Abstract
IMPORTANCE
Tests that predict outcomes for patients with acute myeloid leukemia (AML) are imprecise, especially for those with intermediate risk AML.
OBJECTIVES
To determine whether genomic approaches can provide novel prognostic information for adult patients with de novo AML.
DESIGN, SETTING, AND PARTICIPANTS
Whole-genome or exome sequencing was performed on samples obtained at disease presentation from 71 patients with AML (mean age, 50.8 years) treated with standard induction chemotherapy at a single site starting in March 2002, with follow-up through January 2015. In addition, deep digital sequencing was performed on paired diagnosis and remission samples from 50 patients (including 32 with intermediate-risk AML), approximately 30 days after successful induction therapy. Twenty-five of the 50 were from the cohort of 71 patients, and 25 were new, additional cases.
EXPOSURES
Whole-genome or exome sequencing and targeted deep sequencing. Risk of identification based on genetic data.
MAIN OUTCOMES AND MEASURES
Mutation patterns (including clearance of leukemia-associated variants after chemotherapy) and their association with event-free survival and overall survival.
RESULTS
Analysis of comprehensive genomic data from the 71 patients did not improve outcome assessment over current standard-of-care metrics. In an analysis of 50 patients with both presentation and documented remission samples, 24 (48%) had persistent leukemia-associated mutations in at least 5%of bone marrow cells at remission. The 24 with persistent mutations had significantly reduced event-free and overall survival vs the 26 who cleared all mutations. Patients with intermediate cytogenetic risk profiles had similar findings.
| Digital Sequencing (n=50) | Intermediate Cytogenetic Risk Profile (n=32) |
|||||
|---|---|---|---|---|---|---|
| Persistent Mutations (n=24) |
Cleared Mutations (n=26) |
HR (95% CI) |
Persistent Mutations (n=14) |
Cleared Mutations (n=18) |
HR (95% CI) |
|
| Event-free survival, median (95% CI), mo |
6.0 (3.7–9.6) |
17.9 (11.3–40.4) |
3.67 (1.93–7.11) |
8.8 (3.7–14.6) |
25.6 (11.4-not estimable) |
3.32 (1.44–7.67) |
| Overall survival, median (95% CI), mo |
10.5 (7.5–22.2) |
42.2 (20.6-not estimable) |
2.86 (1.39–5.88) |
19.3 (7.5–42.3) |
46.8 (22.6-not estimable) |
2.88 (1.11–7.45) |
CONCLUSIONS AND RELEVANCE
The detection of persistent leukemia-associated mutations in at least 5%of bone marrow cells in day 30 remission samples was associated with a significantly increased risk of relapse, and reduced overall survival. These data suggest that this genomic approach may improve risk stratification for patients with AML.
Approximately 20% of adult patients with acute myeloid leukemia (AML) fail to achieve remission with initial induction chemotherapy, and approximately 50% ultimately experience relapse after achieving complete remission.1–3 Even though potentially curative therapy (eg, allogeneic hematopoietic stem cell transplantation) is now available for many patients, this therapy is expensive and is associated with significant morbidity. Thus, identifying patients at high risk for relapse would be clinically useful and is the basis of current risk stratification approaches, which include conventional karyotyping, clinical features, and the mutational status of a limited panel of genes.4–9
Genomic approaches have identified somatic mutations in coding genes that are associated with outcomes, however, no study has yet addressed whether mutations in noncoding and regulatory regions may further improve outcome predictions for adults with de novo AML. Further, it is not yet clear whether genomic approaches can be used to assess the clearance of leukemia cells after chemotherapy, which has historically been done by morphologic examination and more recently, by multicolor flow cytometry.10,11 The known prognostic value of persistent clonal cytogenetic abnormalities in remission samples12,13 (which is relevant for the 50%–60% of AML cases with clonal cytogenetic abnormalities at presentation) suggests that higher-resolution genomic approaches that can be applied to all AML samples may provide useful prognostic information.
In this study, we used whole-genome sequencing to determine whether mutations anywhere in the genome (either coding or noncoding regions) detected at presentation were associated with outcomes in patients with AML. We also evaluated an alternative approach: tracking the clearance of leukemia-associated mutations after induction chemotherapy (using approaches that should be informative for nearly all AML cases) to determine whether persistent molecular disease was associated with less-favorable clinical outcomes.
Methods
Patient Selection
All samples from patients with AML were collected as part of a study approved by the Human Research Protection Office at Washington University School of Medicine. All patients provided informed consent explicit for whole-genome or exome sequencing, using a protocol approved by the Washington University School of Medicine Institutional Review Board.
Patients were selected from a larger cohort of more than 500 adult patients (enrolled from March 2002 to December 2013). Previously, we studied 200 of these cases (chosen to reflect the natural distribution of AML cases) for The Cancer Genome Atlas (TCGA) AML study.8 Of the 71 patients described herein, 68 were also included in the TCGA study and the diagnostic material was banked prior to 2010. These samples were selected for further study after excluding patients with good risk cytogenetic abnormalities, lack of intensive induction chemotherapy, or death during induction therapy, in accordance with accepted guidelines (for full description, see eMethods, section A.1.1, in the Supplement).14
All 68 of the cases chosen for this study received intensive chemotherapy with an induction regimen of intravenous cytarabine (100 mg/m2 × 64 patients; 200 mg/m2 × 4 patients) and an anthracycline (either idarubicin or daunorubicin). Patients were then divided into 3 outcome-stratified groups: (1) the refractory group, including patients with primary refractory disease after 2 rounds of induction chemotherapy, and also patients with relapse in less than 6 months; (2) the R6-12 group, including patients with relapse between 6 and 12 months; and (3) the long first remission group (LFR), including patients with chemotherapy only, surviving at least 12 months without an allogeneic stem cell transplant. To identify additional patients fulfilling these criteria, we reanalyzed our updated database and found 3 additional refractory group patients who were not included in the TCGA study. No additional LFR patients were available. We allowed for up to 2 rounds of intensive induction chemotherapy to achieve complete remission, consistent with other recent studies.15 Patients with a documented relapse within the first year were included in the study (in either the refractory or R6-12 groups), regardless of transplantation status.
Thirty-five of these 71 patients had formalin-fixed paraffin-embedded postchemotherapy remission samples at approximately day 30 available for targeted sequencing to follow the clearance of each leukemia-specific mutation in remission; 25 of these samples provided adequate DNA for further testing. We also identified 25 additional cases with cryopreserved cells available from presentation and at first remission at approximately day 30, as well as a bone marrow sample from a first remission that lasted at least 1 year, or at first relapse (banking of all AML patient follow-up samples—including day 30 and remission samples—began at our institution in late 2011; the diagnostic material for the 25 patients selected for this study was therefore banked between 2011 and 2013, allowing for a follow-up period of at least 1 year after presentation). Cases of acute promyelocytic leukemia were excluded from this cohort of 50 cases because these patients received all-trans-retinoic acid as part of induction therapy, but we included 6 cases with translocations producing RUNX1-RUNX1T1 (GenBank 861 and 862) or CBFB-MYH11 (GenBank 865 and 4629) fusions. For all cases, cytogenetic risk was determined by conventional cytogenetics or fluorescence in situ hybridization, unless otherwise stated.
Identification of Recurring Nonprotein Coding Mutations
Using whole-genome sequence data from 110 AML samples sequenced at the McDonnell Genome Institute (including all 58 whole-genome sequencing cases reported in this study), we assessed all genomic regions that were sequenced to adequate depth for variant calling. We identified 8673 regions with recurrent putative mutations, and these sites were then deeply sequenced in all 71 cases in this study, using a NimbleGen custom-capture array. Additional details of this capture, sequencing, and variant validation are described in the eMethods, section A.2.6, in the Supplement.
