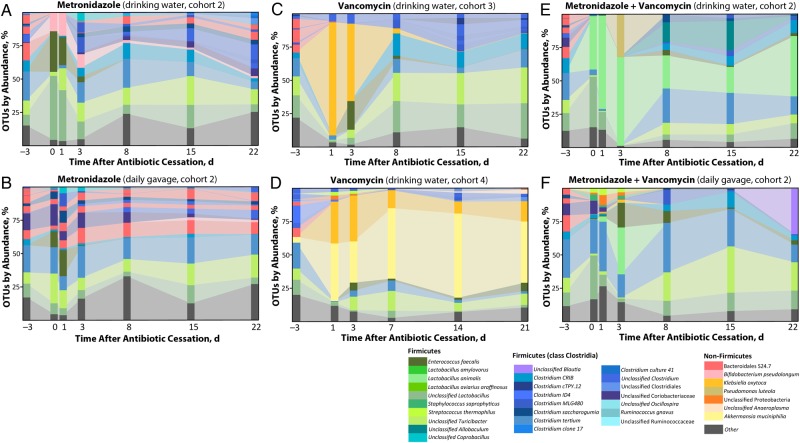
Figure 3.
Impact of starting microbiota and route of antibiotic administration on resulting changes in bacterial composition. The experiment represented in Figure 2 (mouse cohort 1) was repeated with different cohorts of wild-type C57BL/6 Jackson mice. Mice from cohort 2 were subdivided into 2 additional treatment groups, with the first group receiving antibiotics dissolved in water (A, E), and the other receiving antibiotics by daily oral gavage (B, F). Each stacked bar represents a mean of 3 mice per time point per treatment group (exception: F at day 22, where n = 2). Abbreviation: OTUs, operational taxonomic units.
Many patients report that metronidazole has an unpleasant metallic taste, which could affect how much treated water the mice drink during the 3 days of antibiotic administration. We therefore gavaged mice every day for 3 days with metronidazole or metronidazole plus vancomycin (using doses that corresponded to the measured daily intake of water in untreated mice) and then challenged them with C. difficile spores, as described elsewhere [19]. Although the mice administered antibiotics via oral gavage seemed to preserve slightly more diversity, as measured by the inverse Simpson and Bray–Curtis indices, this trend was not statistically significant (Figure 3B, 3F, and Supplementary Figure 2). These results indicate that the timing of antibiotic administration has less of an impact on bacterial shifts than the initial microbial populations present in the mice before treatment begins. The effects of antibiotic treatments on microbial composition and diversity are summarized in Figure 4 and Supplementary Figure 3.
Figure 4.
Summary of effects of metronidazole and vancomycin on fecal microbial populations. Sequences from fecal samples were binned into operational taxonomic units (OTUs), and abundance of the 50 most highly represented OTUs was plotted based on percentage of total sequences. Each horizontal bar represents 1 OTU; each vertical bar represents the mean abundance of individual OTUs within 1 cohort of mice at specified time point and treatment group (n = 3 for each vertical bar).
Correlation of Antibiotic Treatment-Mediated Changes in Commensal Bacterial Populations With C. difficile Colonization Resistance
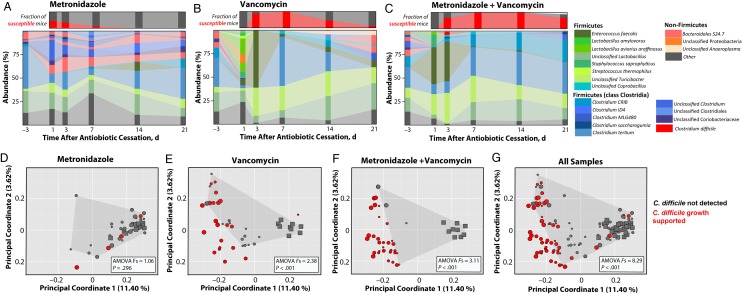
We next asked whether the observed shifts in commensal microbial populations after antibiotic treatment corresponded with susceptibility to C. difficile spore challenge. At 24 hours after infection, colonic content was collected from mice, sequenced, and analyzed as described elsewhere. Mice treated with metronidazole alone were found to maintain a relatively stable microbiota, which corresponded with the rapid recovery of resistance to C. difficile (Figure 5A). In contrast, the colons of mice treated with vancomycin or metronidazole plus vancomycin were found to contain severely disrupted microbiota, which correlated with delays in returning to a colonization-resistant state (Figure 5B and 5C, respectively). Examination of microbial communities with principal coordinate analysis demonstrated that, in the case of vancomycin (Figure 5E) and metronidazole plus vancomycin treatment (Figure 5F), colon samples from mice that were unable to suppress C. difficile growth (susceptible mice) clustered separately from the samples in which C. difficile growth was undetected 24 hours after spore challenge (resistant mice) (analysis of molecular variance F statistic in vancomycin- vs metronidazole plus vancomycin–treated mice, 2.38 vs 3.11; both P < .001). These data demonstrate that not only do C. difficile– targeting antibiotics significantly alter intestinal microbial communities but these disrupted communities are also then more likely to support C. difficile expansion.
