Abstract
An assessment of roles of rhizospheric microbial diversity in plant growth is helpful in understanding plant-microbe interactions. Using random combinations of rhizospheric bacterial species at different richness levels, we analysed the contribution of species richness, compositions, interactions and identity on soil microbial respiration and plant biomass. We showed that bacterial inoculation in plant rhizosphere enhanced microbial respiration and plant biomass with complementary relationships among bacterial species. Plant growth was found to increase linearly with inoculation of rhizospheric bacterial communities with increasing levels of species or plant growth promoting trait diversity. However, inoculation of diverse bacterial communities having single plant growth promoting trait, i.e., nitrogen fixation could not enhance plant growth over inoculation of single bacteria. Our results indicate that bacterial diversity in rhizosphere affect ecosystem functioning through complementary relationship among plant growth promoting traits and may play significant roles in delivering microbial services to plants.
Microbial diversity in soil plays critical roles in ecosystem functioning including a large number of important processes like nutrient cycling1,2,3, promotion of plant growth and health4,5. These services are indispensable in agriculture and may enhance productivity in a sustainable manner. However, the functional role of microbial diversity in relation to plant productivity is poorly understood. Studying the functioning of rhizospheric microbial diversity for enhancing plant productivity will be helpful in understanding plant-microbe interactions. Diverse communities may use resources more efficiently and are more productive and stable under mild environmental fluctuations than monocultures6,7. Positive relationships between microbial diversity and ecosystem functioning have already been established where productivity, stability and resistance to biotic invasion increases at higher levels of diversity8,9,10,11. However, negative interactions in microbial communities are not uncommon12 and increasing frequency of antagonistic interactions at higher levels of species richness may result in the collapse of the whole community13.
Beneficial effects of biodiversity in microbial communities may enhance their survival and fitness and result in an overall increase in microbial biomass. Rhizospheric bacteria enhance plant productivity through various plant growth promoting activities such as nitrogen fixation, increasing the availability of essential nutrients, hormonal modulation and antagonising the growth of deleterious microbes14. Functional diversity is an important determinant of ecosystem processes15. The relationship between phylogenetic diversity and ecosystem functioning indicates that related organisms may share functional traits16,17. However, the distribution of functional traits in bacterial taxa is not fully understood and becomes complicated due to horizontal gene transfer and high evolutionary change18,19.
In a heterogeneous environment, biodiversity can increase ecosystem functioning through niche partitioning, habitat modification and direct facilitation to other organisms. For example, nitrogen fixers may provide extra nitrogen to other microbes, thus increasing their population, which, in turn, may facilitate nitrogen fixation through increasing availability of other essential nutrients like phosphorus. Here we study the functioning of microbial diversity in the rhizosphere and its role in plant growth. Bacterial strains were isolated from rhizospheric soil of Ocimum sanctum L. (Lamiaceae) on the basis of their common occurrence in the plant rhizosphere and their fast growing capability on nutrient agar medium. We hypothesized that the diversity in plant growth promoting traits in a bacterial community will be beneficial to plant growth, and functionally diverse bacterial community will promote more plant growth than simplified community.
