Abstract
Entamoeba histolytica, a microaerophilic protozoan parasite, possesses mitosomes. Mitosomes are mitochondrion-related organelles that have largely lost typical mitochondrial functions, such as those involved in the tricarboxylic acid cycle and oxidative phosphorylation. The biological roles of Entamoeba mitosomes have been a long-standing enigma. We previously demonstrated that sulfate activation, which is not generally compartmentalized to mitochondria, is a major function of E. histolytica mitosomes. Sulfate activation cooperates with cytosolic enzymes, i.e., sulfotransferases (SULTs), for the synthesis of sulfolipids, one of which is cholesteryl sulfate. Notably, cholesteryl sulfate plays an important role in encystation, an essential process in the Entamoeba life cycle. These findings identified a biological role for Entamoeba mitosomes; however, they simultaneously raised a new issue concerning how the reactions of the pathway, separated by the mitosomal membranes, cooperate. Here, we demonstrated that the E. histolytica mitochondrial carrier family (EhMCF) has the capacity to exchange 3′-phosphoadenosine 5′-phosphosulfate (PAPS) with ATP. We also confirmed the cytosolic localization of all the E. histolytica SULTs, suggesting that in Entamoeba, PAPS, which is produced through mitosomal sulfate activation, is translocated to the cytosol and becomes a substrate for SULTs. In contrast, ATP, which is produced through cytosolic pathways, is translocated into the mitosomes and is a necessary substrate for sulfate activation. Taking our findings collectively, we suggest that EhMCF functions as a PAPS/ATP antiporter and plays a crucial role in linking the mitosomal sulfate activation pathway to cytosolic SULTs for the production of sulfolipids.
INTRODUCTION
Entamoeba histolytica is a microaerophilic protozoan parasite causing intestinal and extraintestinal amebiasis in humans. These infectious diseases have spread worldwide and have become a serious public health problem (1). E. histolytica possesses mitosomes, a type of mitochondrion-related organelle (MRO) (2–5). This organelle was originally called crypton when its discovery was reported, and such organelles are currently known as mitosomes, the widely accepted name (6, 7). MROs are derived from canonical mitochondria and are ubiquitously found in anaerobic/microaerophilic eukaryotes (2, 4). MROs display a variety of unique characteristics which are conferred by essentially reduced and/or modified mitochondrial functions and that occasionally result from the addition of new functions acquired through lateral gene transfer (5, 8). It has been postulated that the uniqueness of MROs helps organisms to adapt to various niches for their survival (2–5, 8). Entamoeba mitosomes have largely lost typical mitochondrial functions, such as those involved in the tricarboxylic acid cycle, electron transport, oxidative phosphorylation, and β-oxidation of fatty acids (4, 5). This raises two fundamental questions. Why does Entamoeba maintain mitosomes? What are their biological roles in this organism?
Despite being recognized, these important issues have not been satisfactorily addressed. We previously showed that sulfate activation, which is not generally compartmentalized to mitochondria, is a major function in E. histolytica mitosomes (3, 5, 9). Furthermore, we demonstrated that 3′-phosphoadenosine 5′-phosphosulfate (PAPS), which is synthesized through mitosomal metabolism, acts as an activated sulfur donor mainly to produce sulfolipids by the catalytic actions of putative cytosolic sulfotransferases (SULTs) (5). Cholesteryl sulfate (CS) is one such sulfolipid that plays an important role in encystation, a cell differentiation process necessary for maintaining the Entamoeba life cycle (9). These findings provide not only an explanation for the biological role of Entamoeba mitosomes but also evidence to support the uniqueness of MROs.
Importantly, our findings indicate that in Entamoeba, PAPS, a metabolite synthesized through the mitosomal pathway, needs to be translocated to the cytosol for the production of sulfolipids. Therefore, a molecule that can translocate PAPS from mitosomes to the cytosol must be present. Three candidates which could be responsible for this important function are encoded in the E. histolytica genome. One is a PAPS transporter, and the others are mitochondrial carrier (MC) proteins (EHI_068590, EHI_095150, and EHI_153760, respectively) (AmoebaDB; http://amoebadb.org/amoeba/). However, the PAPS transporter can be ruled out because its nonmitosomal localization has already been demonstrated (5). Hence, we focused on the MC proteins as the most likely candidates.
MC proteins belong to a large family of mitochondrial inner membrane carriers that transport various metabolites between the cytosol and mitochondrial matrix (10, 11). Most MC proteins are localized in mitochondria, but atypical localizations in chloroplasts and peroxisomes have recently been reported (11). The structural features conserved in MC proteins include a size of 30 to 35 kDa, three tandemly repeated homologous domains, each of which has ∼100 amino acid residues, and six transmembrane helices forming an aqueous cavity. Substances transported by MC proteins include nucleotides, amino acids, keto acids, and inorganic phosphate (Pi) (10, 11).
In this study, to address the issue of how the mitosomal sulfate activation pathway and putative cytosolic SULTs cooperate in E. histolytica, we performed biochemical and cell biological characterizations of an E. histolytica MC protein, E. histolytica mitochondrial carrier family (EhMCF), and related sulfate metabolism enzymes, the E. histolytica SULTs (EhSULTs) and E. histolytica 3′(2′),5′-bisphosphate nucleotidases (EhPAPases).
MATERIALS AND METHODS
Materials.
[14C]leucine (stock radioactivity, 100 μCi/ml), [32P]ATP (stock radioactivity, 10 mCi/ml), and [35S]PAPS (stock radioactivity, 1 mCi/ml) were purchased from PerkinElmer Japan (Yokohama, Japan). Asolectin was from Fluka (Buchs, Switzerland).
Culture of E. histolytica.
The E. histolytica HM-1:IMSS cl6 strain was routinely cultured in vitro in Diamond's BI-S-33 medium at 37°C as described previously (3, 5). Transformants were also obtained in Diamond's BI-S-33 medium as described below.
Indirect immunofluorescence analyses of E. histolytica transformants expressing HA-tagged EhSULTs or EhPAPases.
