Abstract
Candida albicans is associated with humans as both a harmless commensal organism and a pathogen. Cph2 is a transcription factor whose DNA binding domain is similar to that of mammalian sterol response element binding proteins (SREBPs). SREBPs are master regulators of cellular cholesterol levels and are highly conserved from fungi to mammals. However, ergosterol biosynthesis is regulated by the zinc finger transcription factor Upc2 in C. albicans and several other yeasts. Cph2 is not necessary for ergosterol biosynthesis but is important for colonization in the murine gastrointestinal (GI) tract. Here we demonstrate that Cph2 is a membrane-associated transcription factor that is processed to release the N-terminal DNA binding domain like SREBPs, but its cleavage is not regulated by cellular levels of ergosterol or oxygen. Chromatin immunoprecipitation sequencing (ChIP-seq) shows that Cph2 binds to the promoters of HMS1 and other components of the regulatory circuit for GI tract colonization. In addition, 50% of Cph2 targets are also bound by Hms1 and other factors of the regulatory circuit. Several common targets function at the head of the glycolysis pathway. Thus, Cph2 is an integral part of the regulatory circuit for GI colonization that regulates glycolytic flux. Transcriptome sequencing (RNA-seq) shows a significant overlap in genes differentially regulated by Cph2 and hypoxia, and Cph2 is important for optimal expression of some hypoxia-responsive genes in glycolysis and the citric acid cycle. We suggest that Cph2 and Upc2 regulate hypoxia-responsive expression in different pathways, consistent with a synthetic lethal defect of the cph2 upc2 double mutant in hypoxia.
INTRODUCTION
Candida albicans is a member of the natural human commensal microbiota found in the oral cavity, the gastrointestinal (GI) tract, and the genitourinary tract (1). However, C. albicans can cause both mucosal and systemic infections. Disseminated candidiasis is one of the most important nosocomial infections, with mortality rates of around 40% (2). Systemic C. albicans infections are believed to originate from the commensal mucosal microbiota disseminating to tissues following damage to the mucosal barrier, alterations in the host immune system, and C. albicans overgrowth following immunosuppressive drug and/or antibiotic treatments (3, 4). In addition to the status of the host immune system, functions of C. albicans genes also contribute to the outcome of the host-C. albicans interaction. While C. albicans genes important for invasive candidiasis have been studied extensively (5), genes necessary for gastrointestinal colonization are less known but are gaining attention (6–11).
The DNA binding domain of C. albicans Cph2 protein is similar to that of mammalian sterol response element binding proteins (SREBPs), which are a family of basic helix-loop-helix transcription factors with a characteristic tyrosine residue (12, 13). SREBPs are master regulators of cellular cholesterol levels and are highly conserved from fungi to mammals (12, 14). Under normal sterol levels, they exist as inactive endoplasmic reticulum (ER) transmembrane proteins. Depletion of cellular sterol results in the translocation of the SREBP to the Golgi apparatus, where it undergoes proteolytic cleavage releasing the N-terminal transcription regulatory domain, which then enters the nucleus to promote expression of genes required for sterol biosynthesis by binding to sterol regulatory elements (SREs) in the promoters of target genes. This regulation of SREBPs in response to sterol level in mammalian cells also exists in Schizosaccharomyces pombe (15, 16), Cryptococcus neoformans, and Aspergillus fumigatus (16, 17), where it is important for hypoxic adaptation, fungal pathogenesis, and resistance to antifungal drugs. In S. pombe, the proteolytic cleavage of the SREBP Sre1 is regulated by ergosterol, which is an indirect measure of oxygen supply, as sterol synthesis is inhibited when oxygen levels are low (15, 18, 19). Furthermore, degradation of the cleaved N-terminal Sre1 is regulated by Ofd1, a prolyl 4-hydroxylase-like 2-oxoglutarate-Fe(II) dioxygenase, in response to oxygen levels (20, 21). Unlike in S. pombe, C. neoformans, and A. fumigatus, ergosterol biosynthesis is regulated by the zinc finger transcription factor Upc2 in Saccharomyces cerevisiae, C. albicans, Candida glabrata, and Candida parapsilosis (22–26). Upc2 is important for growth under anaerobic conditions and for resistance to antifungal drugs in C. albicans (24, 27). The transcriptional response of C. albicans to hypoxia is associated with increased expression of ergosterol synthesis and glycolytic genes and reduced expression of components of the respiratory chain, ATP synthesis, and the citric acid cycle (28–30). The hypoxic induction of the ergosterol pathway is mimicked by treatment with sterol-lowering drugs and requires Upc2. However, Upc2 is not responsible for hypoxia-induced expression of glycolytic genes or decreased expression of genes in respiratory pathways (30). Interestingly, Yarrowia lipolytica, which represents the most divergent lineage of the Saccharomycotina, has both SREBP and Upc2 (31). Both SREBP and Upc2 play a role in response to hypoxic conditions in Y. lipolytica, but Upc2 is the major regulator of sterol genes. Therefore, Upc2 transcription factors displaced SREBPs as the major sterol regulators during Saccharomycotina evolution (31). The SREBP-like protein Cph2 is present throughout the CTG clades (31). However, there is very little evidence that Cph2 proteins in the CTG clades are membrane associated or play a role under hypoxic conditions.
Cph2 was first found as a regulator of hyphal development that is necessary for sustained hyphal growth under certain growth conditions (13). It was later determined to be important for colonization of the murine gastrointestinal tract (9). However, hyphal formation is not necessary for GI colonization (3, 32). Several studies suggest that C. albicans has developed mechanisms that allow it to inhabit the mammalian GI tract as well as the bloodstream. Specific environments in the mammalian GI tract have been shown to promote phenotypic heterogeneity in colonizing C. albicans cells and to produce niche-specific cells that are better adapted for growth in the GI tract (8, 11). In addition, regulatory networks used by C. albicans in different nutritional parameters, such as iron levels of the GI tract and bloodstream, are being identified (7, 8, 10). A recent screen of 72 transcription regulator deletion mutants in mouse models of GI colonization and systemic infection identified an interlocking transcriptional feedback loop, in which some key regulators were found to be important for GI tract colonization and/or systemic infection, regulating many similar genes as well as each other (6). Hms1 and Rtg1/3 of the regulatory circuit bind upstream of the regulatory genes in the circuit as well as upstream of CPH2 (6). Functions of Cph2 in GI colonization are not known.
Here we show that, like SREBPs, Cph2 is a membrane-associated transcription factor that is processed to release the N-terminal transcriptional regulator. Unlike the case for SREBPs or Sre1, the cleavage of Cph2 is not regulated by cellular levels of ergosterol or oxygen. Chromatin immunoprecipitation sequencing (ChIP-seq) of Cph2 shows that Cph2 binds to the promoters of HMS1 and other components of the regulatory circuit for GI tract colonization and systemic infection. Transcriptome sequencing (RNA-seq) identifies differential expression of OFD1 and many hypoxia-responsive genes in glycolysis and the citric acid cycle in cph2. As in Y. lipolytica, we find that Cph2 plays a minor role while Upc2 plays a major role in growth under hypoxic conditions. Conservation and divergence of fungal Cph2 proteins to SREBPs are discussed.
MATERIALS AND METHODS
Media.
Standard techniques and media for growing yeast were used in this study. The media used were YPD (2% Bacto peptone, 1% yeast extract, 0.015% l-Tryptophan, 2% dextrose), SC (0.224% complete amino acid mixture, 0.15% YNB without amino acids or ammonium sulfate, 0.2 mM inositol, 0.5% ammonium sulfate, 2% dextrose), and SD (0.17% YNB without amino acids or ammonium sulfate, 0.5% ammonium sulfate, 2% dextrose, amino acids supplemented for auxotrophs). YPM has 2% maltose instead of the dextrose of YPD. SC–2% maltose has maltose instead of the dextrose in SC. Rapamycin (10 nM) or geldanamycin A (10 μM) was added to YPD for hyphal maintenance.
Plasmid and strain construction.
All strains, plasmids, and primers are listed in Tables S5 to S7 in the supplemental material. Details of plasmid construction are described below.
(i) Mal2p-myc-GFP.
A 700-bp product containing GFP was cloned into the Mal2p-myc-HGC1 vector (33) at the MluI-KpnI site using GFPMluIForward and GFPKpnIReverse primers for PCR from yEGFP3 (pHL339) (34).
(ii) Mal2-GFP-HGC1.
A 700-bp product containing green fluorescent protein (GFP) (pHL339) was inserted into the XbaI-MluI site of Mal2p-myc-HGC1 (33), replacing the myc tag as well as the −800-bp upstream region of Hgc1 with GFP using primers GFPXbaIForward and 842.
(iii) Mal2p-myc-Cph2.
A full-length 2.6-kb Cph2 fragment was amplified by PCR with primers 1052 and 1053 and was cloned into the Mal2p-myc-GFP (p866) plasmid at the MluI-KpnI site, replacing GFP.
(iv) Cph2p-Cph2-myc.
A 2.5-kb PCR product (primers 1 and 2) containing the CPH2 coding region was inserted into the BamHI-MluI site of pPR673 (35). The resulting plasmid was digested with PstI to target integration into its own loci to express Cph2-13Myc.
(v) Cph2p-myc-Cph2.
From plasmid p1021 (Mal2p-myc-Cph2), Mal2p was cut out (0.55 kb) with XhoI and NotI. The Cph2 promoter fragment (2.0 kb) was PCR amplified with primers 1102 and 1103 and digested with SpeI and NotI to clone into the plasmid.
(vi) Mal2p-myc-Cph2N.
The Cph2N fragment was PCR amplified from plasmid p1021 (Mal2mycCph2) with primers 1052 and 1128. The Cph2N fragment (1.2 kb) was inserted into the MluI-KpnI site of p1021, replacing the full-length Cph2 fragment.
(vii) Mal2p-GFP-Cph2.
A 2.6-kb Cph2 product was inserted into the MluI-KpnI site, replacing Hgc1, from p873 (Mal2p-GFP-Hgc1), using primers 1052 and 1053.
(viii) Mal2p-GFP-Cph2-myc.
The Cph2-myc (∼2.5-kb) PCR product from the Cph2p-Cph2-myc plasmid was cloned into the Mal2p-GFP-Cph2 plasmid at the ClaI-KpnI site using primers 1086 and 925.
All plasmids except TET1-Cph2 (SacII-KpnI) and Cph2p-Cph2-myc (PstI) were digested with AscI to integrate into the ADE2 locus in C. albicans strains. ACT1p-6×HIS-FLAG-HMS1 (pLC597) was graciously given by the Cowen lab (36) and linearized with BsrGI before transformation into C. albicans. Homozygous deletion strains generated by the method of Noble and Johnson (37) were made prototrophs by the transformation of the ARG4 gene (1577 and 1578) by PCR of the CAI4 strain. The cph2 upc2 double mutants were made by sequential gene disruption of CPH2 in the upc2 mutant (17).