Identification of Germline Polymorphisms
A total of 518 germline variants that were exclusive to either refractory group or LFR group samples were identified from whole-genome sequencing, and then validated with targeted sequencing. We conducted standard association analysis in PLINK,16,17 version 1.07, by comparing allele frequencies between affected (LFR group, 25 patients) and unaffected (refractory group, 34 patients) patients. We obtained the results of a 1df χ2 with asymptotic significance value. Bonferroni correction was used to account for multiple comparisons.
mRNA and miRNA Analysis
RNA sequencing data was obtained for 45 of 71 samples (42 from the TCGA study, plus the 3 new refractory cases), and differentially expressed transcripts between the refractory group (n = 26) and LFR group (n = 19) were inferred using edgeR (Bioconductor), version 2.6.12.18 Each differentially expressed transcript was required to exceed a fragments per kilobase of transcript per million mapped reads (FPKM) value of 1 in at least 50% of either the refractory or LFR samples or both. Unsupervised hierarchical clustering was performed for the 1000 most variably expressed transcripts across all 45 samples using the heatmap. 2 function (with default clustering parameters) in the gplots package for R (R Foundation), version 2.8.0. MicroRNA (miRNA) expression data was available for 24 refractory group cases and 19 LFR group cases. Differentially expressed miRNAs were detected using edgeR.
Outcomes
Outcomes were assessed according to standard guidelines.14 Event-free survival was defined as the period of time from diagnosis to treatment failure, relapse, or death from any cause. Overall survival was defined as the period of time from diagnosis to death. Patients were required to complete appropriate induction chemotherapy and were then evaluated by bone marrow biopsy approximately 30 days after induction initiation.
Statistical Approach
P values for continuous variables were calculated from the Kruskal-Wallis test, categorical values were compared with the Fisher exact test, and time-to-event comparisons were from the log-rank test with univariate proportional hazards regression used to calculate hazard ratios. Multivariate proportional hazards models were constructed for both event-free survival and overall survival. Covariates considered included cytogenetics, clinical variables such as age, white blood cell count, and the percentage of bone marrow blasts, as well as somatic mutations and mutation clearance. Details are presented in the eMethods, sectionA.7, in the Supplement. All significance tests are 2-tailed, with a P value of .05 used as the threshold for statistical significance. Statistical analyses were performed with SAS (SAS Institute), version 9.3.
Sequencing
The sequence data for all tumors and matched normal samples has been deposited in the database of Genotypes and Phenotypes (dbGaP) under accession number phs000159.v6.p4.
Additional information is available in the Supplement.
Results
Patient Characteristics and Genomic Findings for the AML Cases Analyzed at Diagnosis
Among the 71 patients with de novo AML whose initial diagnostic samples were analyzed, 34 were in the refractory group, 12 in the R6-12 group, and 25 in the LFR group (Figure 1, Table, and eTable 1 and eFigure 1 in the Supplement). Those with intermediate-risk AML (n = 52) were more commonly in the LFR group, but were found in all 3 cohorts. No significant differences were observed in the number of coding mutations, in the number of total genomic mutations in the whole-genome sequencing cases, or in the number of detectable subclones among the groups (eFigures 2–3 and eTables 1–2 in the Supplement). More copy number alterations were detected among patients in the refractory group because more of these patients had unfavorable cytogenetic profiles, which is associated with complex cytogenetic abnormalities (eFigure 4 and eTable 3 in the Supplement).
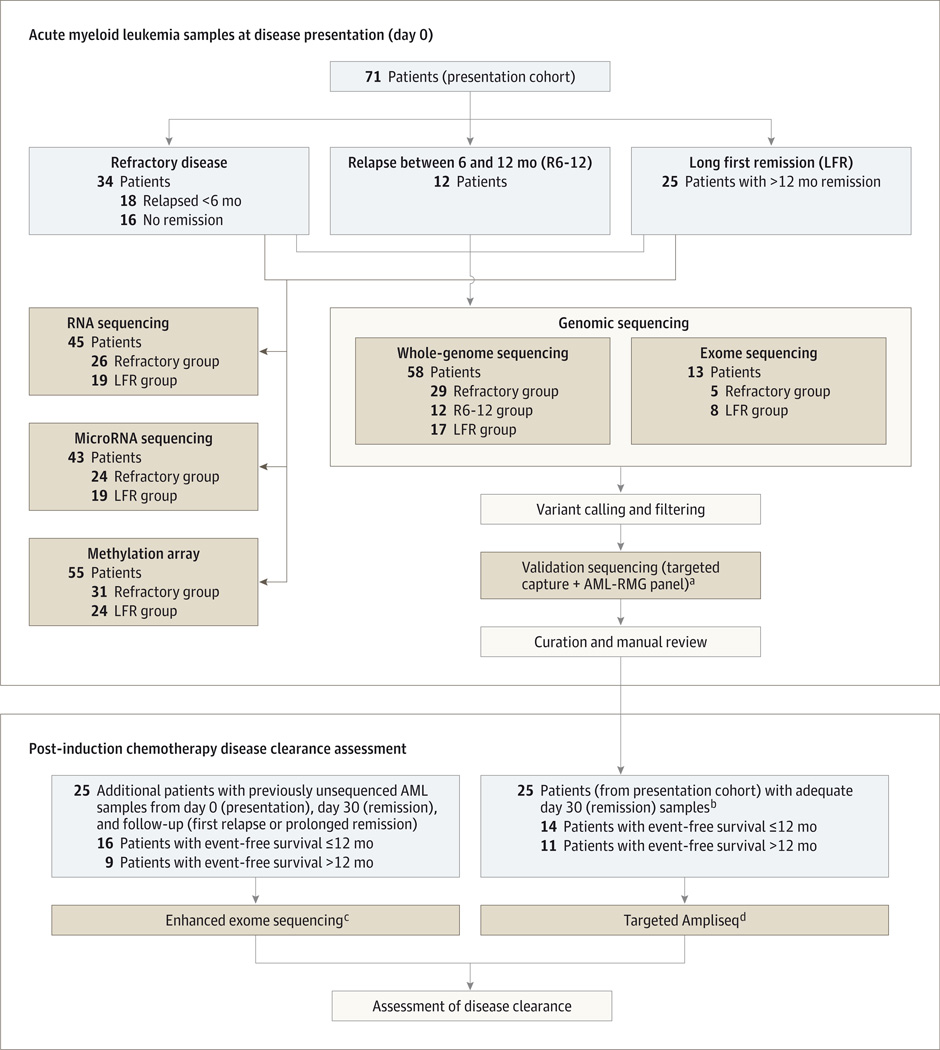
Figure 1.
Flowchart Outlining the Selection of Samples and Sequencing Approaches in the Study
aAML-RMG is a capture reagent consisting of all of the exons of the genes that are currently known to be recurrently mutated in acute myeloid leukemia, based on The Cancer Genome Atlas AML study.8
bThe only samples with sufficient day 30 DNA for sequencing and assessment of disease clearance (refractory group, 6 patients; R6-12 group, 8 patients; LFR group, 11 patients).
c Enhanced exome sequencing is exome capture-based sequencing supplemented with the AML-RMG panel of target genes, to improve coverage of critical regions of the exome.
d Targeted Ampliseq is a polymerase chain reaction–based digital sequencing approach that allows for accurate determination of the frequency of specific mutations in acute myeloid leukemia samples.
Table.