Expression plasmids for hemagglutinin (HA)-tagged EhSULTs and EhPAPases were constructed by PCR amplification of the corresponding open reading frames (ORFs) using appropriate primers sets (Table 1). Amplicons, except for the one containing EhSULT7, were then digested with BglII and inserted into the corresponding sites of the pEhEx-HA plasmid (5). The PCR-amplified EhSULT7 fragment was directly cloned into BglII-digested plasmid pEhEx-HA using an In-Fusion HD cloning kit from TaKaRa Bio (Otsu, Japan) according to the manufacturer's instructions. Lipofection transfection of E. histolytica trophozoites with the constructed plasmids, drug selection, maintenance of selected transformants, and indirect immunofluorescence analysis of independent established transformants were performed as described previously (5).
TABLE 1.
List of primers
| Described section in Materials and Methods | Target gene name | Protein IDa no. | Primer name | Primer sequence | Primer direction |
|---|---|---|---|---|---|
| Production of the rEhMCF | EhMCF | EHI_095150 | srMC-F | 5′-GCTACTAGTAACAATGATACAAGGTATGACTTATAAACGATTTC-3′ | Sense |
| srMC-R | 5′-ACTGTCGACTTACAATTTAAAGAACTTCTTATATTGATCAAAAG-3′ | Antisense | |||
| Expressions of HA-tagged genes in E. histolytica | EhSULT3 | EHI_197340 | Ex-3-F | 5′-GGCAGATCTATGCTTACCCAAAACCCATATTTATATG-3′ | Sense |
| Ex-3-R | 5′-GCGAGATCTAAGGTATTTTCCATATTCAG-3′ | Antisense | |||
| EhSULT4 | EHI_092490 | Ex-4-F | 5′-GGCAGATCTATGGAATTTAAACGAGCAG-3′ | Sense | |
| Ex-4-R | 5′-GCGAGATCTATTAGTATCTTTCTTTCCCC-3′ | Antisense | |||
| EhSULT5 | EHI_090430 | Ex-5-F | 5′-GGCAGATCTATGTCTTTTCCTAATACTGTTC-3′ | Sense | |
| Ex-5-R | 5′-GCGAGATCTTTTCTTTCCCCAGTAATTTGG-3′ | Antisense | |||
| EhSULT6 | EHI_146990 | Ex-6-F | 5′-GGCAGATCTATGGCTGAAGGATTTGAAAATG-3′ | Sense | |
| Ex-6-R | 5′-GCGAGATCTAAACATTGTGGGGAATCTATC-3′ | Antisense | |||
| EhSULT7 | EHI_114190 | Ex-7-F | 5′-AACACATTAACAGATCTATGTCTCTTCCAAATAGTTATCCAG-3′ | Sense | |
| Ex-7-R | 5′-ATATGGATACATAGATCTATTATCTCCCCAATAATTTGGC-3′ | Antisense | |||
| EhSULT8 | EHI_181190 | Ex-8-F | 5′-GGCAGATCTATGAGTTCCCAAGAATATGCTC-3′ | Sense | |
| Ex-8-R | 5′-GCGAGATCTTTTCTTACCATAATAATATGGCATATATTG-3′ | Antisense | |||
| EhSULT9 | EHI_031640 | Ex-9-F | 5′-GGCAGATCTATGAGTGTTGAAAACTATCC-3′ | Sense | |
| Ex-9-R | 5′-GCGAGATCTTTTTTTACCATAAAAGTTTGGCATATATTG-3′ | Antisense | |||
| EhSULT10 | EHI_062680 | Ex-10-F | 5′-GGCAGATCTATGAACATCTCTTTAAATGATTG-3′ | Sense | |
| Ex-10-R | 5′-GCGAGATCTAAGTTTCTTTGAAAAAAATTGTTTGATGTAC-3′ | Antisense | |||
| EhPAPase1 | EHI_193350 | Ex-11-F | 5′-AGATCTATGTCATTTGATAAAGAAC-3′ | Sense | |
| Ex-11-R | 5′-AGATCTTTTTAAATCAGAAAGTAC-3′ | Antisense | |||
| EhPAPase2 | EHI_179820 | Ex-12-F | 5′-AGATCTATGCAAACTTCGTTATTTG-3′ | Sense | |
| Ex-12-R | 5′-AGATCTAAGTAAAATTGTTTTTGAAATAG-3′ | Antisense | |||
| EhPAPase3 | EHI_175410 | Ex-13-F | 5′-AGATCTATGACTGAAGAAGAGTTA-3′ | Sense | |
| Ex-13-R | 5′-AGATCTTTGTTTTAATATATCAAATAATTG-3′ | Antisense |
ID, identification.
Production and purification of rEhMCF protein.
The recombinant EhMCF (rEhMCF) protein that is encoded in the E. histolytica genome (EHI_095150 in AmoebaDB; http://amoebadb.org/amoeba/) was produced using a wheat germ cell-free translation system in the presence of asolectin liposomes, which were freshly prepared just prior to use, as described previously (12). Two plasmids, pYT08-EhMCF and pYT08-codon-optimized EhMCF, were constructed as the templates for in vitro mRNA synthesis. For pYT08-EhMCF, a DNA fragment encoding the EhMCF ORF was amplified from E. histolytica cDNA by PCR with an appropriate primer set (Table 1), digested with SpeI and SalI, and inserted into the corresponding sites of the pYT08 vector (12). For the pYT08-codon-optimized EhMCF, a synthetic DNA encoding an EhMCF ORF in which the codon usage is optimized to that of the wheat germ translation system (GenBank accession number LC036596) was custom synthesized by GenScript Japan (Tokyo, Japan) and cloned into SpeI and SalI sites of the pYT08 vector (12). The mRNAs synthesized from the constructed plasmids were then used in the cell-free translation system in the presence of asolectin liposomes and, when needed, with [14C]leucine (final radioactivity, 2 μCi/ml) as described previously (12). Subsequently, the reaction mixtures were centrifuged at 20,000 × g for 20 min at 4°C, and the supernatant and the precipitate were separately collected. Finally, the precipitate was suspended in 150 μl 10 mM PIPES-NaOH (pH 6.5) and subjected to ultrasonication (Digital Sonifier model 250 D; Branson, Danbury, CT, USA) for 18 s (50% duty cycle). The suspension obtained was used for vesicle preparations for the transporter activity assay as described in the following section. As a blank suspension, the reaction mixture operated in the absence of mRNA was treated in the same way as the rEhMCF protein suspension. To verify the production of rEhMCF and its purity, 1/100 volumes from each fraction were analyzed by SDS-PAGE, followed by visualization through either autoradiography or Coomassie brilliant blue staining.
