Abstract
As a successful commensal and pathogen of humans, Candida albicans encounters a wide range of environmental conditions. Among them, ambient pH, which changes frequently and affects many biological processes in this species, is an important factor, and the ability to adapt to pH changes is tightly linked with pathogenesis and morphogenesis. In this study, we report that pH has a profound effect on white-opaque switching and sexual mating in C. albicans. Acidic pH promotes white-to-opaque switching under certain culture conditions but represses sexual mating. The Rim101-mediated pH-sensing pathway is involved in the control of pH-regulated white-opaque switching and the mating response. Phr2 and Rim101 could play a major role in acidic pH-induced opaque cell formation. Despite the fact that the cyclic AMP (cAMP) signaling pathway does not play a major role in pH-regulated white-opaque switching and mating, white and opaque cells of the cyr1/cyr1 mutant, which is defective in the production of cAMP, showed distinct growth defects under acidic and alkaline conditions. We further discovered that acidic pH conditions repressed sexual mating due to the failure of activation of the Ste2-mediated α-pheromone response pathway in opaque a cells. The effects of pH changes on phenotypic switching and sexual mating could involve a balance of host adaptation and sexual reproduction in C. albicans.
INTRODUCTION
One of the most important environmental factors for all microbes is pH. The human fungal pathogen Candida albicans encounters a wide range of ambient pH values (from <2.0 to >8.5) during its life cycle (1, 2). The ability to sense and respond to pH changes is essential for this pathogen to survive in the host and cause infections (2).
The observation that pH regulates the development of filaments, a key feature of virulence in C. albicans, has been intensively investigated (3–7). A neutral to alkaline pH favors filamentation, while an acidic pH has a repressing effect on this process. The Rim101 pathway mediates pH sensing both in the model yeast Saccharomyces cerevisiae and in C. albicans (8–10). The Rim101 transcription factor itself is regulated by external pH. Deletion of the RIM101 gene causes defects in filamentous growth and in the regulation of pH response genes (11). The full-length form of the Rim101 protein has no activity (8), but in response to alkaline conditions, Rim101 is processed to a short activated form by proteolysis. The function of Rim101 is dependent on the basic helix-loop-helix transcription factor Efg1, which is a key regulator of filamentation downstream of the cyclic AMP (cAMP) signaling pathway (12). The cell surface glycosidases Phr1 and Phr2 function as pH sensors and are transcriptionally regulated by Rim101 in C. albicans (2, 8). PHR1 is induced by a high pH (above 5.5) and filamentation and is required for systemic infections, while PHR2 is induced by a low pH (below 5.5) and is essential for virulence in vaginal infection models (3, 4).
Carbon dioxide (CO2) and pH are tightly linked in the environment and biological systems (13). It has been shown that CO2 regulates both filamentation and white-opaque switching in C. albicans (14, 15). CO2 is sensed via the cAMP signaling pathway and by a currently unidentified pathway in C. albicans (16, 17). Both pathways converge on the Flo8 transcription factor (18). N-Acetylglucosamine (GlcNAc) is another host environmental cue and a potent inducer of both filamentation and the opaque phenotype in C. albicans. GlcNAc functions primarily through the cAMP signaling pathway in the regulation of both filamentation and white-opaque switching (16, 19–21).
In contrast to the yeast-filament transition, white-opaque switching is epigenetically regulated and exhibits heritable and bistable features (22–24). White and opaque cells differ in a number of aspects, including cell shape and surface, gene expression profile, and mating competence (22, 25–27). The transition between white and opaque cell types is controlled by both the master regulator Wor1 (28–30) and the mating type-like locus (26). Only opaque cells can mate efficiently (26). Therefore, in order to mate, diploid C. albicans cells must first undergo homozygosis at the MTL locus and then switch to the opaque mating-competent form (24, 26).
In this study, we investigated the role of pH in the regulation of white-opaque switching and sexual mating in C. albicans. We show that acidic pH induces the opaque phenotype but represses sexual mating. The Rim101-mediated pH-sensing pathway is involved in the regulation of white-opaque switching and cell growth under different pH conditions. We further demonstrate that the activation of the Ste2-mediated pheromone-sensing pathway is essential for efficient mating in C. albicans.
MATERIALS AND METHODS
Growth conditions and strains.
C. albicans cells were routinely cultured at 25°C in YPD (1% yeast extract, 2% peptone, 2% glucose) or Lee's glucose medium (16). Modified Lee's glucose medium (pH 4.0 to 8.0) was adjusted with HCl (37%) or NaOH (10%) solutions and used for white-opaque switching and mating assays. To make Lee's GlcNAc medium, 1.25% GlcNAc was used as the sole carbon source (16). Glucose (2%) was added to synthetic complete (SCD) medium as the sole carbon source.
The C. albicans strains used in this study are listed in Table S1 in the supplemental material. Strain GH1013 (16) was used to generate the null mutants of PHR1 and PHR2. Strain GH1352 was used to generate the cyr1/cyr1 mutant. Fusion PCR strategies (31) were used to delete both alleles of PHR1, PHR2, or CYR1. The nutrient marker genes ARG4, HIS1, and URA3 were amplified from plasmids pSN69 (31), pSN52 (31), and pGEM-URA3 (32), respectively. The primers used in this study are listed in Table S2 in the supplemental material. To construct the PHR1-, PHR2-, and RIM101-reconstituted strains, a fragment containing the promoter, the open reading frame (ORF), and terminal region of each gene, the C. albicans SAT1 (CaSAT1) cassette (from plasmid pNIM1) and a fragment of the 3′ untranslated region (UTR) sequence were first amplified by PCR with the primers listed in Table S2 in the supplemental material. Fusion PCR assays were then performed with these PCR products. To construct the CYR1-reconstituted strain, the promoter region, the region encoding the CYR1-catalyzing domain, the terminator sequence, and a fragment of the 3′ UTR sequence were amplified from the genomic DNA of strain SC5314 with the primers listed in Table S2 in the supplemental material. The PCR products were used for fusion PCR assays with the CaSAT1 cassette. The fusion PCR product was used for transforming the corresponding mutants and generating the reconstituted strains.
