Abstract
The parasite Trypanosoma brucei lives in the bloodstream of infected mammalian hosts, fully exposed to the adaptive immune system. It relies on a very high rate of endocytosis to clear bound antibodies from its cell surface. All endo- and exocytosis occurs at a single site on its plasma membrane, an intracellular invagination termed the flagellar pocket. Coiled around the neck of the flagellar pocket is a multiprotein complex containing the repeat motif protein T. brucei MORN1 (TbMORN1). In this study, the phenotypic effects of TbMORN1 depletion in the mammalian-infective form of T. brucei were analyzed. Depletion of TbMORN1 resulted in a rapid enlargement of the flagellar pocket. Dextran, a polysaccharide marker for fluid phase endocytosis, accumulated inside the enlarged flagellar pocket. Unexpectedly, however, the proteins concanavalin A and bovine serum albumin did not do so, and concanavalin A was instead found to concentrate outside it. This suggests that TbMORN1 may have a role in facilitating the entry of proteins into the flagellar pocket.
INTRODUCTION
Trypanosoma brucei is an important parasite of humans and domestic animals in sub-Saharan Africa, as the causative agent of sleeping sickness and nagana, respectively. Its complex life cycle involves transitions between tsetse fly vectors (its definitive hosts) and mammalian intermediate hosts. This life cycle involves a number of different cell stages, of which the procyclic form (found in the tsetse fly) and the slender bloodstream form (BSF) (found in the mammalian bloodstream) are the best studied in a laboratory setting. The procyclic form and the BSF of T. brucei share similar cytoskeletal architectures (1, 2).
The principal feature of this cytoskeleton is a corset of microtubules that lie directly underneath the plasma membrane and impart to the cell its distinctive shape (3). A single invagination of the plasma membrane, termed the flagellar pocket (FP), constitutes a distinct subdomain and is found at the posterior end of the cell (4). The FP is the site of all endo- and exocytic traffic (5, 6). Abutting the FP membrane is a basal body that nucleates the single flagellum of the trypanosome cell. The flagellum exits the FP and is adhered longitudinally to the cell body along a left-handed helical path (7). Once outside the FP, the axoneme of the flagellum is paralleled by an associated intraflagellar structure called the paraflagellar rod (PFR). The PFR is composed of a paracrystalline lattice and is associated with cellular motility (8). Nucleated adjacent to the basal body is a specialized microtubule quartet that traces around the FP and then underlies the flagellum as far as the anterior end of the cell (4).
The small cylinder of membrane that connects the FP to the rest of the plasma membrane constitutes a third subdomain and is called the flagellar pocket neck (FPN) (4). A number of discrete cytoskeletal structures cluster around the FPN membrane on its cytoplasmic face. Of these, the best characterized is an electron-dense horseshoe-shaped structure named the flagellar pocket collar (FPC) (4). The only known component of the FPC is the protein TbBILBO1, which has been localized to the FPC by immunoelectron microscopy (immuno-EM) and shown to be essential for FP biogenesis (9–12).
Situated on top of the FPC is another multiprotein complex, containing the repeat motif protein T. brucei MORN1 (TbMORN1) (13). The TbMORN1 (40 kDa) molecules in the complex are arranged in a linear macromolecular filament of ∼0.2 by 2 μm whose posterior end is tightly coiled around the FPN, producing an overall fishhook-shaped morphology (13). At least nine other proteins are known to partially or wholly associate with this complex: TbLRRP1, TBCCD1, and seven currently uncharacterized proteins identified in a screen using proximity-dependent biotinylation (14–16). Both the TbMORN1 filament and the FPC are strongly associated with the microtubule-based cytoskeleton.
In the past, the TbMORN1 complex has been described variously as the “bilobe,” “bi-lobe,” or “bi-lobed structure” (14, 17, 18). This bi-lobed structure was originally defined as a centrin-containing complex that was proposed to influence Golgi biogenesis (17). However, recent higher-resolution morphological study has cast doubt on whether the TbMORN1 complex and the centrin-containing complex are indeed associated, and the two structures may be physically distinct (13). To avoid confusion, and to emphasize that the results described here refer solely to the TbMORN1 complex, the term “bilobe” has not been used.
Previous functional work on TbMORN1 focused primarily on procyclic cells but noted that depletion was lethal in BSFs (18). This report describes the phenotypic effects of TbMORN1 depletion in BSFs and the discovery of an unexpected role in facilitating protein entry to the FP.
MATERIALS AND METHODS
Antibodies and reagents.
The anti-TbMORN1 (rabbit polyclonal) and anti-TbBILBO1 (rabbit polyclonal) antibodies have been described previously (13, 16). The anti-BiP antibodies were a gift from Jay Bangs (University at Buffalo, USA) (19). Dextran (10 kDa, lysine fixable, fluorescein conjugated) and concanavalin A (ConA; tetramethylrhodamine-conjugated) were purchased from Life Technologies (Carlsbad, CA). Bovine serum albumin (BSA; conjugated to 5-nm gold beads) was purchased from British Biocell International (Cardiff, United Kingdom).
Cell lines and culture.
Generation of the BSF TbMORN1 RNAi cell line has been described previously (18). The cells were cultured in HMI-9 medium supplemented with 10% heat-inactivated fetal calf serum, 2.5 μg/ml of G418 and 5 μg/ml of hygromycin at 37°C and 5% CO2.
Growth curves and immunoblotting.
Parallel cultures of BSF TbMORN1 RNA interference (RNAi) cells were seeded at 105 cells/ml. RNAi was induced in one population by the addition of 1 μg/ml of tetracycline. Population density was measured every hour, with an average of 18 readings over a 48-h period in a single experiment. Experiment start times were staggered to ensure that each data point was covered in at least 4 independent inductions. Population density was measured using a Z2 Coulter Counter (Beckman Coulter, Krefeld, Germany). For immunoblotting, whole-cell lysates from control and RNAi-induced populations were prepared using standard techniques at a final concentration of 2 × 105 cells/μl. The control lysates were serially diluted using SDS loading buffer. SDS-PAGE and immunoblotting were carried out using standard techniques. For quantitative immunoblots, 6 × 105 cells/lane were loaded; for immunoblots of undiluted (100%) samples, 4 × 105 cells/lane were loaded to avoid signal saturation.
Cell cycle counts and quantification of rounded cells.
Parallel cultures of BSF TbMORN1 RNAi cells were seeded at 105 cells/ml in a total volume of 50 ml, and RNAi was induced in one culture by the addition of 1 μg/ml of tetracycline. At the desired time points, samples of 7 × 105 cells from each population were fixed using isothermal glutaraldehyde according to a previously published protocol (20). Fixed cells were washed with phosphate-buffered saline (PBS) and attached to poly-l-lysine-coated coverslips by centrifugation (1,000 × g for 1 min). The coverslips were mounted on glass slides using DAPI-Fluoromount G purchased from Southern Biotech (Birmingham, AL). 4′,6-Diamidino-2-phenylindole (DAPI) and differential interference contrast (DIC) images were taken using a customized Zeiss Axio Observer Z1 microscope (Jena, Germany) running Visiview software (Visitron Systems, Puchheim, Germany) and equipped with a PCO 1600 camera (PCO, Kelheim, Germany) and a Plan-Apochromat 100×/1.46 oil immersion lens. Image processing was carried out using ImageJ and Adobe Photoshop. Rounded cells and cell cycle counts were quantified from the images obtained and processed using Microsoft Excel.
Immunofluorescence microscopy.
Cell density in control (−Tet) and induced (+Tet) cultures was measured 14 h postinduction, and 106 cells/coverslip were taken. The cells were pelleted by centrifugation (1,800 × g for 2 min at 4°C) and resuspended in 1 ml of ice-cold PBS. This wash step was repeated. The cells were then resuspended in 500 μl of ice-cold PBS and attached to poly-l-lysine-coated coverslips in a 24-well plate by centrifugation (1,000 × g for 1 min at 4°C). The cells were extracted (5 min, ice) using extraction buffer (0.5% NP-40, 0.1 M PIPES-NaOH, 2 mM EGTA, 1 mM MgSO4, 0.1 mM EDTA, cOmplete protease inhibitors [Roche]) and washed twice with 1 ml of ice-cold PBS. The cells were fixed using 4% paraformaldehyde in ice-cold PBS (20 min on ice and then 40 min at room temperature [RT]) and then washed with 1 ml of ice-cold PBS. Blocking and antibody labeling steps have been described previously (13). Image acquisition and processing were as described in the paragraph above. The same camera exposure times were used for control (−Tet) and induced (+Tet) samples, and the images were processed using the same channel levels. The images in the figures are maximum-intensity z-projections. Throughout the protocol, great care was taken to prevent cell damage: all liquid handling was carried out using a P1000 micropipette, and pipetting was done as gently as possible to minimize shear forces.
Fractionation.