Transport activity assay of rEhMCF in vesicles.
Two types of the vesicles, namely, substrate-loaded vesicles and empty vesicles, were prepared and used for the assays. The substrate-loaded vesicles were prepared essentially as described previously (12). In detail, the rEhMCF protein suspension (see the section above) was mixed with an equal volume of solution containing liposomes into which one of the countersubstrates examined was preloaded. To prepare the liposome solution, 200 mM PIPES–NaOH (pH 6.5) containing 40 mM potassium gluconate and 50 or 20 mM countersubstrate was added on acetone-washed asolectin (80 mg lipid/ml), followed by ultrasonication (Digital Sonifier model 250 D; Branson, Danbury, CT, USA) for 5 min on ice. Subsequently, the obtained mixture was frozen in liquid nitrogen, thawed at room temperature, and subjected to ultrasonication (Digital Sonifier model 250 D; Branson, Danbury, CT, USA) for 18 s (50% duty cycle). Finally, the countersubstrate that was not loaded into vesicles was removed by gel filtration with a Dowex AG-1X8 column (Bio-Rad, Tokyo, Japan) using 10 mM PIPES–NaOH (pH 6.5) containing 40 mM potassium gluconate and 100 mM sodium gluconate. The empty vesicles were prepared in a same way as described for the substrate-loaded vesicles except that no countersubstrates were preloaded. Blank controls for each type of vesicle were likewise prepared using the blank (see the section above) suspension in place of the rEhMCF protein suspension.
The transport activity of the rEhMCF in vesicles was measured as described previously (12). Briefly, a quantitative evaluation of the uptake of either [32P]ATP or [35S]PAPS into substrate-loaded or empty vesicles was performed. The final concentration of preloaded countersubstrate in the transport assay of the uptake of [32P]ATP or [35S]PAPS was 25 mM or 10 mM, respectively, and that of added radiolabeled substrate (either [32P]ATP or [35S]PAPS) was 0.5 mM. The reaction was initiated at 25°C upon addition of 5 μl radiolabeled substrate into 100 μl 10 mM PIPES–NaOH (pH 6.5) containing either type of vesicle, [32P]ATP (final radioactivity, 2.7 μCi/ml) or [35S]PAPS (final radioactivity, 1.5 μCi/ml). Reactions were terminated by the addition of 15 μl stop solution (360 mM pyridoxal 5′-phosphate, 64 mM mersalyl acid). After removal of extravesicle radiolabeled substrate by gel filtration performed with a Dowex AG-1X8 column (Bio-Rad, Tokyo, Japan) using 200 mM sodium acetate, the radioactivity associated with each type of vesicle was measured as units of disintegrations per minute using an LSC-6100 liquid-scintillation counter from Aloka (Tokyo, Japan). In parallel, a blank control for each type of vesicle was assayed. Finally, the amount of substrate transported into the vesicles was calculated after subtracting the radioactivity of a blank control from that of the corresponding sample. The amount of protein which was estimated prior to vesicle preparation was used to determine the specific transporter activity. Its estimation was accomplished by measuring the intensity of the corresponding bands in SDS-PAGE gels stained by Coomassie brilliant blue. Bovine serum albumin was used as a standard.
For a time course analysis, a quantitative evaluation of the uptake of [32P]ATP into substrate-loaded vesicles into which nonradiolabeled ATP was preloaded or into empty vesicles was performed at 0, 5, 10, or 20 min after starting the incubation. For a countersubstrate specificity analysis, a quantitative evaluation of the uptake of either [32P]ATP or [35S]PAPS into the substrate-loaded vesicles into which one of the various substances tested was preloaded or into empty vesicles was performed after 10 min of incubation. The obtained activities were statistically analyzed with Student's t test.
Nucleotide sequence accession numbers.
The proteins described in this study (GenBank accession numbers, E. histolytica genome identification numbers) and the accession number of the EhMCF synthetic gene are as follows: MCF (XP_649800, genome identification number EHI_095150), Pi transporter (XP_656350, EHI_153760), PAPS transporter (XP_654175, EHI_068590), SULT1 (XP_654200, EHI_140740), SULT2 (XP_654101, EHI_166030), SULT3 (XP_651675, EHI_197340), SULT4 (XP_655605, EHI_092490), SULT5 (XP_650013, EHI_090430), SULT6 (XP_649714, EHI_146990), SULT7 (XP_651768, EHI_114190), SULT8 (XP_652989, EHI_181190), SULT9 (XP_653539, EHI_031640), SULT10 (XP_650544, EHI_062680), PAPase1 (XP_651950, EHI_193350), PAPase2 (XP_655585, EHI_179820), PAPase3 (XP_650613, EHI_175410), and the synthetic EhMCF cDNA (LC036596).
RESULTS AND DISCUSSION
All EhSULTs are localized in the cytosol.