RNA-seq.
Wild-type (WT) and cph2 mutant strains were grown in YPD overnight to optical densities at 600 nm (OD600) of 14.14 and 11.54 for the wild type and 12.05 and 12.35 for the cph2 mutant (2 samples each). Total RNA was extracted using the RiboPure kit from Ambion along with DNase digestion as described in the manual. Samples were processed for library construction by the Genomics High-Throughput Facility at UC Irvine. mRNA was enriched using the Invitrogen Dynabeads mRNA purification kit. The library was constructed using the NuGen Encore Complete RNA-seq Library Systems kit, and a sequencing run was carried out using Illumina HiSeq 2500 at the UCI Genomics High-Throughput Facility. RNA-seq data preprocessing and alignment were performed using the Illumina CASAVA software package.
ChIP-seq.
For ChIP-seq, the protocol from the Snyder lab was used (38) with some modifications. Wild-type cells carrying Mal2p-Cph2N were grown in 50 ml YEP–2% maltose overnight to an OD600 of ∼10. Fifty milliliters of cells was fixed with 1.4 ml 37% formaldehyde and 2.7 ml 2.5 M glycine for the time indicated. Cells were broken using zirconium beads with a FastPrep 6 times for 60 s at a speed of 6.0 m/s. Lysates were sonicated with a Covaris machine (duty cycle, 20%; intensity, 5; cycles/burst, 200; power mode, frequency sweeping) for 30 min at 4°C and incubated with ∼450 μl Sigma EZ view anti-myc affinity gel overnight, and then 100 to 700 bp of 2 input and the corresponding immunoprecipitation (IP) samples along with a wild-type nontagged control were gel purified for library construction by the Genomics High-Throughput Facility at UC Irvine. The library was generated using the BioScientific NextFlex ChiP-Seq kit. ChIP-seq using Illumina HiSeq 2500 was carried out by the UCI Genomics High-Throughput Facility. The sequencing data were analyzed using QuEST (39) followed by visual confirmation by uploading the data to MochiView (40).
Fluorescence microscopy.
Wild-type cells containing either Mal2p-GFP-Cph2 or Mal2p-GFP-Cph2-myc grown in SC–2% glucose overnight were washed with water and then induced in SC–2% maltose with DAPI (4′,6′-diamidino-2-phenylindole) (2.5 μg/ml) for nuclear localization. Cells were collected over a period of 6 h for Western blotting and fluorescence microscopy on an inverted Zeiss Axio Observer.Z1 microscope fluorescent system (Carl Zeiss MicroImaging) using GFP, DAPI, and differential interference contrast (DIC) channels (41). Two hundred cells from each time point were counted, and localization was determined.
Western blotting.
Protein was extracted using a combination of NaOH and β-mercaptoethanol on ice (42) and precipitated with trichloroacetic acid (TCA). The proteins were resuspended and boiled in urea buffer (8 M urea, 4% SDS, 10% β-mercaptoethanol, 125 mM Tris, pH 6.8). The supernatant was combined with an equal volume of 20% glycerol-bromophenol blue before loading on the appropriate SDS-polyacrylamide gel. This method of protein extraction was used for all Cph2 Western blots because Cph2 is readily degraded with regular extraction buffers and not detectable. After blocking, the blot was incubated with a 1:2,000 dilution of BD Living Color GFP monoclonal antibody (Clontech). Anti-c-myc–peroxidase antibody (Roche) was diluted 1:4,000. The secondary antibody was a 1:2,000 dilution of goat anti-mouse antibody (Bio-Rad). For the control blot, a 1:2,000 dilution of anti-cdc2 (PSTAIRE) from Santa Cruz Biotechnology and a 1:2,000 dilution of goat anti-rabbit antibody (Bio-Rad) were used. For blocking and subsequent antibody incubation, 3% milk–phosphate-buffered saline-Tween 20 (PBST) was used at room temperature. After washing, the blots were exposed to SuperSignal West Pico chemiluminescent substrate for 5 min before detection by film.
qPCR.
cDNA was synthesized using iScript reverse transcription Supermix for reverse transcription-quantitative PCR (qPCR) (Bio-Rad) from 1 μg total RNA extracted either with the RNeasy kit (Qiagen) with on-column DNase digestion or with the RiboPure kit (for RNA used in RNA-seq) and subsequently used for qPCR using iQ SYBR green Supermix (Bio-Rad) using a CFX96 Touch real-time PCR detection system (Bio-Rad). CFX Manager software (Bio-Rad) was used to analyze results. Primers for CPH2 (1124 and 1125) and HMS1 (1501 and 1502) were used. For verification of RNA-seq results, qPCR was performed on cDNA synthesized from the first sample of RNA used for RNA-seq. Primers used for qPCR are listed in Table S6 in the supplemental material. Analysis of each sample was performed in triplicate with genomic DNA standards. All results were transferred to Excel (Microsoft), and means and standard deviations were calculated and normalized to ACT1 (990 and 991).
Microarray data accession numbers.
The GEO (Gene Expression Omnibus) accession number for the RNA-seq data is GSE71902, and that for the ChIP-seq data is GSE71728.
RESULTS
Cph2 is a membrane-bound transcriptional factor whose N-terminal DNA binding domain is processed and localized to the nucleus.
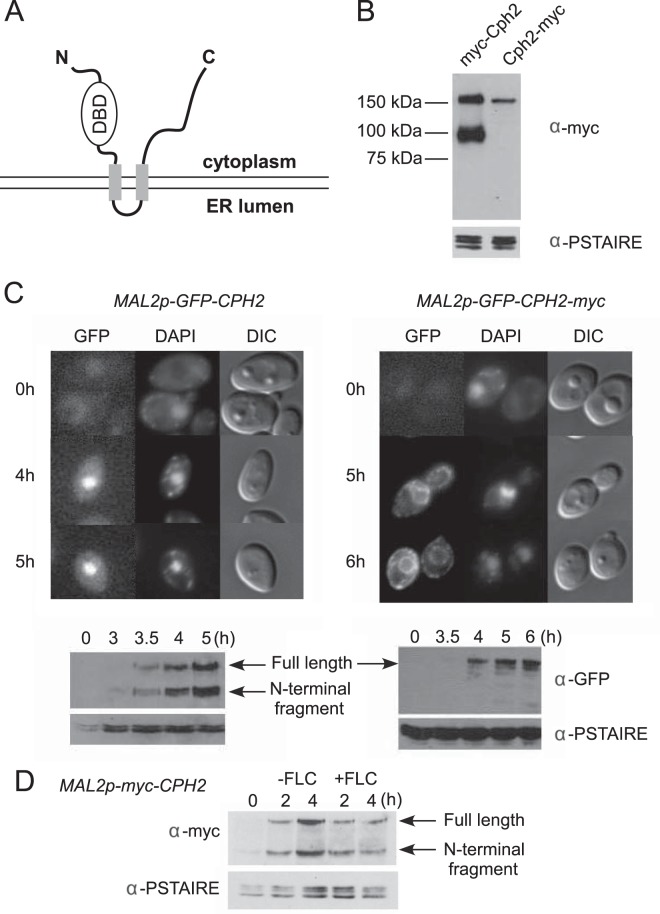
CPH2 encodes a protein of 853 amino acid that has an N-terminal basic helix-loop-helix DNA binding domain similar to that of mammalian sterol response element binding proteins (SREBPs) and two predicted transmembrane segments, like mammalian SREBPs and S. pombe Sre1 (Fig. 1A). The N terminus and C terminus are predicted to be in the cytoplasm. To determine whether Cph2 is proteolytically processed like SREBPs, we tagged the N terminus or the C terminus of Cph2 with myc13 and placed it under control of the CPH2 promoter. By immunoblotting with anti-myc, we were able to detect a full-length (myc-Cph2) product and a processed (myc-Cph2N) product of expected molecular weights with the N-terminally tagged Cph2 (Fig. 1B). In contrast, only a full-length product (Cph2-myc) was detected with the C-terminally tagged Cph2 (Fig. 1B). Therefore, like SREBPs, Cph2 is processed to release the N-terminal transcriptional regulator, but the cleavage of Cph2 can occur under normal growth conditions without the inhibition of sterol biosynthesis or hypoxia.
FIG 1.
Processing and nuclear localization of the Cph2 N-terminal domain from a transmembrane Cph2 protein. (A) Predicted configuration of Cph2 across a membrane. Cph2 has two predicted transmembrane segments (aa 408 to 430 and 501 to 523), with the N terminus and C terminus facing the cytosol. The N terminus contains a DNA binding domain similar to mammalian SREBPs and S. pombe Sre1. (B) Western blot of N- and C-terminally tagged Cph2. Cells of both Cph2p-myc-Cph2 (HLY3927) and Cph2p-Cph2-myc (HLY3829) strains were inoculated into fresh YPD (1:100) at 30°C and grown for 4 h for protein extraction. (C) Cellular localization of GFP-tagged Cph2 and Cph2-myc. Cells carrying Mal2p-GFP-Cph2 (HLY3918) and Mal2p-GFP-Cph2-myc (HLY3919) from overnight in SCD were inoculated into SC–2% maltose. After the indicated hours of growth, cells were collected for fluorescence microscopy and Western analysis. Cells with representative GFP localizations are shown. The C-terminal myc tag seems to have slowed down Cph2 processing and the transition of GFP-Cph2 from the ER to the nucleus. (D) Cph2 processing is not affected by the addition of fluconazole. Wild-type cells containing Mal2p-myc-Cph2 (HLY3968) were grown in YPD overnight at 30°C. Cells were washed 3 times with water, inoculated into YPM at 30°C with or without 10 μM fluconazole (FLC), and collected after 2 and 4 h for Western analysis.