Characteristics for the 71 Adult Patients With Acute Myeloid Leukemiaa
| Refractory Group | R6–12 Group | LFR Group | P Valueb | Additional Cases for Clearance Assessment |
|
|---|---|---|---|---|---|
| Patients in cohort, No. | 34 | 12 | 25 | 25 | |
| Age at study entry, mean (95% CI), y | 51.6 (46.4–56.9) | 48.7 (39.0–58.3) | 50.6 (45.4–55.8) | .82 | 57.4 (53.1–61.8) |
| Male sex, No. (%) | 15 (44.1) | 5 (41.7) | 14 (56.0) | .59 | 12 (48.0) |
| Bone marrow blasts at diagnosis, mean (95% CI), % | 72.5 (65.8–79.2) | 63.1 (50.2–76.0) | 75.4 (69.5–81.3) | .15 | 56.4 (47.5–65.4) |
| White cell count at diagnosis, mean (95% CI), per mm3 | 52 600 (37 400–67 700) | 75 900 (27 600–124 200) | 44 800 (17 700–71 900) | .18 | 33 940 (21 040–46 830) |
| Median (IQR) | 47 100 (12 000–80 500) | 51 900 (15 400–118 200) | 19 800 (5 000–59 300) | 21 700 (7500–21 700) | |
| Event-free survival, median (95% CI), mo | 3.0 (2.2–3.7) | 8.2 (6.4–10.2) | 20.6 (16.6–40.4) | <.001 | 10.7 (9.5–17.3) |
| Overall survival, median (95% CI), mo | 7.5 (5.7–9.3) | 11.7 (7.9–16.4) | 42.3 (24.8-NE) | <.001 | 20.4 (15.1–22.2) |
| Normal cytogenetic profile, No. (%) | 13 (38.2) | 8 (66.7) | 20 (80.0) | .02 | 12 (48.0) |
| Cytogenetic risk group, No. (%) | |||||
| Favorable | 0 | 0 | 0 | 6 (24.0) | |
| Intermediate | 19 (55.9) | 10 (83.3) | 23 (92.0) | 14 (56.0) | |
| Unfavorable | 13 (38.2) | 2 (16.7) | 2 (8.0) | 5 (20.0) | |
| Missing data | 2 (5.9) | 0 | 0 | 0 | |
| Total coding mutations, mean (95% CI)c | 21.8 (17.7–25.9) | 20.8 (14.6–27.1) | 16.0 (12.1–19.9) | .06 | 11.8 (9.7–13.9) |
| Median (IQR) | 20.0 (14.0–27.0) | 19.5 (15.0–25.5) | 14.0 (9.0–20.0) | 12.0 (8.5–14.0) | |
| Common AML somatic mutations, No. (%) | |||||
| NPM1 | 7 (20.6) | 6 (50.0) | 15 (60.0) | .005 | 8 (32.0) |
| DNMT3A | 8 (23.5) | 5 (41.7) | 10 (40.0) | .31 | 5 (20.0) |
| FLT3 | 13 (38.2) | 6 (50.0) | 10 (40.0) | .77 | 10 (40.0) |
| TP53 | 7 (20.6) | 2 (16.7) | 1 (4.0) | .19 | 1 (4.0) |
| IDH1 and/or IDH2 | 10 (29.4) | 1 (8.3) | 10 (40.0) | .13 | 4 (16.0) |
| KRAS and/or NRAS | 6 (17.7) | 2 (16.7) | 5 (20) | >.99 | 11 (44.0) |
| WT1 | 6 (17.7) | 3 (25) | 1 (4.0) | .16 | 1 (4.0) |
| RUNX1 | 5 (14.7) | 0 (0.0) | 0 (0.0) | .08 | 2 (8.0) |
| NF1 | 4 (11.8) | 0 (0.0) | 0 (0.0) | .16 | 0 |
| WGS cases, samples | 29 | 12 | 17 | ||
| Total variants, mean (95% CI) | 411.2 (336.2–486.3) | 422.9 (306.6–539.3) | 375.1 (284.6–465.6) | .78 | |
| Median (IQR) | 435.0 (282.0–509.0) | 414.5 (268.0–583.5) | 378.0 (287.0–439.0) | ||
| Subclones, mean (95% CI) | 1.2 (0.9–1.5) | 1.3 (0.6–2.1) | 1.4 (0.9–1.9) | .78 | |
| Median (IQR) | 1.0 (1.0–2.0) | 1.0 (0.5–2.5) | 1.0 (1.0–2.0) |
Abbreviations: AML, acutemyeloid leukemia; IQR, interquartile range; LFR, long first remission; R6–12, relapsed between 6 and 12 months from the initiation of therapy;WGS, whole-genome sequencing.
Percentages may not total 100 because of rounding.
P values are based on a comparison of the 3 different outcome groups.
Coding variants for the 71 refractory, R6–12, or LFR cases are based on the number of coding variants from the WGS or exome sequencing data; for the 25 additional cases for clearance assessment, this number is based on the total number of variants identified by exome sequencing. Common AML somatic mutations are the numbers of cases with any somatic mutation in DNMT3A, TP53 (including deletions), WT1, RUNX1, or NF1, and canonical mutations in IDH1, IDH2, KRAS, NRAS, or FLT3.
For these 71 cases, FLT3 (GenBank 2322; n = 33 mutations [29 cases]), NPM1 (GenBank 4869; n = 28 mutations [28 cases]), and DNMT3A (GenBank 1788; n = 26 mutations [23 cases]) were the most commonly mutated genes (eFigure 5 in the Supplement). Mutations in genes involved in drug metabolism (eg, components of cytarabine metabolism or solute carrier family members) were uncommon (eFigure 6 in the Supplement). Although mutations in a few genes and functional categories (eTable 4 in the Supplement) were enriched in the different out-come groups, the mutational spectrum at presentation was not significantly associated with outcomes (eMethods, sectionA.2.9 and A.7, and eFigure 7 in the Supplement).
Identification of Recurring Mutations in Nonprotein Coding Regions
Using a genome-wide discovery approach and targeted validation sequencing, we identified 25 regions of 10 kbp or less that contained mutations in 4 or more cases (eFigure 8 and eTable 5 in the Supplement); 13 regions contained mutations in protein-coding genes only, whereas the remaining 12 had at least 1 noncoding mutation (including mutations affecting antisense transcripts within the WT1 gene: WT1-AS [GenBank 51352]). Eight of these regions were characterized exclusively by noncoding variants, but none were common enough to be associated with an outcome group (eTable 6 in the Supplement).
Identification of Germline Polymorphisms and Expression and Methylation Patterns
Neither an analysis of the whole-genome sequencing data nor a focused interrogation of polymorphisms associated with chemotherapeutic response or neoplasia (eFigure 9 and eTable 7 in the Supplement) revealed an association with any polymorphism and an outcome group. Further, using RNA sequencing, miRNA sequencing, and methylation data from 450K arrays,8 we identified only limited signatures associated with outcome groups (eFigures 10–14 and eTables 8–13 in the Supplement).
Genomic Assessment of Mutation Clearance After Induction Chemotherapy
These data suggested that comprehensive genomic characterization of adult AML samples at presentation added little additional prognostic value beyond standard approaches (eg, cytogenetics, age, or identification of coding mutations in genes that are recurrently mutated in AML). We therefore investigated whether digital sequencing could be used to follow the clearance of leukemia-specific mutations after the bone marrow recovered from induction therapy, and whether this approach could improve risk stratification for AML patients. All 50 cases in this part of the study met morphologic criteria for complete remission (<5% blasts) approximately 30 days (median, 34 days [interquartile range, 30–38]) after the initiation of induction chemotherapy (and all but 1 patient also had complete recovery of peripheral counts at the sampling date).19
We first identified 25 previously unsequenced, de novo AML samples from patients who achieved morphologic remission at approximately day 30, with cryopreserved cells banked at presentation, at approximately day 30, and at follow-up (either at first relapse or during a prolonged first remission; eTable 14 in the Supplement). All samples for these 25 cases were sequenced using a modified exome capture reagent with additional probes covering all of the exons of the 264 recurrently mutated genes (RMG) in AML8 (hereafter referred to as the AML-RMG capture set; for additional information about enhanced exome sequencing, see eMethods, section A.6.1, in the Supplement). The mean coverage of exome variants in the day 0 samples from this set was 199X, with 383X coverage for genes in the AML-RMG capture set. In the day 30 samples, the mean coverage of all leukemia-associated variants previously identified on day 0 was 256X, and 543X for those in the AML-RMG capture set (eTable 15 in the Supplement).