The E. histolytica genome contains 10 genes that encode putative SULTs, which can catalyze the production of sulfated molecules, e.g., sulfolipids, from PAPS and sulfur acceptors (AmoebaDB; http://amoebadb.org/amoeba/ [5, 9]). We previously demonstrated the cytosolic localization of EhSULT1 and -2 (5). To fully determine the localization of all EhSULTs, we established independent transformants expressing HA-tagged EhSULT3 to -10 in E. histolytica and analyzed the transformants obtained. The fluorescent signals detected in these transformants were distributed throughout cells, except in organelles such as the nucleus, vacuoles, and small vesicles (Fig. 1). This result, combined with the data corresponding to the cytosolic localization of EhSULT1 and -2 (5), indicates that all EhSULTs are localized in the cytosol (Fig. 1). Importantly, it also confirms the cytosolic localization of EhSULT6, which catalyzes the production of CS, a sulfolipid important for Entamoeba encystation (9). These findings are consistent with the requirement for PAPS, a mitosomal sulfate-activated metabolite, to be transported into the cytosol for it to be a substrate for EhSULTs in E. histolytica.
FIG 1.
Localization of EhSULT3 to -10 in E. histolytica. Indirect immunofluorescence images of transformants expressing HA-tagged EhSULT3 to -10 are shown. Bars, 10 μm.
Production, purification, and characterization of rEhMCF protein.
Two E. histolytica MC proteins (EHI_095150 and EHI_153760, respectively) have been characterized as an ADP/ATP carrier (AAC) and a Pi carrier (AmoebaDB; http://amoebadb.org/amoeba/ [6, 9, 13]). In addition, phylogenetic analysis showed that only the E. histolytica AAC (EhAAC) and not the Pi carrier is a member of a subfamily cluster of MC proteins, which includes carriers transporting adenine nucleotides and coenzyme A (CoA) (13). Recently, a carrier that was originally characterized as an AAC of the thylakoid membrane (14) was shown to possess a capacity for countertransporting PAPS; therefore, it is now known as a PAPS transporter (15). These findings narrow down the mitosomal membrane PAPS transporter candidates and are consistent with our hypothesis that EhAAC acts as an antiporter that can transport PAPS across mitosomal membranes in E. histolytica. EhAAC is more commonly termed E. histolytica mitochondrial carrier family (EhMCF) (3, 5, 6, 16); therefore, unless otherwise stated, we use the name EhMCF here.
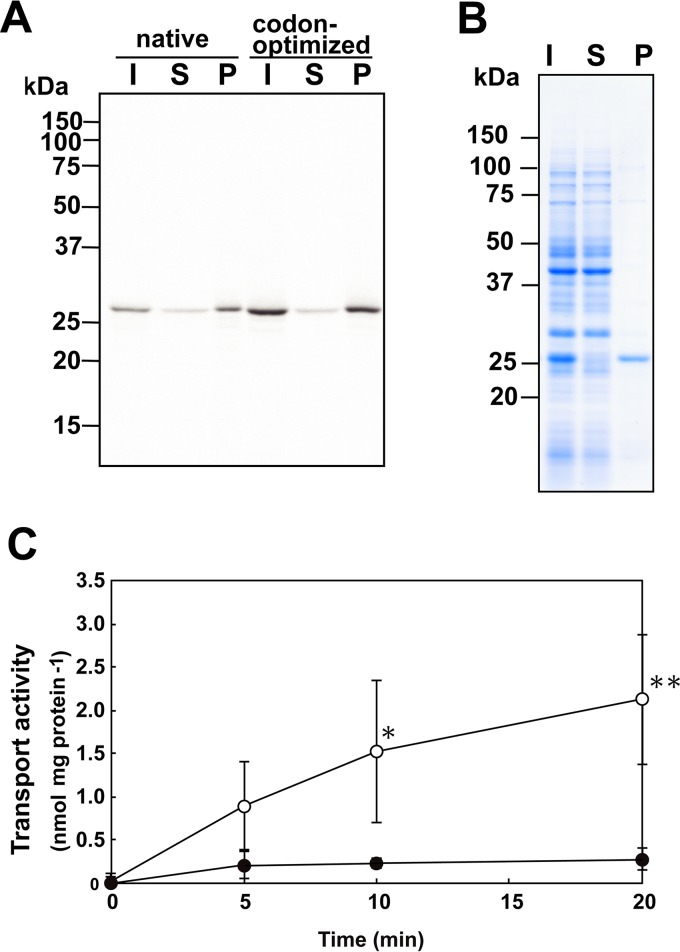
Obtaining functional rEhMCF protein is necessary to address how the mitosomal sulfate activation pathway cooperates with cytosolic SULTs in E. histolytica. To achieve this, we exploited a wheat germ cell-free translation system in the presence of asolectin liposomes because this system circumvents several problems encountered in expressing recombinant membrane proteins in surrogate organisms or cells (17, 18). In this system, we can trace only the target recombinant protein as a nascent protein using a radiolabeled leucine. The molecular size of the synthesized radiolabeled protein was ∼30 kDa, which is close to the deduced molecular mass of EhMCF (30,443 Da), and it was predominantly recovered from the fraction precipitated at 20,000 × g (Fig. 2A). The yield of rEhMCF could be improved by using a synthetic EhMCF cDNA in which the codon usage is optimized to that of the wheat germ translation system (Fig. 2A; GenScript Japan [Tokyo, Japan]). Most importantly, rEhMCF was highly enriched and became a major protein in the precipitated fraction, indicating that centrifugal fractionation is sufficient to purify rEhMCF nearly to homogeneity (Fig. 2B).
FIG 2.
Production, purification, and characterization of rEhMCF. Production (A) and purification (B) of rEhMCF and the assay system for measurement of its transporter activity (C). Autoradiograph (A) and Coomassie brilliant blue-stained (B) SDS-PAGE gels are shown. The loaded samples, which were prepared by centrifugal fractionation of a reaction mixture from the cell-free translation system, were as follows: I, initial material; S, supernatant; P, precipitate. The different DNA fragments carrying an EhMCF ORF in the pYT-08 vector are as follows: native, a PCR-amplified DNA from E. histolytica cDNA; codon-optimized, a synthetic DNA in which the codon usage of the EhMCF ORF is optimized to that of the wheat germ translation system. (C) Time course of the uptake of ATP into vesicles. The uptake of external [32P]ATP into the substrate-loaded vesicles preloaded with nonradiolabeled ATP (open) or into empty vesicles (filled) was measured. The data are shown as means ± standard deviations (SDs) (indicated with error bars) of the results from more than three independent experiments. Asterisks indicate significant differences from empty vesicles (*, P < 0.05; **P < 0.01).