To determine Cph2 localization, we constructed GFP fusions to the N terminus of Cph2 or Cph2-myc and placed the constructs under the control of the MAL2 promoter. The GFP fusions were examined at various times after transferring cells from SCD to SCM medium by immunofluorescence microscopy and Western blotting. In cells carrying the GFP-Cph2 construct, GFP was detected in the nucleus in an increasing number of cells after 4 h in maltose medium (Fig. 1C). By 5 h, 53% of the cells counted had GFP-Cph2 in the nucleus. For cells carrying the GFP-Cph2-myc construct, the GFP signal was seen mostly in the membrane structures surrounding the nucleus and close to the plasma membrane. The subcellular locations of the membrane structures are characteristic of the endoplasmic reticulum (ER). Fifty-four percent of the GFP-Cph2-myc construct was localized to the ER at 5 h. Thus, like mammalian SREBP and S. pombe Sre1, Cph2 protein is a membrane-bound transcriptional factor that is translated on the ER. The GFP signal was seen in the nucleus after overnight culturing in YPM (data not shown). Therefore, having a myc tag at the C terminus seemed to have slowed down the transition of Cph2 through the ER. Western analysis of corresponding cells showed that the timing of GFP-Cph2 detected in the nucleus coincided with the appearance of the processed GFP-Cph2. Moreover, lack of nuclear localization in cells carrying the GFP-Cph2-myc construct was consistent with the lack of processed GFP-Cph2 at 5 to 6 h of induction. Therefore, like SREBPs, Cph2 is a transmembrane protein that is translated on the ER but quickly transitioned into subsequent compartments in the secretion pathway. Like SREBPs, the N-terminal DNA binding domain of Cph2 is processed and translocates to the nucleus, where we believe it functions as a transcriptional regulator. However, unlike the case for Sre1, the N-terminal processing of Cph2 can happen without inhibiting ergosterol biosynthesis or by growth in hypoxia.
Since detection of processed Cph2 in normoxia does not preclude sterol-mediated regulation, we examined whether inhibition of ergosterol biosynthesis with fluconazole could increase the level of Cph2 processing. We found that fluconazole does not increase the level of processed Cph2N (Fig. 1D). Therefore, Cph2 processing in C. albicans is not regulated in response to a change in sterol levels.
ChIP-seq of Cph2 indicates that Cph2 is an integral part of the regulatory circuit with Hms1 and Rtg1/3.
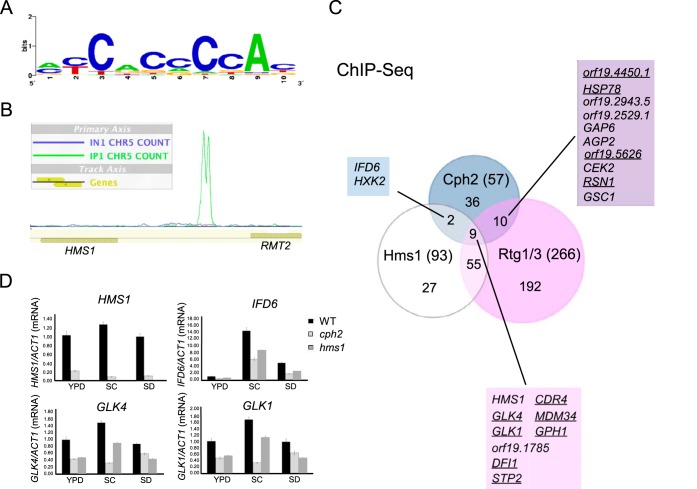
cph2 mutant cells showed reduced levels of colonization in the murine GI tract (9) but were not found to cause reduced virulence in systemic infections (5). To identify direct targets of Cph2, we carried out ChIP-seq in a wild-type strain carrying N-terminal myc-tagged Cph2 (amino acids [aa] 1 to 407) under control of the MAL2 promoter (MAL2-myc-Cph2N). The MAL2-myc-Cph2N construct is functional based on its ability to complement the defect in hyphal development in a cph2 mutant (data not shown). The Cph2N fragment does not contain the transmembrane domains and should be nucleus localized without processing. Cells were grown at 30°C in YEP-maltose medium to an OD600 of around 10, as higher levels of the N-terminal Cph2 were observed in more saturated cells (see Fig. S1 in the supplemental material). IP and input samples from 2 independent experiments, as well as a sample of untagged wild-type control, were sequenced. After initial analysis of the ChIP-seq data using Quest followed by visual confirmation by uploading the data to MochiView, there were a total of 70 peaks identified (see Table S1 in the supplemental material). We have previously shown that Cph2 binds to sterol regulatory element 1 (Sre1)-like elements in vitro (13). With a P value cutoff of 10−3, the Sre1-like motif (Fig. 2A) was found in 56 out of 70 sequences by using the FIMO tool from MEME's suite. For binding at an intergenic region with divergent genes, only one gene is listed in Table S1 in the supplemental material, based on expression data and the closeness of the peak to the coding sequence. After elimination of the intragenic and downstream peaks, 58 genes (83%) showed enrichment in IP versus input in the upstream region (Table 1). orf19.4450.1, which has been shown to be reduced in a cph2 mutant and upregulated in the mouse cecum (9), was identified as a direct target of Cph2 in our ChIP-seq. This validated our ChIP-seq experiment. Of the Cph2 target genes, HMS1 had the highest peak score (Fig. 2B; see Table S1 in the supplemental material). This and the fact that the upstream region of Cph2 is bound by Hms1 and Rtg1/3 (6) prompted us to examine if Cph2 is a part of the regulatory circuit with Hms1 for GI colonization and pathogenesis. Comparison of the Cph2-bound upstream regions to target genes found for Hms1 and other regulators of the regulatory circuit (6) revealed that 50% of Cph2 targets are also bound by Hms1, Rtg1/3, Tye7, or Zcf21 (Table 1). Among them, 19 Cph2 targets are bound by Rtg1/3, 11 by Hms1, 8 by Tye7, and 9 by Zcf21 (Table 1). This suggests that Cph2 is a component of the tightly knit regulatory network. In contrast, Lys144 does not bind any of the Cph2 targets, and Lys14 only binds to only two (IFD6 and orf19.6874). Considering that the DNA binding domains (basic helix-loop-helix domain) of Cph2, Hms1, and Tye7 are all similar to SREBPs with a characteristic tyrosine residue (13), it is possible that they all bind to the same DNA sequences. Unlike SREBPs and Cph2, Hms1 and Tye7 do not contain transmembrane domains. Overlapping genes bound by Cph2, Hms1, and/or Rtg1/3 are shown in Fig. 2C. Interestingly, genes that are upregulated in the mouse gut (9) are all bound by multiple regulators but not Cph2 alone, as shown in Table 1 and by the underlined genes in Fig. 2C. Among the 9 genes bound by all three regulators, 7 are upregulated in the mouse gut. Of the 7 genes, DFI1 encodes a cell surface glycoprotein that is important for colonization of the murine GI tract (6), invasive filamentation into semisolid medium, and virulence in a murine model of disseminated candidiasis (43). GLK1 and GLK4 encode glucokinases that catalyze the phosphorylation of glucose in the first irreversible step of glucose metabolism. HXK2 encodes hexokinase isoenzyme 2, a target of Cph2 and Hms1, that also catalyzes the phosphorylation of glucose. GPH1 encodes a putative glycogen phosphorylase essential for the breakdown of glycogen polysaccharide to glucose-1-phosphate and glucose, which feed into glycolysis. Therefore, carbon metabolism seems to be a common output of Cph2 and the regulatory circuit for GI colonization. This is consistent with the function of Tye7 in regulating carbohydrate metabolism and glycolytic flux (29).
FIG 2.
Hms1 is the top target bound by Cph2 from ChIP-seq. (A) The Cph2 binding motif found in 56 out of 70 Cph2-bound sequences with a P value cutoff of 10−3 when searching with the Sre1-1 motif. (B) The peak of Cph2 binding upstream of HMS1 and RMT2 from MochiView. Green and blue lines show IP and input sequencing counts mapped to the region. (C) Venn diagram to show overlap in genes bound by Cph2 from ChIP-seq of this study and by Hms1 and Rtg1/3 from ChIP-chip determined by Perez et al. (6). The underlined genes are those that are also upregulated in the mouse gut (9). (D) Differential expression of Cph2 targets in the cph2 mutant and in different media. qPCR of the top 4 Cph2-bound genes from the ChIP-seq was performed. Cells of SN250 with ARG4 (HLY4151), cph2 with ARG4 (HLY4152), and hms1 with ARG4 (HLY4153) from a YPD overnight culture were inoculated at 1:100 dilution and grown at 37°C overnight in YPD, SC, and SD. Results were repeated in triplicate and normalized to ACT1.
TABLE 1.