Materials and Methods
Isolation and molecular characterization of rhizospheric bacteria
Seeds of Ocimum sanctum (cv. CIM-Angana) were obtained from the National Gene Bank for Medicinal and Aromatic Plants at CSIR-Central Institute of Medicinal and Aromatic plants (CSIR-CIMAP), Lucknow, India. Surface sterilized seeds were grown in organic farming experimental fields. Six weeks old healthy plants were uprooted and the soil firmly attached to the roots was collected after removing loosely adhered soil by shaking vigorously. One gram of homogenised soil sample was serially diluted in saline water (0.85%) and aliquot of 100 μL was spread on nutrient agar plates (peptic digest of animal tissue, 5 gL−1; beef extract, 1.5 gL−1; yeast extract, 1.5 gL−1; NaCl, 5 gL−1; agar 15 gL−1; HiMedia Laboratories India). After incubation for 2–7 days at 28 °C, the resulting conspicuous colonies were picked and purified by repeatedly streaking on nutrient agar. More than 50 bacterial isolates were purified and stored at −80 °C in 10% glycerol. Twenty-three bacterial strains were selected on the basis of their fast growing habit and with different colony characteristics. Isolates were examined for their plant growth promoting activities such as nitrogen fixation20, phosphate solubilization21 and production of indole acetic acid22, cellulase23 and siderophores24. Sixteen positive isolates showing at least one functional activity were selected for present study (Table 1). The selected bacterial strains were characterized by 16S rRNA gene (rDNA) sequence analysis as done earlier20. These strains were assembled in two groups. Ten phylogenetically diverse nitrogen fixing isolates were used for constructing communities of nitrogen-fixing bacteria (hereafter called as nitrogen-fixing bacteria). Twelve phylogenetically diverse but possessing different functional activities (six nitrogen fixers, two phosphate solubilizers, two indole acetic acid producers, one cellulose producer and one siderophore producer) were considered for constituting communities in case of phylogenetically as well as functionally diverse communities of rhizospheric bacteria (hereafter called as rhizospheric bacteria; Table S1a and S1b in Supplementary Information).
Table 1. Bacterial species used in the study, as identified through 16 s rRNA gene sequence analysis.
| Sp. Id | Closest NCBI match | *NCBI Accession number | Functional activity |
|---|---|---|---|
| An | Klebsiella pneumoniae | JX512025 | Nitrogen fixer |
| B | Staphylococcus pasteuri | JX512027 | IAA Producer |
| Cn | Bacillus cereus | JX512029 | Nitrogen fixer |
| D | Brevibacillus agri | JX512031 | Phosphate solubilizer |
| E | Bacillus subtilis | JX512032 | Phosphate solubilizer |
| Fn | Pseudomonas putida | JX512033 | Nitrogen fixer |
| Gn | Bacillus megaterium | JX512035 | Nitrogen fixer |
| Hn | Enterobacter sp. | JX512036 | Nitrogen fixer |
| I | Cronobacter sakazakii | JX512037 | Siderophore producer |
| J | Pantoea agglomerans | JX512038 | Cellulase producer |
| Kn | Alcaligenes sp. | JX512039 | Nitrogen fixer |
| L | Micrococcus sp. | JX512040 | IAA Producer |
| Mn | Bacillus thuringiensis | JX512028 | Nitrogen fixer |
| Nn | Bacillus firmus | JX512034 | Nitrogen fixer |
| On | Pseudomonas rhizosphaerae | JX512030 | Nitrogen fixer |
| Pn | Flexibacter sp. | JX512026 | Nitrogen fixer |
Sequences have been submitted to NCBI GenBank. Please refer to Supplementary Information for separate list of species used in two experiments.
n denotes the strains which were included in nitrogen fixing bacterial group.
Experimental design and treatments
One-month-old seedlings obtained from germinating surface sterilized seeds under sterile conditions were taken for study. The potting material consisted of soil: sand (2:1) mixture sieved through 2.5 mm mesh and then autoclaved for 3 h at 121 °C in three consecutive cycles within 48 hours. The soil was sandy loam (Ustifluvent) with pH, 7.19; electrical conductivity, 0.41 dS m−1; organic carbon, 4.47 g kg−1; available N (alkaline permanganate extractable), 123 kg ha−1; available P (0.46 M NaHCO3 extractable), 9.8 kg ha−1; and available K (1 N NH4OAc extractable), 95 kg ha−1.
Bacterial species were assembled in various combinations with different diversity gradients (1, 2, 3, 4, 6, 12 for rhizospheric bacteria and 1, 2, 5, 10 for nitrogen-fixing bacteria) through random samplings17. At all richness levels, species combinations were made randomly without replacement such that each species is selected equally at each of the richness level (Supplementary Information Tables S2 and S3). The process of constructing a set of experimental units was carried out independently three times at each richness level, and each of the combination was replicated three times. In addition, un-inoculated control with three replications (without any bacterial inoculation) was also included.