We then examined whether the rEhMCF purified as the precipitated fraction was functional by measuring its ability to transport ATP, a standard substrate for MC proteins. Uptake of 32P-labeled ATP into substrate-loaded vesicles proceeded in a time-dependent manner, while uptake of 32P-labeled ATP into empty vesicles could not be detected (Fig. 2C). These results validate the biochemical methodology used in this study. Furthermore, they indicate that rEhMCF is properly reconstituted into the lipid bilayer of asolectin liposomes, indicating that rEhMCF can function as an antiporter.
The EhMCF functions as a PAPS/adenosine 3′,5′-bisphosphate (PAP) and PAPS/PAPS antiporter in vitro.
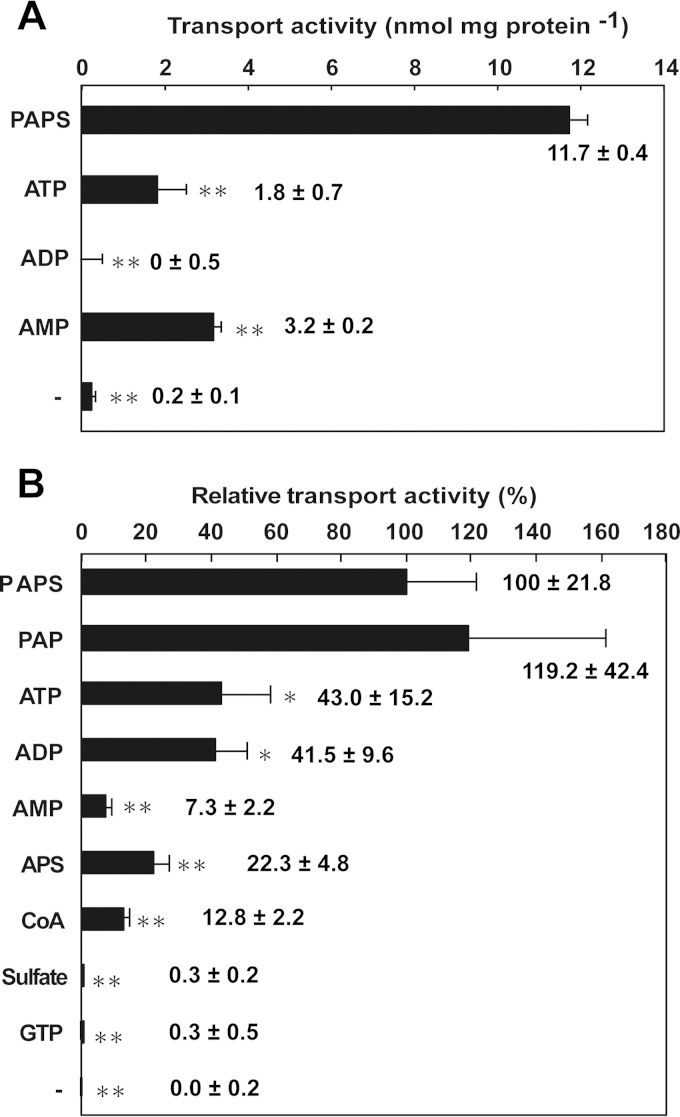
Our primary question—does EhMCF, a mitosomal protein, participate in the translocation of PAPS across mitosomal membranes in E. histolytica?—can be now addressed. To address this question, we measured the countertransport activity of the reconstituted rEhMCF for ATP with PAPS, ATP, ADP, or AMP. The uptake of 32P-labeled ATP into the substrate-loaded vesicles was significantly higher when PAPS was used as a countersubstrate (11.7 ± 0.4 nmol/mg protein) than when adenosine mono-, di-, or triphosphates were used (3.2 ± 0.2, 0.0 ± 0.5, or 1.8 ± 0.7 nmol/mg protein, respectively). The uptake of 32P-labeled ATP into empty vesicles could not be detected (Fig. 3A). These data are partly inconsistent with those from a previous study owing to differences in the ATP/ADP exchange activity (6). In this study, no uptake of 32P-labeled ATP into the ADP-loaded vesicles or the empty vesicles could be detected, while in the previous study, a significant uptake of 14C-labeled ATP into ADP-loaded vesicles was observed. Assessment of this difference is hampered by the lack of demonstration of uptake of 14C-labeled ATP into the empty vesicles in the former work. They also showed a significant uptake of 14C-labeled ADP into ATP-loaded vesicles, a result which was well supported by showing competitive inhibition by excess cold ADP as well as the absence of uptake into the empty vesicles. This inconsistency in the reported ATP/ADP exchange activity may be due to differences in proteoliposome preparations, in components in the reactions (e.g., the use of radiolabeled materials as a substrate or of preloaded molecules as a countersubstrate), and/or in assay conditions (e.g., incubation time or concentrations of substrate added and of countersubstrate preloaded). Another explanation is possible differences in the lipid compositions because of different membrane sources having been used for protein reconstitutions (this study and reference 6). More importantly, the data (Fig. 3A) clearly indicate that EhMCF indeed has the capacity for countertransport of ATP using PAPS as a preferred countersubstrate.
FIG 3.