Cph2-bound genes from ChIP-seqa
| No. | Systematic name | Gene name | Bound by: |
Upregulated in gut | Description | |||
|---|---|---|---|---|---|---|---|---|
| Rtg1/3 | Hms1 | Tye7 | Zcf21 | |||||
| 1 | orf19.921 | HMS1 | Y | Y | Y | Y | Transcriptional regulator required for morphogenesis | |
| 2 | orf19.1048 | IFD6 | Y | Y | Aldo-keto reductase family member | |||
| 3 | orf19.6116 | GLK4 | Y | Y | Y | Putative glucokinase | ||
| 4 | orf19.734 | GLK1 | Y | Y | Y | Putative glucokinase | ||
| 5 | orf19.1785 | UNK | Y | Y | Hap43p-repressed gene | |||
| 6 | orf19.7084 | DFI1 | Y | Y | Y | Cell surface-associated glycoprotein | ||
| 7 | orf19.4961 | STP2 | Y | Y | Y | Amino acid-regulated transcription factor | ||
| 8 | orf19.5079 | CDR4 | Y | Y | Y | Y | Putative transporter of ATP binding cassette superfamily | |
| 9 | orf19.542 | HXK2 | Y | Y | Hexokinase II | |||
| 10 | orf19.1826 | MDM34 | Y | Y | Y | Putative transcription factor with zinc finger DNA binding motif | ||
| 11 | orf19.7021 | GPH1 | Y | Y | Y | Putative glycogen phosphorylase with a role in glycogen metabolism | ||
| 12 | orf19.4450.1 | UNK | Y | Y | Protein conserved among the fungal CTG clade | |||
| 13 | orf19.882 | HSP78 | Y | Y | Putative heat shock protein | |||
| 14 | orf19.2943.5 | UNK | Y | Y | ORF, dubious | |||
| 15 | orf19.2529.1 | UNK | Y | Spider biofilm repressed | ||||
| 16 | orf19.6659 | GAP6 | Y | Y | Y | Broad-specificity amino acid permease | ||
| 17 | orf19.4679 | AGP2 | Y | Amino acid permease | ||||
| 18 | orf19.5626 | UNK | Y | Y | Y | Biofilm-induced gene | ||
| 19 | orf19.460 | CEK2 | Y | Mitogen-activated protein kinase required for wild-type efficiency of mating | ||||
| 20 | orf19.347 | RSN1 | Y | Y | Hap43p-repressed gene | |||
| 21 | orf19.2929 | GSC1 | Y | Subunit of beta-1,3-glucan synthase | ||||
| 22 | orf19.727 | UNK | Y | Predicted ORF overlapping major repeat sequence on chromosome R | ||||
| 23 | orf19.6734 | TCC1 | Y | Protein involved in regulation of filamentous growth and virulence | ||||
| 24 | orf19.4476 | UNK | Protein with NADP-dependent oxidoreductase domain, biofilm-induced gene | |||||
| 25 | orf19.5191 | FGR6-1 | Y | Protein lacking an ortholog in S. cerevisiae | ||||
| 26 | orf19.3682 | CWH8 | Putative dolichyl pyrophosphate phosphatase | |||||
| 27 | orf19.6874 | UNK | Putative helix-loop-helix transcription factor with a role in filamentous growth | |||||
| 28 | orf19.7436 | AAF1 | Y | Possible regulatory protein | ||||
| 29 | orf19.3549 | CDC21 | Putative thymidylate synthase | |||||
| 30 | orf19.3231 | CDC27 | Putative ubiquitin-protein ligase | |||||
| 31 | orf19.5005 | OSM2 | Putative mitochondrial fumarate reductase | |||||
| 32 | orf19.2809 | CTN3 | Predicted peroxisomal carnitine acetyltransferase | |||||
| 33 | orf19.5604 | MDR1 | Y | Plasma membrane multidrug efflux pump | ||||
| 34 | orf19.1659 | ALG8 | Putative glucosyltransferase involved in cell wall mannan biosynthesis | |||||
| 35 | orf19.88 | ILV5 | Ketol-acid reductoisomerase | |||||
| 36 | orf19.5762 | PGA61 | Putative glycosylphosphatidylinositol-anchored protein | |||||
| 37 | orf19.33 | LPF1 | Similar to orf19.7550 | |||||
| 38 | orf19.4669 | AAT22 | Protein similar to S. cerevisiae Aat2p | |||||
| 39 | orf19.434 | PRD1 | Putative proteinase | |||||
| 40 | orf19.251 | GLX3 | Glutathione-independent glyoxylase, Spider biofilm induced | |||||
| 41 | orf19.5952 | UNK | Sef1p-, Sfu1p-, and Hap43p-induced protein of unknown function | |||||
| 42 | orf19.6869 | BZD98 | Putative lipid raft-associated protein, Spider biofilm induced | |||||
| 43 | orf19.1313 | CDR3 | Y | Transporter of the Pdrp/Cdrp family of the ATP binding cassette superfamily | ||||
| 44 | orf19.3981 | MAL31 | Y | Putative high-affinity maltose transporter | ||||
| 45 | orf19.6738 | VAN1 | Member of Mnn9p family of mannosyltransferases | |||||
| 46 | orf19.4702 | UNK | Biofilm-induced gene | |||||
| 47 | orf19.4081 | UNK | Dubious ORF | |||||
| 48 | orf19.3490 | FGR6-4 | Y | Protein lacking an ortholog in S. cerevisiae | ||||
| 49 | orf19.6986 | MSB3 | Has domain(s) with predicted Rab GTPase activator activity | |||||
| 50 | orf19.2468 | TMT1 | Ortholog(s) has trans-aconitate 3-methyltransferase activity | |||||
| 51 | orf19.2825 | UNK | Putative cytosolic Fe-S protein assembly protein | |||||
| 52 | orf19.3405 | ZCF18 | Putative transcription factor with zinc finger DNA binding motif | |||||
| 53 | orf19.1690 | TOS1 | Protein similar to alpha agglutinin anchor subunit | |||||
| 54 | orf19.837 | GNA1 | Glucosamine-6-phosphate acetyltransferase | |||||
| 55 | orf19.515 | UNK | Has domain(s) with predicted nucleic acid binding, zinc ion binding activity | |||||
| 56 | orf19.396 | EAF6 | Y | Subunit of the NuA4 histone acetyltransferase complex | ||||
| 57 | orf19.4064 | GPI7 | Protein involved in attachment of glycosylphosphatidylinositol-linked proteins to cell wall | |||||
The list only includes the peaks found in the upstream regions of genes (83% of the total peaks found). Y, yes.
To determine if Cph2-bound genes are transcriptionally regulated by Cph2, expression of the top 4 genes from the ChIP-seq, HMS1, IFD6, GLK4, and GLK1, was confirmed by qPCR, which found them to be differentially regulated in the wild type and the cph2 mutant when grown in YPD, SC, and SD at 37°C for 24 h (Fig. 2D). Whereas HMS1 was highly regulated by Cph2 in all three media, IFD6 was regulated but expressed at a very low level in YPD, more highly expressed in SD, and much more highly expressed in SC medium. Both GLK4 and GLK1 were similarly regulated by Cph2. They were differentially expressed in the wild type versus the cph2 mutant only in SC medium but were not regulated as dramatically in YPD or SD. The expression of IFD6, GLK4, and GLK1 was also impaired to various levels in the hms1 mutant, consistent with their being direct targets of Hms1.
Expression of CPH2 and HMS1 is higher at 30°C and above, and both function downstream of Hsp90 in sustained hyphal elongation.
The HMS1 promoter had the highest peak in the ChIP-seq of Mal2-myc-Cph2N, and HMS1 expression was also dependent on Cph2 (Fig. 2). Interestingly, both Hms1 and Cph2, along with Stp2, are found to be important for filamentation at high temperature or when Hsp90 activity is compromised by geldanamycin A (36). We were interested in the relationship of the expression of CPH2 and HMS1 to temperature and to each other, so their expression was examined by qPCR experiments at 25°C, 30°C, 37°C, and 42°C in YPD in WT, cph2, and hms1 strains (Fig. 3A). We discovered that the expression of both CPH2 and HMS1 was affected by temperature, as their expression was lower at 25°C and highest at 30°C. At 37°C and 42°C, their expression was also elevated in comparison to that at 25°C. HMS1 expression was almost completely dependent on Cph2, as the transcript level was greatly reduced in the cph2 mutant compared to the wild type. There may be Cph2-independent regulation of HMS1 transcription, as there was still some transcript in the cph2 mutant, which increased with temperature (Fig. 3A). In contrast, despite the fact that Hms1 also binds to the upstream region of CPH2 (6), CPH2 expression was not dependent on Hms1 under the in vitro conditions assayed, as the hms1 mutant had only slightly lowered expression of CPH2 (Fig. 3A). Thus, both CPH2 and HMS1 show increased expression at 30°C and above, and for the most part HMS1 expression requires Cph2.
FIG 3.
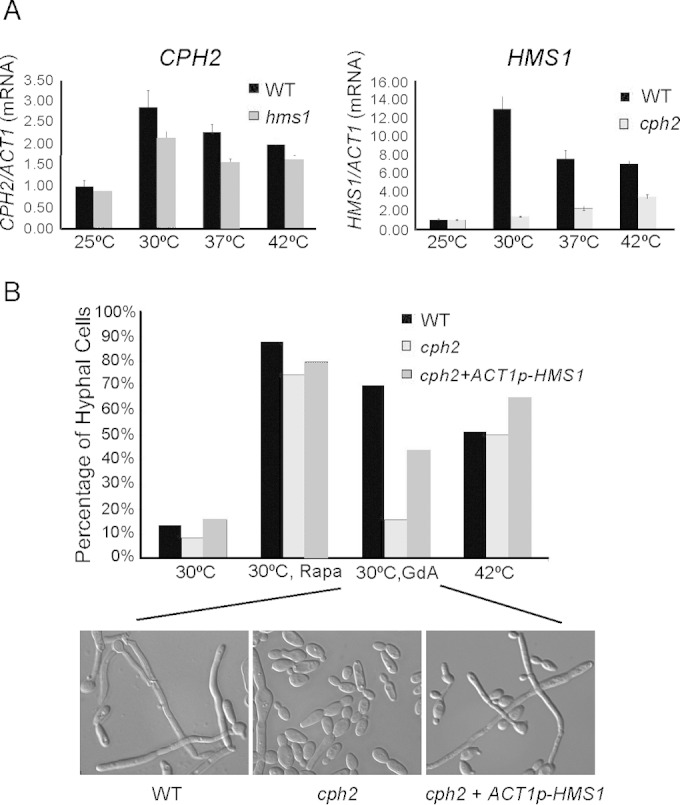
Temperature affects expression of CPH2 and HMS1 and their roles in hyphal maintenance. (A) qPCR of CPH2 and HMS1 at various temperatures and in hms1 and cph2 mutants. SC5314, cph2 (HLY1928), and hms1 (54) strains were inoculated from overnight cultures grown in YPD at 30°C (1:50 dilution) into YPD prewarmed to 25°C, 30°C, 37°C, and 42°C and grown for 4 h. qPCR results were repeated in triplicate and normalized to ACT1. (B) Geldanamycin A-induced filamentation is partially Cph2 dependent. SC5314, cph2 (HLY1928), and cph2 + ACT1p-HMS1 (HLY4149) strains were inoculated into YPD at 37°C (1:100 dilution) for 1 h to initiate hyphae and then switched to 30°C, 30°C with 10 nM rapamycin, 30°C with 10 μM geldanamycin A, or 42°C and grown for 5 h. The percentage of hyphae was determined by counting 200 cells. Pictures were taken with a Zeiss inverted microscope. Cell morphology of only strains grown in 30°C plus geldanamycin A is shown.
Our previous work has demonstrated that Cph2 is involved in regulating hyphal development and that the defect is seen after a long period of time in hyphal growth (13). Therefore, Cph2 is expected to play a role primarily in hyphal maintenance and not in hyphal initiation (44). Consistent with this, the cph2 mutant formed germ tubes in YPD at 37°C for 1 h, just like wild-type cells. To determine if Cph2 is involved in hyphal elongation at high temperature, germ tubes of the wild type and the cph2 mutant were then switched to either 30°C, 30°C with 10 nM rapamycin, 30°C with 10 μM geldanamycin A, or 42°C for an additional 5 h of growth. The wild-type cells converted to yeast growth in YPD at 30°C by 5 h, and rapamycin was able to sustain hyphae under this condition, as shown previously (44, 45). Interestingly, rapamycin also sustained hyphal elongation in the cph2 mutant (Fig. 3B), suggesting that Cph2 does not function downstream of the Tor1 pathway. In contrast, the cph2 mutant displayed a dramatic reduction in the percentage of hyphae compared to the wild type at 30°C with geldanamycin A (Fig. 3B), consistent with the previous notion that the cph2 mutant is impaired in hyphal morphogenesis induced by Hsp90 compromise (36). However, cph2 displayed no impairment in hyphal maintenance at 42°C (Fig. 3B).