Bacterial cultures were prepared by growing them in 10 ml nutrient broth for 24 h in an orbital incubator shaker (200 rpm at 28 °C), harvesting cells by centrifuging at 8000 × g for 5 min at 4 °C and diluting their pellets with sterile saline water (0.85%) to a CFU (colony forming units) of ∼108 mL−1. CFU was determined by plating serially diluted 0.1 ml culture on nutrient agar plates after incubating for 24 hours. For preparing inoculum of different combinations, an equal quantity of each bacterial culture was mixed in a sterile vial so that total individual density in each combination remains similar. Roots of one-month-old seedlings were dipped into bacterial culture for one hour and transplanted into the pots (20 cm high and 20 cm internal diameter). Additionally, 2 ml of culture (108 CFU mL−1) containing appropriate bacterial combinations in equal amounts was placed into the planting hole. To maintain uniformity, control plants were inoculated with autoclaved saline water. The pots were kept in a glass house and were irrigated with sterilized water as and when required.
Measurements of plant biomass, soil microbial respiration
Plants were harvested 60 days after transplanting (at the initiation of flowering), and total biomass of oven dried plant material was recorded. Microbial respiration was estimated from the potting soil sieved through 2 mm mesh after manually removing plant residues. A sample of 50 g soil was kept in airtight glass jars with 1N NaOH in a separate tube. Jars were incubated for 10 days at 28 °C. CO2 evolution by microbial respiration was estimated through titrating the NaOH with 1N HCl and expressed as g CO2 kg−1 day−1, based on dry weight25.
Statistical analyses
The relationship between response variables (plant biomass, soil microbial respiration) and predicting variables (species richness, functional group richness) was analyzed through linear regression. Pairwise differences between individual species and between different levels of species richness were determined with one-way ANOVA with Tukey’s post hoc test. In a particular composition, if monocultures didn’t differ in performance, transgressive overyielding was not calculated and was reported as zero. When differences among monocultures were significant at P < 0.05 level, transgressive overyielding was calculated as the difference between mixture yield and yield of most performing species present in the mixture.
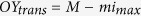 |
where OYtrans is transgressive overyielding and mimax is the maximum monoculture yield.
The effects of species richness (linear and fixed factor), species identity and compositions were analysed with general linear models in R version 3.1.0 (http://www.r-project.org) following Bell et al.26. The advantage of the separation of species richness into linear and fixed factor is that once linear richness has been entered into the model, the effect of species identity and the effect of nonlinear species richness are orthogonal (i.e., do not share sums of squares). In this analysis, the nonlinear richness explains the effect of species interaction on the response variable. For avoiding the problem of pseudoreplication, F-statistic was calculated with specific error terms. The effects of species richness were tested against partitioned species pools and the effects of species identities against compositions. Diagnostic plots of residuals versus fitted values revealed the lack of significant heterogeneity of variance and Q-Q plots showed that assumptions of normality were justified. Cook’s distances were tested to examine the level of influence of extreme data points. No data point was noticed influential to change the output of analyses and interpretation of results.
Results
Plant biomass varied on inoculations with different bacteria in monocultures (Fig. 1a). Although all the bacterial species were found to express plant growth promoting activity in vitro, only 50% of them significantly increased the plant biomass as compared to control. Two bacteria i.e. Klebsiella pneumoniae and Brevibacillus agri were found to be better than others in increasing plant growth in monoculture. On the other hand, 60% of nitrogen-fixing bacteria were found to be plant growth promoters in pot experiments (Fig. 1a). Soil microbial respiration was not significantly different among species monocultures, and it was also not different from control suggesting the species individually couldn’t increase the microbial respiration over control as analysed through our protocol (Fig. 1b).
Figure 1.