White-opaque switching assays.
White-opaque switching assays were performed as previously described (16, 33). Cells were plated on Lee's glucose medium plates (pH 6.8) and grown at 25°C for 5 days. Homogeneous white or opaque colonies were then suspended in double-distilled H2O and replated onto modified Lee's glucose or GlcNAc medium plates (with different pHs). Phloxine B (5 μg/ml), which exclusively stains opaque colonies red, was added to the solid media. For quantitative assays, C. albicans cells were normally grown on plates for 5 or 7 days in air or 5% CO2. For the cyr1/cyr1 mutant, cells were grown on plates for 12 days.
Mating assays.
Quantitative mating assays were performed as described in our previous publication with slight modifications (18, 34). Lee's glucose solid, Lee's glucose liquid, SCD solid, and SCD liquid media were used for these assays. The pH of the medium was adjusted to 4.0 to 8.0. Opaque a cells (1 × 106; strain SZ306u [34]) and opaque α cells (1 × 106) were mixed and then spotted onto nutrient plates or inoculated into fresh liquid medium. After 48 h of incubation on solid medium plates or in liquid medium at 25°C, cells were collected and replated onto SCD medium selectable plates (SCD medium-arginine-uridine, SCD medium-arginine, and SCD medium-uridine) for phototrophic growth. Mating efficiencies were calculated as previously described (26, 34).
Pheromone response assays.
A 14-mer α-pheromone peptide (GFRLTNFGYFEPGK) was chemically synthesized and used for induction of the pheromone response. Opaque cells of the green fluorescent protein (GFP) reporter strain (strain SZ306a PMFa-GFP [34]) were cultured in liquid Lee's glucose medium (pH 6.8) at 25°C for 24 h and then inoculated into fresh Lee's glucose medium of different pHs (5.0 and 7.0) at a concentration of 1 × 106 cells/ml. The α-pheromone was added to the cultures twice at an interval of 12 h. The final concentration of the α-pheromone was 1 × 10−4 mol/liter. MFA1 expression induced by α-pheromone was indicated by the GFP fluorescence.
qRT-PCR assay.
Opaque cells were grown in liquid Lee's glucose medium (pH 6.8) at 25°C for 24 h. Opaque a cells were then inoculated into fresh liquid media of different pHs. For pheromone response assays, synthesized α-pheromone peptide was added to the single-strain cultures of SZ306aPMFa-GFP twice at an interval of 12 h. The final concentration of α-pheromone was 1 × 10−4 mol/liter. Alternatively, 2 × 106 opaque a and α cells were mixed, inoculated into fresh medium, and incubated at 25°C for 24 h. Cells were collected, and total RNA was extracted for quantitative real-time PCR (qRT-PCR) assays. qRT-PCR assays were performed as previously described (34) with slight modifications. A total of 0.6 mg of total RNA per sample was used to synthesize cDNA with RevertAid H Minus reverse transcriptase (Thermo Scientific). The relative expression level of each gene was normalized to that of ACT1.
RESULTS
Acidic pH promotes white-to-opaque switching.
It has been demonstrated that CO2 can induce white-to-opaque switching in C. albicans (15). We suspected that the hydrolysis of CO2, which results in the production of protons and intracellular acidification, could contribute to the induction of the opaque phenotype. We therefore tested whether acidic pH culture conditions had a similar effect on white-opaque switching. White cells of WO-1 (FC4, a stock of WO-1 [15]) were plated onto Lee's glucose medium in air at 25°C. This stock was used because it is sensitive to the stimulation of environmental factors and exhibits high white-to-opaque switching frequencies. As shown in Fig. 1, the majority of colonies underwent white-to-opaque switching and carried one or multiple opaque sectors at pH 4, while colonies with opaque sectors were much less frequent under neutral to alkaline pH conditions (pH 6 to 8). To confirm the inducing effect of acidic pH on the opaque phenotype, we performed quantitative white-to-opaque switching assays in four genetically independent strains. As demonstrated in Table 1, acidic pH notably promoted white-to-opaque switching in both WO-1 and SN250α in air. However, this promoting effect was not observed in the other two strains (12C and P37005) due to their low switching frequencies (<1%) under all pH conditions. These results suggest that acidic pH has an inducing effect on the opaque phenotype, but this appeared to be dependent on the genetic background of the strain.
FIG 1.