To increase the number of cases available for this analysis, genomic DNA was extracted from day 30 remission formalin-fixed, paraffin-embedded bone marrow biopsy samples from 35 of the 71 cases described in the first part of this study; these cases did not have cryopreserved cells available from day 30. Although the DNA obtained from these samples was highly degraded (and not suitable for standard targeted deep sequencing), we were able to generate short amplicons (using Ampliseq [Thermo Fisher Scientific]) for deep sequencing on the Ion Torrent platform from 25 of the 35 cases. We selected 6 to 15 somatic mutations (mean, 12.1) identified at day 0 from each case, and assessed their presence in the day 30 remission bone marrow (mean coverage of these leukemia-associated variants on day 30, 14 780X).
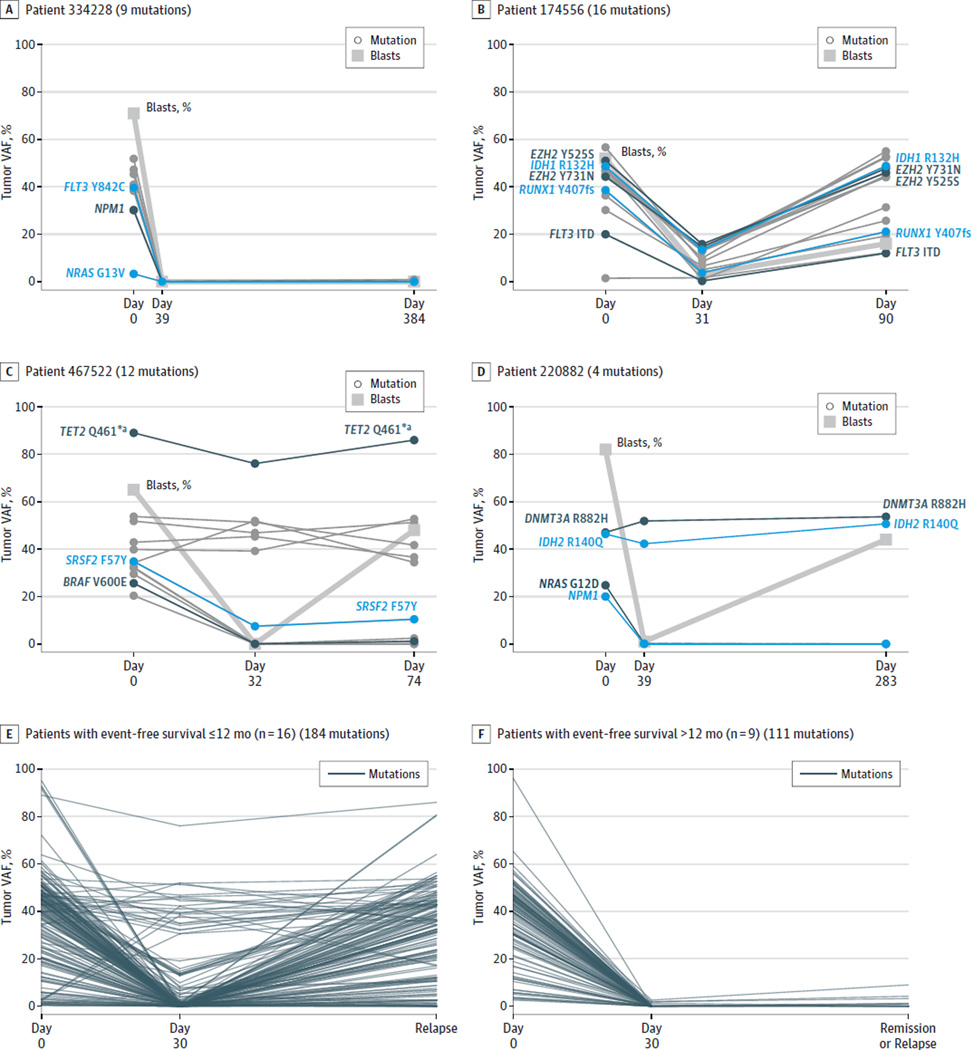
A total of 597 variants (eTable 16 in the Supplement) identified in the day 0 samples were deeply sequenced in the 50 remission samples, with a mean of 11.9 variants per case (range, 5–30). Even though all 50 patients were in morphologic remission on day 30, the clearance patterns of leukemia-associated mutations varied substantially among samples (Figure 2, panels A–D). Some patients essentially cleared all variants on day 30 (Figure 2A), whereas among other patients, only a few of the mutations were cleared and the same mutations returned at relapse (Figure 2B). In other cases, remission at day 30 was associated with clearance of a subset of the mutations (probably representing subclones), whereas the founding clone mutations persisted in virtually all cells, despite the normal morphology of the samples (Figure 2C and 2D). We separately evaluated the cases with follow-up exome sequencing with an event-free survival of 12 months or less (n = 16 cases) vs an event-free survival of more than 12 months (n = 9 cases) (Figure 2, panel E vs F). Many more patients with an event-free survival of 12 months or less had persistent disease (as measured by the maximum day 30 variant allele frequency [VAF] for each patient), as compared with those with an event-free survival of more than 12 months (mean maximum VAF, 18.39 for ≤12 months vs 0.56 for >12 months; P = .01). Importantly, the same persistent day 30 variants often increased at relapse, as cells containing these mutations reexpanded.
Figure 2.
Clearance Patterns of Acute Myeloid Leukemia–Associated Mutations Detected by Exome Sequencing
VAF indicates variant allele frequency. Panels A through D show examples of different patterns of clearance of leukemia-associated mutations after induction therapy in 4 acute myeloid leukemia cases. Key leukemic variants are highlighted by labels and color. All patients had a morphologic complete remission in the approximately day 30 sample (thick gray line shows that blast counts declined to <5%on approximately day 30 for all cases). The patterns include complete clearance of all variants at day 39 that remain undetectable during an extended remission (>1 year) (panel A); incomplete clearance of most variants at day 31, with subsequent return of these mutations at relapse on day 90 (panel B); clearance of subclonal variants at day 32 (panel C) or day 39 (panel D), with persistence of founding clone variants that remain present at relapse (eg, TET2 for panel C and DNMT3A and IDH2 for panel D). Panels E and F, for the 25 samples with follow-up exome sequencing, the clearance patterns of all leukemia-associated variants detected on day 0 are separately shown for patients with an event-free survival of 12 months or less (n = 16, panel E) or more than 12 months (n = 9, panel F).
a The high TET2 VAF levels (75%–90%) in panel C suggest that 1 copy of TET2 was probably deleted in the founding clone of this acute myeloid leukemia sample.
The determination of whether a mutation is present at levels above background sequencing error depends not only on sequencing depth, but also on sequence context, and the error rate of the sequencing platform. Because different sequencing platforms and levels of coverage were used in the two 25-sample cohorts (and to ensure that these data would be broadly applicable to other studies using different sequencing approaches), we applied VAF thresholds for each variant in the day 30 remission sample. Because nearly all somatic mutations in AML genomes are heterozygous,8 a mutation with a VAF of 2.5% on day 30 (day 30-VAF2.5%) suggests that 5% of the cells in that sample contained that mutation. In addition to being a conservative threshold that is robust across different genomic contexts, coverage levels, and sequencing platforms, the day 30-VAF2.5% threshold is congruent with previous studies of leukemia clearance using karyotypic analysis (ie, 1 in 20 metaphases), and can easily be extended to other molecular technologies.