To examine whether the primary function of Cph2 in hyphal elongation is to express Hms1, ACT1p-HMS1 was introduced into a cph2 strain to see if ectopically expressed HMS1 would bypass the cph2 defect in geldanamycin A-induced hyphal maintenance. Whereas the wild type maintained 70.3% hyphae and the cph2 mutant only had 15.75% hyphae when induced with geldanamycin A, the cph2 mutant with ACT1p-HMS1 showed restored hyphal maintenance to 44% (Fig. 3B). Thus, HMS1 is able to partially restore the hyphal maintenance defect of the cph2 mutant at 30°C, consistent with the requirement of Cph2 for HMS1 expression at 30°C. Cph2 was not required for hyphal maintenance at 42°C, and HMS1 expression was also less dependent on Cph2 at 42°C (Fig. 3A and B). Unlike the cph2 mutant, the hms1 mutant showed a similar hyphal defect at 30°C with geldanamycin A and at 42°C (36). Therefore, we suggest that Cph2 functions downstream of independent regulation of Hms1 expression and hyphal elongation at 42°C.
RNA-seq links Cph2 function to the expression of OFD1 and other genes differentially regulated in hypoxia.
To identify genes differentially expressed in the cph2 mutant, we sequenced the poly(A) fraction of RNA extracted from replicate wild-type and cph2 mutant cells grown in YPD to an OD600 of ∼10 (see Materials and Methods), expecting to find genes that were differentially regulated in the cph2 mutant versus the wild-type control. Sequence reads from each sample were aligned to the Candida albicans genome sequence (assembly 21) from CGD (46) that were removed of introns (47). Sequence counts for each of the open reading frames (ORFs) were obtained, and the fold change difference between the wild type and mutant were calculated, as well as the P value. Only genes with a P value of less than 0.01 and whose actual wild-type expression value is greater than 20 were included for further analysis. There are a total of 183 genes that met the criteria (see Table S2 in the supplemental material). To validate the RNA-seq results, qPCR was done on 12 genes of the wild-type and cph2 samples from cDNA generated from the original RNA used for RNA-seq as well as additional RNA samples from cells grown for 4 h in YPD at 30°C. All 12 genes were verified by qPCR to be differentially regulated between the wild-type and cph2 strains in YPD (see Table S3 in the supplemental material), which validated our RNA-seq data. Gene ontology (GO) analysis of the 183 differentially regulated genes indicated enrichment in the processes of oxidation-reduction, glycolysis, single-organism metabolism and biosynthesis, generation of precursor metabolites and energy, cofactor metabolism, and thioester and acyl coenzyme A (acyl-CoA) metabolism (see Table S4 in the supplemental material). Eight genes were found by both RNA-seq (183 genes in Table S2 in the supplemental material) and ChIP-seq (58 genes from Table 1). These genes are HMS1, IFD6, orf19.4450.1, orf19.4476, ILV5, orf19.2529.1, CDR4, and CTN3 (shown in bold in Table S2 in the supplemental material). The ChIP-seq represents the genes that are directly regulated by Cph2, but some of the Cph2-targeted genes may not be expressed at a high enough level or display sufficient differential expression in YPD to meet the cutoffs in our data analysis, such as GLK1 and GLK4 (Fig. 2D). It is possible that Cph2-dependent expression of additional Cph2 target genes will be detected if cells are grown under a different growth condition, such as early-log-phase cultures, a different carbon source, or SC medium.
OFD1 is one the genes differentially regulated by Cph2 from the RNA-seq experiment. Ofd1 is a prolyl hydroxylase-like 2-oxoglutarate-Fe(II) dioxygenase (20, 48). This family of enzymes uses oxygen as a substrate to perform hydroxylation of proline residues of a protein, thus providing a means of oxygen-level dependent regulation of their functions. C. albicans Ofd1 acts as an oxygen sensor and regulates hyphal development by promoting the degradation of the hyphal regulator Ume6, in a mechanism similar to that of S. pombe Ofd1 (20, 48). OFD1 expression is higher under hypoxia conditions in both organisms (20, 48). This and the fact that the GI tract is also hypoxic prompted us to compare genes that are differentially regulated by Cph2 and hypoxia. Among the 183 genes differentially regulated by Cph2, 51 are differentially regulated in response to hypoxia based on a previous report of hypoxia-regulated genes (30). These 51 genes are listed in Table 2. The study by Synnott et al. (30) showed that hypoxic induction of the ergosterol pathway is mimicked by treatment with sterol-lowering drugs and requires Upc2. However, increased expression of glycolytic genes and reduced expression of respiratory pathway genes in hypoxia are not regulated by sterol-lowering drugs and Upc2 (30). A comparison of genes differentially regulated by Cph2, Upc2, and hypoxia identified 41 genes that are differentially regulated by both Cph2 and hypoxia but not by Upc2 (Table 2). Eighteen out of 41 genes (43%) are annotated as encoding oxidoreductase by GO analysis. OFD1 belongs to this group. Many of the 41 genes are glycolytic genes induced by hypoxia or genes in the respiratory pathway that are repressed by hypoxia (Table 2). Genes induced by hypoxia showed reduced expression in the cph2 mutant, whereas genes repressed by hypoxia showed higher levels of expression in the cph2 mutant. For example, TDH3, PGK1, GPM1, ENO1, ADH1, ADH5, and ALD5 are upregulated by Cph2 and hypoxia, whereas CIT1 and FUM12 are downregulated by Cph2 and hypoxia (Fig. 4A). In contrast, UPC2 and genes in the ergosterol biosynthesis pathways, such as ERG1, ERG3, ERG5, and ERG11, are among the 56 genes regulated by Upc2 in response to hypoxia or sterol-lowering drugs (30) (Fig. 4A). Cph2 has little effect on the expression of these 56 genes. ERG251 is the only ERG gene upregulated by Cph2 and hypoxia, but it is not regulated by Upc2 (Table 2). Ten genes are regulated by Cph2, hypoxia, and Upc2. Four of them are involved in pathogenesis, biofilm formation, and filamentation, such as BRG1. Four are uncharacterized ORFs. Only 11 genes are differentially expressed in both cph2 and upc2 mutants, from 183 of Cph2- and 532 of Upc2-regulated genes, and with no enrichment in a specific functional class by GO. In conclusion, our RNA-seq data suggest that Cph2 participates in the transcriptional upregulation of glycolytic genes and downregulation of genes in the respiratory pathway that are differentially regulated in response to hypoxia.
TABLE 2.
Cph2-regulated genes from RNA-seq that overlap genes regulated by hypoxia and Upc2
| Condition | Systematic name | Gene name | Description | Fold change, WT/cph2 | Mean log fold changea |
|
|---|---|---|---|---|---|---|
| WT, −O2/+O2 | WT/upc2, −O2 | |||||
| Cph2 + hypoxia | orf19.6882 | OSM1 | Putative flavoprotein subunit of fumarate reductase | −4.709 | 1.74 | |
| orf19.2608 | ADH5 | Putative alcohol dehydrogenase | −4.688 | 2.39 | ||
| orf19.2023 | HGT7 | Putative glucose transporter | −4.133 | 0.90 | ||
| orf19.2344 | ASR1 | Putative heat shock protein | −4.098 | 1.56 | ||
| orf19.7284 | ASR2 | Gene regulated by cyclic AMP and by osmotic stress | −3.912 | 3.62 | ||
| orf19.2241 | PST1 | Putative 1,4-benzoquinone reductase | −3.675 | 1.95 | ||
| orf19.4044 | MUM2 | Protein similar to S. cerevisiae Mum2p | −3.264 | 0.97 | ||
| orf19.3822 | SCS7 | Putative ceramide hydroxylase | −3.248 | 1.67 | ||
| orf19.2803 | HEM13 | Coproporphyrinogen III oxidase | −3.187 | 2.48 | ||
| orf19.1395 | Ortholog(s) has inorganic phosphate transmembrane transporter activity | −3.084 | 1.73 | |||
| orf19.5158 | Protein with similarity to product of a human gene associated with colon cancer and to orf19.5158 | −3.029 | 0.99 | |||
| orf19.4631 | ERG251 | C-4 sterol methyl oxidase with a role in ergosterol biosynthesis | −2.947 | 1.21 | ||
| orf19.3651 | PGK1 | Phosphoglycerate kinase, enzyme of glycolysis | −2.861 | 2.36 | ||
| orf19.5437 | RHR2 | Glycerol 3-phosphatase | −2.736 | 2.85 | ||
| orf19.6321 | PGA48 | Putative glycosylphosphatidylinositol-anchored, adhesin-like protein | −2.605 | 1.08 | ||
| orf19.3707 | YHB1 | Nitric oxide dioxygenase, acts in nitric oxide scavenging/detoxification | −2.604 | −1.61 | ||
| orf19.6770 | ENT4 | Ortholog of Candida dubliniensis CD36, CD36_87190 | −2.575 | 1.11 | ||
| orf19.1862 | Possible stress protein | −2.569 | 3.30 | |||
| orf19.1048 | IFD6 | Aldo-keto reductase family member | −2.560 | 3.27 | ||
| orf19.7027 | Ortholog of C. parapsilosis CDC317, CPAR2_301430 | −2.557 | 0.98 | |||
| orf19.3997 | ADH1 | Alcohol dehydrogenase complements S. cerevisiae adh1 adh2 adh3 mutation; biofilm, fluconazole, and farnesol induced | −2.4875 | 2.22 | ||
| orf19.395 | ENO1 | Enolase, enzyme of glycolysis and gluconeogenesis | −2.3912 | 2.55 | ||
| orf19.5079 | CDR4 | Putative transporter of ATP binding cassette superfamily | −2.3527 | 2.45 | ||
| orf19.1802 | OFD1 | Putative termination and polyadenylation protein | −2.2979 | 1.54 | ||
| orf19.903 | GPM1 | Phosphoglycerate mutase | −2.2705 | 2.55 | ||
| orf19.3139 | Hap43p-repressed gene | −2.0063 | 1.04 | |||
| orf19.1034 | Hap43p-repressed gene | −1.9417 | 1.60 | |||
| orf19.6814 | TDH3 | NAD-linked glyceraldehyde-3-phosphate dehydrogenase | −1.918 | 2.90 | ||
| orf19.3104 | YDC1 | Protein with Mob2p-dependent hyphal regulation | −1.9012 | 1.18 | ||
| orf19.5205 | Hap43p-repressed gene | −1.8474 | 0.83 | |||
| orf19.5801 | RNR21 | Ribonucleoside-diphosphate reductase | −1.6451 | 1.06 | ||
| orf19.339 | NDE1 | Putative NADH dehydrogenase | 1.74861 | −1.08 | ||
| orf19.685 | YHM1 | Putative mitochondrial carrier protein | 1.85585 | −1.01 | ||
| orf19.9 | Ortholog of C. parapsilosis CDC317, CPAR2_204280 | 2.00352 | 0.53 | |||
| orf19.6916 | Ortholog(s) has unfolded protein binding activity | 2.26845 | −1.02 | |||
| orf19.6724 | FUM12 | Putative fumarate hydratase, enzyme of citric acid cycle | 2.30815 | −2.08 | ||
| orf19.5806 | ALD5 | NAD-aldehyde dehydrogenase | 2.97536 | −2.30 | ||
| orf19.183 | HIS3 | Imidazole glycerol-phosphate dehydratase | 3.32823 | −1.86 | ||
| orf19.7112 | FRP2 | Putative ferric reductase | 4.10759 | 1.02 | ||
| orf19.4773 | AOX2 | Alternative oxidase | 4.43443 | −2.22 | ||
| orf19.4393 | CIT1 | Citrate synthase | 5.18316 | −2.10 | ||
| Cph2 + Upc2 + hypoxia | orf19.4450.1 | Protein conserved among the fungal CTG clade | −41.01 | −1.15 | 1.84 | |
| orf19.3548.1 | WH11 | Cytoplasmic protein expressed specifically in white phase yeast cells | −3.577 | 5.64 | 1.36 | |
| orf19.4943 | PSA2 | Mannose-1-phosphate guanyltransferase | −2.951 | 2.13 | 1.22 | |
| orf19.4530.1 | Biofilm- and planktonic growth-induced gene | −2.4962 | 2.16 | 1.35 | ||
| orf19.2770 | Ortholog of C. parapsilosis CDC317, CPAR2_500340 | −2.3585 | 1.08 | 1.48 | ||
| orf19.4476 | Predicted ORF in Assemblies 19, 20 and 21 | −2.0498 | 0.78 | 1.76 | ||
| orf19.4056 | BRG1 | DNA binding transcription factor involved in control of biofilm formation | −1.9173 | 1.62 | 1.16 | |
| orf19.260 | SLD1 | Sphingolipid delta-8 desaturase | −1.7096 | 1.94 | −1.10 | |
| orf19.4506 | LYS22 | Putative homocitrate synthase | 1.9834 | −0.99 | 1.39 | |
| orf19.5741 | ALS1 | Adhesin; ALS family of cell surface glycoproteins | 3.0932 | −1.03 | 0.93 | |
Data are from reference 30.