Figure 5.
Antibiotic-induced disruptions of microbial communities contribute to Clostridium difficile susceptibility. A–C, Colon samples were collected from mice 24 hours after C. difficile infection and assessed for abundance of individual bacterial operational taxonomic units (large panels). Each stacked bar represents mean microbiota composition of 3 independently housed mice from cohort 1. Small panels in A–C represent the fraction of mice found susceptible to C. difficile 24 hours after infection in all cohorts (red bar; n = 9 mice per time point). D–G, Principal coordinate analysis of colon samples from all cohorts 24 hours after infection. Squares represent preantibiotic samples; circles, postantibiotic treatment samples. Circle sizes represent the time point of each posttreatment sample, with large circles representing earliest time points. Analysis of molecular variance (AMOVA) F statistics were used to compare samples in which C. difficile was not detected (gray points bounded by shaded region) with samples that supported C. difficile growth (red points).
Impact of Metronidazole and Vancomycin Treatment on Susceptibility to Infection With Other Nosocomial Pathogens
We next asked whether the observed shifts in microbial composition after metronidazole or vancomycin treatment also contribute to heightened risk of colonization with other nosocomial infections. Of growing concern in many hospitals are VRE species, KPC, and E. coli infections [4]. We therefore treated a cohort of wild-type C57BL/6 Jackson mice with metronidazole or vancomycin for 3 days, followed by challenge with approximately 50 000 CFUs of VRE, KPC, or E. coli at 1, 7, or 14 days after antibiotic cessation (Figure 6). In all cases we saw a similar pattern of intestinal colonization. First, the untreated cohort was colonized at very low densities (or not at all) throughout the time course. Second, the metronidazole cohort displayed moderate bacterial burden at early time points, then recovered colonization resistance resembling that in the control mice. Finally, the vancomycin cohort remained highly susceptible to colonization throughout the duration of the experiment. Only in the VRE-infected group did the vancomycin-treated mice display a trend toward recovery of colonization resistance 2 weeks after antibiotic removal (Figure 6A).
Figure 6.
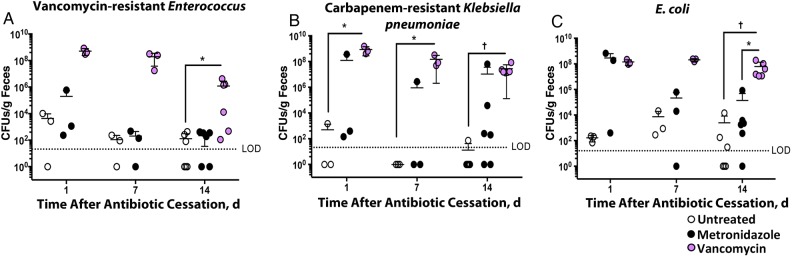
Exposure to metronidazole and vancomycin affects the colonization ability of vancomycin-resistant Enterococcus, carbapenem-resistant Klebsiella pneumoniae, and Escherichia coli. Mice were treated with indicated antibiotics for 3 days and then allowed to recover. After cessation of antibiotics, mice were challenged with approximately 50 000 colony-forming units (CFUs) of vancomycin-resistant Enterococcus (A), carbapenem-resistant K. pneumoniae (B), or E. coli (C). Fecal pellets were collected from mice 24 hours after infection and assessed for the corresponding pathogen burden. *P < .05, †P < .01. Horizontal lines represent means; error bars standard deviations (n = 3 [days 1 and 7] or n = 6 [day 14]). Abbreviation: LOD, limit of detection.