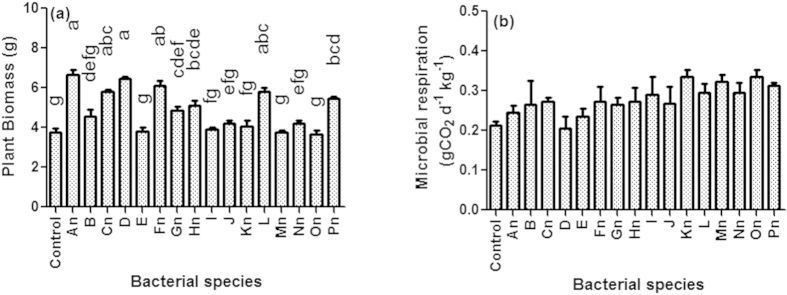
Effect of bacterial species monocultures, inoculated in the rhizosphere of Ocimum sanctum, on plant biomass (a) and soil microbial respiration (b). Bars followed by different letters were significantly different at P < 0.05 level (Tukey’s test). Error bars represent ± 1SEM. ndenotes the strains which were included in nitrogen fixing bacterial group.
Plant biomass increased linearly with inoculation of increasing diversity of rhizospheric bacterial diversity (Fig. 2; F1, 172 = 133.4, P < 0.0001), on the other hand, no relationship was found between plant biomass and nitrogen-fixing bacterial species richness (F1, 94 = 1.055, P = 0.3071). Species richness, however, greatly increased soil microbial respiration (F1, 172 = 5.27, P < 0.0001 for rhizospheric bacteria and F1, 94 = 60.51, P < 0.0001 for nitrogen-fixing bacteria). Interestingly, some species-poor compositions performed similar to some compositions of higher richness levels indicating the positive effects of the presence of particular species in the compositions in both the cases.
Figure 2. Effect of bacterial communities varying in species richness, inoculated in the rhizosphere of Ocimum sanctum, on plant biomass and soil microbial respiration.
Effect of rhizospheric bacterial communities varying from one to twelve species richness on plant biomass (a) and soil microbial respiration (b). Effect of nitrogen-fixing bacterial communities varying from one to ten species richness on plant biomass (c) and soil microbial respiration (d).
As species richness was found to be an important predictor of improved plant biomass and respiration, we tried to establish if the functional diversity of plant growth promoting traits within species richness further enhances the performance of a community. A strong positive relationship was also observed between functional group richness and dependent variables (plant biomass and soil microbial respiration, Fig. 3). In our experiments, though species richness and functional group richness were observed to be important but found to be highly correlated (r = 0.84), and therefore it is difficult to establish their exclusive effects.
Figure 3.

Effect of functional group richness in rhizospheric bacterial communities, inoculated in the rhizosphere of Ocimum sanctum, on plant biomass (a) and soil microbial respiration (b).
The presence of transgressive overyielding indicates the positive biodiversity effects may be due to niche complementation or mutualistic interactions. Transgressive overyielding was calculated only for plant biomass in case of rhizospheric bacteria where monocultures differed in their performance. At lower levels of species richness transgressive underyielding was also observed, however, at higher levels overyielding was prevalent. A significant relationship between transgressive overyielding and species richness was observed (Fig. 4; F1, 44 = 29.81, P < 0.0001). A similar relationship was observed between transgressive overyielding and functional group richness. As monocultures within nitrogen-fixing bacterial group did not differ among themselves in their ability to promote plant growth and microbial respiration in the soil, no such analyses were performed in this case.
Figure 4.

Transgressive over yield in plant biomass at different levels of species richness (a) and functional group richness (b) in rhizospheric bacterial communities inoculated in the rhizosphere of Ocimum sanctum.
In linear model analyses, the linear effect of species richness on plant biomass and respiration was significant. However, in case of nitrogen-fixing bacteria the relation between species richness and plant biomass (Tables 2 and 3) was not noticed. The significant linear relationship between species richness and ecosystem functioning indicates complementary effects of biodiversity where the presence of any new species has some additional increase in ecosystem functioning. The linear effect of species richness was significant in the case of rhizospheric bacteria only, and the absence of this effect in the case of nitrogen-fixing bacterial group shows the redundancy of functional traits in this group. Fixed effects of species richness on plant biomass were not significant at all, in any case. The fixed effect shows the effect of species richness above the complementarity, where the deviation from linearity indicates the strength of species interactions. Interestingly, the effects of species identities were found significant in the case of soil respiration in nitrogen-fixing bacteria. The effect of particular species compositions on plant biomass and soil microbial respiration was consistently significant in all the cases. It shows that regardless of the general pattern of diversity-functioning relationship, specific compositions of some species, at all richness levels, have a large effect.