FIG 4.
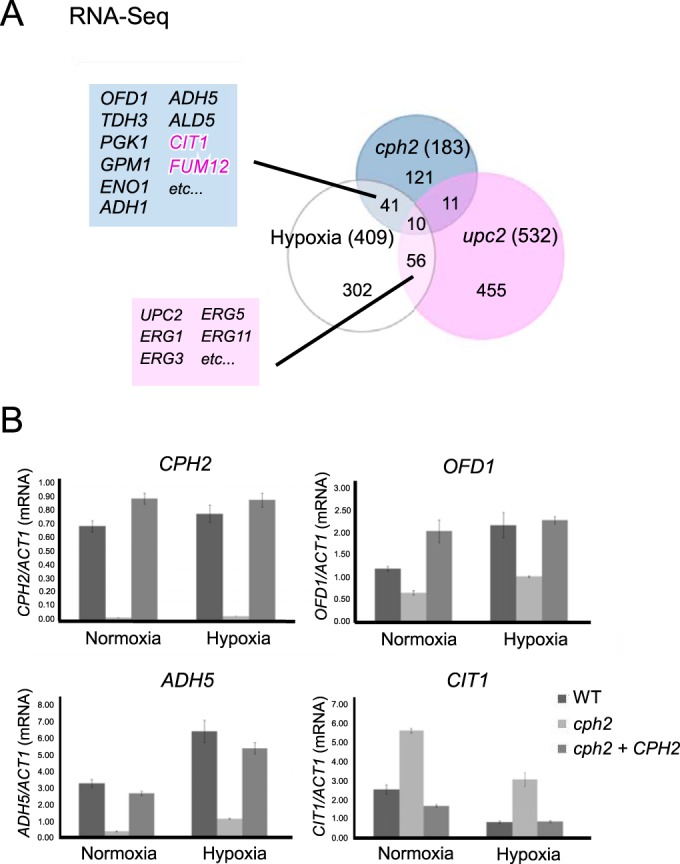
Cph2 is important for the expression of OFD1 and other genes differentially expressed in hypoxia. (A) Venn diagram of Cph2-regulated, Upc2-regulated, and hypoxia-responsive genes. Genes controlled by Cph2 are from RNA-seq in this study (see Table S2 in the supplemental material). Gene sets regulated by Upc2 and hypoxia-responsive genes are from genome-wide ORF microarray analysis (30). A complete list of genes regulated by Cph2 and hypoxia is shown in Table 2. Genes negatively regulated by Cph2 are shown in pink. (B) qPCR analysis of genes regulated by Cph2 in normoxia and hypoxia. SC5314, cph2 (HLY1928), and cph2 + CPH2 (HLY1929) cells were grown in YPD at 37°C for 6 h in air or in 0.2% O2 and 5% CO2 before collection for RNA extraction. CPH2 was used as a control. Results were repeated in triplicate and normalized to ACT1.
We then asked if the hypoxia-responsive expression is mediated through Cph2. CPH2 transcript levels were similar in normoxia and hypoxia (Fig. 4B). In addition, levels of Cph2 protein were similar for cells grown under normoxia and hypoxia (see Fig. S2 in the supplemental material). Therefore, the level of Cph2 under control of its own promoter is not differentially regulated by oxygen levels. Furthermore, Cph2 activity was not responsible for the hypoxia-responsive changes in gene expression, as similar fold changes in gene expression between normoxia and hypoxia were still observed in the cph2 mutant (Fig. 4B). For example, OFD1 and ADH5 expression was upregulated in hypoxia by around 2-fold, as reported previously (30). Their expression was lower in the cph2 mutant, but the hypoxia-induced expression was still observed in the cph2 mutant, with a fold induction similar to that in wild-type cells (Fig. 4B). Similarly, Cph2 was not required for the hypoxia-induced repression of CIT1 expression (Fig. 4B). Therefore, the hypoxia-induced changes in gene expression are not mediated through Cph2.
cph2 upc2 double mutants are unable to grow in hypoxia.
Since Cph2 affected sets of hypoxia-regulated metabolic genes different from those for Upc2 in C. albicans, we then investigated whether the double deletion cph2 upc2 mutant shows a more severe growth defect in hypoxia than any of the single deletion mutants. The cph2 single mutant did not display detectable growth impairment in hypoxia, while the upc2 mutant showed reduced growth in hypoxia (Fig. 5). In contrast, the cph2 upc2 double mutants did not grow at all in hypoxia, and they were able to grow under normoxia conditions (Fig. 5). The genetic interaction between the cph2 and upc2 mutants for growth in hypoxia is consistent with their roles in controlling different sets of hypoxia-regulated metabolic genes. This is also similar to the finding that both Upc2 and SREBP-like Cph2 play a role in response to hypoxic conditions in Y. lipolytica (31).
FIG 5.
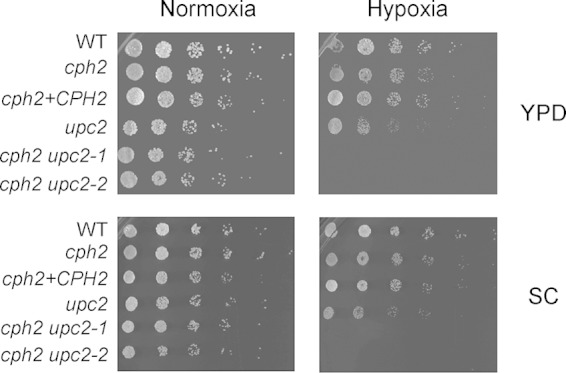
The cph2 upc2 double mutant is completely defective in hypoxic growth. WT (HLY4151), cph2 (HLY4152), cph2+CPH2 (HLY4195), upc2 (54), and two independently obtained cph2 upc2 double mutant strains (HLY4007 and HLY4008) were grown in YPD to an OD600 of ∼1. Cells were diluted to an OD600 of 0.1 and then serially diluted 5-fold, spotted (1.25 μl) onto YPD and SC plates, and grown at 37°C in normoxia or 0.2% O2 and 5% CO2 for a day before pictures were taken.
Ofd1 does not regulate Cph2N stability.
Ofd1 regulates the degradation of the cleaved Sre1 N-terminal domain (Sre1N) in response to oxygen in S. pombe. The Ofd1 N-terminal dioxygenase domain of Ofd1 acts as an oxygen sensor that represses the activity of the C-terminal degradation-promoting domain of Ofd1 in hypoxia (20). Sre1N is rapidly degraded and not detectable in cells grown under normoxia when the Ofd1 C-terminal domain is active. Unlike Sre1N in S. pombe, Cph2N is readily detectable in C. albicans grown under normoxia. However, we observed that the level of Cph2N (expressed under control of its own promoter) is higher in cells from overnight cultures than in cells just inoculated into fresh medium for a few hours of growth (see Fig. S1 and S2 in the supplemental material). To determine if Cph2N is unstable after inoculation into a fresh medium, we used a promoter shutdown experiment with cells carrying the MAL2-myc-CPH2 construct. myc-Cph2 was expressed and processed to myc-Cph2N in cells grown in maltose medium. After cells were transferred into glucose medium, Cph2N rapidly disappeared and was not detected by 60 min (Fig. 6A). Therefore, Cph2N is unstable right after inoculation when cell density is low. Importantly, adding farnesol to the medium blocked Cph2N degradation. Therefore, farnesol stabilizes Cph2N.
FIG 6.