DISCUSSION
The high incidence of C. difficile infection in select, highly vulnerable patient populations has led to the consideration of using metronidazole or vancomycin prophylactically [15, 26]. Although it is not recommended by the Infectious Disease Society of America, clinicians caring for highly immunocompromised patients undergoing cancer treatment or organ or stem cell transplantation are increasingly administering these antibiotics to prevent C. difficile infections. Both metronidazole and vancomycin are known to effectively treat active infections, but their impact on commensal bacterial populations has not been extensively studied. We found that metronidazole treatment disrupts the microbiota initially, but the effect is transient and mice are able to regain colonization resistance to C. difficile relatively quickly. In contrast, vancomycin treatment causes severe shifts in commensal microbial species that correspond with prolonged susceptibility to C. difficile, an effect magnified when vancomycin is administered concurrently with metronidazole. The continued disruption of commensal bacterial species can help explain the high relapse rate associated with C. difficile disease, which most commonly occurs within a few weeks of successful treatment [reviewed in 27].
In addition to recurrent C. difficile episodes, a disordered commensal microbiota also leaves patients susceptible to other nosocomial infections. Of chief concern in hospitals are infections with VRE species, carbapenem-resistant Enterobacteriaceae, such as K. pneumoniae, and E. coli. These pathogens are found with increased prevalence in hospital settings, and many have acquired resistance to remaining available antibiotics [4]. We found that in addition to its effect of prolonged susceptibility to C. difficile infection, brief treatment with vancomycin opens a niche in the intestinal environment that allows dense colonization with VRE, KPC, and E. coli. Metronidazole treatment, however, has a more transient effect. When prescribing metronidazole or vancomycin for active C. difficile infection (or as empiric therapy for suspected cases), clinicians must take into account the collateral damage to protective bacterial species and take steps to prevent patient exposure to nosocomial pathogens long after treatment has been completed. One alternative is fidaxomicin, an antibiotic recently approved by the Food and Drug Administration for the treatment of C. difficile infections. In contrast to vancomycin and metronidazole, fidaxomicin has been demonstrated to have a narrower spectrum of activity against obligate anaerobes of the colon [28, 29]. Given its specificity, it would probably have less impact on colonization resistance, but this still requires further study.
More broadly speaking, our experiments reveal an important challenge regarding any investigation of commensal bacterial populations. Concordant with previous studies [30], we discovered that wild-type mice purchased from the same vendor harbored different microbes depending on the date and barrier facility from which they were obtained (Supplementary Figure 1). The differences in starting commensal species were magnified after conditioning antibiotic treatments (Figure 3C and 3D). This finding highlights the need to actively monitor the microbiota in laboratory animals even before starting experimental treatments.
Although vancomycin treatment is known to improve C. difficile symptoms, studies in mice demonstrate that it also increases recurrence rates more than metronidazole treatment [31]. Our experiments were specifically designed to study the impact of these 2 agents on the normal microbiota and their relative ability to destroy colonization resistance. Our findings demonstrate that oral vancomycin, and to a lesser extent metronidazole, leave the host far more vulnerable to infection with C. difficile but also to additional antibiotic-resistant bacterial species. Another antibiotic occasionally used to treat C. difficile infections (tigecycline) has been shown to disrupt colonization resistance in a similar manner [32]. In clinical circumstances where the risk of exposure to and acquisition of C. difficile spores is high, the short-term potential benefits of antibiotic prophylaxis with either metronidazole or vancomycin needs to be weighed against the increased long-term risk for infection with C. difficile, VRE, carbapenem-resistant Enterobacteriaceae, and E. coli.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank all members of the Eric Pamer Lab, Memorial Sloan Kettering Cancer Center, for their valuable suggestions.
Financial support. This work was supported by the National Institutes of Health (NIH) (grants R01 AI42135 and AI95706) and the Tow Foundation. B. B. L. and C. G. B. were supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences, NIH (award T32GM07739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-institutional MD-PhD Program).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Bartlett JG. Historical perspectives on studies of Clostridium difficile and C. difficile infection. Clin Infect Dis 2008; 46(suppl 1):S4–11. [DOI] [PubMed] [Google Scholar]
- 2.McDonald LC, Killgore GE, Thompson A et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med 2005; 353:2433–41. [DOI] [PubMed] [Google Scholar]
- 3.He M, Miyajima F, Roberts P et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 2013; 45:109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States. http://www.cdc.gov/drugresistance/threat-report-2013/ Accessed 16 January 2015.