The fractionation protocol was adapted from a previously published method (21). Cell density in control (−Tet) and induced (+Tet) cultures (14-h point) was measured, and equal amounts (∼4 × 107 cells) were taken from each. The cells were pelleted by centrifugation (800 × g for 10 min at 4°C), resuspended in 1 ml of wash buffer (0.1 M PIPES-NaOH, 2 mM EGTA, 1 mM MgSO4, 0.1 mM EDTA, cOmplete protease inhibitors), and transferred to 1.5-ml Eppendorf tubes. The cells were pelleted by centrifugation (1,800 × g for 2 min at 4°C), resuspended in 200 μl of extraction buffer (see paragraph above), and incubated (15 min at RT, with orbital mixing). A 10-μl sample (5%) of this input (I) fraction was then taken. Detergent-soluble and -insoluble fractions were separated by centrifugation (3,400 × g for 2 min at RT). The detergent-soluble supernatant (SN) fraction was transferred to a fresh tube and the exact volume noted. The tube containing the pellet was centrifuged for a second time to bring down material sticking to the tube wall; this material was discarded. The detergent-insoluble cytoskeletal pellet (P) was then resuspended in 200 μl of extraction buffer. Samples (5% by volume) of the SN and P fractions were taken. Equal fractions (percent) of the I, SN, and P samples were separated by SDS-PAGE and analyzed by immunoblotting. In the immunoblots shown in Fig. 2, the samples are all 2.5%, equivalent to ∼106 cells for the I fraction.
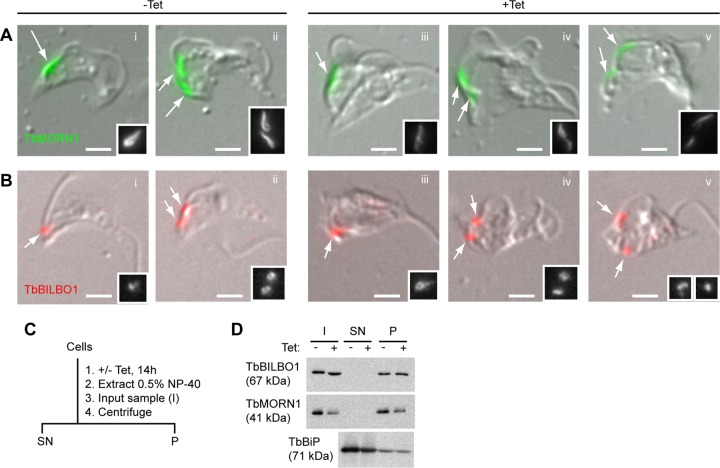
FIG 2.
Imaging and biochemical analysis of TbMORN1-depleted cells. (A and B) TbMORN1 RNAi cells from uninduced (−Tet) and induced (+Tet, 14-h point) populations were extracted with detergent, fixed, and labeled with the indicated antibodies. Immunofluorescence images were acquired using identical exposure times and are shown superimposed on differential interference contrast (DIC) images. Greyscale insets reproduce the structures indicated by arrows. Scale bars, 2 μm. (A) TbMORN1 labeling is reduced in +Tet populations. (B) TbBILBO1 localization and signal intensity seem unchanged in +Tet populations. (C) Schematic of the biochemical fractionation protocol. After extraction in 0.5% NP-40, an input sample (I) was taken. Centrifugation was used to separate the soluble cytoplasmic supernatant (SN) from the insoluble cytoskeletal pellet (P). (D) Immunoblotting of samples from biochemical fractionation (14-h point). I, SN, P samples are as defined for panel C. Equal fractions (2.5% of total; equal to ∼106 cells in I sample) were loaded in each lane. Three independent experiments were carried out; a representative immunoblot is shown.
Electron microscopy and BSA-gold uptake assays.
Cell density in control (−Tet) and induced (+Tet) cultures was measured and equal amounts (∼2 × 108 cells) were taken from each. The cells were sedimented and fixed with 2.5% glutaraldehyde–4% paraformaldehyde in 0.1 M cacodylate buffer for 2 h. After washes with 0.1 M cacodylate buffer, pellets were incubated with 1% OsO4 in cacodylate buffer for 1.5 h. Following dehydration in a graded ethanol series, samples were embedded in Epon resin. Sixty-nanometer-thick sections were cut using a Reichert Ultracut S ultramicrotome (Leica, Vienna, Austria) and contrasted with 4% uranyl acetate and 1% lead citrate. Images were acquired at 80 kV using a Tecnai 20 electron microscope (FEI, Eindhoven, the Netherlands) equipped with an Eagle 4K charge-coupled-device (CCD) camera (FEI) and processed with Adobe Photoshop. For BSA-gold uptake, the cells were pelleted by centrifugation (800 × g for 10 min at 4°C), washed twice in ice-cold serum-free HMI9, and transferred to Eppendorf tubes. The cells were again pelleted (1,800 × g for 2 min at 4°C) and resuspended in ice-cold PBS supplemented with 20 mM glucose to a final density of 2 × 108 cells/ml. The cells were cooled (10 min on ice) and incubated with BSA-gold at ∼5 × 1012 particles/ml (15 min on ice and then 30 min at 37°C). Cooling and incubation on ice were carried out to ensure parity with the other uptake assays. The cells were then pelleted, washed once with serum-free HMI9 (37°C), and fixed by the addition of isothermal glutaraldehyde to a final concentration of 2.5% (10 min at 37°C). Further fixation and processing steps were the same as those described above.
Uptake assays (dextran and ConA).
Cell density in control (−Tet) and induced (+Tet) cultures was measured 14 h postinduction, and 4 samples (2 from each population) of 2 × 106 cells were taken. The cells were pelleted by centrifugation (800 × g for 10 min at 4°C), resuspended in 1 ml of ice-cold serum-free HMI9, and transferred to Eppendorf tubes. They were then pelleted by centrifugation (1,800 × g for 2 min at 4°C), resuspended in 100 μl of ice-cold serum-free HMI9, and chilled (10 min on ice). Dextran (final concentration, 5 mg/ml) and/or ConA (final concentration, 10 μg/ml) was added and mixed by flicking. The cells were then incubated to allow FP loading (15 min on ice). Half the samples were then fixed (time [t] = 0 min). The other half (t = 30 min) were shifted to higher temperature and incubated to allow uptake (37°C for 30 min). For both sets of samples (t = 0 and 30 min), uptake was stopped by the addition of 1 ml of ice-cold serum-free HMI9, and the cells were pelleted and then resuspended and incubated in 4% paraformaldehyde–0.1% glutaraldehyde in ice-cold PBS (20 min on ice and then 40 min at RT). The cells were washed twice in PBS, attached to poly-l-lysine-coated coverslips, and mounted on slides with DAPI-Fluoromount G. The slides were briefly dried (tissue culture hood for 45 min) to stabilize the coverslips and then imaged directly. The same acquisition settings and exposure times were used for −Tet and +Tet samples. The images displayed in Fig. 4 and 5 are maximum-intensity z-projections, although where necessary, signal overlap was confirmed in single z-slices. The images shown in Fig. 6 are all single z-slices.
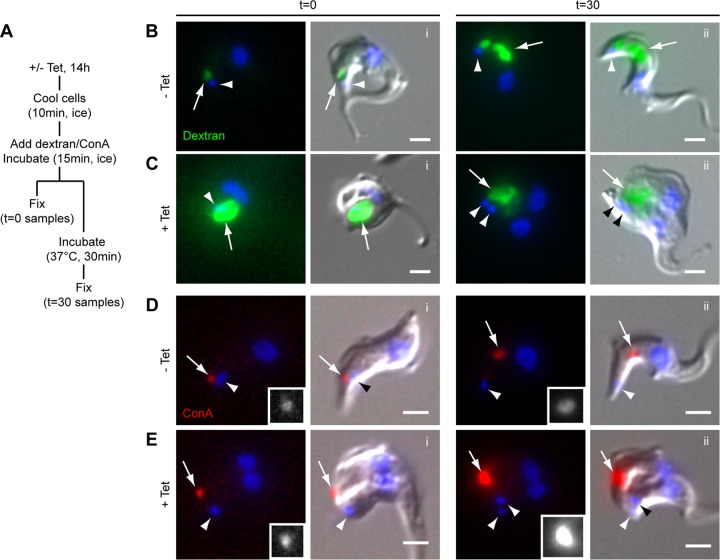
FIG 4.
Dextran (20-Å diameter) appears to accumulate in the enlarged flagellar pocket of TbMORN1-depleted cells, while ConA (80-Å diameter) does not. (A) Schematic of the uptake protocol. (B) Uptake of fluorophore-conjugated dextran (green) in control cells. DNA is labeled with DAPI (blue). The left side of each pair shows the fluorescence image; the right side shows the DIC overlay. At 0 min, dextran (arrow) exhibited a round labeling pattern adjacent to the kinetoplast (arrowhead). At 30 min, robust uptake of dextran into the region of the cell corresponding to the endosomal-lysosomal system was observed. (C) Accumulation of fluorophore-conjugated dextran in TbMORN1-depleted cells. At 0 min, a large accumulation of dextran could be observed in the region of the cell corresponding to the enlarged FP. The label was observed to persist in this region at 30 min. Data were obtained from three independent experiments. (D) Uptake of fluorophore-conjugated ConA (red) in control cells. The labeling pattern is similar to that seen for dextran. The ConA signal (arrows) is shown enlarged in greyscale in the insets. (E) Accumulation of ConA after TbMORN1 depletion. At 0 min, ConA was found in a punctate accumulation (arrow) some distance from the kinetoplast (arrowhead). The distribution did not change at 30 min. Data were obtained from three independent experiments, and cells were imaged using the same exposure times for −Tet and +Tet samples. Scale bars, 2 μm.