Countersubstrate specificities of rEhMCF reconstituted in vesicles. The substrate-loaded vesicles (preloaded substances are indicated by names) and the empty vesicles (indicated by a minus sign) were assayed for the uptake of either [32P]ATP (A) or [35S]PAPS (B). The specific transport activity (A) and the transport activity relative to that of PAPS as the control (set as 100%) (B) are shown. The data are presented as the means ± SDs calculated from the results of three independent experiments. The raw data used to calculate the relative activity levels are as follows: PAPS, 32.8 ± 7.2; PAP, 39.1 ± 13.9; ATP, 14.1 ± 5.0; ADP, 13.6 ± 3.2; AMP, 2.4 ± 0.7; APS, 7.3 ± 1.6; CoA, 4.2 ± 0.7; sulfate, 0.1 ± 0.1; GTP, 0.1 ± 0.2; −, 0.0 ± 0.1 (nmol/mg protein/min). Asterisks indicate significant differences between PAPS and other countersubstrates (*, P < 0.05; **P < 0.01).
To investigate further the countersubstrate specificity, we measured the activity of PAPS countertransport with various substances. Among potential countersubstrates examined, adenosine 3′,5′-bisphosphate (PAP) gave the highest activity for the uptake of 35S-labeled PAPS into the substrate-loaded vesicles (119.2 ± 42.4% relative to that of PAPS as 100% control). Adenosine mono-, di-, and triphosphates all gave moderate activities, but a preference for ATP and ADP over AMP (43.0 ± 15.2% and 41.5 ± 9.6% over 7.3 ± 2.2%, respectively) was observed. Similarly, adenosine 5′-phosphosulfate (APS) and coenzyme A (CoA) also gave moderate activities (22.3 ± 4.8% and 12.8 ± 2.2%, respectively). Sulfate and GTP gave nearly untraceable activities (0.3 ± 0.2% and 0.3 ± 0.5%, respectively). Consistent with the assays using 32P-labeled ATP, uptake of 35S-labeled PAPS into empty vesicles could not be detected at all (Fig. 3B). These data (Fig. 3) indicate that EhMCF is different from archetypal AACs regarding countersubstrate specificity and are in agreement with the previous finding that carboxyatractyloside and bongkrekic acid, specific inhibitors of most AACs, are not effective against EhAAC (6). More importantly, these data indicate that EhMCF functions mainly as a PAPS/PAP and PAPS/PAPS antiporter in vitro.
Evidence that EhMCF functions mainly as a PAPS/ATP antiporter in vivo.
Generally, PAP is produced together with a sulfated molecule through the catalytic action of SULT with respect to PAPS and a sulfur acceptor. PAP is then degraded to adenosine 5′-phosphate and Pi by 3′(2′),5′-bisphosphate nucleotidase [19, 20; 3′(2′),5′-bisphosphate nucleotidase is described here as PAPase for ease of reading]. In E. histolytica, all the EhSULTs are localized in the cytosol (Fig. 1) (5), and EhMCF has a high capacity for countertransport of PAPS with PAP or PAPS in vitro (Fig. 3). Therefore, we inferred that knowing the localization of EhPAPase is important for predicting the availability of the substrates for EhMCF in vivo; this will help unravel its precise role.
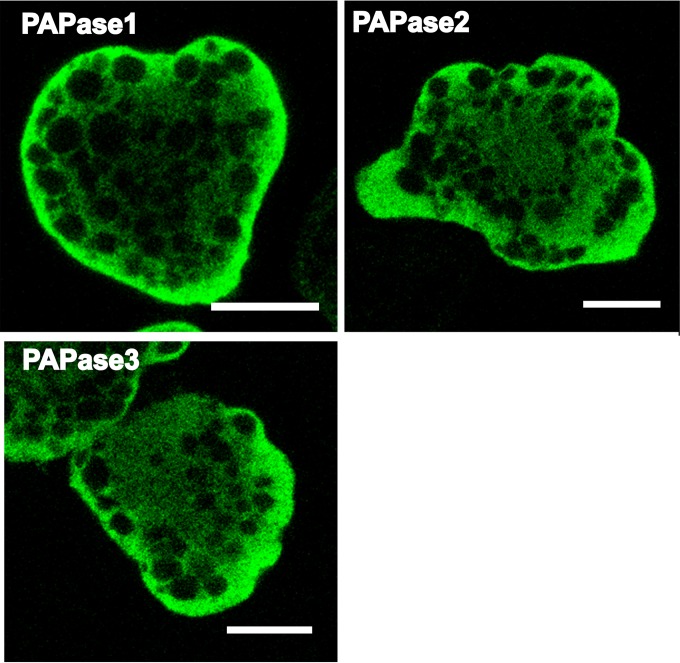
In the E. histolytica genome, three genes encoding putative PAPase1 to -3 (EHI_193350, EHI_179820, and EHI_175410, respectively) (AmoebaDB; http://amoebadb.org/amoeba/) are present. To determine the localization of all the PAPases in E. histolytica, we established independent transformants expressing each HA-tagged EhPAPase. In all the transformants analyzed, except in organelles such as the nucleus, vacuoles, and small vesicles, the fluorescent signals were evenly distributed throughout cells, indicating that EhPAPase1 to -3 are localized in the cytosol (Fig. 4). Consistent with these observations, the cytosolic localizations of EhPAPase1 and -2 have been recently reported (21, 22). Moreover, EhPAPase1 has been biochemically characterized (21) and active transcription of EhPAPase1 to -3 has been reported (23). Collectively, these findings indicate that all the EhPAPases are functional in the E. histolytica cytosol.
FIG 4.
Localization of EhPAPase1 to -3 in E. histolytica. Indirect immunofluorescence images of transformants expressing HA-tagged EhPAPase1 to -3 are shown. Bars, 10 μm.
This notion, together with the cytosolic localization of all the EhSULTs (Fig. 1) (5), suggests that PAP produced by the catalytic action of EhSULTs is sequentially degraded by EhPAPases; therefore, the PAP concentration in the cytosol is maintained at a low level in E. histolytica. This is consistent with the general idea that PAP is toxic to cells (24) but contradicts another interpretation of the results (Fig. 3), i.e., that EhMCF functions mainly as a PAPS/PAP and PAPS/PAPS antiporter in vitro. However, the results (Fig. 3) also showed that the reconstituted rEhMCF had high activity for the exchange of ATP with PAPS. ATP, which is mainly synthesized through cytosolic pathways, is a crucial molecule for the sulfate activation pathway to produce PAPS in E. histolytica mitosomes, whereas PAPS is a necessary substrate for EhSULTs to produce sulfolipids in the cytosol. Hence, maintaining a molecule such as EhMCF that has a high capacity for exchanging ATP with PAPS across the mitosomal membrane could be beneficial for E. histolytica.