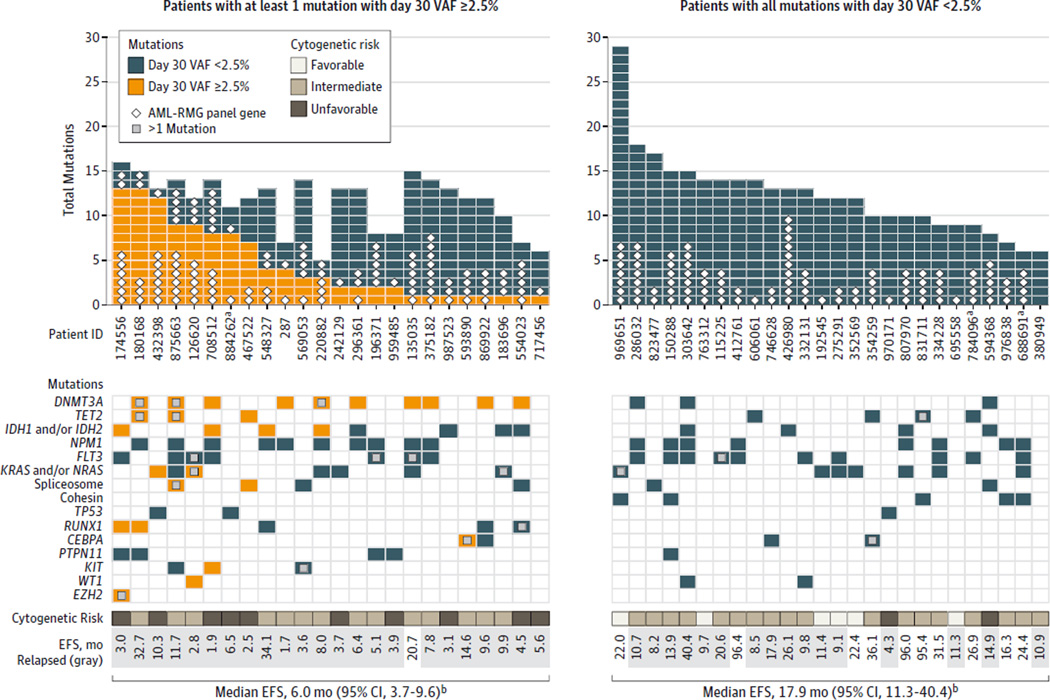
Of the 597 variants followed at day 30, 109 variants (18.3%) had a day 30 VAF of 2.5%or more. To determine whether day 30 clearance was associated with relapse or survival, we stratified patients based on the presence or absence of leukemia-associated variants at this level in the day 30 samples. Twenty-four of the 50 cases had at least 1 persistent (noncleared) mutation at day 30 (Figure 3, left side); many cases had several variants that persisted (mean, 4.5). The number of variants above this threshold varied widely among the cases, but having more persistent variants was not significantly associated with a shortened time to relapse (eFigure 15 in the Supplement). Of the 24 cases that failed to clear all mutations on approximately day 30, 10 cases were in the cohort of 25 cases assessed with exome sequencing, and 14 cases were in the Ampliseq cohort (P = .40).
Figure 3.
Day 30 Mutation Clearance Patterns by Patient for 50 Acute Myeloid Leukemia Cases
AML indicates acute myeloid leukemia; AML-RMG, recurrently mutated AML genes; EFS, event-free survival; VAF, variant allele frequency. Top: bar plots showing the number of mutations assessed at day 30, color coded according to whether they exceeded the day 30 VAF threshold of 2.5%. Mutations that occurred in AML-RMG are labeled with white diamonds. The panel is divided into samples with at least 1 variant with a day 30 VAF of 2.5%or more (left) and samples in which the day 30 VAF for all mutations was less than 2.5% (right). Bottom: key AML genes and pathways, showing patterns of mutations and clearance.
a Three cases that received an allogeneic transplant in the first complete remission. Exclusion of these 3 cases from the analysis did not significantly alter the outcome results.
b The median event-free survival of cases with a day 30 VAF of less than 2.5% for all mutations was 17.9 months vs 6.0 months for the cases in which at least 1 variant persisted with a VAF threshold of 2.5%or more (P < .001).
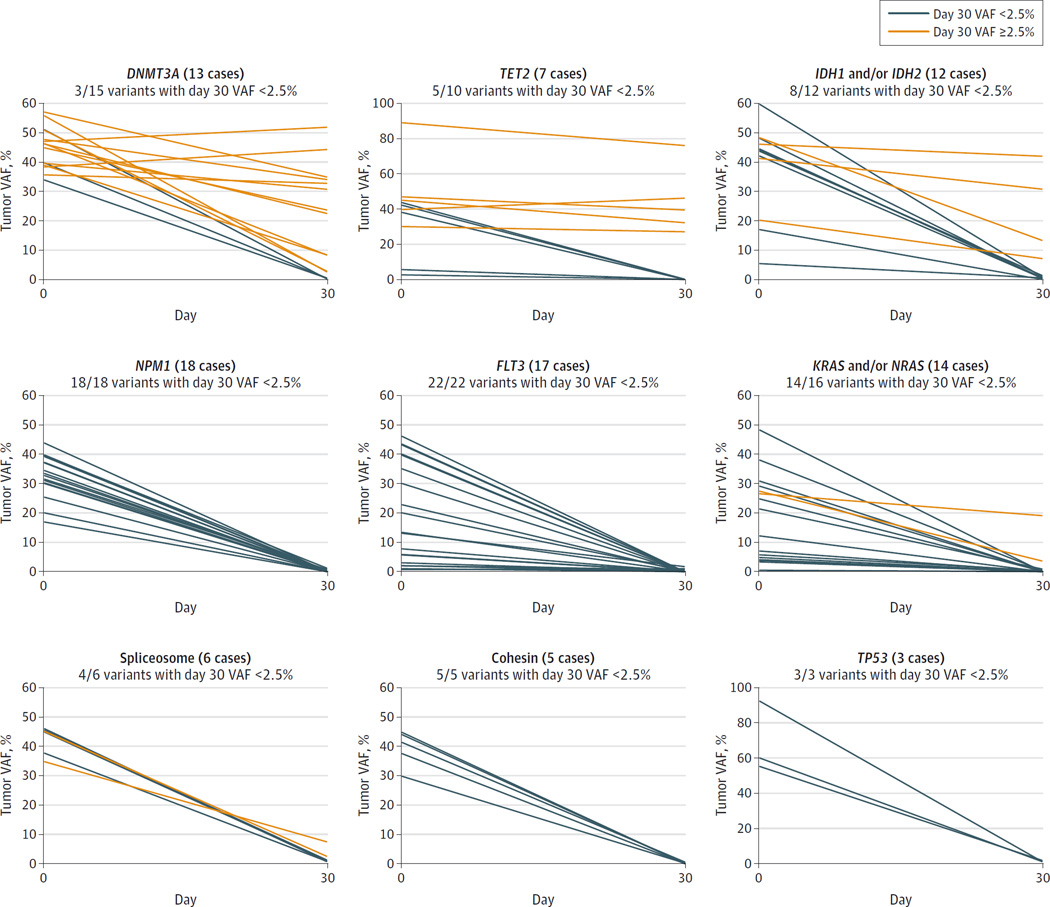
Only a subset of the persistent mutations were in recurrently mutated AML genes (diamonds in Figure 3), with the remainder occurring elsewhere in the genome. Eight cases had only 1 persistent mutation at day 30, with 4 of these occurring in DNMT3A (GenBank 1788). Overall, of the 16 DNMT3A mutations (in 13 cases), only 3 were cleared on day 30. Mutations in TET2 (GenBank 54790) were often persistent in remission, whereas mutations in FLT3, NRAS, and KRAS (GenBank 2322, 4893, and 3845) were usually cleared below the VAF threshold (Figure 4).