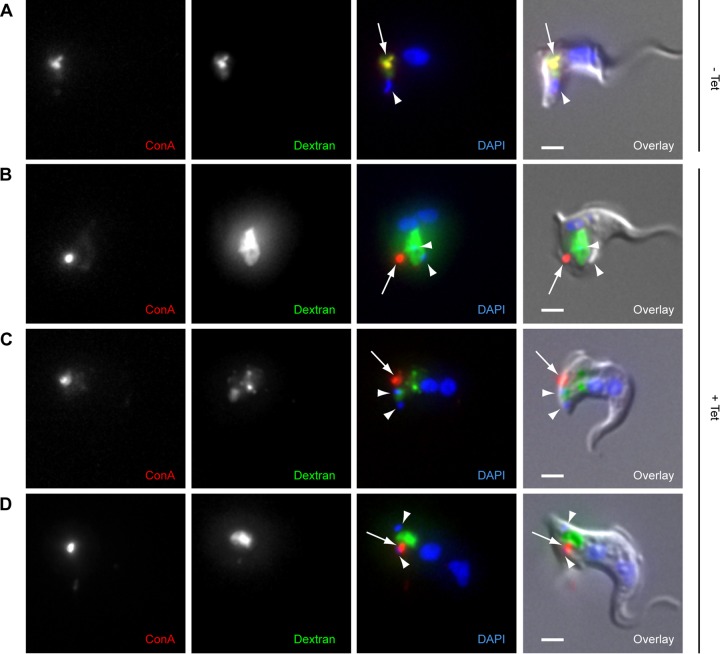
FIG 5.
Loss of overlap between dextran (20-Å diameter) and ConA (80-Å diameter) in TbMORN1-depleted cells. TbMORN1 RNAi cells from uninduced (−Tet) or induced (+Tet, 14-h point) populations were incubated simultaneously with fluorophore-conjugated ConA (red) and dextran (green) for 30 min at 37°C and then fixed. DNA was labeled with DAPI. (A) Uninduced cells showed strong overlap between the two signals in the region of the cell corresponding to the endosomal-lysosomal system. (B to D) TbMORN1-depleted cells show a loss in overlap between the two signals. ConA (arrows) showed a small punctate accumulation, dextran a larger and more amorphous accumulation. Kinetoplasts (sometimes partially obscured by the dextran signal) are indicated with arrowheads. Data were obtained from three independent experiments. Scale bars, 2 μm.
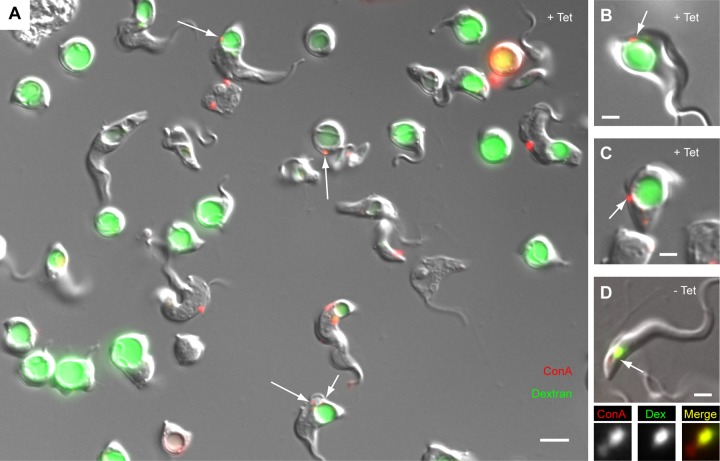
FIG 6.
Loss of overlap between dextran (20-Å diameter) and ConA (80-Å diameter) in unfixed TbMORN1-depleted cells. TbMORN1 RNAi cells from uninduced or induced (14-h point) populations were incubated simultaneously with fluorophore-conjugated ConA (red) and dextran (green) for 30 min at 37°C and analyzed directly. Immunofluorescence images are shown superimposed on DIC images. (A) Representative field of view from a population of TbMORN1-depleted cells. The labeling patterns recapitulate those seen in fixed cells. Dextran was found inside the enlarged FP. The ConA signal could frequently be observed in punctate accumulations (arrows) as seen in fixed cells. Scale bar, 5 μm. (B and C) Two more examples from the induced population. The ConA accumulation (arrow) matches that seen in fixed cells. (D) A cell from the uninduced population. Both labels show a good degree of overlap. The labeled area indicated with the arrow is shown in the insets below. Scale bars: 5 μm (A) and 2 μm (B to D).
ss-YFP assays.
Generation of the signal sequence-yellow fluorescent protein (ss-YFP) plasmid has been described previously (22). The plasmid was linearized by NotI digestion and introduced into TbMORN1 RNAi cells by electroporation according to standard techniques. Clones were selected by limiting dilution in culture medium containing 0.1 μg/ml of puromycin. Intracellular ss-YFP was detected by immunoblotting and immunofluorescence with anti-GFP antibodies following induction of TbMORN1-directed RNAi (14-h point).
RESULTS
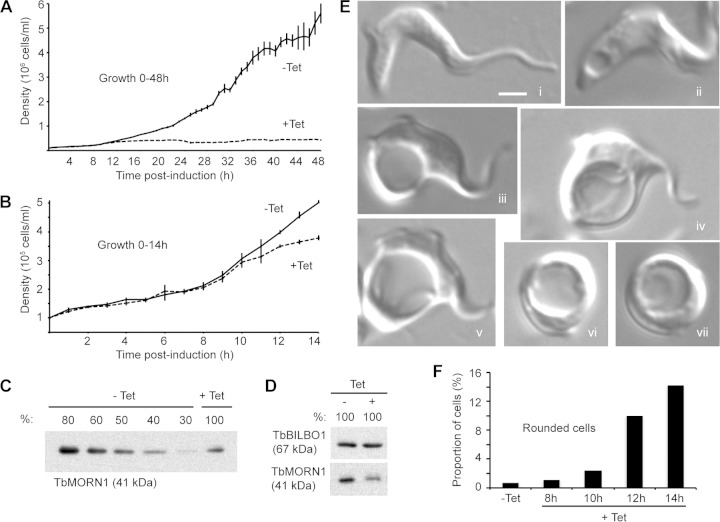
Depletion of TbMORN1 using RNAi has previously been shown to be lethal in BSF T. brucei (18). To determine the critical time window in which to analyze the cells for phenotypic effects, the BSF TbMORN1 RNAi cells were examined at a higher time resolution. As previously reported, induction of TbMORN1-directed RNAi by addition of tetracycline caused a rapid cessation of population growth (Fig. 1A). Cell death in the induced population could be confirmed visually, as the cells ultimately lysed. Analysis of the early stages of the time course (0 to 14 h) revealed a clear difference in the population densities of uninduced (−Tet) and induced (+Tet) populations as early as 12 h after RNAi induction (Fig. 1B). Depletion of TbMORN1 protein was confirmed by immunoblotting of whole-cell lysates from uninduced and induced populations. The immunoblots showed that levels of TbMORN1 protein in the induced population were reduced to ∼50% of preinduction levels in the 12- to 14-h window (Fig. 1C). This matched previously reported depletion kinetics in this cell line (18). To check that the whole-cell lysates used in the quantitative immunoblots were at the same concentration, undiluted samples were also blotted. Immunoblotting with antibodies specific for TbBILBO1 confirmed that the samples were at approximately the same concentration; TbMORN1 levels were again shown to be depleted (Fig. 1D). In all subsequent RNAi experiments, TbMORN1 depletion was confirmed by immunoblotting, and the results were always consistent with those shown here (data not shown).
FIG 1.
Depletion of TbMORN1 is rapidly lethal and is associated with distinct morphological alterations. (A) Growth curves of TbMORN1 RNAi cells in the first 48 h after induction. Uninduced control (−Tet) cells exhibited robust growth. Cells in which TbMORN1-directed RNAi had been induced (+Tet) rapidly cease growth. Cell death (lysis) was confirmed visually. Population density was measured every hour, and each data point was obtained in at least four independent inductions. Data are presented as means ± SEM. (B) Magnification of the data shown in panel A in the 0- to 14-h range. A clear difference in the population densities of uninduced (−Tet) and induced (+Tet) cells was visible 12 h after induction of TbMORN1-directed RNAi. (C) Immunoblotting using anti-TbMORN1 antibodies confirmed depletion of TbMORN1. A representative immunoblot (13-h point) is shown. Diluted whole-cell lysates of uninduced (−Tet) cells were compared with an undiluted (100%) sample from induced (+Tet) cells. TbMORN1 levels were reduced by approximately 50% relative to the uninduced controls. (D) Confirmation that whole-cell lysates from uninduced (−Tet) and induced (+Tet) cells were comparable in protein concentration. Lysates were probed with anti-TbBILBO1 and anti-TbMORN1 antibodies. TbMORN1 levels show depletion; TbBILBO1 levels confirm equivalent concentrations. (E) Examples of morphological alterations observed in live TbMORN1-depleted (+Tet) cells (14-h point). All images are shown at the same magnification. Scale bar, 2 μm. (F) The number of rounded cells increased rapidly following TbMORN1 depletion. Quantification of rounded cells was carried out after fixation using isothermal glutaraldehyde at the indicated time points and is expressed as a fraction of the total population. Data were obtained from three independent experiments (n > 250 cells for each time point).
To determine the morphological consequences of TbMORN1 depletion, live cells were analyzed in the critical 8- to 14-h window. At 8 h postinduction, virtually all cells displayed a normal morphology (Fig. 1Ei). From ∼10 h postinduction, cells with a vacuole at the posterior end of the cell became increasingly common (Fig. 1Eii and iii). The vacuole often achieved a diameter of several micrometers (Fig. 1Eiv and v). Given the position of this vacuole near the posterior end of the cell, it was likely that this was an enlarged FP (a “BigEye” phenotype) (23). The final stage of morphological aberration seemed to be a rounding up of the cells (Fig. 1Evi and vii), after which there was a complete loss of cell integrity, leading to lysis. It should be stressed that as the cells were asynchronous, all these morphological states were comingled even at the later time points (12 h and 14 h). The TbMORN1-depleted population did not progress uniformly through these states. Rather, the morphologically abnormal cells accounted for a larger and larger fraction of the total population. At very late time points (24 h postinduction), virtually all intact cells were of the rounded type (data not shown).