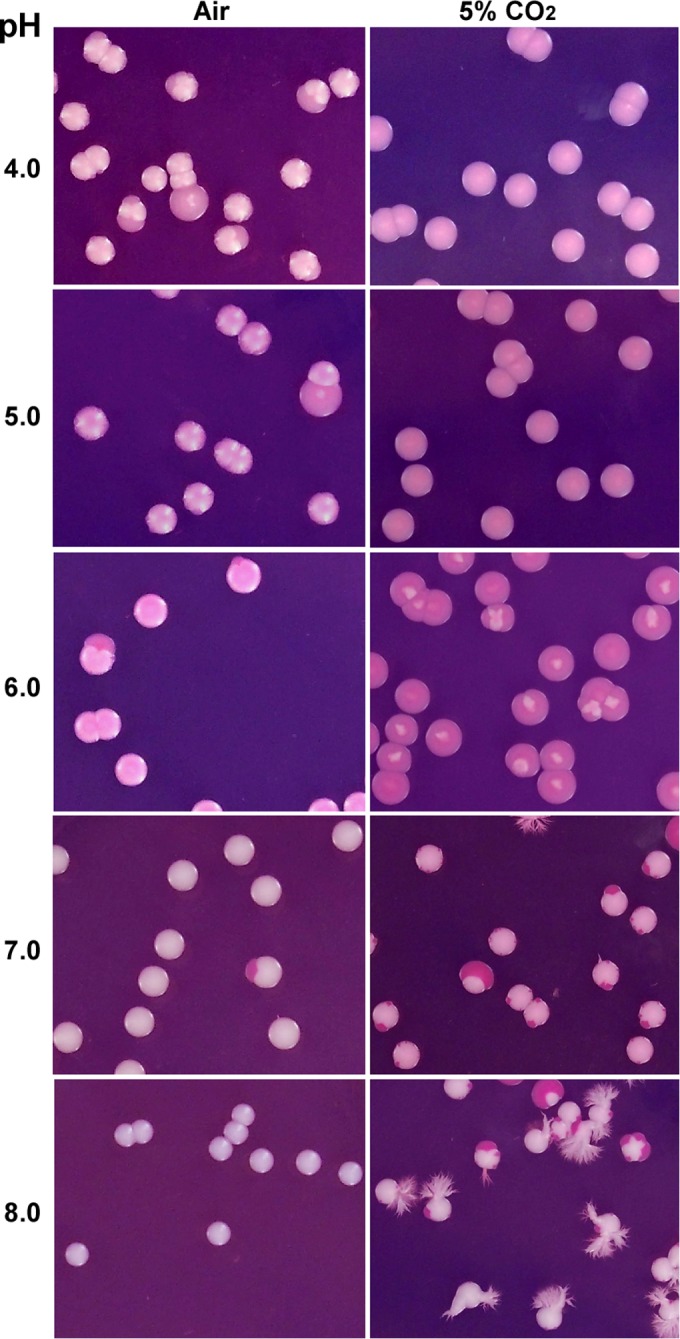
pH regulates white-to-opaque switching in air and in 5% CO2. An α/α strain (FC4, a stock of WO-1 [15]) was used for this experiment. White cells were plated onto Lee's glucose medium plates and incubated at 25°C for 7 days. Quantitative assays of the switching frequencies are shown in Table 1.
TABLE 1.
pH regulates white-to-opaque switching in strains with different backgroundsa
| Strain | Culture condition | White-to-opaque switching frequency (%) at pH: |
||||
|---|---|---|---|---|---|---|
| 5.0 | 6.0 | 7.0 | 7.5 | 8.0 | ||
| WO-1 (FC4, α/α) | Air | 29.1 ± 4.8* | 2.9 ± 0.9 | 2.3 ± 0.2 | 3.0 ± 0.5 | 3.5 ± 0.2 |
| 5% CO2 | 100.0 | 100.0* | 69.4 ± 7.5 | 49.5 ± 13.4 | 66.8 ± 3.4 | |
| SN250α (α/α) | Air | 19.7 ± 8.4 | 15.9 ± 2.2 | 15.3 ± 8.4 | 12.8 ± 7.6 | NA |
| 5% CO2 | 90.3 ± 7.8 | 82.4 ± 25.7* | 27.7 ± 21.5 | 18.3 ± 2.4 | NA | |
| 12C (a/a) | Air | <0.5 | <1.0 | <0.5 | <0.5 | <0.4 |
| 5% CO2 | 22.3 ± 5.9* | 13.0 ± 12.2* | <1.0 | <1.5 | 1.0 ± 1.8 | |
| P37005 (a/a) | Air | <0.3 | <1.0 | <0.4 | <0.5 | <1.0 |
| 5% CO2 | 2.7 ± 0.6* | 1.3 ± 0.5 | <0.5 | <0.4 | <0.4 | |
Two α/α strains (strain FC4, a stock of WO-1, and strain SN250α) and two a/a strains were used. White cells were plated onto Lee's glucose medium plates and incubated in air or 5% CO2 at 25°C for 7 days. Assays for the quantitation of the white-to-opaque switching frequencies were performed. <, no opaque colonies or colonies with opaque sectors were observed. *, the switching rate is significantly (Student's t test, P < 0.05) higher than that under the condition at the next higher pH. For example, the switching rate of strain WO-1 at pH 5.0 (29.1 ± 4.8) was significantly higher than that at pH 6.0 (2.9 ± 0.9). NA, not available.
pH conditions affect CO2- and GlcNAc-induced white-to-opaque switching.
CO2 and GlcNAc are potent inducers of the opaque phenotype in C. albicans (15, 16). We next tested the effect of pH changes on CO2- and GlcNAc-induced white-to-opaque switching in C. albicans. As shown in Fig. 1 and Table 1, CO2 and acidic pH had a synergistic effect on the induction of white-to-opaque switching in the four different strains. Interestingly, CO2 promoted the formation of branched colonies at pH 8.0. These colonies contained very few filamentous cells but showed slightly invasive growth (not shown). Similarly, GlcNAc and acidic pH also had an obvious synergistic effect on the induction of the opaque phenotype (Table 2). Despite the differences among the four different strains, all of them showed higher white-to-opaque switching frequencies under lower-pH conditions than higher-pH conditions. Some strains exhibited higher switching frequencies at pH 8.0 than at pH 7.5. This effect could be due to an indirect effect since extreme pH (8.0) reduced the solubility of some metal ions (such as copper and iron ions), which are important for the maintenance of the original phenotype.