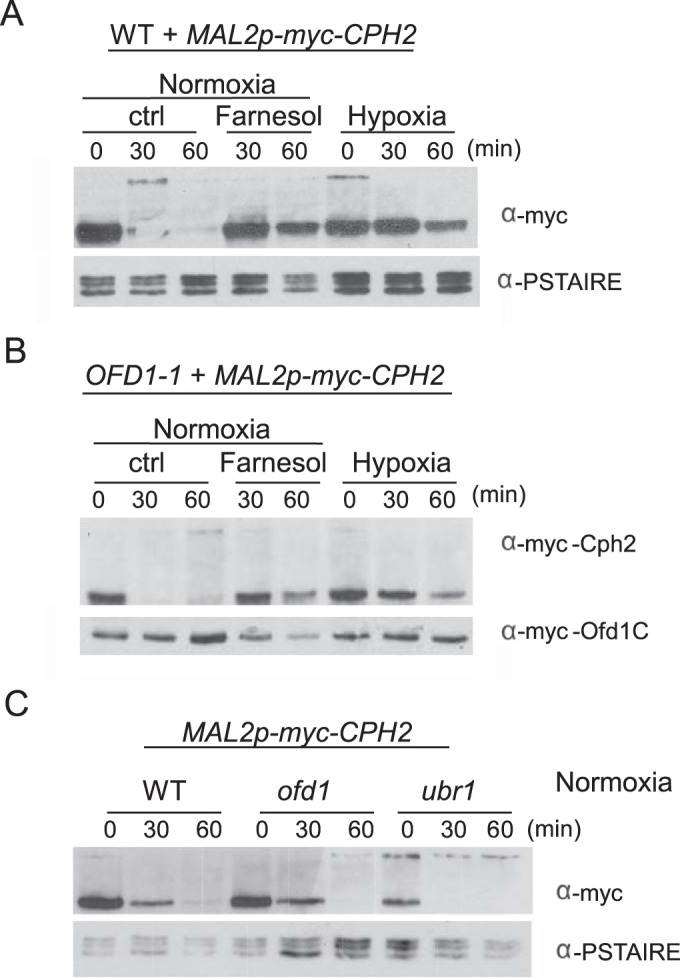
Ofd1 does not regulate Cph2N stability. (A) The N-terminal fragment of Cph2 is stabilized by farnesol. Cells of the WT strain carrying Mal2p-myc-CPH2 (HLY3968) from overnight cultures were inoculated 1:50 into YPM and grown at 30°C for 5 h. The log-phase cells were washed twice with water and inoculated into YPD at 30°C either with or without 40 μM farnesol. For hypoxia (0.2% O2, 5% CO2), cells from overnight culture were washed twice and inoculated 1:50 into YPM and grown for 5 h under 0.2% O2 and 5% CO2 at 30°C before glucose was added. Cells were collected at 30 and 60 min for Western blotting. (B). Ofd1C does not promote Cph2 degradation. WT cells carrying OFD1-1 and MAL2p-myc-CPH2 (HLY4225) were used to determine Cph2 stability under the same conditions as for panel A. (C) The ofd1 and ubr1 deletions do not block Cph2N degradation. WT, ofd1, or ubr1 cells containing Mal2p-myc-CPH2 (HHLY3968, HLY4208, and HLY4224) were used to determine Cph2 stability in air without farnesol as described for panel A.
We also examined whether Cph2 is stable in hypoxia. Cph2 was degraded by 60 min as in normoxia (Fig. 6A, normoxia panel) when cells carrying MAL2-myc-CPH2 were inoculated from a maltose medium into YPD and placed right away into a hypoxic chamber (time zero) (data not shown). However, there is a technical limitation with this procedure, as it takes about 20 min for the hypoxic chamber to reach the O2 level of 0.2. As an alternative approach, cells were inoculated into a maltose medium and grown in hypoxia for 5 h to express myc-Cph2. Cph2N stability in hypoxia was examined by adding glucose to shut down the MAL2 expression. Cph2N was not degraded by 60 min with this procedure (Fig. 6A, hypoxia panel), suggesting that Cph2N was stable in hypoxia. Although C. albicans cells grown anaerobically do not produce farnesol (49), farnesol could still be produced in 0.2% O2 during the 5 h of growth in maltose, which could then prevent Cph2N degradation. Therefore, two different experimental designs gave opposite results. Considering that no difference in the levels of Cph2N was observed when wild-type cells were inoculated into air or hypoxia (see Fig. S2 in the supplemental material), we suggest that Cph2N stability is not regulated by hypoxia.
In parallel, we determined if Ofd1 is involved in Cph2N degradation. We examined Cph2 stability in cells carrying the constitutively active OFD1 (OFD1-1), encoding the C-terminal domain of Ofd1 (48). Because OFD1-1 is active in promoting degradation regardless of oxygen levels, Cph2 is expected to be unstable in hypoxia or in the presence of farnesol if Ofd1 is responsible for promoting Cph2 degradation. However, Cph2N was still detectable after 60 min of growth in farnesol-containing medium or in hypoxia in cells carrying OFD1-1 (Fig. 6B). Furthermore, deletion of OFD1 did not block the degradation of Cph2N in normoxia (Fig. 6C). Therefore, Ofd1 does not regulate Cph2N stability in C. albicans.
In C. albicans, the transcriptional inhibitors Cup9 and Nrg1 are degraded rapidly during inoculation, and both are stabilized by farnesol (50). The Ubr1 ubiquitin ligase mediates the degradation of Cup9 when farnesol is removed (50). The loss of Cup9 allows the expression of Sok1 protein, which is responsible for the degradation of Nrg1 during hyphal initiation (50). We find that Cph2N degradation was not blocked in the ubr1 mutant (Fig. 6C). Since SOK1 is not expressed in the ubr1 mutant because of the stabilization of the Cup9 transcriptional repressor (50), Cph2N degradation is not dependent on the Ubr1 ubiquitin ligase or Sok1. The mechanism and ubiquitin ligase responsible for Cph2 degradation remain to be determined.
DISCUSSION
Cph2 processing is not regulated by the ergosterol pathway.
The N-terminal basic helix-loop-helix DNA binding domain of Cph2 is similar to those of mammalian SREBPs and fungal Sre1 proteins (12). Also, like the SREBPs and Sre1s, Cph2 is a membrane-bound transcription factor that is processed to release the N-terminal DNA binding domain, which translocates to the nucleus. The proteolytic activation/processing of SREBPs and Sre1 proteins is regulated in response to cellular levels of sterols (12). Membrane sterol levels are monitored by the membrane-embedded protein Scap (SREBP cleavage-activating protein) of the ER through its sterol-sensing domain, a polytopic intramembrane sequence (14). When cells are depleted of sterol, Scap dissociates from the ER membrane protein Insig and escorts SREBPs from the ER to the Golgi apparatus, where the SREBPs are proteolytically processed to yield active fragments that enter the nucleus. Sterol blocks the entry of SREBP-Scap into ER transport vesicles, as the ER membrane protein Insig associates with Scap in the presence of high sterol levels and retains the SREBP-Scap in the ER. We show that Cph2 processing is not regulated by cellular ergosterol levels in C. albicans. This is consistent with the lack of a Scap-like protein in C. albicans (12), and ergosterol biosynthesis genes are regulated by Upc2. Like Cph2, fission yeast Sre2 contains the conserved tyrosine residue in its DNA binding domain and two predicted transmembrane segments, and Sre2 is cleaved constitutively in a sterol-independent manner (15). The sterol-independent cleavage raises questions as to why Cph2 and Sre2 are associated with the ER membrane when their release from the membrane is not regulated in response to ergosterol levels. Further studies will be required to determine what cellular signals can inhibit/regulate their processing.
Cph2 functions as an integral part of the regulatory circuit with Hms1 and Rtg1/3 for GI tract colonization.
Cph2 has been shown to be important for GI colonization (9). By ChIP-seq we find that the HMS1 upstream region is the highest-ranked Cph2 target, and HMS1 expression is Cph2 dependent under our assay conditions. The expression of both CPH2 and HMS1 is under Hsp90 regulation and higher at body temperature than at room temperature. Fifty percent of Cph2 targets from our ChIP-seq are also targets of Hms1, Rtg1/3, Tye7, or Zcf21 of the GI regulatory circuit, and the upstream region of Cph2 is also bound by Hms1 and Rtg1/3 (6). Most of the genes bound by Cph2, Hms1, and Rtg1/3 are upregulated in the mouse gut. Most of the genes are involved in glycolysis, consistent with the reported function of Tye7 in regulating carbohydrate metabolism and glycolytic flux (29). Therefore, we suggest that Cph2 is an integral part of the regulatory circuit with Hms1 for GI tract colonization. By RNA-seq, we find that a high percentage of Cph2-regulated genes are differentially regulated in response to hypoxia, and many belong to oxidoreductases by GO analysis. OFD1 is one of them. Therefore, Cph2 participates in the expression of glycolytic genes induced by hypoxia or genes in the respiratory pathway that are repressed by hypoxia. The involvement of Cph2 in hypoxia-responsive expression of genes in glycolysis and respiration explains its importance for C. albicans colonization in the GI tract.
Oxygen sensing in C. albicans.
Molecular mechanisms for how C. albicans senses levels of oxygen and adjusts its transcriptional program for growth under hypoxia are not clear despite the identification of several transcription factors as being involved in hypoxia-responsive expression of genes for glycolysis and ergosterol biosynthesis (28–30, 51). For example, Upc2 is responsible for the increased expression of ergosterol biosynthesis genes under hypoxia (24, 26, 30). However, Upc2 may not directly sense the oxygen level; instead it is activated by ergosterol depletion in hypoxia, as many steps of ergosterol biosynthesis are oxygen dependent. The C-terminal domain of Upc2 has a hydrophobic pocket for ergosterol binding; dissociation from ergosterol leads to its nuclear localization and transcription activation (52). Alternatively, the hypoxic response was suggested to be initiated by a drop in ATP production from the oxidative phosphorylation chain (51). In S. pombe, Sre1N, the regulator of ergosterol biosynthesis, is stabilized in cells grown under hypoxia but is rapidly degraded under normoxia. Sre1N degradation is regulated by Ofd1, which is a prolyl 4-hydroxylase-like 2-oxoglutarate-Fe(II) dioxygenase and can sense oxygen levels directly (20). C. albicans Ofd1 also acts as an oxygen sensor. Instead of regulating the expression of ergosterol biosynthesis genes, it regulates hyphal development by promoting the degradation of the hyphal regulator Ume6, in a mechanism similar to that of the S. pombe Ofd1 (20, 48). Stability of Cph2N is not regulated by Ofd1, consistent with our conclusion that hypoxia-induced changes in gene expression are not mediated through Cph2. However, we do not know if Ofd1 regulates hypoxia-responsive gene expression. Further studies need to elucidate if Ofd1 and/or other 2-oxoglutarate-dependent oxygenases regulate hypoxic gene expression in C. albicans as in higher eukaryotes (53).
Supplementary Material
ACKNOWLEDGMENTS
We thank Suzanne Noble, Alexander Johnson, the Fungal Genetics Stock Center, and Leah Cowen for plasmids and C. albicans strains. We thank members of the Liu lab for helpful discussions and Melanie Oakes for help with sequencing.
This work was supported by National Institutes of Health grants R01GM/AI55155 and R01AI099190 to H.L. This work was made possible, in part, through access to the Genomics High Throughput Facility Shared Resource of the Cancer Center Support Grant (CA-62203) at the University of California, Irvine.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00102-15.