To determine whether TbMORN1 depletion could be correlated with a particular cell cycle stage, cells were fixed with isothermal glutaraldehyde at 8, 10, 12, and 14 h postinduction and stained with DAPI. Quantification of kinetoplast (mitochondrial genome [K]) and nucleus (N) numbers did not reveal a significant block in division at any specific cell cycle stage (see Fig. S1A in the supplemental material). Rounded cells (Fig. 1Evi and vii) could readily be identified in the images of fixed cells, and their numbers were quantified. As expected from the observations of live cells, their numbers rose rapidly in the 8- to 14-h window (Fig. 1F). It should be noted, however, that these quantifications represent an underestimate of the true numbers, due to the fragility of the cells and consequent losses during fixation. The extreme fragility of cells with enlarged FPs has been noted in the past (23).
To verify that the morphological changes in the TbMORN1-depleted population could be correlated with loss of TbMORN1 protein, immunofluorescence assays using anti-TbMORN1 antibodies were carried out. Immunofluorescence assays on paraformaldehyde-fixed whole cells proved difficult, as the morphologically abnormal cells were extremely fragile. Despite immunoblots confirming that TbMORN1 depletion had occurred, in early experiments the TbMORN1 signal from the morphologically abnormal cells was actually higher than from uninduced control cells (see Fig. S1B and C in the supplemental material). Extraction of the cytoplasm prior to fixation (for example, with nonionic detergent) is a common method for improved visualization of cytoskeletal proteins, such as TbMORN1 (13). The removal of cytoplasmic proteins reduces epitope masking and allows better antibody access to the cytoskeletal structures. Therefore, mechanical damage during the prefixation steps of the protocol might have resulted in some cytoplasmic extraction and an anomalously strong TbMORN1 signal relative to controls. To remove this potential source of variability, detergent-extracted cells were analyzed by immunofluorescence assay. In uninduced controls, TbMORN1 labeling described a fishhook-shaped structure close to the point of flagellum entry into the cell (Fig. 2Ai, arrow). Cells that were replicating had two such structures (Fig. 2Aii, arrows).
In TbMORN1-depleted cells (14 h postinduction), the TbMORN1 labeling was now noticeably weaker than in controls, consistent with the immunoblotting data. Although the detergent extraction meant that assigning morphological abnormality was not easy, some of the highest levels of depletion were seen in cells that appeared to exhibit morphological alteration (Fig. 2Aiii, arrow). The morphological effects in Fig. 1 can therefore be correlated with protein levels of TbMORN1. In TbMORN1-depleted cells that were attempting to replicate, both TbMORN1 structures showed decreased signal relative to that of the controls (Fig. 2Aiv and v, arrows). Galleries of additional images are provided in the supplemental material (see Fig. S2).
Immunoblotting had already shown that protein levels of the FPC protein TbBILBO1 were unaffected by TbMORN1 depletion (Fig. 1D). However, in some ways this was unexpected given the close spatial proximity between the FPC and TbMORN1 (13). To investigate this, detergent-extracted cells were labeled with anti-TbBILBO1 antibodies. In uninduced controls, TbBILBO1 labeling was found close to the flagellum base, consistent with the position of the FPC (Fig. 2Bi, arrow). Two structures were observed in replicating cells (Fig. 2Bii, arrows). In TbMORN1-depleted cells (14 h postinduction), there appeared to be no gross change in either the signal intensity or morphology of the anti-TbBILBO1 labeling pattern (Fig. 2Biii, arrow). Duplication and segregation of TbBILBO1 were also apparently unaffected, with many morphologically abnormal cells displaying two structures (Fig. 2Biv and v, arrows).
Although the immunoblots of whole-cell lysates had shown that total protein levels of TbBILBO1 do not change following TbMORN1 depletion (Fig. 1D), this did not exclude the possibility that there was a change in its degree of cytoskeletal association. To test this, biochemical fractionation was carried out. Control and TbMORN1-depleted cells were incubated with nonionic detergent, and after an input (I) sample was taken, the cells were separated by centrifugation into a detergent-soluble supernatant (SN) fraction (containing the cytoplasm) and a detergent-insoluble pellet (P) fraction (containing the cytoskeleton) (Fig. 2C). Equal fractions (percent) of the I, SN, and P were then analyzed by immunoblotting (Fig. 2D). Comparison of TbMORN1 protein levels in control and induced samples showed that depletion had occurred, and in both cases the protein remained wholly associated with the cytoskeletal (P) fraction. As expected, TbBILBO1 levels showed no change between the control and induced cells. There was also no obvious change in the cytoskeletal association of the protein, with the amounts of TbBILBO1 in the cytoskeletal (P) fraction being the same in both populations. Immunoblotting with antibodies specific for the endoplasmic reticulum protein TbBiP was used as a positive control for detergent extraction of the cytoplasm, with the protein found almost entirely in the SN fractions.
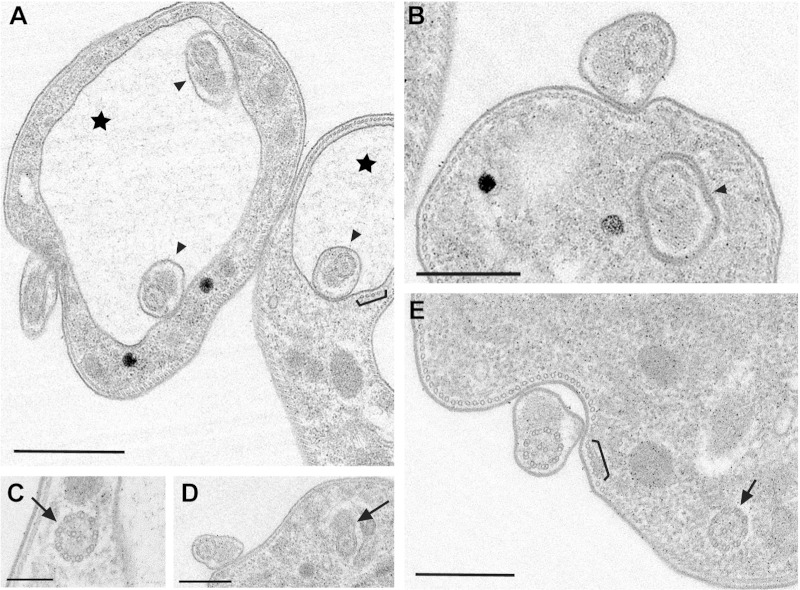
To gain ultrastructural insight into the morphological changes in the TbMORN1-depleted cells, transmission electron microscopy (TEM) was carried out. TEM micrographs supported the assignment of the large vacuole in the TbMORN1-depleted cells as an enlarged FP, due to the presence of flagellum cross sections within it (Fig. 3A, stars). Further data, including longitudinal sections showing the flagellum exiting an enlarged FP, are provided in the supplemental material (see Fig. S3 and S4B and C). Multiple (≥2) axoneme cross sections could frequently be observed within a single FP. These flagellum cross sections also revealed the presence of the PFR inside the FP (Fig. 3A and B, arrowheads, and data not shown). Axoneme cross sections (with or without associated PFR) were also frequently found within the cytoplasm of TbMORN1-depleted cells (Fig. 3C to E, arrows). There did not appear to be any gross abnormality in the microtubule quartet in TbMORN1-depleted cells (Fig. 3A and E, brackets). It was not possible to determine definitively at this level if there was any alteration in the FPC, but gross abnormality seemed unlikely (see Fig. S3; other data are not shown). This was consistent with the observations made using immunofluorescence.
FIG 3.
Ultrastructural phenotype of TbMORN1-depleted cells. Shown are common morphological characteristics of TbMORN1-depleted cells (14-h point). Images were taken from 60-nm-thick resin sections contrasted with uranyl acetate and lead citrate. (A) Enlarged vacuoles (stars) and multiple axonemes with PFRs inside the pocket (arrowheads). The microtubule quartet seems unaffected (bracket; also in panel E). (B) Axoneme with PFR inside the flagellar neck region (arrowhead). (C to E) Intracellular axonemes with and without PFR (arrows). Scale bars: 1 μm (A and D), 500 nm (B and E), and 200 nm (C).
The main morphological changes were quantified. Wild-type (427), control (−Tet), and TbMORN1-depleted cells (+Tet) were scored for the presence of intracellular flagella, PFR(s) inside the FP, an enlarged FP, and multiple axoneme cross sections inside the FP. These traits were all overwhelmingly present only in TbMORN1-depleted cells (Table 1).
TABLE 1.