TABLE 2.
pH regulates GlcNAc-induced white-to-opaque switchinga
| Strain | White-to-opaque switching frequency (%) at pH: |
||||
|---|---|---|---|---|---|
| 5.0 | 6.0 | 7.0 | 7.5 | 8.0 | |
| WO-1 (FC4) | 100.0* | 95.0 ± 4.1* | 28.3 ± 5.9 | 21.1 ± 1.6 | 23.0 ± 4.3 |
| JSM-167 | 99.4 ± 0.5* | 79.4 ± 6.4* | 31.1 ± 3.5 | 40.0 ± 7.0 | 45.2 ± 5.0 |
| 12C | 96.4 ± 2.1* | 75.7 ± 13.1* | 8.5 ± 5.9 | 14.0 ± 3.0 | 23.2 ± 5.6 |
| P37005 | 14.5 ± 4.4* | 2.9 ± 0.1 | 1.4 ± 1.5 | 1.7 ± 1.3 | 1.3 ± 1.1 |
White cells were plated onto Lee's GlcNAc medium plates and incubated in air at 25°C for 7 days. Assays for the quantitation of the white-to-opaque switching frequencies were performed. *, statistically significant difference (Student's t test, P < 0.05; see footnote a of Table 1).
pH has no obvious effect on opaque-to-white switching.
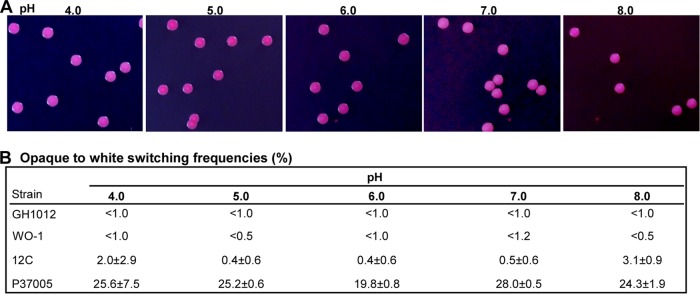
White-opaque switching is a heritable cellular transition (22). We next asked whether pH affects the stability of the opaque phenotype and performed quantitative opaque-to-white switching assays in different strains. As shown in Fig. 2, opaque cells of strains GH1012, WO-1, and 12C were very stable under different pH conditions. Although the opaque-to-white switching frequencies of P37005 were higher than those of the other three strains, there was no significant variation of this frequency under the different pH conditions.
FIG 2.
pH has no obvious effect on opaque-to-white switching. (A) Images of strain GH1012 (a/a) colonies at different pHs. (B) Quantitation of the frequencies of opaque-to-white switching. Four genetically independent strains were used. Opaque cells were plated onto Lee's glucose medium plates and incubated at 25°C in air for 7 days.
Role of cAMP signaling pathway in pH-regulated white-to-opaque switching.
The cAMP signaling pathway is involved in a range of biological processes in C. albicans, such as CO2- and GlcNAc-induced white-to-opaque switching and filamentation (6, 15, 16, 35). As mentioned earlier, the hydrolysis of CO2 leads to acidification. We tested whether this pathway plays a role in acidic pH-induced white-to-opaque switching. As shown in Table 3, deletion of CYR1 had no obvious effect on white-to-opaque switching under different pH conditions. Consistent with a previous report (16), deletion of PDE2 led to mass conversion to the opaque phenotype at pH 5.0 to 7.0, while the switching frequencies under alkaline pH conditions (pH 7.5 and 8.0) were relatively low. Therefore, despite the difference in switching frequencies between strains with the wild-type (WT) and mutant cAMP signaling pathway, the mutant strains had a similar tendency for opaque cell formation under different pH conditions. However, white and opaque cells of the cyr1/cyr1 mutant exhibited a distinct growth defect under different pH conditions. White cells of the mutant grew better under acidic pH conditions (4.0 to 6.0) than under alkaline conditions, while opaque cells exhibited better growth under acidic to neutral pH conditions (6.0 to 7.0). These results suggest that the cAMP signaling pathway may not play a major role in acidic pH-induced white-to-opaque switching in C. albicans but may indirectly affect the cell population through the regulation of cell growth.
TABLE 3.
Roles of cAMP signaling and pH response pathways in white-to-opaque switchinga
| Strain | White-to-opaque switching frequency (%) at pH: |
|||||
|---|---|---|---|---|---|---|
| 4.0 | 5.0 | 6.0 | 7.0 | 7.5 | 8.0 | |
| WT | 18.4 ± 2.9* | 12.5 ± 0.9 | 13.8 ± 0.3* | 5.4 ± 1.0* | <1.0 | <1.3 |
| cyr1/cyr1b | 23.5 ± 3.3* | 7.1 ± 4.1* | 4.8 ± 1.3 | 2.4 ± 3.4 | Growth defect | Growth defect |
| pde2/pde2 | 100.0 | 100.0 | 100.0* | 93.8 ± 2.3 | 74.2 ± 1.8* | 67.5 ± 1.2 |
| phr1/phr1 | 2.7 ± 1.8* | 0.2 ± 0.4 | <0.2 | 0.2 ± 0.4 | <0.3 | <0.3 |
| PHR1 reconstituted | 9.4 ± 0.1* | 5.9 ± 0.4* | 2.9 ± 1.1* | 0.9 ± 0.8* | <0.5 | 0.6 ± 1.0 |
| phr2/phr2 | Growth defect | Growth defect | 0.9 ± 1.2 | 3.8 ± 3.2* | 0.8 ± 1.1 | <0.9 |
| PHR2 reconstituted | 5.8 ± 0.3 | 10.4 ± 2.3* | 4.7 ± 0.8* | 0.5 ± 0.8* | <0.5 | <0.6 |
| rim101/rim101 | 2.4 ± 0.7 | 6.0 ± 0.8* | 1.7 ± 0.8 | 2.6 ± 0.6 | 3.8 ± 2.3 | 4.2 ± 0.2 |
| RIM101 reconstituted | 10.0 ± 2.8* | 6.4 ± 1.3 | 4.6 ± 1.4* | 0.7 ± 1.2* | <0.4 | <0.4 |
The WT was strain GH1013, an SC5314 background strain. White cells were plated onto Lee's glucose medium plates and incubated in air at 25°C for 7 days. Assays for the quantitation of white-to-opaque switching frequencies were performed. <, no opaque or opaque-sectored colonies were observed. PHR1, PHR2, or RIM101 reconstituted, a copy of the PHR1, PHR2, or RIM101 gene, respectively, was integrated into the original locus. *, statistically significant difference (Student's t test, P < 0.05; see footnote a of Table 1).