Figure 4.
Day 30 Mutation Clearance by Gene for 50 Acute Myeloid Leukemia Cases
VAF indicates variant allele frequency. Serial VAF measurements demonstrating the clearance patterns of several recurrently mutated acute myeloid leukemia genes in the set of 50 cases. Orange lines indicate a day 30 VAF of 2.5%or more; blue lines indicate a day 30 VAF of less than 2.5%.
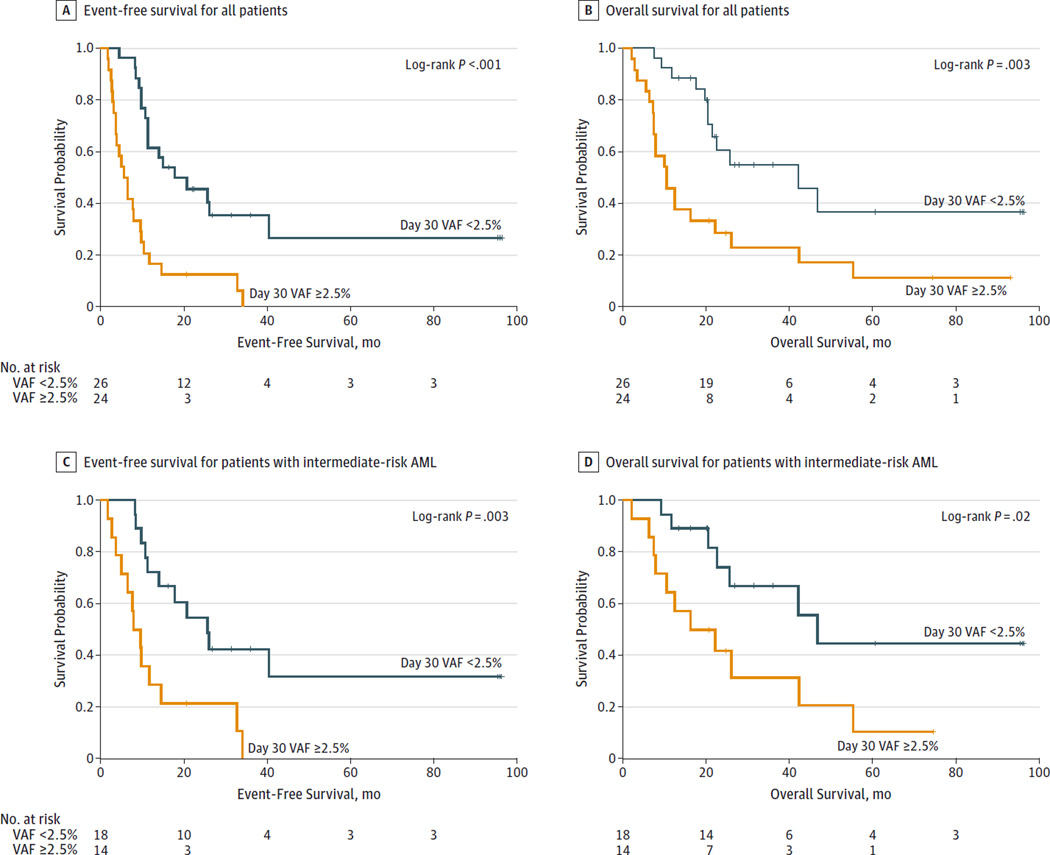
The 24 cases with at least 1 persistent day 30-VAF2.5% mutation had significantly reduced event-free survival (Figure 5A; median, 6.0 months [95% CI, 3.7–9.6]) compared with the 26 cases that cleared all mutations at the day 30-VAF2.5% threshold (median, 17.9 months [95% CI, 11.3–40.4], log-rank P < .001; hazard ratio [HR], 3.67 [95% CI, 1.93–7.11], P < .001). A similar reduction in overall survival was also found (median, 10.5 months [95% CI, 7.5–22.2] for persistent mutations vs 42.2 months [95% CI, 20. 6-not estimable] for cleared mutations, log-rank P = .003; HR, 2.86 [95% CI, 1.39–5.88], P = .004) (Figure 5B). A day 30-VAF threshold of ≥1.0%resulted in more persistent variants (169 variants [28.3%]), but differences in event-free survival were still significant (7.9 months [95% CI, 4.5–9.9] for persistent mutations vs 25.6 months [95% CI, 11.4-not estimable] for cleared mutations, log-rank P = .001; HR, 3.33 [95% CI, 1.61–6.89], P = .001; eFigure 16 in the Supplement). In multivariate proportional hazards regression models, both thresholds were significant with respect to event-free survival in multivariate models: the HR for day 30-VAF1.0% was 2.58 (95% CI, 1.17–5.69, P = .02); for day 30-VAF2.5%, 3.52 (95% CI, 1.68–7.39, P = .001) (section A7 in the Supplement). Neither the day 30-VAF1.0% nor the day 30-VAF2.5% was significant in the multivariate model for overall survival when considering all 50 patients, most likely due to the contribution of cytogenetic findings in the 6 favorable risk cases (for the multivariate model for overall survival: day 30-VAF1.0%, HR, 1.90 [95% CI, 0.70–5.18], P = .21; day 30-VAF2.5%, HR, 2.14 [95% CI, 0.93–4.99], P = .07).
Figure 5.
Association Between Mutation Clearance and Outcomes
AML indicates acute myeloid leukemia; VAF, variant allele frequency. Data were censored at last contact or at January 21, 2015.
Analysis of the 32 intermediate-risk cases (Figure 5C and Figure 5D) revealed that the detection of at least 1 persistent mutation (n = 14 patients) compared with cleared mutations (n = 18 patients) was also associated with reduced event-free survival (median, 8.8 months [95% CI, 3.7–14.6] for persistent mutations vs 25.6 months [95% CI, 11.4-not estimable] for cleared mutations, log-rank P = .003; HR, 3.32 [95% CI, 1.44–7.67], P = .005) and reduced overall survival (median, 19.3 months [95% CI, 7.5–42.3] for persistent mutations vs 46.8 months [95% CI, 22.6-not estimable] for cleared mutations, log-rank P = .02; HR, 2.88 [95% CI, 1.11–7.45], P = .03). In contrast, there was no association of event-free survival with the mutation status of DNMT3A, NPM1, or FLT3 (eFigure 17 in the Supplement). Associations with adverse outcomes were also significant when the day 30-VAF1.0% threshold was applied (eMethods, section A.7, and eFigure 18 in the Supplement).
Discussion
In this study of adult patients with de novo AML treated at a single academic center, novel risk-assessment biomarkers were not identified by extensively evaluating the presentation tumor sample from 71 similarly treated patients with very different outcomes. However, the persistence of leukemia-associated variants approximately 30 days after initiation of chemotherapy was associated with an increased risk of relapse, using a genomic approach that should be informative for nearly all AML cases. Using data from 50 patients from 2 separate AML patient cohorts and different sequencing platforms, we found a statistically significant association between the detection of persistent disease-specific variants at day 30, and a shorter event-free survival and overall survival. These associations also were found among the 32 AML cases with intermediate risk cytogenetics, which are difficult to classify using information available at diagnosis.