Quantification of morphological phenotypes seen by EM
| Phenotypic feature | Cell line (no. of profiles) |
||
|---|---|---|---|
| 427 (52) | TbMORN1 RNAi |
||
| −Tet (48) | +Tet (58) | ||
| Cells with intracellular flagella | 0 | 0 | 12 |
| PFR inside FP | 0 | 0 | 34 |
| Enlarged FP | 1 | 1 | 24 |
| Multiple axonemes in one FP | 0 | 0 | 19 |
Gross enlargement of the FP (a “BigEye” phenotype) is not an unprecedented observation. It is most commonly associated with defects in endocytosis and has been documented following depletion of clathrin (23). To test if depletion of TbMORN1 resulted in an endocytosis defect, dextran uptake experiments were carried out. Dextran is a polysaccharide that is commonly used as a marker for fluid phase uptake. There are well-established protocols for assaying endocytosis in trypanosomes using temperature blocks (6, 24, 25). At 0°C, material can enter the FP but endocytosis is blocked. Shifting the cells to 37°C allows uptake to resume. The uptake protocol can be summarized as follows. Control or TbMORN1-depleted cells (14 h postinduction) were cooled on ice to stop endocytic uptake. They were then incubated with fluorophore-conjugated dextran (molecular mass, ∼10 kDa; hydrodynamic radius, ∼20 Å) on ice. Samples of each population were fixed at this point (t = 0 min). The remaining cell populations were then shifted to 37°C and incubated for a further 30 min to allow endocytosis, and then fixed (t = 30 min) (Fig. 4A).
In control cells, at 0 min the dextran accumulated in a small volume (Fig. 4Bi, arrow) adjacent to the kinetoplast (Fig. 4Bi, arrowhead). This corresponds to the location of the FP. At 30 min, a large amount of dextran had been internalized (Fig. 4Bii, arrow). The bulk of this internalized material was now considerably anterior to the kinetoplast (Fig. 4Bii, arrowhead), consistent with the location of the endosomal-lysosomal system. In TbMORN1-depleted cells, at 0 min, the dextran appeared to accumulate within the enlarged FP (Fig. 4Ci, arrow). At a 30 min, the TbMORN1-depleted cells showed no change in dextran distribution, with the signal still apparently coming from the enlarged FP (Fig. 4Cii, arrow).
As an additional test of endocytic activity, uptake of fluorophore-conjugated concanavalin A (ConA) was assayed (26). ConA is a lectin (∼80 Å across) that can bind to glycoproteins in and adjacent to the FP (26, 27). The glycoprotein-ConA complexes are then taken up by endocytosis. The protocol for ConA uptake was essentially the same as that followed for dextran uptake (Fig. 4A). In control cells, ConA behaved in a similar way to dextran: at 0 min, it was found in a small volume (Fig. 4Di, arrow) adjacent to the kinetoplast (Fig. 4Di, arrowhead), and at 30 min, it was localized predominantly to the region occupied by the endosomal-lysosomal system (Fig. 4Dii, arrow).
Surprisingly, in TbMORN1-depleted cells the behavior of ConA did not recapitulate what had previously been observed with dextran. At 0 min, it was found in a small, intensely bright spot (Fig. 4Ei, arrow). This spot was some distance from the kinetoplast, in contrast to dextran (compare arrowheads in Fig. 4Ci and Fig. 4Ei). At 30 min, there was no change in the localization of ConA, but the signal strength further increased (Fig. 4Eii).
This difference in the behavior of dextran and ConA was unexpected and has not been observed in morphological phenotypes of this type. For example, in clathrin-depleted BigEye cells, ConA accumulates within the enlarged FP in the same way as dextran (23). To check that these different behaviors were real and not an experimental artifact, levels of uptake of both markers were analyzed simultaneously. In control cells, after a 30-min incubation with ConA and dextran at 37°C, there was robust accumulation of both markers in the region of the cell corresponding to the endosomal-lysosomal system. Good overlap was observed between both signals (Fig. 5A, arrow).
In contrast, TbMORN1-depleted cells recapitulated the behavior seen in the single-marker assays. There was no overlap between the two signals, and while dextran accumulated in a large volume, ConA was found next to it in an intensely bright spot (Fig. 5B to D, arrows).
Given the early difficulties encountered during the immunofluorescence assays (see Fig. S1B and C in the supplemental material), a further concern was that the dextran accumulation might have been a fixation artifact caused by membrane damage. To exclude this possibility, the double-uptake protocol was repeated but instead of fixing the cells after the 30-min incubation, live cells were imaged directly (Fig. 6A). More than half of the cells in the TbMORN1-depleted population now had a visibly enlarged FP, compared to less than 4% in controls (n > 400). Quantification of the data indicated that 92% of cells with an enlarged FP showed dextran accumulation within that pocket (n > 400). Imaging of ConA in the live TbMORN1-depleted cells was difficult due to cell movement and the small size of the signal. In a number of examples, however, the ConA punctae were in the focal plane and visible at the same point as previously observed in fixed samples (Fig. 6A to C, arrows). These cells accounted for 26% of all cells with enlarged FP (n > 400). Control cells showed robust uptake and overlap of both markers (Fig. 6D, arrow).
What might account for this difference in the behavior of the two endocytic markers? One possibility was that the discrepancy between the ConA and dextran labeling patterns was due to impaired access of ConA to the FP owing to its greater size. To test this, TbMORN1-depleted cells were incubated with 5 nm gold-conjugated BSA. The BSA-gold, like dextran, traffics in the fluid phase but the particle size (>100 Å) is larger than ConA.
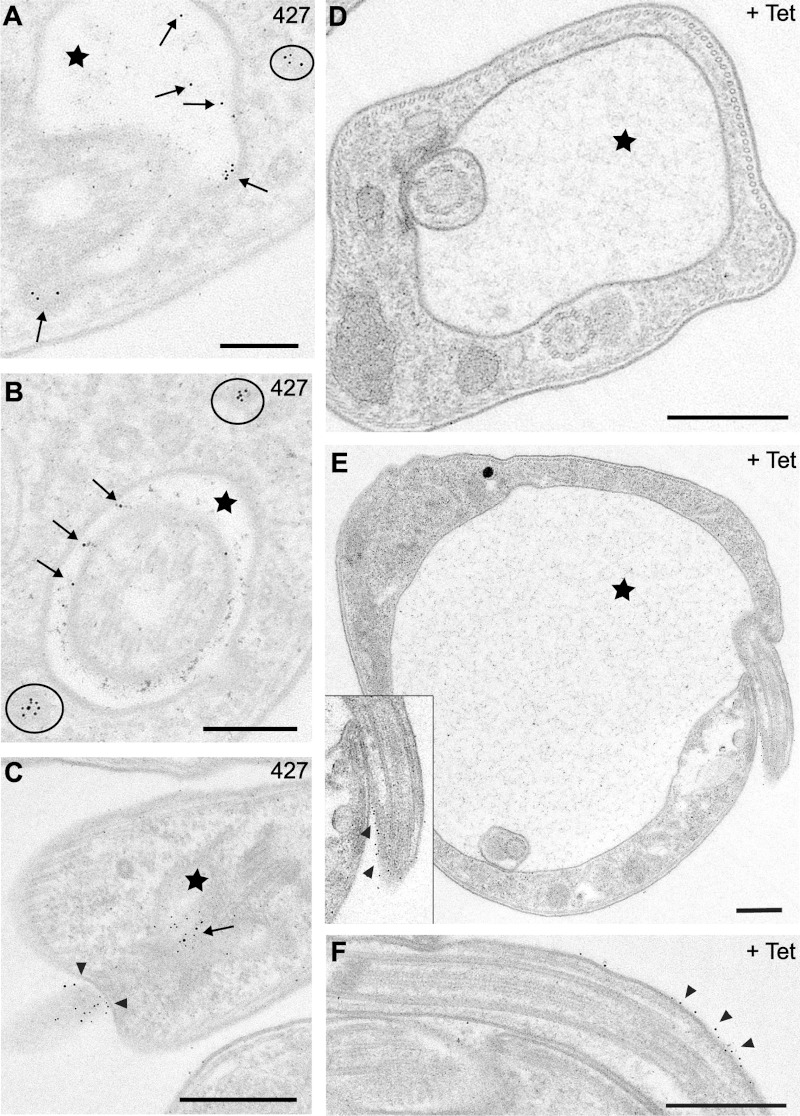
In wild-type (427) cells, BSA-gold was primarily found in three locations: (i) within the FP (Fig. 7A to C, arrows); (ii) outside the FP, near the point of flagellum entry (Fig. 7C, arrowheads); (iii) inside intracellular vesicles (Fig. 7A and B, circles). The same distributions were observed in uninduced control cells (data not shown). In contrast, very few sections from TbMORN1-depleted cells showed BSA-gold within the enlarged FP (Fig. 7D and E, stars; see also Fig. S4A and C in the supplemental material). BSA-gold could, however, be observed on the flagellum and frequently close to the point of flagellum entry into the FP (Fig. 7E and F, arrowheads; see also Fig. S3A' and D in the supplemental material).
FIG 7.
Reduced uptake of BSA-gold (>100-Å diameter) in TbMORN1-depleted cells. Images were taken from 60-nm-thick resin sections contrasted with uranyl acetate and lead citrate of trypanosome cells after incubation with BSA-gold (5 nm; 14-h point). Arrows indicate particles of BSA-gold inside the FP, circles show vesicles containing gold particles, and arrowheads point at gold particles at the FP entrance and flagellum; FPs are labeled with stars. (A to C) Efficient BSA-gold uptake and accumulation within the FP was observed in 427 wild-type cells. (D and E) Enlarged FPs of +Tet cells rarely contained any BSA-gold. (E and F) Extracellular gold particles (arrowheads) were often found just outside the pocket at the flagellum. The relevant area of panel E is enlarged in the inset. Scale bars: 200 nm (A and B) and 500 nm (C to F).
Quantification of the distribution of the gold grains supported these observations. In TbMORN1-depleted cells, only 7 FPs out of a total of 53 (13%) showed one or more BSA-gold grains (Table 2).
TABLE 2.