The cyr1/cyr1 mutant grew extremely slowly. The switching frequencies were examined after 12 days of growth.
Efg1 is a downstream transcription factor of the cAMP signaling pathway and plays a critical role in the regulation of white-opaque switching (36–38). Although the difference in white-opaque switching frequency under different pH conditions was not observed, we found two major colony phenotypes for the efg1/efg1 mutant: smooth and rough. Smooth colonies contained only typical opaque cells, while rough colonies contained both typical opaque cells and more elongated and pseudohypha-like cells. Under acidic pH conditions (<6.5), most colonies of the mutant exhibited the smooth phenotype, while most were rough when cultured at pH 7.0 or higher (data not shown).
Roles of Phr1, Phr2, and Rim101 in white-opaque switching.
As mentioned earlier, the Rim101 pathway governs pH responses and filamentation in C. albicans (4, 8, 9). Deletion of the pH response gene PHR1 had no significant effect on white-to-opaque switching, while deletion of PHR2 led to growth defects under lower-pH conditions (Table 3). The rim101/rim101 mutant exhibited comparable switching frequencies under different pH conditions. Therefore, deletion of RIM101 resulted in the loss of the pH-dependent switching response. Both white and opaque cells of the phr2/phr2 mutant had a growth defect under acidic pH conditions (4.0 to 6.0; see Fig. S1 and S2 in the supplemental material). Deletion of RIM101 or PHR1 had no obvious effect on opaque-to-white switching (not shown), while opaque cells of the phr2/phr2 mutant were extremely unstable at pH 4.0 to 5.0 and underwent mass conversion to the white phenotype. These results indicate that Rim101 and Phr2 may play a major role in the regulation of white-opaque switching in C. albicans.
pH regulates sexual mating in C. albicans.
White-opaque switching and sexual mating are two tightly linked processes in C. albicans (26, 27). To test whether pH conditions affect sexual mating in C. albicans, we performed mating assays under four different culture conditions (shown in Table 4). In SCD media (both solid and liquid), opaque cells mated most efficiently at pH 7.0. On solid Lee's medium, opaque cells mated most efficiently at pH 6.0, while in liquid Lee's medium, opaque cells mated most efficiently at pH 8.0 (of note, the growth of C. albicans cells resulted in acidification of the medium; the final pH of this medium was about 6.9). These results suggest that acidic pH (<5.0) represses sexual mating in C. albicans.
TABLE 4.
pH regulates sexual mating in C. albicansa
| Medium | Mating efficiency at pH: |
||||
|---|---|---|---|---|---|
| 4.0 | 5.0 | 6.0 | 7.0 | 8.0 | |
| Lee's glucose | |||||
| Solid | (6.3 ± 1.2) × 10−2 | (1.0 ± 0.2) × 10−1 | (1.7 ± 0.3) × 10−1* | (5.9 ± 0.4) × 10−2 | (3.1 ± 0.8) × 10−2 |
| Liquid | (5.6 ± 3.9) × 10−5 | (3.5 ± 1.5) × 10−4 | (1.9 ± 1.2) × 10−3 | (5.2 ± 1.4) × 10−2 | (2.4 ± 0.2) × 10−1* |
| SCD | |||||
| Solid | (2.6 ± 0.7) × 10−1 | (1.6 ± 0.3) × 10−1 | (3.4 ± 0.7) × 10−1 | (7.8 ± 1.7) × 10−1* | (5.5 ± 3.0) × 10−1 |
| Liquid | (3.9 ± 4.6) × 10−5 | (5.1 ± 1.0) × 10−2 | (1.3 ± 0.2) × 10−1 | (1.9 ± 0.4) × 10−1* | (5.8 ± 1.6) × 10−2 |
Opaque cells of SZ306u (an a strain) and GH1349 (an α strain) were mixed and used for quantitative mating assays (see the Materials and Methods section). *, the mating efficiency was statistically significantly (Student's t test, P < 0.05) higher than that under the lower-pH conditions.
Acidic pH represses α-pheromone-induced STE2 and MFA1 expression in opaque a cells.