Table 2. Results of general linear models showing the effects of species richness, species identities and compositions of rhizospheric bacterial communities inoculated in the rhizposphere of Ocimum sanctum on plant biomass and microbial respiration.
| Factors | df | Plant biomass |
Microbial respiration |
||||
|---|---|---|---|---|---|---|---|
| SS | F | P | SS | F | P | ||
| Sp. Richness | |||||||
| Linear | 1, 12 | 120.3 | 42.09 | 2.99 × 10−4 | 0.1263 | 8.982 | 0.0111 |
| Fixed | 6, 8 | 14.59 | 0.99 | 0.492 | 0.07988 | 1.2 | 0.394 |
| Sp. ID | 12, 46 | 14.47 | 0.60 | 0.8339 | 0.01973 | 0.438 | 0.9389 |
| Compositions | 58, 116 | 93.08 | 5.84 | 4.14 × 10−16 | 0.17248 | 2.846 | 8.5 × 10−7 |
Sp. Richness, species richness; Sp. ID, species identity; df, degrees of freedom; SS, sum of squares; F, F-statistic; P, P-value.
Table 3. Results of general linear models showing the effects of species richness, species identities and compositions of nitrogen fixing bacterial communities inoculated in the rhizposphere of Ocimum sanctum on plant biomass and microbial respiration.
| Factors | df | Plant biomass |
Microbial respiration |
||||
|---|---|---|---|---|---|---|---|
| SS | F | P | SS | F | P | ||
| Sp. Richness | |||||||
| Linear | 1, 6 | 0.552 | 0.487 | 0.511 | 0.153 | 60.508 | 9.27 × 10−12 |
| Fixed | 4, 4 | 1.003 | 0.173 | 0.941 | 0.005 | 0.4945 | 0.7398 |
| Sp. ID | 10, 22 | 2.712 | 0.296 | 0.975 | 0.023 | 0.94 | 0.51 |
| Compositions | 32, 64 | 20.118 | 1.586 | 0.058 | 0.110 | 2.196 | 0.004 |
Sp. Richness, species richness; Sp. ID, species identity; df, degrees of freedom; SS, sum of squares; F, F-statistic; P, P-value.
Discussion
Functional diversity is assumed a strong predictor of ecosystem functioning27 because it can approximate niche heterogeneity in a community. We found that functionally diverse rhizospheric bacterial communities enhance plant productivity while functionally redundant nitrogen fixing community did not; the plant biomass could not increase over monocultures. Linear increase in ecosystem functioning and presence of transgressive overyielding in several combinations indicate that bacterial functional traits may work in a complementary way and enhance plant biomass. These results fall in the line of several earlier reports, where increasing bacterial richness increased ecosystem functioning of the system8,9,17. However, in these reports as well as in ours, some species combinations at lower richness levels were also observed to be as productive as the higher richness levels, indicating the effects of species identities in these compositions. We found significant antagonistic interactions at lower levels of species richness in case of rhizospheric bacteria, however, these were infrequent and non-significant at higher levels of species richness. It might be due to the modification of species interactions such as direct competition in the presence of other species. An increase in soil respiration with species richness in case of both experiments indicates that bacterial diversity positively affects the functioning of the microbial community, however, its impact on plant growth depends on diverse plant growth promoting activities.