In agreement with this interpretation, we previously demonstrated that MCFgs, an E. histolytica G3 strain in which EhMCF was knocked down by gene silencing, showed a significant reduction in sulfate activation activity and a marked growth defect (3). This finding can be now explained by that the shortage of ATP in the mitosomes as well by as a shortage of PAPS in the cytosol. This would impair the synthesis of sulfolipids, which plays important roles in E. histolytica growth. However, a study to confirm the importance of sulfolipids in E. histolytica is needed.
Mechanistically, the EhMCF knockdown causes an evident reduction in the exchange activity of PAPS with ATP across the mitosomal membrane in E. histolytica; this reduction lowers the flow of cytosolic ATP into mitosomes, leading to a mitosomal ATP shortage. This shortage severely impairs mitosomal sulfate activation activity; therefore, PAPS production significantly decreases. In addition, even the PAPS produced cannot be efficiently translocated into the cytosol because of the defect in EhMCF activity. Consequently, PAPS levels in the cytosol become remarkably low, resulting in the halting of almost all EhSULT sulfolipid synthesis.
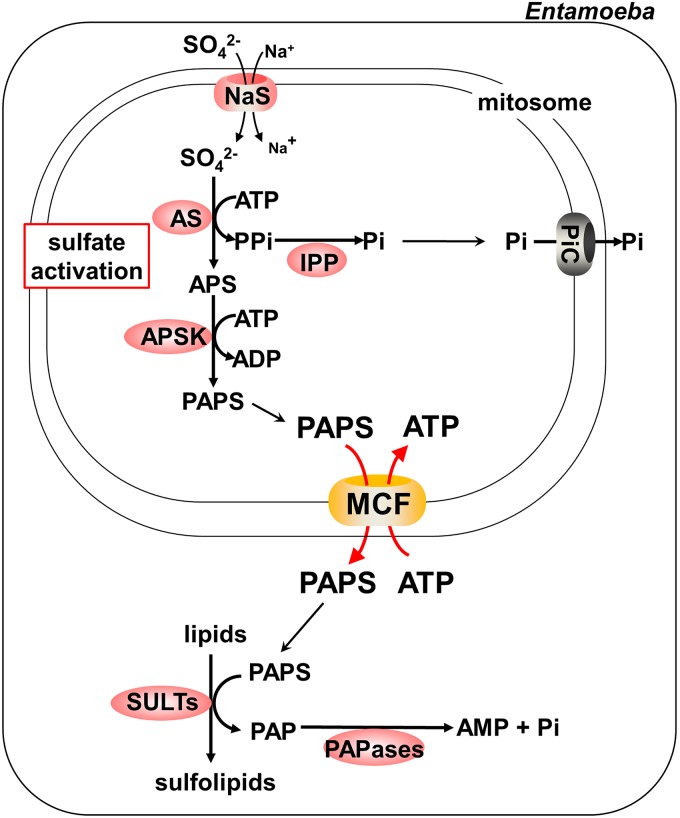
In conclusion, we suggest that EhMCF functions mainly as a PAPS/ATP antiporter and links the mitosomal sulfate activation pathway to the cytosolic chain reaction that is composed of EhSULTs and EhPAPases in E. histolytica (Fig. 5).
FIG 5.
A scheme for sulfate metabolism in E. histolytica. The flow of metabolites and the enzymes involved are depicted, based on evidence from previous studies (3, 5, 13, 21, 22) as well as from the present study. APS, adenosine 5′-phosphosulfate; APSK, APS kinase; AS, ATP sulfurylase; IPP, inorganic pyrophosphatase; MCF, mitochondrial carrier family; NaS, sodium/sulfate symporter; PAP, adenosine 3′,5′-bisphosphate; PAPase, 3′(2′),5′-bisphosphate nucleotidase; PAPS, 3′-phosphoadenosine 5′-phosphosulfate; Pi, inorganic phosphate; PiC, Pi carrier; PPi, pyrophosphate; SULT, sulfotransferase.
ACKNOWLEDGMENTS
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan to F.M. (22890136, 24117517, and 26117719), to A.N. (24580094, 24589511, and 15K07006), to H.Y. (25460594), to Y.T. (24117516 and 26117717), and to T.N. (23117001, 23117005, and 26293093) and from the Research Program on Emerging and Re-emerging Infectious Diseases from Japan Agency for Medical Research and Development (AMED) to T.N. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Shouko Takao, Ritsuko Yoshida, and Shizuko Furukawa for technical assistance.
F.M. and A.N. designed and performed the experiments; F.M., A.N., H.Y., Y.T., and T.N. analyzed the data and wrote the paper.
We declare that we have no conflict of interest.