Quantification of distribution of BSA-5nm gold grains
| Trafficking feature | Cell line (no. of profiles) |
||
|---|---|---|---|
| 427 (52) | TbMORN1 RNAi |
||
| −Tet (48) | +Tet (58) | ||
| Access to FP | |||
| FP profiles | 42 | 43 | 53 |
| FPs containing gold grains | 31 | 21 | 7 |
| Total no. of gold grains | 160 | 106 | 59 |
| No. of grains at pocket entrance | 44 | 49 | 45 |
| Endocytosis | |||
| Total no. of vesicles with endocytosed grains | 46 | 43 | 16 |
| Total no. of endocytosed grains | 150 | 124 | 48 |
The total number of vesicles with endocytosed grains and the total number of endocytosed grains were both reduced in TbMORN1-depleted cells relative to controls (Table 2). In a number of cross sections, the BSA-gold particles could be observed within the FPN (see Fig. S3F and S4B and C).
The enlarged FP observed following depletion of clathrin or other endocytic mediators is thought to arise from compromised endocytosis and concomitant unimpaired secretion: the secretory pathway now delivers membrane to the FP faster than the impaired endocytic pathway can remove it (23). In support of this, export of variant surface glycoprotein (VSG) has been shown to be unaffected following depletion of either clathrin or actin (23, 28). The behavior of signal sequence-YFP (ss-YFP) was monitored in the TbMORN1 RNAi background. ss-YFP is a reporter carrying the signal sequence from the procyclin surface coat protein fused to YFP (22). In wild-type cells, ss-YFP is released into the medium (29). Immunoblotting and immunofluorescence analyses indicated that there was no gross increase in intracellular ss-YFP following TbMORN1 depletion, suggesting that there was no major effect on its trafficking (see Fig. S5A and B).
DISCUSSION
TbMORN1 depletion is rapidly lethal in BSF T. brucei and is accompanied by an enlargement of the FP (a BigEye phenotype) (Fig. 1). Other morphological effects include the presence of the PFR inside the enlarged FP, and the appearance of intracellular axonemes (sometimes with an associated PFR) (Fig. 3; see also Fig. S3 in the supplemental material). These additional morphological effects can be observed in other, but not all, documented BigEye phenotypes (30–35). It is not clear whether these are direct effects following from specific protein loss or secondary effects on biogenesis precipitated by the presence of a massively enlarged FP.
In replicating cells, TbMORN1 depletion results in a loss from both the old and new structures (Fig. 2; see also Fig. S2). Most cytoskeletal structures in T. brucei are replicated by de novo assembly, for example, the FAZ (36, 37). However, the observation that both old and new TbMORN1 complexes are affected suggests that during replication, the old structure makes a physical contribution to the composition of the new structure. This may explain why the progression of the TbMORN1 depletion phenotype is so rapid and penetrant.
The FPC protein TbBILBO1 was found to be unaffected upon TbMORN1 depletion, by both immunofluorescence and biochemical fractionation (Fig. 2). EM images also support the idea that the FPC is intact (see Fig. S3). This lack of effect on TbBILBO1 is also consistent with the fact that the published RNAi phenotypes of TbMORN1 and TbBILBO1 in procyclic T. brucei are very different (9, 18).
BigEye phenotypes are usually associated with a defect in endocytosis, exemplified by the clathrin RNAi phenotype (23). Consistent with this, dextran accumulated inside the enlarged FP of TbMORN1-depleted cells (Fig. 4 to 6). Dextran is a coiled polysaccharide with a hydrodynamic radius of ∼20 Å that traffics in the fluid phase (38).
What was unexpected was the behavior of ConA. Accumulation of ConA inside the enlarged FP has previously been documented in other BigEye phenotypes (23, 34, 39). However, here, instead of accumulating inside the enlarged FP, ConA accumulated outside the FP in an intense spot or patch (Fig. 4). This resulted in a loss of overlap between the dextran and ConA in cells incubated with both reporters (Fig. 5 and 6). ConA is a lectin with a diameter of ∼80 Å (40). It should be noted that the lectin tetramer exists in equilibrium with its dimeric form, which may affect its hydrodynamic radius (41). The size calculations for both the dextran and ConA assume that the contribution from the conjugated fluorophore is negligible.
ConA has been shown to bind a number of T. brucei glycoproteins in lectin blots, including the variant surface glycoprotein (VSG) that comprises the bulk of surface protein in the organism (42, 43). Accordingly, ConA labels the entire surface of fixed T. brucei cells (44). However, surface labeling by ConA is not seen in experiments with live cells (26, 27). Fixation presumably makes the glycan groups on VSG more accessible to ConA. This is supported by the observation that antibodies directed against the C-terminal domain of VSG (which is close to the outer leaflet of the plasma membrane) label the entire surface of fixed cells but do not label live cells (45). Trypsinization of live cells removes VSG and results in ConA labeling of the entire surface, probably due to recognition of other glycoproteins that were previously shielded (26, 27). In live cells, the only ConA labeling is observed in and adjacent to the FP (26, 27).
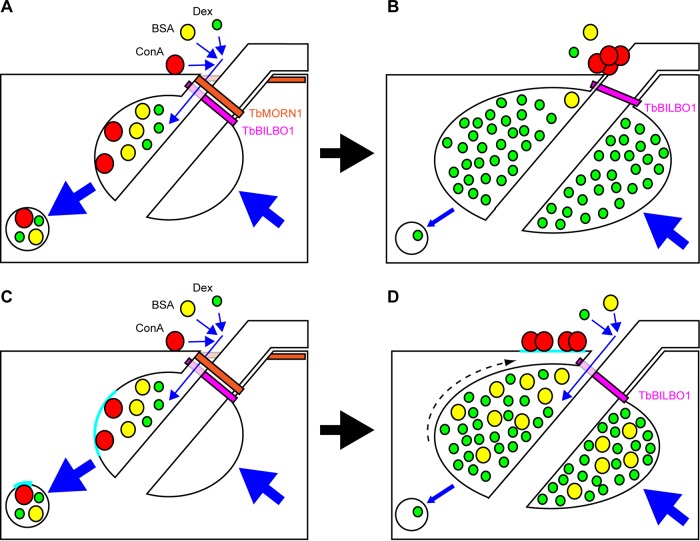
These observations, and those described in Results, lead to two possible interpretations that are presented as working models to guide future enquiry (Fig. 8). These interpretations are referred to as the size exclusion model (Fig. 8A and B) and the membrane subdomain model (Fig. 8C and D). In the size exclusion model, TbMORN1 is responsible for holding open the FPN to permit the entry of macromolecular traffic. Dextran enters in the fluid phase, while ConA binds to VSG at sites of high positive curvature at the top of the FPN and to other glycoproteins at and within the FP (Fig. 8A). Loss of TbMORN1 prevents the larger ConA molecules from accessing the FP, and they accumulate outside it. The smaller dextran molecules can still enter and accumulate inside the enlarged FP owing to an additional endocytosis defect (Fig. 8B).
FIG 8.
Summary of observations and two possible interpretations. Shown are schematic representations of the posterior end of a trypanosome cell in longitudinal cross-section. The FPC is depicted in magenta (TbBILBO1). The TbMORN1 macromolecular complex is shown as an orange fishhook above the FPC. Dextran, BSA, and ConA are depicted, respectively, as green, yellow, red circles. Blue arrows indicate movement. Endocytosis and secretion are represented by large blue arrows. (A) Model 1. In uninduced cells, dextran, BSA, and ConA enter the FP (small blue arrows) and are taken up by endocytosis and trafficked to the endosomal-lysosomal system in transport vesicles. The secretory pathway returns membrane to the FP. (B) Following TbMORN1-directed RNAi, the FPC is intact but TbMORN1 is largely absent. BSA (fluid phase) and ConA (membrane bound) can no longer efficiently access the FP, and ConA accumulates close to the point of flagellum entry. Dextran can still access the FP and accumulates inside it. The rate of endocytosis is reduced (small blue arrow), while secretion is unaffected. (C) Model 2. The situation is the same as that depicted in model 1, except for the behavior of ConA. Here, ConA binds exclusively to glycoproteins present either within or adjacent to the FP in a defined patch (cyan, here shown inside the FP). (D) Following depletion of TbMORN1, the maintenance of subdomains is lost and the ConA-binding glycoprotein patch is displaced outside the FP (dashed arrow), where it accumulates ConA. The rate of endocytosis is reduced as in model 1, leading to an enlargement of the FP and an accumulation of both dextran and BSA within it.
The initial situation in the membrane subdomain model is similar to the size exclusion model, except that the glycoproteins bound by ConA within the FP are present within a defined patch of membrane. TbMORN1 is responsible for corralling this patch within the FP (Fig. 8C). Loss of TbMORN1, and the enlargement of the FP due to the endocytosis defect, results in a displacement of this patch onto the cell surface, where it continues to be bound by ConA (Fig. 8D).
The experiments with BSA-gold act as a first test of these models. Like ConA, it is a protein, but like dextran, it traffics in the fluid phase. BSA is a barrel-shaped molecule with the dimensions 140 by 40 Å (46). It is not clear how many BSA molecules are attached to each 5-nm/50-Å gold grain (company technical support, BBI Solutions, personal communication). Regardless of the number and orientation of the BSA molecules attached to the gold grain, the total particle size will be greater than that of ConA, and generally above 100 Å. Robust uptake of BSA-gold (5 nm and larger) by T. brucei has been demonstrated in the past (20, 24). The size exclusion model predicts that it should be harder for the BSA-gold to enter following TbMORN1 depletion (Fig. 8B). The membrane subdomain model predicts that BSA should accumulate inside the enlarged FP, in the same way as dextran (Fig. 8D).