Diversity effects are overall outcomes of several types of negative and positive interactions such as facilitation and competition in microbial communities. Bacterial strains, which lack a particular activity when tested alone, may act synergistically in promoting ecosystem functioning when included as a part of microbial consortia5. These interactions were reflected in transgressive underyielding in case of plant biomass, where the yield of plants inoculated with species mixture was significantly lower than maximum yielding monocultures. Similar negative biodiversity- ecosystem functioning relationships were obtained by Becker et al.13. Besides direct antagonism among species such as toxin production28, competition for root exudates and space29 may also play a significant role in underyielding in plant biomass. Accumulation and/or dominance of underproductive species may also exert considerable influence in underyielding. The overall effect of diversity is affected by species interactions in which the positive and negative effects may neutralize each other, and the net diversity effect may be zero30. However, at higher richness levels, this deleterious effect may be compensated by the presence of many highly productive species indicating the sampling effect of biodiversity31. Our observations indicate that role of bacterial diversity in the rhizosphere ranges from positive to negative interactions that may also counteract each other at some instances. It is also found that plant actively recruits beneficial microorganisms in rhizosphere probably for increasing its fitness5 through antagonising harmful microorganisms. The probability of including beneficial microorganisms increases with diversity compensating the deleterious effects caused by opportunists. It is supported by our results also where increasing trends in diversity effects (overyielding) with species richness were observed. At higher levels of richness, the beneficial effects superimpose deleterious one and overall improvement in ecosystem services may be achieved.
Several limitations should be considered before generalising the results of present and all similar type of studies. The diversity of our system was very low, and the observations are based on short-term response to initial diversity. The competitive relationships among species may, however, change with long-term observations due to the extirpation of some species. In the present experiment, only a minor part of the variation in plant growth could be explained by bacterial richness, however, this would remain an active question of research that may provide better understanding of rhizosphere ecosystem functioning. In conclusion, we have shown that complementarity among different plant growth promoting traits within the community is useful in increasing plant productivity. These observations would be helpful in understanding plant-microbe interactions in the rhizosphere.
Additional Information
How to cite this article: Singh, M. et al. Complementarity among plant growth promoting traits in rhizospheric bacterial communities promotes plant growth. Sci. Rep. 5, 15500; doi: 10.1038/srep15500 (2015).
Supplementary Material
Acknowledgments
The authors are grateful to Director CSIR-CIMAP for facilities and encouragement, CSIR for the funding of study under network project BSC-203 and University Grant Commission, New Delhi for providing Senior Research Fellowship to M.S. Technical assistance of Mr. Kundan Wasnik is acknowledged.
Footnotes
Author Contributions M.S. and A.K. conceived and planned the experiment, M.S., S.K.S. and R.P.S. done experiment and collected data, A.A. and R.K.V. helped in statistical analyses and interpretation of results, M.S. and A.K. wrote the paper, and all authors reviewed the manuscript.
References
- Smith S. E. & Read. D. J. Mycorrhizal Symbiosis 2nd edn. Academic Press, London, UK, 1997 [Google Scholar]
- Kowalchuk G. A. & Stephen J. R. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Ann. Rev. Microbiol. 55, 485–529 (2001). [DOI] [PubMed] [Google Scholar]
- Duchicela J. et al. Non-native plants and soil microbes: potential contributors to the consistent reduction in soil aggregate stability caused by the disturbance of North American grasslands. New Phytol. 196, 212–222 (2012). [DOI] [PubMed] [Google Scholar]
- Bever J. D. et al. Rooting theories of plant community ecology in microbial interactions. Trend. Ecol. Evol. 25, 468–478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R. et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 332, 1097–1100 (2011). [DOI] [PubMed] [Google Scholar]