REFERENCES
- 1.Ralston KS, Petri WA Jr. 2011. Tissue destruction and invasion by Entamoeba histolytica. Trends Parasitol 27:254–263. doi: 10.1016/j.pt.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makiuchi T, Nozaki T. 2014. Highly divergent mitochondrion-related organelles in anaerobic parasitic protozoa. Biochimie 100:3–17. doi: 10.1016/j.biochi.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 3.Mi-ichi F, Makiuchi T, Furukawa A, Sato D, Nozaki T. 2011. Sulfate activation in mitosomes plays an important role in the proliferation of Entamoeba histolytica. PLoS Negl Trop Dis 5:e1263. doi: 10.1371/journal.pntd.0001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Giezen M. 2009. Hydrogenosomes and mitosomes: conservation and evolution of functions. J Eukaryot Microbiol 56:221–231. doi: 10.1111/j.1550-7408.2009.00407.x. [DOI] [PubMed] [Google Scholar]
- 5.Mi-ichi F, Abu Yousuf Nakada-Tsukui M, Nozaki K, T. 2009. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc Natl Acad Sci U S A 106:21731–21736. doi: 10.1073/pnas.0907106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan KW, Slotboom DJ, Cox S, Embley TM, Fabre O, van der Giezen M, Harding M, Horner DS, Kunji ER, Leon-Avila G, Tovar J. 2005. A novel ADP/ATP transporter in the mitosome of the microaerophilic human parasite Entamoeba histolytica. Curr Biol 15:737–742. doi: 10.1016/j.cub.2005.02.068. [DOI] [PubMed] [Google Scholar]
- 7.Mai Z, Ghosh S, Frisardi M, Rosenthal B, Rogers R, Samuelson J. 1999. Hsp60 is targeted to a cryptic mitochondrion-derived organelle (“crypton”) in the microaerophilic protozoan parasite Entamoeba histolytica. Mol Cell Biol 19:2198–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Müller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu RY, van der Giezen M, Tielens AG, Martin WF. 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev 76:444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mi-ichi F, Miyamoto T, Takao S, Jeelani G, Hashimoto T, Hara H, Nozaki T, Yoshida H. 2015. Entamoeba mitosomes play an important role in encystation by association with cholesteryl sulfate synthesis. Proc Natl Acad Sci U S A 112:E2884–E2890. doi: 10.1073/pnas.1423718112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monné M, Palmieri F. 2014. Antiporters of the mitochondrial carrier family. Curr Top Membr 73:289–320. doi: 10.1016/B978-0-12-800223-0.00008-6. [DOI] [PubMed] [Google Scholar]
- 11.Palmieri F, Pierri CL, De Grassi A, Nunes-Nesi A, Fernie AR. 2011. Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J 66:161–181. doi: 10.1111/j.1365-313X.2011.04516.x. [DOI] [PubMed] [Google Scholar]
- 12.Nozawa A, Fujimoto R, Matsuoka H, Tsuboi T, Tozawa Y. 2011. Cell-free synthesis, reconstitution, and characterization of a mitochondrial dicarboxylate-tricarboxylate carrier of Plasmodium falciparum. Biochem Biophys Res Commun 414:612–617. doi: 10.1016/j.bbrc.2011.09.130. [DOI] [PubMed] [Google Scholar]
- 13.Dolezal P, Dagley MJ, Kono M, Wolynec P, Likic VA, Foo JH, Sedinova M, Tachezy J, Bachmann A, Bruchhaus I, Lithgow T. 2010. The essentials of protein import in the degenerate mitochondrion of Entamoeba histolytica. PLoS Pathog 6:e1000812. doi: 10.1371/journal.ppat.1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thuswaldner S, Lagerstedt JO, Rojas-Stutz M, Bouhidel K, Der C, Leborgne-Castel N, Mishra A, Marty F, Schoefs B, Adamska I, Persson BL, Spetea C. 2007. Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis. J Biol Chem 282:8848–8859. doi: 10.1074/jbc.M609130200. [DOI] [PubMed] [Google Scholar]
- 15.Gigolashvili T, Geier M, Ashykhmina N, Frerigmann H, Wulfert S, Krueger S, Mugford SG, Kopriva S, Haferkamp I, Flugge UI. 2012. The Arabidopsis thylakoid ADP/ATP carrier TAAC has an additional role in supplying plastidic phosphoadenosine 5′-phosphosulfate to the cytosol. Plant Cell 24:4187–4204. doi: 10.1105/tpc.112.101964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, Embley TM. 2010. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos Trans R Soc Lond B Biol Sci 365:713–727. doi: 10.1098/rstb.2009.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nozawa A, Tozawa Y. 2014. Modifications of wheat germ cell-free system for functional proteomics of plant membrane proteins. Methods Mol Biol 1072:259–272. doi: 10.1007/978-1-62703-631-3_19. [DOI] [PubMed] [Google Scholar]
- 18.Bernhard F, Tozawa Y. 2013. Cell-free expression—making a mark. Curr Opin Struct Biol 23:374–380. doi: 10.1016/j.sbi.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Murguía JR, Belles JM, Serrano R. 1995. A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science 267:232–234. doi: 10.1126/science.7809627. [DOI] [PubMed] [Google Scholar]
- 20.Ramaswamy SG, Jakoby WB. 1987. (2′)3′,5′-Bisphosphate nucleotidase. J Biol Chem 262:10044–10047. [PubMed] [Google Scholar]
- 21.Faisal Tarique K, Arif Abdul Rehman S, Gourinath S. 2014. Structural elucidation of a dual-activity PAP phosphatase-1 from Entamoeba histolytica capable of hydrolysing both 3′-phosphoadenosine 5′-phosphate and inositol 1,4-bisphosphate. Acta Crystallogr D Biol Crystallogr 70:2019–2031. doi: 10.1107/S1399004714010268. [DOI] [PubMed] [Google Scholar]
- 22.Faisal Tarique K, Arif Abdul Rehman S, Betzel C, Gourinath S. 2014. Structure-based identification of inositol polyphosphate 1-phosphatase from Entamoeba histolytica. Acta Crystallogr D Biol Crystallogr 70:3023–3033. doi: 10.1107/S1399004714021245. [DOI] [PubMed] [Google Scholar]
- 23.Husain A, Jeelani G, Sato D, Nozaki T. 2011. Global analysis of gene expression in response to L-Cysteine deprivation in the anaerobic protozoan parasite Entamoeba histolytica. BMC Genomics 12:275. doi: 10.1186/1471-2164-12-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toledano E, Ogryzko V, Danchin A, Ladant D, Mechold U. 2012. 3′-5′ phosphoadenosine phosphate is an inhibitor of PARP-1 and a potential mediator of the lithium-dependent inhibition of PARP-1 in vivo. Biochem J 443:485–490. doi: 10.1042/BJ20111057. [DOI] [PMC free article] [PubMed] [Google Scholar]