REFERENCES
- 1.Odds FC. 1988. Candida and candidosis, 2nd ed Bailliere Tindall, Philadepphia, PA. [Google Scholar]
- 2.Zaoutis TE, Argon J, Chu J, Berlin JA, Walsh TJ, Feudtner C. 2005. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis 41:1232–1239. doi: 10.1086/496922. [DOI] [PubMed] [Google Scholar]
- 3.Koh AY, Kohler JR, Coggshall KT, Van Rooijen N, Pier GB. 2008. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog 4:e35. doi: 10.1371/journal.ppat.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odds FC, Davidson AD, Jacobsen MD, Tavanti A, Whyte JA, Kibbler CC, Ellis DH, Maiden MC, Shaw DJ, Gow NA. 2006. Candida albicans strain maintenance, replacement, and microvariation demonstrated by multilocus sequence typing. J Clin Microbiol 44:3647–3658. doi: 10.1128/JCM.00934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noble SM, French S, Kohn LA, Chen V, Johnson AD. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet 42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez JC, Kumamoto CA, Johnson AD. 2013. Candida albicans commensalism and pathogenicity are intertwined traits directed by a tightly knit transcriptional regulatory circuit. PLoS Biol 11:e1001510. doi: 10.1371/journal.pbio.1001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Pande K, French SD, Tuch BB, Noble SM. 2011. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 10:118–135. doi: 10.1016/j.chom.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pande K, Chen C, Noble SM. 2013. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet 45:1088–1091. doi: 10.1038/ng.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenbach A, Dignard D, Pierce JV, Whiteway M, Kumamoto CA. 2010. Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryot Cell 9:1075–1086. doi: 10.1128/EC.00034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pierce JV, Dignard D, Whiteway M, Kumamoto CA. 2013. Normal adaptation of Candida albicans to the murine gastrointestinal tract requires Efg1p-dependent regulation of metabolic and host defense genes. Eukaryot Cell 12:37–49. doi: 10.1128/EC.00236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierce JV, Kumamoto CA. 2012. Variation in Candida albicans EFG1 expression enables host-dependent changes in colonizing fungal populations. mBio 3:e00117-12. doi: 10.1128/mBio.00117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bien CM, Espenshade PJ. 2010. Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryot Cell 9:352–359. doi: 10.1128/EC.00358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lane S, Zhou S, Pan T, Dai Q, Liu H. 2001. The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1. Mol Cell Biol 21:6418–6428. doi: 10.1128/MCB.21.19.6418-6428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstein JL, DeBose-Boyd RA, Brown MS. 2006. Protein sensors for membrane sterols. Cell 124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 15.Hughes AL, Todd BL, Espenshade PJ. 2005. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA Jr. 2008. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog 4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. 2007. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol Microbiol 64:614–629. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- 18.Hughes BT, Nwosu CC, Espenshade PJ. 2009. Degradation of sterol regulatory element-binding protein precursor requires the endoplasmic reticulum-associated degradation components Ubc7 and Hrd1 in fission yeast. J Biol Chem 284:20512–20521. doi: 10.1074/jbc.M109.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porter JR, Burg JS, Espenshade PJ, Iglesias PA. 2010. Ergosterol regulates sterol regulatory element binding protein (SREBP) cleavage in fission yeast. J Biol Chem 285:41051–41061. doi: 10.1074/jbc.M110.144337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes BT, Espenshade PJ. 2008. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J 27:1491–1501. doi: 10.1038/emboj.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee CY, Stewart EV, Hughes BT, Espenshade PJ. 2009. Oxygen-dependent binding of Nro1 to the prolyl hydroxylase Ofd1 regulates SREBP degradation in yeast. EMBO J 28:135–143. doi: 10.1038/emboj.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies BS, Wang HS, Rine J. 2005. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: similar activation/regulatory domains but different response mechanisms. Mol Cell Biol 25:7375–7385. doi: 10.1128/MCB.25.16.7375-7385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vik A, Rine J. 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol 21:6395–6405. doi: 10.1128/MCB.21.19.6395-6405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silver PM, Oliver BG, White TC. 2004. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot Cell 3:1391–1397. doi: 10.1128/EC.3.6.1391-1397.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagi M, Nakayama H, Tanabe K, Bard M, Aoyama T, Okano M, Higashi S, Ueno K, Chibana H, Niimi M, Yamagoe S, Umeyama T, Kajiwara S, Ohno H, Miyazaki Y. 2011. Transcription factors CgUPC2A and CgUPC2B regulate ergosterol biosynthetic genes in Candida glabrata. Genes Cells 16:80–89. doi: 10.1111/j.1365-2443.2010.01470.x. [DOI] [PubMed] [Google Scholar]
- 26.Guida A, Lindstadt C, Maguire SL, Ding C, Higgins DG, Corton NJ, Berriman M, Butler G. 2011. Using RNA-seq to determine the transcriptional landscape and the hypoxic response of the pathogenic yeast Candida parapsilosis. BMC Genomics 12:628. doi: 10.1186/1471-2164-12-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacPherson S, Akache B, Weber S, De Deken X, Raymond M, Turcotte B. 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob Agents Chemother 49:1745–1752. doi: 10.1128/AAC.49.5.1745-1752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Setiadi ER, Doedt T, Cottier F, Noffz C, Ernst JF. 2006. Transcriptional response of Candida albicans to hypoxia: linkage of oxygen sensing and Efg1p-regulatory networks. J Mol Biol 361:399–411. doi: 10.1016/j.jmb.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 29.Askew C, Sellam A, Epp E, Hogues H, Mullick A, Nantel A, Whiteway M. 2009. Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog 5:e1000612. doi: 10.1371/journal.ppat.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Synnott JM, Guida A, Mulhern-Haughey S, Higgins DG, Butler G. 2010. Regulation of the hypoxic response in Candida albicans. Eukaryot Cell 9:1734–1746. doi: 10.1128/EC.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maguire SL, Wang C, Holland LM, Brunel F, Neuveglise C, Nicaud JM, Zavrel M, White TC, Wolfe KH, Butler G. 2014. Zinc finger transcription factors displaced SREBP proteins as the major Sterol regulators during Saccharomycotina evolution. PLoS Genet 10:e1004076. doi: 10.1371/journal.pgen.1004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White SJ, Rosenbach A, Lephart P, Nguyen D, Benjamin A, Tzipori S, Whiteway M, Mecsas J, Kumamoto CA. 2007. Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog 3:e184. doi: 10.1371/journal.ppat.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang A, Lane S, Tian Z, Sharon A, Hazan I, Liu H. 2007. Temporal and spatial control of HGC1 expression results in Hgc1 localization to the apical cells of hyphae in Candida albicans. Eukaryot Cell 6:253–261. doi: 10.1128/EC.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cormack BP, Bertram G, Egerton M, Gow NA, Falkow S, Brown AJ. 1997. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 35.Lu Y, Su C, Mao X, Raniga PP, Liu H, Chen J. 2008. Efg1-mediated recruitment of NuA4 to promoters is required for hypha-specific Swi/Snf binding and activation in Candida albicans. Mol Biol Cell 19:4260–4272. doi: 10.1091/mbc.E08-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shapiro RS, Sellam A, Tebbji F, Whiteway M, Nantel A, Cowen LE. 2012. Pho85, Pcl1, and Hms1 signaling governs Candida albicans morphogenesis induced by high temperature or Hsp90 compromise. Curr Biol 22:461–470. doi: 10.1016/j.cub.2012.01.062. [DOI] [PubMed] [Google Scholar]
- 37.Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lefrancois P, Zheng W, Snyder M. 2010. ChIP-Seq using high-throughput DNA sequencing for genome-wide identification of transcription factor binding sites. Methods Enzymol 470:77–104. doi: 10.1016/S0076-6879(10)70004-5. [DOI] [PubMed] [Google Scholar]
- 39.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. 2008. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods 5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Homann OR, Johnson AD. 2010. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol 8:49. doi: 10.1186/1741-7007-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mochon AB, Liu H. 2008. The antimicrobial peptide histatin-5 causes a spatially restricted disruption on the Candida albicans surface, allowing rapid entry of the peptide into the cytoplasm. PLoS Pathog 4:e1000190. doi: 10.1371/journal.ppat.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kornitzer D, Raboy B, Kulka RG, Fink GR. 1994. Regulated degradation of the transcription factor Gcn4. EMBO J 13:6021–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zucchi PC, Davis TR, Kumamoto CA. 2010. A Candida albicans cell wall-linked protein promotes invasive filamentation into semi-solid medium. Mol Microbiol 76:733–748. doi: 10.1111/j.1365-2958.2010.07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Y, Su C, Wang A, Liu H. 2011. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol 9:e1001105. doi: 10.1371/journal.pbio.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Y, Su C, Liu H. 2012. A GATA transcription factor recruits Hda1 in response to reduced Tor1 signaling to establish a hyphal chromatin state in Candida albicans. PLoS Pathog 8:e1002663. doi: 10.1371/journal.ppat.1002663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van het Hoog M, Rast TJ, Martchenko M, Grindle S, Dignard D, Hogues H, Cuomo C, Berriman M, Scherer S, Magee BB, Whiteway M, Chibana H, Nantel A, Magee PT. 2007. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol 8:R52. doi: 10.1186/gb-2007-8-4-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De La Vega FM, Johnson AD. 2010. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet 6:e1001070. doi: 10.1371/journal.pgen.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y, Su C, Solis NV, Filler SG, Liu H. 2013. Synergistic regulation of hyphal elongation by hypoxia, CO2, and nutrient conditions controls the virulence of Candida albicans. Cell Host Microbe 14:499–509. doi: 10.1016/j.chom.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dumitru R, Hornby JM, Nickerson KW. 2004. Defined anaerobic growth medium for studying Candida albicans basic biology and resistance to eight antifungal drugs. Antimicrob Agents Chemother 48:2350–2354. doi: 10.1128/AAC.48.7.2350-2354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu Y, Su C, Unoje O, Liu H. 2014. Quorum sensing controls hyphal initiation in Candida albicans through Ubr1-mediated protein degradation. Proc Natl Acad Sci U S A 111:1975–1980. doi: 10.1073/pnas.1318690111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sellam A, van het Hoog M, Tebbji F, Beaurepaire C, Whiteway M, Nantel A. 2014. Modeling the transcriptional regulatory network that controls the early hypoxic response in Candida albicans. Eukaryot Cell 13:675–690. doi: 10.1128/EC.00292-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang H, Tong J, Lee CW, Ha S, Eom SH, Im YJ. 2015. Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2. Nat Commun 6:6129. doi: 10.1038/ncomms7129. [DOI] [PubMed] [Google Scholar]
- 53.Ploumakis A, Coleman ML. 2015. Oh, the places you'll go! Hydroxylation, gene expression and cancer. Mol Cell 58:729–741. doi: 10.1016/j.molcel.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 54.Homann OR, Dea J, Noble SM, Johnson AD. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet 5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.