In TbMORN1-depleted cells, some BSA-gold was still observed within the enlarged FP, but this may simply reflect the greater sensitivity of the assay compared to that of the ConA experiments. An important caveat is that the enlargement of the FP means that the number of sections through it will increase. As the experiments did not involve serial sections, it means that it is therefore not possible to formally conclude anything from the totals of gold grains. There are overall fewer grains in randomly chosen sections from TbMORN1-depleted cells, however, which offers some support for the conclusion that less BSA-gold enters the enlarged FP. If one assumes that the gold grains within the FP are evenly distributed, then analysis of single sections provides a meaningful comparison between conditions. In this light, it is significant that in TbMORN1-depleted cells there are far more empty FPs than in controls, again suggesting that less BSA-gold accesses the enlarged FP. It is hard to see how this effect would be produced by an accumulation of BSA-gold, which is the prediction made by the membrane subdomain model (Fig. 8D).
Therefore, although the results of the BSA-gold experiments do not clinch the argument, they do lead us to favor the size exclusion model for now. It is also not clear how the patch described for the membrane subdomain model retains its cohesion once outside the FP, or why it seems to remain in the vicinity of the FP. And unless all the ConA-binding glycoproteins are present in the patch, it is not clear why there is no ConA labeling within the enlarged FP of TbMORN1-depleted cells. It should be noted that the existence of a patch of ricin-binding glycoproteins within the FP has been documented in the past (27).
The two models prompt a number of testable predictions, some of which are outlined below. First, the transferrin receptor has been shown to localize exclusively within the FP (47). Does labeled transferrin then accumulate inside the enlarged FP of TbMORN1-depleted cells (as predicted by the size exclusion model) or on a patch outside (as predicted by the membrane subdomain model, and assuming that the transferrin receptor is present in the patch)? Second, does anti-VSG IgM still enter the enlarged FP of TbMORN1-depleted cells? IgM has a mass of roughly 970 kDa and a hydrodynamic radius of around 130 Å but is still efficiently internalized (48–50). Third, is the neck channel still open following TbMORN1 depletion, or has it collapsed? The neck channel is a recently described corridor through the FPN that connects the FP lumen to the extracellular space (20). The position of this channel alongside the specialized microtubule quartet puts it in close proximity to the position of TbMORN1. The possibility of the neck channel being involved in a size-exclusion mechanism has already been noted (20). Fourth, although it would not constitute a direct test of the two models, it would be very interesting to assay the uptake of low-density lipoprotein. Low-density lipoprotein particles have an approximately 3-MDa mass and are around 220 Å across (51). The low-density lipoprotein receptor in T. brucei is present within and just outside the FP, similar to the pattern of ConA binding (52).
Finally, it should be stressed that the two models are not entirely mutually exclusive: it is possible to envisage a situation in which TbMORN1 both facilitates macromolecule entry into the FP and works to establish and maintain membrane subdomains. For instance, it has recently been shown that a phosphatidylinositol 4-phosphate 5-kinase, TbPIPKA, is present in the FPN (53). This kinase manufactures the phosphatidylinositol 4,5-bisphosphate that is essential for endocytosis. If TbPIPKA depended on TbMORN1 for its correct localization, then this would additionally explain why there is an apparent endocytosis defect. These and other predictions raised by the models should provide fascinating avenues for future enquiry.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Graham Warren, in whose laboratory this work was conducted. Financial support was primarily from Austrian Science Fund (FWF) grant P22265-B12 to Graham Warren; support subsequently came from FWF grant P27016-B21 to Brooke Morriswood.
We thank Catarina Gadelha (University of Nottingham) for advice on fixation protocols and Susanne Kramer, Ines Subota, and Tim Krüger (all from the University of Würzburg) and Mélanie Bonhivers (University of Bordeaux) for critical reading of the manuscript. We also thank colleagues at the University of Oxford and the University of Nottingham for valuable discussions. Ivanna Fedorenko provided technical assistance with the EM.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00094-15.
REFERENCES
- 1.Gull K. 1999. The cytoskeleton of trypanosomatid parasites. Annu Rev Microbiol 53:629–655. doi: 10.1146/annurev.micro.53.1.629. [DOI] [PubMed] [Google Scholar]
- 2.Wheeler RJ, Gluenz E, Gull K. 2013. The limits on trypanosomatid morphological diversity. PLoS One 8:e79581. doi: 10.1371/journal.pone.0079581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson DR, Sherwin T, Ploubidou A, Byard EH, Gull K. 1995. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J Cell Biol 128:1163–1172. doi: 10.1083/jcb.128.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacomble S, Vaughan S, Gadelha C, Morphew MK, Shaw MK, McIntosh JR, Gull K. 2009. Three-dimensional cellular architecture of the flagellar pocket and associated cytoskeleton in trypanosomes revealed by electron microscope tomography. J Cell Sci 122:1081–1090. doi: 10.1242/jcs.045740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grünfelder CG, Engstler M, Weise F, Schwarz H, Stierhof YD, Morgan GW, Field MC, Overath P. 2003. Endocytosis of a glycosylphosphatidylinositol-anchored protein via clathrin-coated vesicles, sorting by default in endosomes, and exocytosis via RAB11-positive carriers. Mol Biol Cell 14:2029–2040. doi: 10.1091/mbc.E02-10-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engstler M, Thilo L, Weise F, Grunfelder CG, Schwarz H, Boshart M, Overath P. 2004. Kinetics of endocytosis and recycling of the GPI-anchored variant surface glycoprotein in Trypanosoma brucei. J Cell Sci 117:1105–1115. doi: 10.1242/jcs.00938. [DOI] [PubMed] [Google Scholar]
- 7.Heddergott N, Kruger T, Babu SB, Wei A, Stellamanns E, Uppaluri S, Pfohl T, Stark H, Engstler M. 2012. Trypanosome motion represents an adaptation to the crowded environment of the vertebrate bloodstream. PLoS Pathog 8:e1003023. doi: 10.1371/journal.ppat.1003023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastin P, Pullen TJ, Sherwin T, Gull K. 1999. Protein transport and flagellum assembly dynamics revealed by analysis of the paralysed trypanosome mutant snl-1. J Cell Sci 112(Part 21):3769–3777. [DOI] [PubMed] [Google Scholar]
- 9.Bonhivers M, Nowacki S, Landrein N, Robinson DR. 2008. Biogenesis of the trypanosome endo-exocytotic organelle is cytoskeleton mediated. PLoS Biol 6:e105. doi: 10.1371/journal.pbio.0060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vidilaseris K, Morriswood B, Kontaxis G, Dong G. 2014. Structure of the TbBILBO1 protein N-terminal domain from Trypanosoma brucei reveals an essential requirement for a conserved surface patch. J Biol Chem 289:3724–3735. doi: 10.1074/jbc.M113.529032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vidilaseris K, Shimanovskaya E, Esson HJ, Morriswood B, Dong G. 2014. Assembly mechanism of Trypanosoma brucei BILBO1, a multidomain cytoskeletal protein. J Biol Chem 289:23870–23881. doi: 10.1074/jbc.M114.554659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Florimond C, Sahin A, Vidilaseris K, Dong G, Landrein N, Dacheux D, Albisetti A, Byard EH, Bonhivers M, Robinson DR. 2015. BILBO1 is a scaffold protein of the flagellar pocket collar in the pathogen Trypanosoma brucei. PLoS Pathog 11:e1004654. doi: 10.1371/journal.ppat.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esson HJ, Morriswood B, Yavuz S, Vidilaseris K, Dong G, Warren G. 2012. Morphology of the trypanosome bilobe, a novel cytoskeletal structure. Eukaryot Cell 11:761–772. doi: 10.1128/EC.05287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Q, Gheiratmand L, Chen Y, Lim TK, Zhang J, Li S, Xia N, Liu B, Lin Q, He CY. 2010. A comparative proteomic analysis reveals a new bi-lobe protein required for bi-lobe duplication and cell division in Trypanosoma brucei. PLoS One 5:e9660. doi: 10.1371/journal.pone.0009660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andre J, Harrison S, Towers K, Qi X, Vaughan S, McKean PG, Ginger ML. 2013. The tubulin cofactor C family member TBCCD1 orchestrates cytoskeletal filament formation. J Cell Sci 126:5350–5356. doi: 10.1242/jcs.136515. [DOI] [PubMed] [Google Scholar]
- 16.Morriswood B, Havlicek K, Demmel L, Yavuz S, Sealey-Cardona M, Vidilaseris K, Anrather D, Kostan J, Djinovic-Carugo K, Roux KJ, Warren G. 2013. Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryot Cell 12:356–367. doi: 10.1128/EC.00326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He CY, Pypaert M, Warren G. 2005. Golgi duplication in Trypanosoma brucei requires Centrin2. Science 310:1196–1198. doi: 10.1126/science.1119969. [DOI] [PubMed] [Google Scholar]