Previous studies have shown a strong association between persistent clonal cytogenetic markers in first remission samples, and an increased risk of relapse.12,13 Although the sensitivity of this technique is low—only 1 in 20 metaphases (ie, 5% of cells)—it still has considerable prognostic value when it is applicable (approximately 50%–60% of AML cases). Although this approach holds no value for patients with normal karyotype AML, it provides an important scientific foundation, and a benchmark for the sensitivity threshold used in this study. Direct comparison of the 2 methods was possible for the 14 cases with clonal cytogenetic abnormalities at presentation: using the day 30-VAF2.5% threshold, all 6 cases with persistent cytogenetic abnormalities at day 30 also had persistent mutations. Of the 8 cases with complete cytogenetic clearance at day 30, 5 had persistent mutations. These data suggest the methods correlate well, but that digital sequencing is more sensitive, probably because it samples many more cells, and more mutations, than standard karyotyping.
Although this study was too small to establish an optimal sensitivity threshold for mutation clearance and outcome predictions, our data (and previous cytogenetic clearance studies) suggest that ultrasensitive methods after induction therapy may not be required to improve risk assessment. In fact, these methods may overestimate the risk of relapse and cause patients to be overtreated. For example, RUNX1-RUNX1T1 fusions can often be detected by ultrasensitive polymerase chain reaction methods in patients who are in long, durable remissions.20,21 Further studies are needed to fully understand the levels of sequencing sensitivity that will be required for optimal risk assessment.
These data also provide biological information regarding the clonal structure of AML cases, which may be relevant for clinical testing. For example, NPM1 mutations were cleared below the day 30-VAF2.5% threshold in all 18 cases; in contrast, 12 of 15 DNMT3A mutations (in 13 cases) persisted on day 30, and 12 of these 13 cases have relapsed. There were 8 doubly mutant AML cases with persistent DNMT3A mutations in remission despite clearance of NPM1, consistent with the hypothesis that DNMT3A mutations often precede NPM1 mutations.22,23 In 4 cases, only DNMT3A mutations remained on day 30 (3 cases have relapsed, but 1 remains in remission at 25 months), consistent with other reports of persistent DNMT3A mutations found in remission samples.22,24–26 Mutations in TET2 were also likely to persist at day 30, which corroborates recent studies that identified DNMT3A, TET2 and other mutations in the peripheral blood of elderly individuals with clonally skewed hematopoiesis, but no overt myeloid disease.27–30 Collectively, these data support the hypothesis that these mutations are initiating events for AML, but are not sufficient to directly cause this disease. In contrast, somatic mutations that activate signaling pathways (eg, mutations in FLT3, KRAS, or NRAS) were usually cleared on day 30, suggesting that subclones containing these mutations may be more sensitive to induction chemotherapy.
Although the optimal method for measuring clearance of AML cells following induction chemotherapy is currently unknown, this study suggests that monitoring a small subset of genes may have limited usefulness for many AML cases. Even testing with the panel of 264 genes that are recurrently mutated in AML will be inadequate for assessing clearance for some patients (eg, 5 cases from this set of 50). Flow cytometry–based assessment of residual disease in pediatric acute leukemias is becoming well established,11,21,31,32 and recent data suggest that the same will apply to adult AML cases.10 However, this test was not available for the patients studied in this report, because they were enrolled in the study approximately 2 to 10 years ago, and because it is not yet a standard-of-care test for adult patients with AML. The detection of residual disease by flow cytometry and the measurement of clonal clearance by digital sequencing are very different assays with different sensitivities and specificities, and potentially different strengths and weaknesses, and will need to be compared in future prospective studies.
Some of the conclusions of this study are limited in scope, primarily because of the relatively small sizes of the sample cohorts. For example, recurrent mutations in noncoding regions of AML genomes were identified in this study, but were not common enough to be associated with outcomes. Larger studies will be needed to further investigate the importance of these regions for AML pathogenesis, as well as any rare polymorphisms that may influence AML development or drug resistance. Likewise, the lack of a strong association with mutational status and patient outcomes may be related to the relatively small size of this study. However, another recent study has also shown that the mutational status of NPM1 and FLT3 are not as strongly associated with outcomes as previously suggested.5 The lack of the predictive value of individual mutations in presentation AML samples also reflects the now well-recognized combinatorial complexity of AML-associated mutations: most AML cases have at least 3 to 5 driver mutations, occurring in the more than 250 genes that are recurrently mutated in this disease; this yields an extraordinary number of mutation combinations that could be relevant for pathogenesis and outcomes.8 This study was not designed to make definitive statements about such mutation combinations because a very large number of samples would be needed to identify all significant co-associations. For example, we only had 4 cases in this study with an NPM1 mutation along with an IDH1 (GenBank 3417) or IDH2 (GenBank 3418) mutation that did not have a coexisting FLT3 or DNMT3A mutation; this combination of mutations is associated with a favorable outcome.6 Finally, virtually all AML samples are clonally complex at presentation; subclones can have unique functional properties that are sometimes critical for relapse.33,34 Therefore, we suspect that the mutational and clonal complexity of this disease will confound efforts to classify risk, if assessment is based only on the mutations that are detected at presentation.
The data presented in this report begin to define a genomic method for the risk stratification of patients with AML that places greater emphasis on the clearance of somatic mutations after chemotherapy than the identification of specific mutations at the time of presentation. It builds on previous observations that utilized the clearance of cytogenetic markers to predict relapse risk,12,13 but extends these findings considerably because it appears to be broadly applicable for intermediate risk patients, which usually have a normal karyotype. Although this study was not designed to determine the optimal clearance threshold for the association with outcomes, it represents a foundation for prospective trials focused on the role of digital sequencing to improve risk stratification for AML patients, and perhaps other cancer types as well.
Conclusions
The detection of persistent leukemia-associated mutations in at least 5% of bone marrow cells in day 30 remission samples was associated with a significantly increased risk of relapse, and reduced overall survival. These data suggest that this genomic approach may improve risk stratification for patients with AML.
Supplementary Material
Acknowledgments
Dr Spencer reports receiving personal fees from Cofactor Genomics. Dr Duncavage reports receiving personal fees from Cofactor Genomics and nonfinancial support from Agilent Technologies. Dr Ozenberger reports receiving grant funding from the National Cancer Institute. No other disclosures were reported.
Funder/Sponsor: This work was supported by grants KO8 HL116605 (Dr Klco), PO1 CA101937 (Dr Ley), and U54 HG003079 (Dr Wilson) from the National Institutes of Health and grant 00335-0505-02 (Dr Ley) from the Barnes–Jewish Hospital Foundation. Technical assistance was provided by the Alvin J. Siteman Cancer Center Tissue Procurement Core, the High Speed Cell Sorting Core, and the Molecular and Genomic Analysis Core at Washington University School of Medicine and Barnes-Jewish Hospital in St Louis, Missouri, which are all supported by grant P30CA91842 from the National Cancer Institute Cancer Center. Dr Klco reports holding a Career Award for Medical Scientists from the Burroughs Wellcome Fund.
Role of the Funder/Sponsor: The funding agencies had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Additional Contributions: We thank Josh McMichael, BS, for graphical support, and Nancy Reidelberger, CRA, for administrative support (both from Washington University). Neither were compensated for their contributions.
Glossary
- AML
acute myeloid leukemia
- LFR
long first remission (ie, an initial remission of >12 months with chemotherapy only)
- R6-12
patients who relapsed between 6 and 12 months from the initiation of therapy
- VAF
variant allele frequency
Footnotes
Author Contributions: Drs Klco and Ley had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Drs Klco and Miller contributed equally to this work.