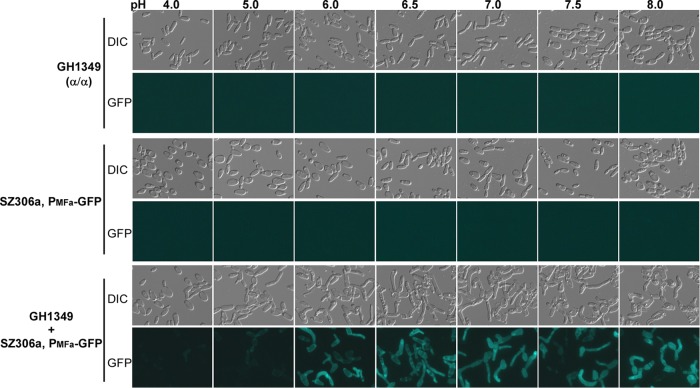
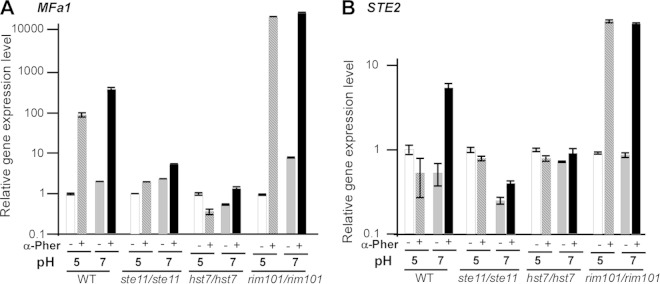
In C. albicans, the α-pheromone precursor, MFα1, is constitutively expressed in α opaque cells (34, 39–41). However, the a-pheromone precursor, MFa1, is expressed only in opaque a cells in response to α-pheromone or opaque α cells (34, 42). We suspected that opaque α cells failed to induce the expression of MFA1 in opaque a cells under acidic pH conditions. Thus, pheromone and mating responses could not occur efficiently. To test this hypothesis, the expression of MFA1 was examined using an MFA1 promoter-controlled GFP reporter a strain (34). As shown in Fig. 3, the GFP fluorescence of the reporter strain was extremely weak at pH 4.0 and 5.0, while the signal was much stronger at pHs over 6.0 when the reporter strain was coincubated with opaque α cells. The relative expression levels of MFA1 in cells treated with chemically synthesized α-pheromone at pH 5.0 and 7.0 were then determined using qRT-PCR assays. As expected, the expression of MFA1 was about five times higher at pH 7.0 than at pH 5.0 in α-pheromone-treated cells (Fig. 4A). Of note, the expression of MFα1 in opaque α cells was not affected by different pH conditions (not shown). The α-pheromone receptor gene STE2 is a pheromone-responsive gene in opaque a cells (39). The induction of STE2 expression in opaque a cells was observed only at pH 7.0 when the cells were treated with α-pheromone and not at pH 5.0 (Fig. 4B). These results indicate that the induction of the Ste2-mediated pheromone response pathway and expression of MFA1 are dependent on pH conditions.
FIG 3.
pH regulates the induction of MFA1 expression in response to α-pheromone (produced by opaque α cells). A GFP-encoding sequence was integrated into the MFA locus and used as a reporter of the MFA1 promoter. Opaque α cells of GH1349, which served as an α-pheromone producer, were mixed with opaque a cells of the PMFa1-GFP reporter strain. Single-strain cultures (GH1349 and the reporter strain) served as controls. Cells were incubated in liquid Lee's glucose medium with shaking at 25°C for 24 h. DIC, differential interference contrast.
FIG 4.
Relative expression levels of MFA1 (A) and STE2 (B) in response to α-pheromone under different pH conditions. The WT (GH1013) control and the ste11/ste11, hst7/hst7, and rim101/rim101 mutants are all SC5314 derivatives. To generate the MFA1 reporter strain SZ306a PMFa-GFP, a GFP-encoding sequence was integrated into the MFA locus. Opaque cells of SZ306a PMFa-GFP were initially grown in liquid Lee's glucose medium (pH 6.8) to stationary phase. Cells were then reinoculated into fresh Lee's glucose medium (pH 5.0 and 7.0) and treated with α-pheromone at 25°C for 24 h.
Since deletion of RIM101 led to the loss of pH-regulated white-to-opaque switching, we examined the relative expression levels of MFA1 and STE2 in the rim101/rim101 mutant. As shown in Fig. 4, the expression levels of both MFA1 and STE2 in α-pheromone-treated cells were comparable, suggesting that the expression of the two genes is independent of pH conditions in this mutant. The pheromone response is transduced from Ste2 to the STE11- mitogen-activated protein kinase (MAPK) pathway (43, 44). We next tested whether this MAPK pathway is essential for α-pheromone-activated expression of MFA1 and STE2. As expected, the expression levels of these two genes in the ste11/ste11 and hst7/hst7 mutants were very low in all treatments under both pH conditions.
DISCUSSION
To better survive and cause infections in the human host, C. albicans must adapt to broadly fluctuating pH conditions. Despite an ambient pH that is relatively stable in human blood and tissues, pH variations frequently occur in the major niches for the commensal lifestyle of C. albicans (such as the digestive tract and vaginal cavity of human hosts). The two related processes, white-opaque switching and mating, are thought to play important roles in the regulation of pathogenesis in C. albicans (23, 24, 27). A variety of environmental cues have been reported to regulate these two processes (7, 23, 24, 27). In this study, we report that changes of the pH conditions affect not only white-opaque switching but also sexual mating in C. albicans. Despite the difference in the response to pH changes, acidic pH conditions favor the opaque phenotype in all clinically dependent strains tested (Table 1 and Fig. 2). Since acidic pH-induced opaque cell formation is highly dependent on strain backgrounds and exhibits a relatively minor effect (compared to the effects of CO2 and GlcNAc), we consider that pH may play an additive role in the environmental regulation of this phenotypic switching in C. albicans.
The Rim101 pH-sensing pathway is highly conserved in pathogenic fungi (2, 45). It has been shown that the rim101/rim101 mutant exhibits the defect of alkaline-induced hyphal growth (2, 8). Given the central role of Rim101 in the regulation of pH adaptation, it is reasonable that the deletion of RIM101 led to the loss of the pH-dependent switching response (reduced switching under higher-pH conditions). Consistent with this observation, α-pheromone-activated expression of MFA1 and STE2 was also independent of pH conditions in the rim101/rim101 mutant. Inactivation of the pH response gene PHR1 had no obvious effect on white-opaque switching (Table 3), but deletion of PHR2 led to instability of the opaque cell type at acidic pHs (see Fig. S2 in the supplemental material). Phr2 may play a major role in acidic pH-induced white-to-opaque switching. Consistent with our data, it has been demonstrated that PHR2 is expressed only at a pH below 5.5 (4).