- 18.Morriswood B, He CY, Sealey-Cardona M, Yelinek J, Pypaert M, Warren G. 2009. The bilobe structure of Trypanosoma brucei contains a MORN-repeat protein. Mol Biochem Parasitol 167:95–103. doi: 10.1016/j.molbiopara.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Bangs JD, Uyetake L, Brickman MJ, Balber AE, Boothroyd JC. 1993. Molecular cloning and cellular localization of a BiP homologue in Trypanosoma brucei. Divergent ER retention signals in a lower eukaryote. J Cell Sci 105(Part 4):1101–1113. [DOI] [PubMed] [Google Scholar]
- 20.Gadelha C, Rothery S, Morphew M, McIntosh JR, Severs NJ, Gull K. 2009. Membrane domains and flagellar pocket boundaries are influenced by the cytoskeleton in African trypanosomes. Proc Natl Acad Sci U S A 106:17425–17430. doi: 10.1073/pnas.0909289106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson D, Beattie P, Sherwin T, Gull K. 1991. Microtubules, tubulin, and microtubule-associated proteins of trypanosomes. Methods Enzymol 196:285–299. doi: 10.1016/0076-6879(91)96027-O. [DOI] [PubMed] [Google Scholar]
- 22.Ho HH, He CY, de Graffenried CL, Murrells LJ, Warren G. 2006. Ordered assembly of the duplicating Golgi in Trypanosoma brucei. Proc Natl Acad Sci U S A 103:7676–7681. doi: 10.1073/pnas.0602595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen CL, Goulding D, Field MC. 2003. Clathrin-mediated endocytosis is essential in Trypanosoma brucei. EMBO J 22:4991–5002. doi: 10.1093/emboj/cdg481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brickman MJ, Cook JM, Balber AE. 1995. Low temperature reversibly inhibits transport from tubular endosomes to a perinuclear, acidic compartment in African trypanosomes. J Cell Sci 108(Part 11):3611–3621. [DOI] [PubMed] [Google Scholar]
- 25.Hall BS, Pal A, Goulding D, Field MC. 2004. Rab4 is an essential regulator of lysosomal trafficking in trypanosomes. J Biol Chem 279:45047–45056. doi: 10.1074/jbc.M407271200. [DOI] [PubMed] [Google Scholar]
- 26.Balber AE, Frommel TO. 1988. Trypanosoma brucei gambiense and T. b. rhodesiense: concanavalin A binding to the membrane and flagellar pocket of bloodstream and procyclic forms. J Protozool 35:214–219. doi: 10.1111/j.1550-7408.1988.tb04326.x. [DOI] [PubMed] [Google Scholar]
- 27.Brickman MJ, Balber AE. 1990. Trypanosoma brucei rhodesiense bloodstream forms: surface ricin-binding glycoproteins are localized exclusively in the flagellar pocket and the flagellar adhesion zone. J Protozool 37:219–224. doi: 10.1111/j.1550-7408.1990.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 28.Nolan DP, Garcia-Salcedo JA. 2008. Loss of actin does not affect export of newly synthesized proteins to the surface of Trypanosoma brucei. Mol Biochem Parasitol 157:233–235. doi: 10.1016/j.molbiopara.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Sealey-Cardona M, Schmidt K, Demmel L, Hirschmugl T, Gesell T, Dong G, Warren G. 2014. Sec16 determines the size and functioning of the Golgi in the protist parasite, Trypanosoma brucei. Traffic 15:613–629. doi: 10.1111/tra.12170. [DOI] [PubMed] [Google Scholar]
- 30.García-Salcedo JA, Perez-Morga D, Gijon P, Dilbeck V, Pays E, Nolan DP. 2004. A differential role for actin during the life cycle of Trypanosoma brucei. EMBO J 23:780–789. doi: 10.1038/sj.emboj.7600094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall B, Allen CL, Goulding D, Field MC. 2004. Both of the Rab5 subfamily small GTPases of Trypanosoma brucei are essential and required for endocytosis. Mol Biochem Parasitol 138:67–77. doi: 10.1016/j.molbiopara.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Morgan GW, Denny PW, Vaughan S, Goulding D, Jeffries TR, Smith DF, Gull K, Field MC. 2005. An evolutionarily conserved coiled-coil protein implicated in polycystic kidney disease is involved in basal body duplication and flagellar biogenesis in Trypanosoma brucei. Mol Cell Biol 25:3774–3783. doi: 10.1128/MCB.25.9.3774-3783.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheader K, Vaughan S, Minchin J, Hughes K, Gull K, Rudenko G. 2005. Variant surface glycoprotein RNA interference triggers a precytokinesis cell cycle arrest in African trypanosomes. Proc Natl Acad Sci U S A 102:8716–8721. doi: 10.1073/pnas.0501886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price HP, Stark M, Smith DF. 2007. Trypanosoma brucei ARF1 plays a central role in endocytosis and golgi-lysosome trafficking. Mol Biol Cell 18:864–873. doi: 10.1091/mbc.E06-08-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barquilla A, Crespo JL, Navarro M. 2008. Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proc Natl Acad Sci U S A 105:14579–14584. doi: 10.1073/pnas.0802668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunter JD, Varga V, Dean S, Gull K. 2015. A dynamic coordination of flagellum and cytoplasmic cytoskeleton assembly specifies cell morphogenesis in trypanosomes. J Cell Sci 128:1580–1594. doi: 10.1242/jcs.166447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Q, Hu H, He CY, Li Z. 2015. Assembly and maintenance of the flagellum attachment zone filament in Trypanosoma brucei. J Cell Sci 128:2361–2372. doi: 10.1242/jcs.168377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armstrong JK, Wenby RB, Meiselman HJ, Fisher TC. 2004. The hydrodynamic radii of macromolecules and their effect on red blood cell aggregation. Biophys J 87:4259–4270. doi: 10.1529/biophysj.104.047746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manna PT, Gadelha C, Puttick AE, Field MC. 2015. E/ANTH domain proteins participate in AP2-independent clathrin-mediated endocytosis. J Cell Sci 128:2130–2142. doi: 10.1242/jcs.167726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hardman KD, Ainsworth CF. 1972. Structure of concanavalin A at 2.4-A resolution. Biochemistry 11:4910–4919. doi: 10.1021/bi00776a006. [DOI] [PubMed] [Google Scholar]
- 41.Senear DF, Teller DC. 1981. Thermodynamics of concanavalin A dimer-tetramer self-association: sedimentation equilibrium studies. Biochemistry 20:3076–3083. doi: 10.1021/bi00514a014. [DOI] [PubMed] [Google Scholar]
- 42.Frommel TO, Balber AE. 1987. Trypanosoma brucei brucei, T brucei gambiense, and T brucei rhodesiense: common glycoproteins and glycoprotein oligosaccharide heterogeneity identified by lectin affinity blotting and endoglycosidase H treatment. Exp Parasitol 63:32–41. [DOI] [PubMed] [Google Scholar]
- 43.Frommel TO, Kohler MF, Balber AE. 1987. Trypanosoma brucei brucei and Trypanosoma brucei gambiense: stage specific differences in wheat germ agglutinin binding and in endoglycosidase H sensitivity of glycoprotein oligosaccharides. Exp Parasitol 64:104–110. doi: 10.1016/0014-4894(87)90014-2. [DOI] [PubMed] [Google Scholar]
- 44.Gadelha C, Zhang W, Chamberlain JW, Chait BT, Wickstead B, Field MC. 2015. Architecture of a host-parasite interface: complex targeting mechanisms revealed through proteomics. Mol Cell Proteomics 14:1911–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwede A, Jones N, Engstler M, Carrington M. 2011. The VSG C-terminal domain is inaccessible to antibodies on live trypanosomes. Mol Biochem Parasitol 175:201–204. doi: 10.1016/j.molbiopara.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Majorek KA, Porebski PJ, Dayal A, Zimmerman MD, Jablonska K, Stewart AJ, Chruszcz M, Minor W. 2012. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol Immunol 52:174–182. doi: 10.1016/j.molimm.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salmon D, Geuskens M, Hanocq F, Hanocq-Quertier J, Nolan D, Ruben L, Pays E. 1994. A novel heterodimeric transferrin receptor encoded by a pair of VSG expression site-associated genes in T. brucei. Cell 78:75–86. doi: 10.1016/0092-8674(94)90574-6. [DOI] [PubMed] [Google Scholar]
- 48.Czajkowsky DM, Shao Z. 2009. The human IgM pentamer is a mushroom-shaped molecule with a flexural bias. Proc Natl Acad Sci U S A 106:14960–14965. doi: 10.1073/pnas.0903805106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engstler M, Pfohl T, Herminghaus S, Boshart M, Wiegertjes G, Heddergott N, Overath P. 2007. Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell 131:505–515. doi: 10.1016/j.cell.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 50.Müller R, Grawert MA, Kern T, Madl T, Peschek J, Sattler M, Groll M, Buchner J. 2013. High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation. Proc Natl Acad Sci U S A 110:10183–10188. doi: 10.1073/pnas.1300547110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furuya D, Yagihashi A, Nasu S, Endoh T, Nakamura T, Kaneko R, Kamagata C, Kobayashi D, Watanabe N. 2000. LDL particle size by gradient-gel electrophoresis cannot be estimated by LDL-cholesterol/apolipoprotein B ratios. Clin Chem 46:1202–1203. [PubMed] [Google Scholar]
- 52.Coppens I, Baudhuin P, Opperdoes FR, Courtoy PJ. 1988. Receptors for the host low density lipoproteins on the hemoflagellate Trypanosoma brucei: purification and involvement in the growth of the parasite. Proc Natl Acad Sci U S A 85:6753–6757. doi: 10.1073/pnas.85.18.6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Demmel L, Schmidt K, Lucast L, Havlicek K, Zankel A, Koestler T, Reithofer V, de Camilli P, Warren G. 2014. The endocytic activity of the flagellar pocket in Trypanosoma brucei is regulated by an adjacent phosphatidylinositol phosphate kinase. J Cell Sci 127:2351–2364. doi: 10.1242/jcs.146894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.