Study concept and design: Klco, Miller, Wartman, Shen, Fulton, Ozenberger, Walter, Graubert, Radich, Link, DiPersio, Wilson, Ley.
Acquisition, analysis, or interpretation of data: Klco, Miller, Griffith, Petti, Spencer, Ketkar-Kulkarni, Wartman, Christopher, Lamprecht, Helton, Duncavage, Payton, Baty, Heath, Griffith, Shen, Hundal, Chang, Fulton, O’Laughlin, Fronick, Magrini, Demeter, Larson, Kulkarni, Welch, Westervelt, Mardis, Ley.
Drafting of the manuscript: Klco, Miller, Petti, Ketkar-Kulkarni, O’Laughlin, Fronick, Wilson, Ley.
Critical revision of the manuscript for important intellectual content: Klco, Miller, Griffith, Petti, Spencer, Wartman, Christopher, Lamprecht, Helton, Duncavage, Payton, Baty, Heath, Griffith, Shen, Hundal, Chang, Fulton, Magrini, Demeter, Larson, Kulkarni, Ozenberger, Welch, Walter, Graubert, Westervelt, Radich, Mardis, DiPersio, Wilson, Ley.
Statistical analysis: Klco, Miller, Petti, Ketkar-Kulkarni, Baty, Shen, Hundal, Chang, Link, Ley.
Obtained funding: Wilson, Ley.
Administrative, technical, or material support: Klco, Wartman, Lamprecht, Helton, Duncavage, Payton, Heath, Fulton, O’Laughlin, Fronick, Demeter, Larson, Kulkarni, Ozenberger, Welch, Walter, Graubert, Westervelt, Radich, Link, Ley.
Study supervision: Klco, Miller, Fulton, Magrini, Larson, Mardis, DiPersio, Wilson, Ley.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
REFERENCES
- 1.Breems DA, Van Putten WL, Huijgens PC, et al. Prognostic index for adult patients with acute myeloid leukemia in first relapse. J Clin Oncol. 2005;23(9):1969–1978. doi: 10.1200/JCO.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 2.Ravandi F, Cortes J, Faderl S, et al. Characteristics and outcome of patients with acute myeloid leukemia refractory to 1 cycle of high-dose cytarabine-based induction chemotherapy. Blood. 2010;116(26):5818–5823. doi: 10.1182/blood-2010-07-296392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walter RB, Kantarjian HM, Huang X, et al. Effect of complete remission and responses less than complete remission on survival in acute myeloid leukemia: a combined Eastern Cooperative Oncology Group, Southwest Oncology Group, and M.D. Anderson Cancer Center Study. J Clin Oncol. 2010;28(10):1766–1771. doi: 10.1200/JCO.2009.25.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrd JC, Mrózek K, Dodge RK, et al. Cancer and Leukemia Group B (CALGB 8461). Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100(13):4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 5.Kihara R, Nagata Y, Kiyoi H, et al. Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia. 2014;28(8):1586–1595. doi: 10.1038/leu.2014.55. [DOI] [PubMed] [Google Scholar]
- 6.Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlenk RF, Döhner K, Krauter J, et al. German-Austrian Acute Myeloid Leukemia Study Group. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter RB, Othus M, Borthakur G, et al. Prediction of early death after induction therapy for newly diagnosed acute myeloid leukemia with pretreatment risk scores: a novel paradigm for treatment assignment. J Clin Oncol. 2011;29(33):4417–4423. doi: 10.1200/JCO.2011.35.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Xie H, Wood BL, et al. Relation of clinical response and minimal residual disease and their prognostic impact on outcome in acute myeloid leukemia. J Clin Oncol. 2015;33(11):1258–1264. doi: 10.1200/JCO.2014.58.3518. [DOI] [PubMed] [Google Scholar]
- 11.Loken MR, Alonzo TA, Pardo L, et al. Residual disease detected by multidimensional flow cytometry signifies high relapse risk in patients with de novo acute myeloid leukemia: a report from Children’s Oncology Group. Blood. 2012;120(8):1581–1588. doi: 10.1182/blood-2012-02-408336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Cortes J, Estrov Z, et al. Persistence of cytogenetic abnormalities at complete remission after induction in patients with acute myeloid leukemia: prognostic significance and the potential role of allogeneic stem-cell transplantation. J Clin Oncol. 2011;29(18):2507–2513. doi: 10.1200/JCO.2010.34.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcucci G, Mrózek K, Ruppert AS, et al. Abnormal cytogenetics at date of morphologic complete remission predicts short overall and disease-free survival, and higher relapse rate in adult acute myeloid leukemia: results from cancer and leukemia group B study 8461. J Clin Oncol. 2004;22(12):2410–2418. doi: 10.1200/JCO.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 14.Cheson BD, Bennett JM, Kopecky KJ, et al. International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol. 2003;21(24):4642–4649. doi: 10.1200/JCO.2003.04.036. [published correction appears in J Clin Oncol. 2004;22(3):576]. [DOI] [PubMed] [Google Scholar]
- 15.Rowe JM, Kim HT, Cassileth PA, et al. Adult patients with acute myeloid leukemia who achieve complete remission after 1 or 2 cycles of induction have a similar prognosis: a report on 1980 patients registered to 6 studies conducted by the Eastern Cooperative Oncology Group. Cancer. 2010;116(21):5012–5021. doi: 10.1002/cncr.25263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.PLINK: whole genome association analysis toolset. [Accessed July 28, 2015]; http://pngu.mgh.harvard.edu/~purcell/plink/anal.shtml. [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hourigan CS, Karp JE. Minimal residual disease in acute myeloid leukaemia. Nat Rev Clin Oncol. 2013;10(8):460–471. doi: 10.1038/nrclinonc.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamoto T, Weissman IL, Akashi K. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc Natl Acad Sci U S A. 2000;97(13):7521–7526. doi: 10.1073/pnas.97.13.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inaba H, Coustan-Smith E, Cao X, et al. Comparative analysis of different approaches to measure treatment response in acute myeloid leukemia. J Clin Oncol. 2012;30(29):3625–3632. doi: 10.1200/JCO.2011.41.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shlush LI, Zandi S, Mitchell A, et al. HALT Pan-Leukemia Gene Panel Consortium. Identification of preleukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–333. doi: 10.1038/nature13038. [published correction appears in Nature. 2014;508(7496):420]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krönke J, Bullinger L, Teleanu V, et al. Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood. 2013;122(1):100–108. doi: 10.1182/blood-2013-01-479188. [DOI] [PubMed] [Google Scholar]
- 24.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A. 2014;111(7):2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindsley RC, Mar BG, Mazzola E, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125(9):1367–1376. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pløen GG, Nederby L, Guldberg P, et al. Persistence of DNMT3A mutations at long-term remission in adult patients with AML. Br J Haematol. 2014;167(4):478–486. doi: 10.1111/bjh.13062. [DOI] [PubMed] [Google Scholar]
- 27.Busque L, Patel JP, Figueroa ME, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44(11):1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paietta E. Minimal residual disease in acute myeloid leukemia: coming of age. Hematology Am Soc Hematol Educ Program. 2012;2012:35–42. doi: 10.1182/asheducation-2012.1.35. [DOI] [PubMed] [Google Scholar]
- 32.Terwijn M, van Putten WL, Kelder A, et al. High prognostic impact of flow cytometric minimal residual disease detection in acute myeloid leukemia: data from the HOVON/SAKK AML 42A study. J Clin Oncol. 2013;31(31):3889–3897. doi: 10.1200/JCO.2012.45.9628. [DOI] [PubMed] [Google Scholar]
- 33.Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klco JM, Spencer DH, Miller CA, et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 2014;25(3):379–392. doi: 10.1016/j.ccr.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.