The cAMP signaling pathway plays a central role in phenotypic transitions and environmental sensing in C. albicans (6, 46, 47). The adenylyl cyclase of C. albicans, Cyr1, is a large protein that contains several functional domains (35, 47). Cyr1 functions as a signal sensor for a variety of environmental cues. Consistent with previous studies, inactivation of the cAMP signaling pathway decreases the frequency of white-to-opaque switching under neutral pH conditions, while activation of this pathway by deletion of the high-affinity cyclic nucleotide phosphodiesterase gene, PDE2, induces the opaque phenotype. However, acidic pH favors the development of the opaque cell type in both CYR1 and PDE2 mutants (Table 3), suggesting that acidic pH-induced opaque cell formation is not dependent on the cAMP signaling pathway. There could be other pathways involved in the regulation of this process. Interestingly, white and opaque cells of the cyr1/cyr1 mutant showed distinct growth defects under different pH conditions (Fig. 3), suggesting that the cAMP signaling pathway could play critical but distinct roles in the regulation of the growth of white and opaque cells. Thus, the two cell types may have different abilities to adapt to extreme pH conditions.
pH affects multiple biological processes in fungi, including metabolism, morphogenesis, meiosis, and the virulence of pathogens (48). Here we observed that acidic pH conditions repress sexual mating in C. albicans (Table 4). This repressing effect is due to the failure of activation of the Ste2-mediated pheromone response pathway and the induction of a-pheromone expression. Since only opaque cells can mate efficiently, acidic pH conditions favor opaque cell formation, which indirectly promotes mating in this species in nature. The repressing effect of acidic pH conditions on the mating process would thus play a role in balancing the sexual and asexual lifestyles in this species. The situation in S. cerevisiae is similar (49). The optimum pH for mating-specific adhesion is 6 to 7, while pretreatment of S. cerevisiae cells at pH 3.5 significantly increases mating efficiencies.
The roles of pH regulation in white-opaque switching and sexual mating may support the adaptation of C. albicans to environmental variations in the host. However, the host environment is very complex. There are many effectors in every niche, and a lot of questions remain to be investigated. For example, what is the relationship between CO2- and pH-sensing pathways? Are there any Rim101-independent pH-sensing pathways? Does the pH signaling pathway function directly or indirectly (for example, through the regulation of cellular metabolisms) on the induction of the opaque cell type? How do pH-sensing pathways cross talk with regulatory pathways controlling phenotypic switching and sexual mating?
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to David Soll, Bill Fonzi, and Suzanne Noble for the generous gifts of plasmids and strains and thank Malcolm Whiteway for helpful comments on the manuscript.
This work was supported by grants from the Chinese National Natural Science Foundation (31370175, 31170086, and 81322026 to G.H.; 31200115 to L.T.) and a 100 Talent Program grant from the Chinese Academy of Sciences (to G.H.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00123-15.
REFERENCES
- 1.Kararli TT. 1995. Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharm Drug Dispos 16:351–380. doi: 10.1002/bdd.2510160502. [DOI] [PubMed] [Google Scholar]
- 2.Davis D. 2003. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr Genet 44:1–7. doi: 10.1007/s00294-003-0415-2. [DOI] [PubMed] [Google Scholar]
- 3.De Bernardis F, Muhlschlegel FA, Cassone A, Fonzi WA. 1998. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun 66:3317–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fonzi WA. 1999. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of beta-1,3- and beta-1,6-glucans. J Bacteriol 181:7070–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramon AM, Porta A, Fonzi WA. 1999. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J Bacteriol 181:7524–7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas S, Van Dijck P, Datta A. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev 71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang G. 2012. Regulation of phenotypic transitions in the fungal pathogen Candida albicans. Virulence 3:251–261. doi: 10.4161/viru.20010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis D, Wilson RB, Mitchell AP. 2000. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol Cell Biol 20:971–978. doi: 10.1128/MCB.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Barkani A, Kurzai O, Fonzi WA, Ramon A, Porta A, Frosch M, Muhlschlegel FA. 2000. Dominant active alleles of RIM101 (PRR2) bypass the pH restriction on filamentation of Candida albicans. Mol Cell Biol 20:4635–4647. doi: 10.1128/MCB.20.13.4635-4647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeda T. 2012. The signaling mechanism of ambient pH sensing and adaptation in yeast and fungi. FEBS J 279:1407–1413. doi: 10.1111/j.1742-4658.2012.08548.x. [DOI] [PubMed] [Google Scholar]
- 11.Bensen ES, Martin SJ, Li M, Berman J, Davis DA. 2004. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol Microbiol 54:1335–1351. doi: 10.1111/j.1365-2958.2004.04350.x. [DOI] [PubMed] [Google Scholar]
- 12.Nie X, Liu X, Wang H, Chen J. 2010. Deletion of EFG1 promotes Candida albicans opaque formation responding to pH via Rim101. Acta Biochim Biophys Sin (Shanghai) 42:735–744. doi: 10.1093/abbs/gmq076. [DOI] [PubMed] [Google Scholar]
- 13.Hetherington AM, Raven JA. 2005. The biology of carbon dioxide. Curr Biol 15:R406–R410. doi: 10.1016/j.cub.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 14.Klengel T, Liang WJ, Chaloupka J, Ruoff C, Schroppel K, Naglik JR, Eckert SE, Mogensen EG, Haynes K, Tuite MF, Levin LR, Buck J, Muhlschlegel FA. 2005. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol 15:2021–2026. doi: 10.1016/j.cub.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang G, Srikantha T, Sahni N, Yi S, Soll DR. 2009. CO2 regulates white-to-opaque switching in Candida albicans. Curr Biol 19:330–334. doi: 10.1016/j.cub.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. 2010. N-Acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog 6:e1000806. doi: 10.1371/journal.ppat.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cottier F, Raymond M, Kurzai O, Bolstad M, Leewattanapasuk W, Jimenez-Lopez C, Lorenz MC, Sanglard D, Vachova L, Pavelka N, Palkova Z, Muhlschlegel FA. 2012. The bZIP transcription factor Rca1p is a central regulator of a novel CO2 sensing pathway in yeast. PLoS Pathog 8:e1002485. doi: 10.1371/journal.ppat.1002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du H, Guan G, Xie J, Cottier F, Sun Y, Jia W, Muhlschlegel FA, Huang G. 2012. The transcription factor Flo8 mediates CO2 sensing in the human fungal pathogen Candida albicans. Mol Biol Cell 23:2692–2701. doi: 10.1091/mbc.E12-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cassone A, Sullivan PA, Shepherd MG. 1985. N-Acetyl-d-glucosamine-induced morphogenesis in Candida albicans. Microbiologica 8:85–99. [PubMed] [Google Scholar]