- Langenheder S., Bulling M. T., Solan M. & Prosser J. I. Bacterial biodiversity-ecosystem functioning relations are modified by environmental complexity. PLoS ONE 5, e10834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreau M. Biodiversity and ecosystem functioning: recent theoretical advances. Oikos. 91, 3–17 (2000). [Google Scholar]
- Bell T., Newman J. A., Silverman B. S., Turner S. L. & Lilley A. K. The contribution of species richness and composition to bacterial services. Nature 436, 1157–1160 (2005). [DOI] [PubMed] [Google Scholar]
- Salles J. F., Poly F. & Roux X. L. Community niche predicts the functioning of denitrifying bacterial assemblages. Ecology 90, 3324–3332 (2009). [DOI] [PubMed] [Google Scholar]
- Wittebolle L. et al. Initial community evenness favours functionality under selective stress. Nature 458, 623–626 (2009). [DOI] [PubMed] [Google Scholar]
- Eisenhauer N., Schulz W., Scheu S. & Jousset A. Niche dimensionality links biodiversity and invisibility of microbial communities. Funct. Ecol. 27, 282–288 (2013). [Google Scholar]
- Foster K. R. & Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 22, 1845–1850 (2012). [DOI] [PubMed] [Google Scholar]
- Becker J., Eisenhauer N., Scheu S. & Jousset A. Increasing antagonistic interactions cause bacterial communities to collapse at high diversity. Ecol. Lett. 15, 468–474 (2012). [DOI] [PubMed] [Google Scholar]
- Glick B. R. Plant Growth-Promoting Bacteria: Mechanisms and Applications. http://dx.doi.org/10.6064/2012/963401 (2012). [DOI] [PMC free article] [PubMed]
- Tilman D. Functional diversity. In: Levin S. A. (Ed.), Encyclopedia of Biodiversity Academia Press, San Diego. pp.109–120 (2001). [Google Scholar]
- Cadotte M. W. et al. Phylogenetic diversity metrics for ecological communities: integrating species richness, abundance and evolutionary history. Ecol. Lett. 13, 96–105 (2010). [DOI] [PubMed] [Google Scholar]
- Awasthi A., Singh M., Soni S. K., Singh R. & Kalra A. Biodiversity acts as insurance of productivity of bacterial communities under abiotic perturbations. ISME J 8, 2445–52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle W. F. Phylogenetic classification and the universal tree. Science 284, 2124–2129 (1999). [DOI] [PubMed] [Google Scholar]
- Snel B., Bork P. & Huynen M. A. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Gen. Res. 12, 17–25 (2002). [DOI] [PubMed] [Google Scholar]
- Awasthi A. et al. Synergistic effect of Glomus mosseae and nitrogen fixing Bacillus subtilis strain Daz26 on artemisinin content in Artemisia annua L. Appl. Soil Ecol. 49, 125–130 (2011). [Google Scholar]
- Mehta S. & Nautiyal C. S. An efficient method for qualitative screening of phosphate solubilizing bacteria. Curr. Microbiol. 43, 57–58 (2001). [DOI] [PubMed] [Google Scholar]
- Gordan S. A. & Weber R. P. Colorimetric estimation of indole acetic acid. Plant Physiol. 26, 192–195 (1951). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendels M. & Reese E. T. Induction of cellulases in fungi trichoderma virde as influencing carbon source. J. Bactriol. 37, 265–275 (1957). [Google Scholar]
- Schwyn B. & Neilds J. B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56 (1987). [DOI] [PubMed] [Google Scholar]
- Bekku Y., Koizumi H., Oikava T. & Iwaki H. Examination of four methods for measuring soil respiration. Appl. Soil Ecol. 5, 247–254 (1997). [Google Scholar]
- Bell T. et al. A linear model method for biodiversity-ecosystem functioning experiments. Am Nat. 174, 836–849 (2009). [DOI] [PubMed] [Google Scholar]
- Flynn D. F. B. et al. Functional and phylogenetic diversity as predictors of biodiversity–ecosystem-function relationships. Ecology 92, 1573–1581 (2011). [DOI] [PubMed] [Google Scholar]
- Czaran T. L., Hoekstra R. F. & Pagie L. Chemical warfare between microbes promotes biodiversity. Proc. Natl. Acad. Sc. USA, 99, 786–790 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais H. P., Weir T. L., Perry L. G., Gilroy S. & Vivanco J. M. The role of root exudates in rhizosphere interations with plants and other organisms. Ann. Rev. Plant Biol. 57, 233–266 (2006). [DOI] [PubMed] [Google Scholar]
- Kirwan L. et al. Diversity-interaction modeling: estimating contributions of species identities and interactions to ecosystem function. Ecology. 90, 2032–2038 (2009). [DOI] [PubMed] [Google Scholar]
- Huston M. A. Hidden treatments in ecological experiments: re-evaluating the ecosystem function of biodiversity. Oecologia. 110, 449–460 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