- 5.Pépin J, Valiquette L, Alary M-E et al. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. Can Med Assoc J 2004; 171:466–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feuerstadt P, Das R, Brandt LJ. The evolution of urban C. difficile infection (CDI): CDI in 2009–2011 is less severe and has better outcomes than CDI in 2006–2008. Am J Gastroenterol 2014; 109:1265–76. [DOI] [PubMed] [Google Scholar]
- 7.Bauer MP, Notermans DW, van Benthem BH et al. Clostridium difficile infection in Europe: a hospital-based survey. Lancet 2011; 377:63–73. [DOI] [PubMed] [Google Scholar]
- 8.Slimings C, Riley TV. Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis. J Antimicrob Chemother 2014; 69:881–91. [DOI] [PubMed] [Google Scholar]
- 9.Garey KW, Sethi S, Yadav Y, DuPont HL. Meta-analysis to assess risk factors for recurrent Clostridium difficile infection. J Hosp Infect 2008; 70:298–304. [DOI] [PubMed] [Google Scholar]
- 10.Perras C, Tsakonas E, Ndegwa S, Conly J, Valiquette L, Farrah K. Vancomycin or metronidazole for treatment of Clostridium difficile infection: clinical and economic analyses. http://www.ncbi.nlm.nih.gov/pubmedhealth/PMH0063523/. Accessed 15 December 2014. [PubMed] [Google Scholar]
- 11.Drekonja DM, Butler M, MacDonald R et al. Comparative effectiveness of Clostridium difficile treatments: a systematic review. Ann Intern Med 2011; 155:839–47. [DOI] [PubMed] [Google Scholar]
- 12.Zar FA, Bakkanagari SR, Moorthi KM, Davis MB. A comparison of vancomycin and metronidazole for the treatment of Clostridium difficile-associated diarrhea, stratified by disease severity. Clin Infect Dis 2007; 45:302–7. [DOI] [PubMed] [Google Scholar]
- 13.Johnson S, Louie TJ, Gerding DN et al. Vancomycin, metronidazole, or tolevamer for Clostridium difficile infection: results from two multinational, randomized, controlled trials. Clin Infect Dis 2014; 59:345–54. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez S, Hernandez MB, Tarchini G et al. Risk of Clostridium difficile infection in hospitalized patients receiving metronidazole for a non-C difficile infection. Clin Gastroenterol Hepatol 2014; 12:1856–61. [DOI] [PubMed] [Google Scholar]
- 15.Saade E, Deshpande A, Kundrapu S et al. Appropriateness of empiric therapy in patients with suspected Clostridium difficile infection. Curr Med Res Opin 2013; 29:985–8. [DOI] [PubMed] [Google Scholar]
- 16.Abujamel T, Cadnum JL, Jury LA et al. Defining the vulnerable period for re-establishment of Clostridium difficile colonization after treatment of C. difficile infection with oral vancomycin or metronidazole. PLoS One 2013; 8:e76269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buffie CG, Jarchum I, Equinda M et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun 2012; 80:62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ubeda C, Taur Y, Jenq RR et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 2010; 120:4332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buffie CG, Bucci V, Stein RR et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015; 517:205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schloss PD, Westcott SL, Ryabin T et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009; 75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeSantis TZ, Hugenholtz P, Larsen N et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheneman L, Evans J, Foster JA. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics 2006; 22:2823–4. [DOI] [PubMed] [Google Scholar]
- 23.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 2005; 71:8228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology 2001; 26:32–46. [Google Scholar]
- 25.Chen X, Katchar K, Goldsmith JD et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology 2008; 135:1984–92. [DOI] [PubMed] [Google Scholar]
- 26.Owens RC. Clostridium difficile-associated disease: an emerging threat to patient safety: insights from the society of infectious diseases pharmacists. Pharmacotherapy 2006; 26:299–311. [DOI] [PubMed] [Google Scholar]
- 27.Aslam S, Hamill RJ, Musher DM. Treatment of Clostridium difficile-associated disease: old therapies and new strategies. Lancet Infect Dis 2005; 5:549–57. [DOI] [PubMed] [Google Scholar]
- 28.Louie TJ, Miller MA, Mullane KM et al. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med 2011; 364:422–31. [DOI] [PubMed] [Google Scholar]
- 29.Eyre DW, Babakhani F, Griffiths D et al. Whole-genome sequencing demonstrates that fidaxomicin is superior to vancomycin for preventing reinfection and relapse of infection with Clostridium difficile. J Infect Dis 2014; 209:1446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ubeda C, Lipuma L, Gobourne A et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med 2012; 209:1445–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warren CA, van Opstal EJ, Riggins MS et al. Vancomycin treatment's association with delayed intestinal injury, clostridial overgrowth, and recurrence of Clostridium difficile infection in mice. Antimicrob Agents Chemother 2013; 57:689–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bassis CM, Theriot CM, Young VB. Alteration of the murine gastrointestinal microbiota by tigecycline leads to increased susceptibility to Clostridium difficile infection. Antimicrob Agens Chemother 2014; 58:2767–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.