- 20.Konopka JB. 2012. N-Acetylglucosamine (GlcNAc) functions in cell signaling. Scientifica (Cairo) 2012:489208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naseem S, Parrino SM, Buenten DM, Konopka JB. 2012. Novel roles for GlcNAc in cell signaling. Commun Integr Biol 5:156–159. doi: 10.4161/cib.19034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol 169:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohse MB, Johnson AD. 2009. White-opaque switching in Candida albicans. Curr Opin Microbiol 12:650–654. doi: 10.1016/j.mib.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soll DR. 2009. Why does Candida albicans switch? FEMS Yeast Res 9:973–989. doi: 10.1111/j.1567-1364.2009.00562.x. [DOI] [PubMed] [Google Scholar]
- 25.Lan CY, Newport G, Murillo LA, Jones T, Scherer S, Davis RW, Agabian N. 2002. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc Natl Acad Sci U S A 99:14907–14912. doi: 10.1073/pnas.232566499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller MG, Johnson AD. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293–302. doi: 10.1016/S0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 27.Soll DR, Lockhart SR, Zhao R. 2003. Relationship between switching and mating in Candida albicans. Eukaryot Cell 2:390–397. doi: 10.1128/EC.2.3.390-397.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. 2006. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci U S A 103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, Gerstein M, Yi S, Snyder M, Soll DR. 2006. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell 5:1674–1687. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zordan RE, Galgoczy DJ, Johnson AD. 2006. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci U S A 103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol 181:1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie J, Tao L, Nobile CJ, Tong Y, Guan G, Sun Y, Cao C, Hernday AD, Johnson AD, Zhang L, Bai FY, Huang G. 2013. White-opaque switching in natural MTLa/alpha isolates of Candida albicans: evolutionary implications for roles in host adaptation, pathogenesis, and sex. PLoS Biol 11:e1001525. doi: 10.1371/journal.pbio.1001525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao L, Cao C, Liang W, Guan G, Zhang Q, Nobile CJ, Huang G. 2014. White cells facilitate opposite- and same-sex mating of opaque cells in Candida albicans. PLoS Genet 10:e1004737. doi: 10.1371/journal.pgen.1004737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rocha CR, Schroppel K, Harcus D, Marcil A, Dignard D, Taylor BN, Thomas DY, Whiteway M, Leberer E. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell 12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonneborn A, Tebarth B, Ernst JF. 1999. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect Immun 67:4655–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srikantha T, Tsai LK, Daniels K, Soll DR. 2000. EFG1 null mutants of Candida albicans switch but cannot express the complete phenotype of white-phase budding cells. J Bacteriol 182:1580–1591. doi: 10.1128/JB.182.6.1580-1591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bockmuhl DP, Ernst JF. 2001. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 157:1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennett RJ, Uhl MA, Miller MG, Johnson AD. 2003. Identification and characterization of a Candida albicans mating pheromone. Mol Cell Biol 23:8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lockhart SR, Zhao R, Daniels KJ, Soll DR. 2003. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot Cell 2:847–855. doi: 10.1128/EC.2.5.847-855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panwar SL, Legrand M, Dignard D, Whiteway M, Magee PT. 2003. MFalpha1, the gene encoding the alpha mating pheromone of Candida albicans. Eukaryot Cell 2:1350–1360. doi: 10.1128/EC.2.6.1350-1360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dignard D, El-Naggar AL, Logue ME, Butler G, Whiteway M. 2007. Identification and characterization of MFA1, the gene encoding Candida albicans a-factor pheromone. Eukaryot Cell 6:487–494. doi: 10.1128/EC.00387-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bensen ES, Filler SG, Berman J. 2002. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot Cell 1:787–798. doi: 10.1128/EC.1.5.787-798.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi S, Sahni N, Daniels KJ, Pujol C, Srikantha T, Soll DR. 2008. The same receptor, G protein, and mitogen-activated protein kinase pathway activate different downstream regulators in the alternative white and opaque pheromone responses of Candida albicans. Mol Biol Cell 19:957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornet M, Gaillardin C. 2014. pH signaling in human fungal pathogens: a new target for antifungal strategies. Eukaryot Cell 13:342–352. doi: 10.1128/EC.00313-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whiteway M, Bachewich C. 2007. Morphogenesis in Candida albicans. Annu Rev Microbiol 61:529–553. doi: 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y. 2013. Fungal adenylyl cyclase acts as a signal sensor and integrator and plays a central role in interaction with bacteria. PLoS Pathog 9:e1003612. doi: 10.1371/journal.ppat.1003612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Penalva MA, Tilburn J, Bignell E, Arst HN Jr. 2008. Ambient pH gene regulation in fungi: making connections. Trends Microbiol 16:291–300. doi: 10.1016/j.tim.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 49.Soellick TR, Uhrig JF. 2001. Development of an optimized interaction-mating protocol for large-scale yeast two-hybrid analyses. Genome Biol 2:RESEARCH0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.